Abstract
Background and Aims
Leaf growth is a complex developmental process controlled by genetic and environmental factors and is determined by a proliferation, expansion and maturation phase. Mutational analysis in Arabidopsis thaliana showed that leaf size and shape is dependent on cell division and cell expansion activity. An investigation was made at the cytophysiological and ultrastructural level of the elo1 mutant of Arabidopsis thaliana, which is defective in one of the components of the histone acetyl transferase Elongator complex and displays a distinct ‘narrow leaves’ phenotype, owing to a reduced cell number and no transition between petiole and lamina. Relative expression levels of three sucrose metabolism/transport-related genes were also investigated. The aim was to determine the physiological basis of leaf morphology in this mutant, by investigating the modulatory role of sucrose.
Methods
The elo1 mutant was taken as representative of all the elo mutations and investigated at cytophysiological level. A germination test and growth assays were performed on seedlings grown for 21 d at different sucrose concentrations. Leaf morphometric and ultrastructural features were also investigated by image analysis and electron microscopy, respectively. Finally, a quantitative PCR (qPCR) analysis was performed with three sucrose metabolism/transport-related genes that were investigated under different sucrose concentrations.
Key Results
elo1 plants at high sucrose concentrations exhibited an enhancement of germination and inhibition of leaf growth as compared with wild-type plants. qPCR experiments with three sucrose metabolism/transport-related genes showed an interaction between sucrose availability and the elo1 mutation. Furthermore, electron microscopy analysis provided the first ultrastructural description of an elo mutant, which showed a hypotonic vacuole, alterations in the size of grana and starch grains in the chloroplasts, and the massive presence of Golgi vesicles in the cytoplasm.
Conclusions
Based on the results obtained it is proposed that mechanisms producing carbon assimilates or importing sucrose could be affected in elo1 plants and could account for the observed differences, implying a role for Elongator in the regulation of these processes.
Key words: Elongator complex, elo1, leaf development, germination, cell division, cell expansion, morphometric analysis, electron microscopy, qPCR, Arabidopsis thaliana, sucrose
INTRODUCTION
Leaf morphogenesis is a complex developmental process, and is controlled by genetic and environmental factors. It ultimately produces a functional photosynthetic organ that is able to capture light, produce carbon metabolites, exchange gasses, and transpire water for plant cooling and circulation (Pozzi et al., 2001; Tsukaya, 2002, 2005; Micol and Hake, 2003; Kessler and Sinha, 2004; Fleming, 2005). Leaf growth is determined by cell division and expansion. According to the ‘neo cell theory’ (Tsukaya, 2005), the size and shape of leaves is secondarily affected by the size and shape of each leaf cell, which is considered the unit of all tissues and organs. Both final cell number and cell polarity have a genetic basis, thus controlling tissue organization and, finally, leaf shape. Indeed, the manipulation of cell cycle or cell-wall extensibility resulted in different leaf shapes (Fleming, 2002). Peculiarly, in plant leaf an inter-reliant cellular compensatory mechanism can deliver the same morphological output in different ways: for instance, a specific lamina width can be achieved with many small palisade cells or vice versa with a smaller number of large palisade cells. However, this so-called compensation mechanism does not always occur. Furthermore, environmental variables, such as light, temperature and nutrients, play a modulatory role and can modify an established morphogenetic programme (Van Volkenburgh, 1999).
In this context, an important role is played by sucrose, which is an important metabolite for plant growth, tissue differentiation and maturation. Namely, its import and cleavage, via invertases and sucrose synthases, into signal hexoses (glucose, fructose, UDP-glucose) is known to act at the cellular level, controlling cell division and expansion (Weber et al., 1997; Sturm, 1999; Lemoine, 2000; Weschke et al., 2003; Koch, 2004; Roitsch and González, 2004).
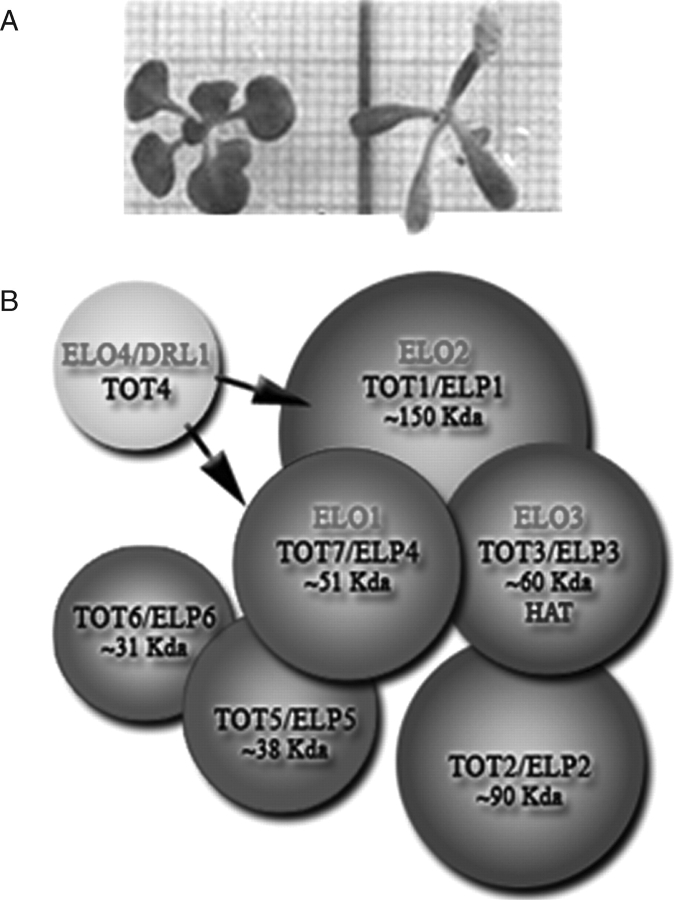
In the present work, a ‘narrow leaf’ elo1 mutant of Arabidopsis thaliana, which displays a slim lamina (Fig. 1A), the so-called ‘narrow leaves’ phenotype, was taken as representative of all the elo mutations and investigated at cytophysiological level. elo1 belongs to the elongata class of leaf mutants (Berná et al., 1999) and has a mutation in the At3g11220 gene, which is homologous to the yeast ELP4 component of the Elongator complex (Fig. 1B) (Otero et al., 1999; Fellows et al., 2000; Frohloff et al., 2001; Jablonowski et al., 2001; Krogan and Greenblatt, 2001; Jablonowski and Schaffrath, 2002; Kim et al., 2002; Slaugenhaupt and Gusella, 2002; Fichtner et al., 2003; Nelissen et al., 2003, 2005; Gilbert et al., 2004).
Fig. 1.
(A) Wild-type Ler (left) and mutant elo1 (right) plantlets grown under standard condition 21 DAV. (B) The holo-Elongator complex. Red, accessory complex; green, core-Elongator; yellow, DRL1, which interacts with the holo-Elongator complex. Red codes (ELO1 to ELO4) refer to plant elongata genes (Berna et al., 1999), while black codes refer to yeast Elongator genes (TOT1 to TOT7) and their plant homologues (ELP1 to ELP6) (Nelissen et al., 2005).
In yeast, the holo-Elongator complex, which contains histone acetyltransferase (HAT) activity (Winkler et al., 2002), consists of two subcomplexes: ELP1, ELP2 and ELP3 (HAT) that compose the core-Elongator and ELP4, ELP5 and ELP6 that constitute the accessory subcomplex. The ELP genes are the homologues of the Toxin Target (TOT) genes of Saccharomyces cerevisiae that, upon mutation, slowly adapt growth to changing conditions and resistance to the zymocin toxin (Fig. 1B) (Frohloff et al., 2001). KTI12 is a putative regulator of Elongator and its respective mutants display a similar phenotype as the elp/tot mutants. DRL1 is the homologue of KTI12 in Arabidopsis and its mutant alleles also show a narrow leaves phenotype (Nelissen et al., 2003). A scheme of the Elongator complex, based on epistatic interactions (Nelissen et al., 2005) and components in plants (ELO), is shown in Fig. 1B; the yeast nomenclature (ELP/TOT) is also included.
The aim of the present study was to determine the effect of different sucrose concentrations on seed germination, leaf growth and morphogenesis of the elo1 mutant. In addition, the level of expression of three sucrose metabolism/transport-related genes was also evaluated. The elo1 mutation is shown here to affect the sucrose metabolism.
MATERIALS AND METHODS
Plant material and in vitro growth conditions
Seeds of Arabidopsis thaliana (L.) Heynh. ecotype Landsberg erecta (Ler) and elongata1 (elo1) have been described previously (Nelissen et al., 2003, 2005). Seed stocks from both Ler and elo1 were harvested at the same time and stored in the same conditions in order to avoid differences in seed germination and in vitro growth.
For standard growth conditions, seeds were strongly sterilized by incubation for 2 min in 100 % ethanol and for 12 min in 1·75 % hypochlorite solution (NaClO). Thereafter, seeds were germinated on plates with germination medium (GM) at 1 % sucrose (Valvekens et al., 1988) and 0·7 % plant cell culture agar (Sigma-Aldrich). Plates were kept for 3 d overnight at 4 °C for seed vernalization prior to be transferred to the germination chamber, where plants were grown under sterile conditions under a 16-h light/8-h dark regime at 22 °C, with light intensity of 150 µmol m−2 s−1 and 60 % relative humidity. Plant age was estimated after completion of the 3-d vernalization period and the first day of the plant corresponds to 1 d after vernalization (DAV). Different growth conditions were tested as described below.
Germination analysis
Germination tests performed under standard growth conditions were used as control. Different sterilization (water, 100 % ethanol, 7, 5·25, 3·5 % sodium hypochlorite solution) and nutrient (0–3 % sucrose concentration) conditions were independently tested. Three replicates were performed; for each replica, n > 50 seeds were germinated for each sample.
Morphological analysis
Analysis was performed on first and third expanded leaves of 24-d-old (21 DAV) Ler and elo1 plantlets, grown under standard conditions on GM, supplemented with three different sucrose concentrations (0·5, 1 or 2 %). Entire leaves were excised at the basis of the petiole, mounted onto a microscope slide, put on a millimetre paper and photographed with a fixed 6·3-megapixel Finepix S7000 digital camera (Fuji). Image analysis was performed with the ImageJ 1·032j software (Wayne Rasband, National Institutes of Health, Bethesda, MD, USA) after pixel/mm conversion. Four parameters were measured: lamina length, width and area, and petiole length. Three different replicas were made and n > 10 leaves were utilized for each sample.
Histological analysis
Analysis was performed on first and third fully expanded leaves of 24-d-old (21 DAV) Ler and elo1 seedlings, grown under standard conditions on GM, supplemented with three different sucrose concentrations (0·5, 1 or 2 %). From each collected leaf a median sector was excised under a stereomicroscope and immediately fixed either in 4 % paraformaldeyde overnight or in a 3 % glutaraldehyde/0·5 % paraformaldehyde/phosphate-buffered saline solution at 4 °C, for 3 h. After dehydration, samples were embedded in Tecnovit 8100 resin and cross-sectioned at 4 µm with an RM 2155 Microtome (Leica); other samples were embedded in paraplast embedding media (Sigma-Aldrich) and cross-sectioned at 10 µm. Sections were stained for 10 min with either toluidine blue (0·05 % in 0·1 m phosphate buffer, pH 6·7) or periodic acid Schiff (PAS) and mounted with Canada balsam. Screen shots of transverse sections were consecutively acquired with a DMRB Microscope (Leica) and the IM50 software under a 200 × magnification (Leica). Palisade cell number (PCN) (Cnops et al., 2004) and palisade cell/gap number (PCGN) were estimated on the three widest sections of each leaf. PCGN analysis takes into account gaps present in the palisade layer. One gap corresponds to one ‘virtual’ palisade cell with the same mean area of the two cells that are adjacent to the gap. PCGN analysis was necessary in the case of elo1 mutants, characterized by high number of gaps in the palisade layer (>5 %), in order to obtain a more significant correlation with lamina width. Stomatal chambers were not taken into account. The screen shots of the transverse sections were merged together with the ‘photomerge’ function (Adobe Photoshop CS2) and cell area measurements on digital images were made with the freely available IMAGEJ software, after pixel/μm conversion. Three different replicas were done and n > 20 leaves were taken for each sample. Cell area analysis (n > 600 for each sample) was performed on the same sections. In addition, cell area was also measured from the adaxial side through differential interference contrast optics. Cleared first leaves were used to measure the cell area from the adaxial side. The cells seen under the microscope were digitalized and analysed with the IMAGEJ software, after pixel/μm conversion. Other parameters, such as cell length (along the proximal–distal axis of the leaf) and width (along the lateral axis of the leaf), were measured on the same pictures.
Electron microscopy
Analysis was performed on first and third fully expanded leaves of 24-d-old (21 DAV) Ler and elo1 seedlings, grown under standard conditions on GM, supplemented with three different sucrose concentrations (0·5, 1 or 2 %). Slices (n > 5 for each sample) were fixed overnight at 4 °C in 3 % glutaraldehyde in 0·1 m cacodylate buffer (pH 6·9) and post-fixed for 2 h in 15 % osmium tetroxide in the same buffer and at the same temperature. Specimens were dehydrated in a graded series of ethyl alcohol and propylene oxide solutions and embedded in araldite. Staining with uranyl acetate was carried out while dehydrating with 75 % alcohol. Ultrathin sections (0·06 µm) were cut with an Ultracut UCT (Leica), stained with lead citrate and observed with a transmission electron microscope (EM900; Zeiss) operating at 50 kV.
qPCR
For the quantitative PCR (qPCR) experiment, plants were grown for 15 d on medium containing 0·5, 1 or 2 % sucrose. RNA was extracted from the first two leaves (RNeasy, Qiagen Benelux B.V, The Netherlands). The analysed genes were sucrose synthase (At4g02280), sucrose transporter (At1g71890), sugar transporter (At5g18840) and actin (At3g60830). The primers were designed using the Beacon designer 4 program (Premier BioSoft International). The qPCR was performed on the LightCycler TM 480 (Roche). Data points in the exponential phase of the qPCR were linearly interpolated after log correction (0·5) with a perl script using the Statistics::Regression module. This script is available from the authors upon request. The actin gene was found to be stable in all samples by using the GeNorm program (Vandesompele et al., 2002) and was used to normalize the data in Qbase (Hellemans et al., 2007) taking into account the efficiencies of the PCR reactions, calculated by log-linear regression.
Statistical analysis
For morphometric analyses all data were evaluated statistically with the SPSS software (Statistical Package for the Social Sciences, version 11·0·0; SPSS, Chicago, IL, USA). In order to verify whether the distribution was normal or left/right skewed, a ‘descriptive statistics’ was performed. In the case of a skewed distribution, a logarithmic transformation (lnX) of the data was applied, thus transforming it into a normal distribution. Thereafter, a Student's t-test between two sets of data was perfromed to obtain a significance value P of the mean differences. In our case the null hypothesis (H0) of ‘equality of means’ was rejected if P = 0·05.
For qPCR the experimental design comprised two biological replicates and the measurement was run four times. An analysis of variance (ANOVA) was performed on the qPCR data that were normalized with the actin gene using the SPSS software. The ANOVA model was a three-factor model with runs as random factor and genotype and medium as fixed factor and with medium × genotype interaction. The equality of variance error was controlled by Levene's test (P > 0·05).
RESULTS
Under sucrose depletion elo1 seed germination is more efficient
In order to assess the best sterilization conditions, different solutions were tested. As expected, washing and only ethanol sterilization were ineffective in avoiding contamination for both elo1 and Ler seeds. By contrast, 14 min treatment with NaClO solutions suppressed any contamination whatever concentration was applied. Surprisingly, after treatment with 7 % NaClO solution, seed germination was completely inhibited for Ler seeds (0 %) and drastically reduced for elo1 seeds (<10 % germination) (Fig. 2A). Treatments with serial dilutions (25, 50 and 75 %) of NaClO solution showed that elo1 seeds were more resistant to hypochlorite than Ler seeds, being able to maintain good germination rates (>60 %) at higher bleach concentrations (5·25 %) compared with wild-type seeds (3·5 %) (Fig. 2A).
Fig. 2.
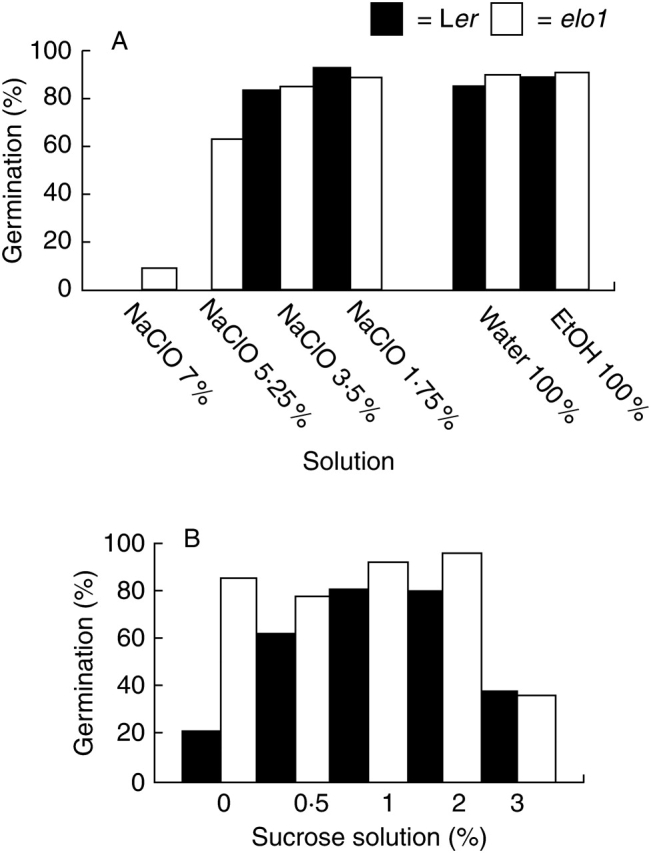
Seed germination rates under different environmental conditions: (A) sodium hypochlorite solution concentration, and (B) sucrose concentration.
Thereafter, the nutrient test was performed by using the lowest NaClO concentration for sterilization and using sucrose concentration ranging from 0 to 3 % (Fig. 2B). It was apparent that germination of Ler seeds was always affected when standard sucrose concentrations varied from sucrose depletion to sucrose abundance. By contrast, elo1 seeds exhibited a reduced germination rate only at the highest sucrose concentrations (3 %).
Sucrose-dependent differences in growth stage exist between elo1 and Ler plants
Growth stage-based analysis (Boyes et al., 2001) of Ler and elo1 plants (Table 1) cultured for 24 d under standard in vitro conditions on GM supplied with different sucrose concentrations (0·5, 1 and 2 %) was undertaken. Seeds were synchronized by means of a 3-DAV period, but radicle appearance and cotyledon emergence from the seed coat were not synchronized (data not shown). However, 7 DAV, both wild-type and mutant plantlets reached stage 1·00 (Boyes et al., 2001), with the two cotyledons fully open. Thereafter, plant growth proceeded dissimilarly between mutants and wild-types. At 12 DAV, sucrose starvation (0·5 %) had clearly a macroscopic and visible effect on elo1 plants, which displayed a one-leaf delay as compared with those grown under standard conditions (1 % sucrose) and in sucrose abundance (2 %). Namely, the former exhibited only three rosette leaves larger than 1 mm (stage 1·03), while the latter two were able to produce four rosette leaves larger than 1 mm (stage 1·04). This delay was maintained until 21 DAV where stages 1·05 and 1·06 were reached, respectively. No visible differences were induced by sucrose treatment in Ler plants until 15 DAV. From this point onward, sucrose abundance transiently accelerated leaf emergence (stage 1·07) compared with standard conditions (stage 1·06). More precisely, at 21 DAV, Ler plants grown under standard conditions and in sucrose abundance reached the same stage (stage 1·07, with seven rosette leaves larger than 1 mm), because of a faster growth under standard conditions than under sucrose abundance. In addition, at 18 DAV, Ler plants cultured either under sucrose starvation or standard conditions displayed no growth differences and the one-leaf delay was observed only at 21 DAV.
Table 1.
Growth stage-based analysis of Ler and elo1 plants grown in standard in vitro conditions on germination medium supplied with different sucrose concentrations
| Phenotype | Sucrose (%) | 7 DAV | 12 DAV | 15 DAV | 18 DAV | 21 DAV |
|---|---|---|---|---|---|---|
| Ler | 0·5 | 1 | 1·04 | 1·05 | 1·06 | 1·06 |
| Ler | 1 | 1 | 1·04 | 1·05 | 1·06 | 1·07 |
| Ler | 2 | 1 | 1·04 | 1·05 | 1·07 | 1·07 |
| elo 1 | 0·5 | 1 | 1·02 | 1·03 | 1·04 | 1·05 |
| elo 1 | 1 | 1 | 1·02 | 1·04 | 1·05 | 1·06 |
| elo 1 | 2 | 1 | 1·02 | 1·04 | 1·05 | 1·06 |
DAV= days after vernalization; growth stages: 1 = two cotyledons fully open, 1·02–1·07 = plantlets with 2–7 rosette leaves larger than 1 mm.
Taken together, these observations showed that elo1 plants had a final one-stage retardation compared with Ler plants grown under the same nutrient conditions, but also that growth dynamics of wild-type and mutant plants are differentially driven by sucrose availability. Namely, sucrose abundance had no effect on elo1 plants and early effects of sucrose starvation were observed in elo1 plants with respect to wild-type, in such a way that the delay in leaf emergence was anticipated.
Under different sucrose concentrations Ler and elo1 leaves exhibit opposite lamina growth dynamics
Fully expanded leaves can be considered as standardized material in which cell size and cell number are representative of total cell expansion and cell division contributing to final leaf size (Cnops et al., 2004).
Leaf morphometric parameters were measured of the fully expanded first and third leaves of 21 DAV plants grown on media supplemented with 0·5, 1 and 2 % sucrose. Four parameters were taken into account: petiole length, and lamina length, width and area (Fig. 3). In elo1 plants grown at 2 % sucrose, petiole and lamina length were not evaluated, owing to the absence of a transition between lamina and petiole; in this case the whole leaf length was measured.
Fig. 3.
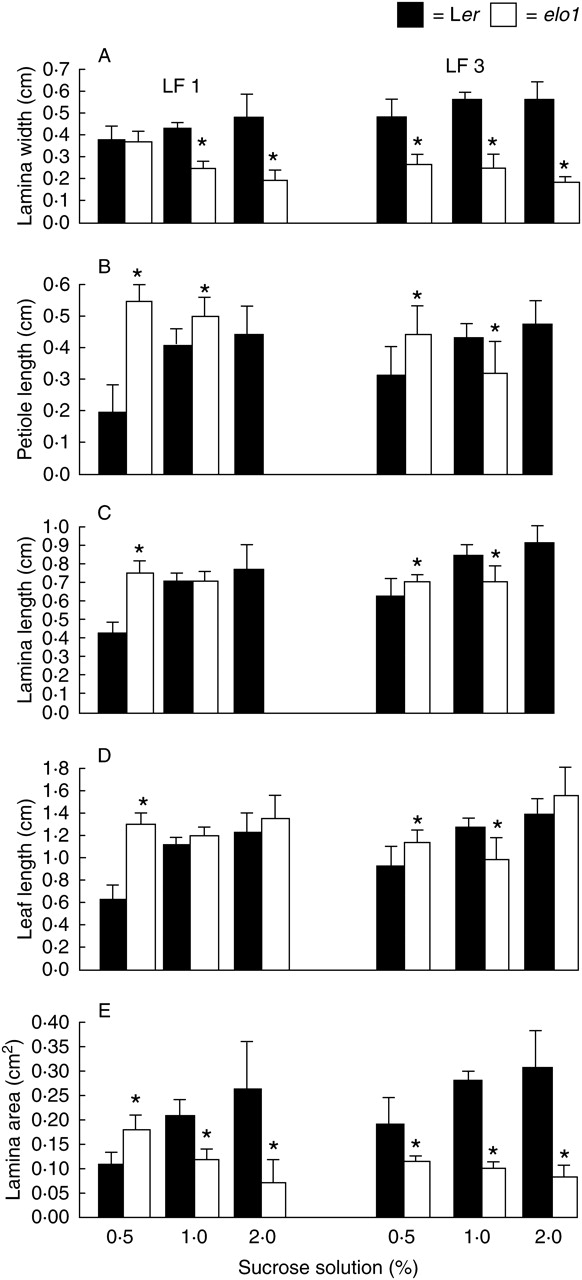
Mean and s.d. of (A) lamina width, (B) petiole length, (C) lamina length, (D) leaf length, and (E) lamina area of 21-DAV Ler and elo1 first and third leaves (LF1 and LF3, respectively), grown under standard in vitro conditions, but with different sucrose concentrations (0·5, 1 and 2 %) in the medium. Asterisks indicate a statistically significant difference between elo1 and Ler values (t-test, P < 0·05).
The data showed that, with the exception of the first leaf at 0·5 % sucrose concentration, elo1 leaves were always significantly narrower than Ler leaves (Fig. 3A). Secondarily, in Ler and elo1 plants leaf growth responded to sucrose concentration in opposite ways: high sucrose concentrations enlarged Ler leaves, namely lamina width and area, but narrowed elo1 leaves (Fig. 3A, E). Notably, the differences in lamina width and area were statistically significant at 2 % sucrose concentration and especially for the third leaf.
PC(G)N analyses show different cell division number at increasing sucrose concentrations
In order to investigate whether sucrose treatments induced variations in lamina width by cell division number, the numbers of cells and gaps were evaluated in the palisade layer, PC(G)N, of leaf mesophyll. The analysis was performed on transverse sections of fully expanded first and third leaves of Ler and elo1 plants grown for 21 DAV on media supplemented with 0·5, 1 and 2 % sucrose (Fig. 4A).
Fig. 4.
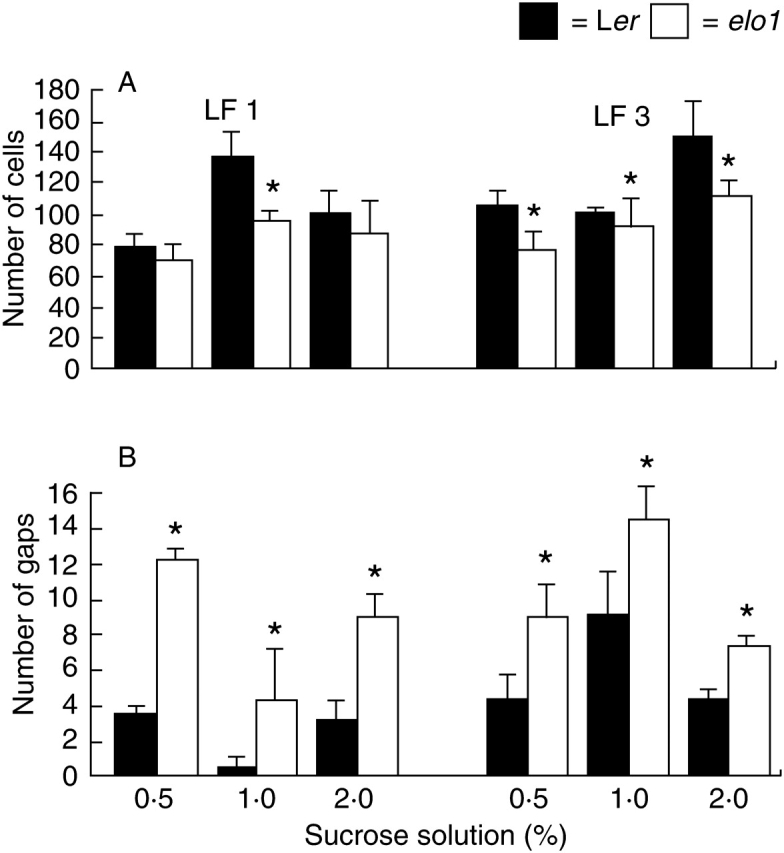
(A) Mean and s.d. palisade cell number and (B) gaps of 21-DAV Ler and elo1 first and third leaves, grown under standard in vitro conditions, but with different sucrose concentrations (0·5, 1 and 2 %) in the medium. Asterisks indicate a statistically significant difference between elo1 and Ler values (t-test, P < 0·05).
Mutants had a significantly reduced PCN compared with Ler plants, while PCGN was reduced under almost all sucrose conditions, with the exception of first leaves in sucrose starvation and abundance. As expected, PCN increased for Ler first leaves from 0·5 to 1 % sucrose and for Ler third leaves from 1 to 2 %. These results are consistent with a clear effect of sucrose concentrations on cell division (Wobus and Weber, 1999). On the other hand, PCGN values showed that the reduction of cell number was accompanied by the presence of several gaps (Fig. 4B) that, at 0·5 and 2 % sucrose concentrations, for first leaves partially compensate for the reduction of cells in the mutant.
Sucrose concentrations differentially affected cell expansion in elo1 and wild-type plants
In order to gain a complete overview of lamina growth, the area of palisade cells was evaluated on paradermal and transverse leaf sections (Fig. 5A). Under standard conditions, from paradermal microscopic observations, elo1 cells showed an increased area compared with Ler cells. Furthermore, wild-type cells increased their area when sucrose concentration was enhanced, while under the same conditions cell expansion was reduced in the mutant. By contrast, under sucrose starvation, elo1 cells showed increased area whereas Ler cells showed reduced area.
Fig. 5.
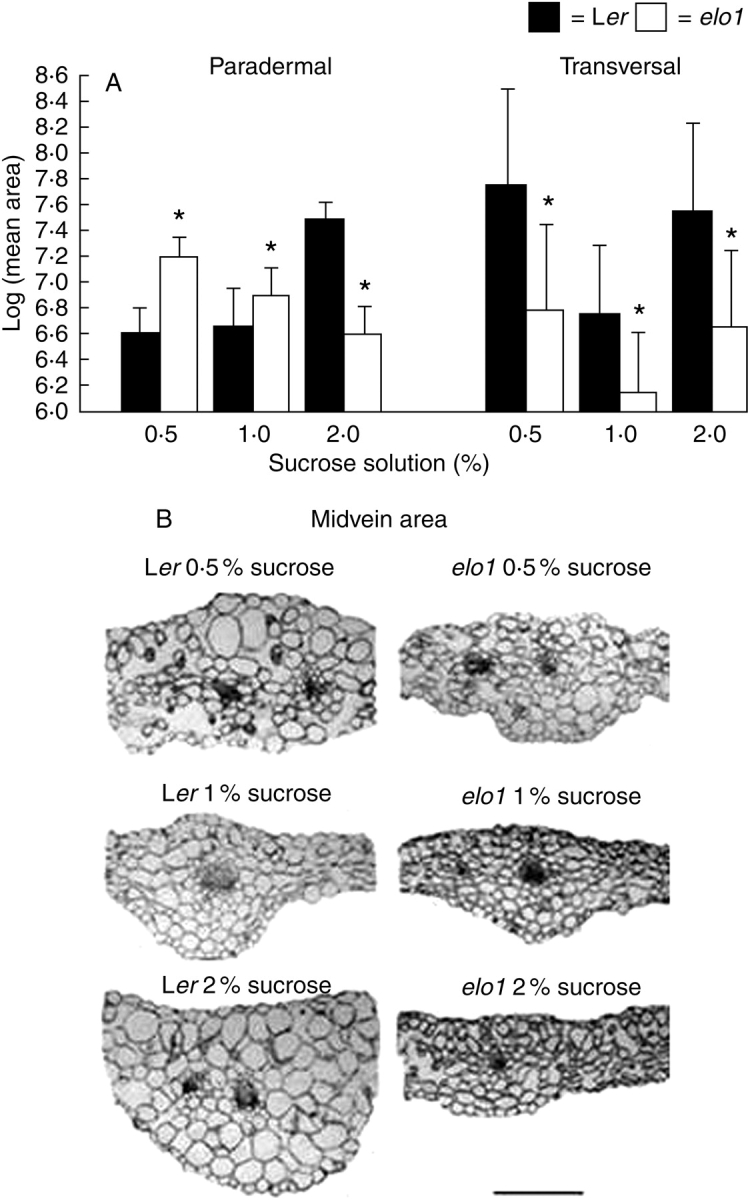
(A) Logarithmic mean and s.d. of paradermal (measured from the adaxial face of the leaf) and transverse cell area of 21-DAV Ler and elo1 first leaves, grown under standard in vitro conditions, but with different sucrose concentrations (0·5, 1 and 2 %) in the medium. Asterisks indicate a statistically significant difference between elo1 and Ler values (t-test, P < 0·05). (B) Transverse sections of 21-DAV Ler and elo1 first leaves grown on different germination media with 0·5, 1 and 2 % sucrose showing the midvein and cell area differences. All sections are shown at the same magnification (scale bar = 500 µm).
Cell dimensions were also measured along the two main axes of the leaf, the proximal–distal x-axis and the lateral y-axis. No significant polarity was found in cell growth along these two axes (data not shown) and such conditions were not affected by sucrose concentration.
The palisade cell area was also estimated on transverse sections. Surprisingly, compared with controls grown under different sucrose concentrations, mutants exhibited an overall reduced cell area (Fig. 5B). When the effects of different sucrose concentrations were analysed, it was evident that high sucrose concentration induced a significant increase of cell area in Ler leaves, whereas elo1 cell area stayed relatively constant and small.
These results suggest clearly that cell expansion along the dorsal–ventral axis (z-axis) is affected in mutants and that an opposite behaviour is active in elo1 and Ler plants, in a sucrose-dependent manner. Finally, in the elo1 mutant, sucrose starvation induced cells to become smaller along the z-axis and larger in the xy lamina, thus showing the establishment of a compensatory polarity in cell growth.
elo1 plants have less stacked grana, a hypotonic vacuole and an active exocytosis
Transmission electron microscopy was carried out on ultrathin sections of Ler and elo1 first leaves harvested at 21 DAV (Fig. 6). Plants were grown on germination media with different sucrose concentrations.
Fig. 6.
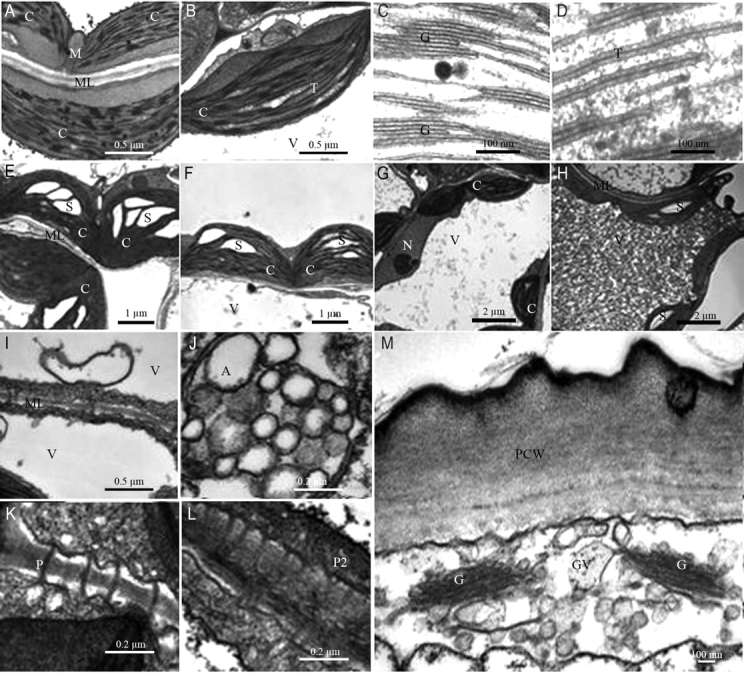
Chloroplasts of (A) Ler and (B) elo1 cells under standard conditions. Chloroplast thylakoids of Ler (C) and elo1 (D). Chloroplasts of Ler (E) and elo1 (F) plants grown on 2 % sucrose medium, showing a strong starch accumulation in the stroma. elo1 cells of plants grown on 0·5 % (G) and 2 % (H) sucrose concentration medium. Autophagy in Ler (I) cells grown under sucrose starvation and elo1 (J) cells grown under standard conditions. Plasmodesmata of elo1 cells of plants grown under standard conditions (K) and in sucrose abundance (L). (M) Golgi of elo1 cells of plants grown under standard conditions. Magnifications: (A, B, I) 20 000 ×; (C, D) 85 00 ×; (E, F) 7000 ×; (G, H) 4400 ×; (J–L) 50,000 ×; (M) 30 000 ×. C, chloroplast; M, mitochondrion; N, nucleus; S, starch; ML, median lamella; V, vacuole; A, autophagic products; P, simple plasmodesma; P2, Y plasmodesma; G, Golgi apparatus; GV, Golgi vesicles; PCW, primary cell wall.
Comparing wild-type and mutant plants grown under standard conditions (1 % sucrose), it was evident that chloroplasts were well differentiated in Ler (Fig. 6A), with abundant grana and thylakoids distributed in the stroma (Fig. 6C), whereas elo1 chloroplasts (Fig. 6B) had long double interconnecting thylakoids and fewer stacked granal thylakoids (Fig. 6D). Furthermore, chloroplasts of Ler preferentially accumulated lipid/protein, as evidenced by the presence of numerous plastoglobules (Fig. 6A), as compared with the reduced number or complete absence of primary starch. By contrast, in elo1 primary starch was frequently visible in addition to few plastoglobules (Fig. 6B). In both Ler and elo1 seedlings grown on GM at 2 % sucrose concentration, cells exhibited chloroplasts with large starch grains (Fig. 6E, F). In addition, a peculiar feature of elo1 (Fig. 6G) cells, as compared with control (Fig. 6H), was a hypotonic vacuole, probably related to a reduced turgor pressure. The vacuole consisted of a dense matrix in elo1 cells grown on 2 % sucrose media (Fig. 6H). Such a matrix is possibly a mixture of intravacuolar components and glycosides, as supported by eosine staining (data not shown). As a consequence of a lower vacuole/cytoplasm ratio, nuclei were more easily detectable in elo1 cells. Mitochondria and peroxisomes were normally distributed in both mutant and wild-type cells, attached to chloroplast (data not shown). Autophagy was observed in Ler cells grown under sucrose starvation (Fig. 6I) and in elo1 cells grown under standard conditions (Fig. 6J). Dense plasmodesmata were present in elo1 cells of plants grown on 1 % (Fig. 6K, simple plasmodesmata) and 2 % (Fig. 6L, Y plasmodesmata) sucrose media, indicating functional and active symplastic co-operation between cells. Finally, in these elo1 cells the Golgi apparatus (Fig. 6M) showed a high number of exocytosis vesicles and a large primary cell-wall formation.
qPCR shows differential expression of sucrose-related genes in elo1 mutant
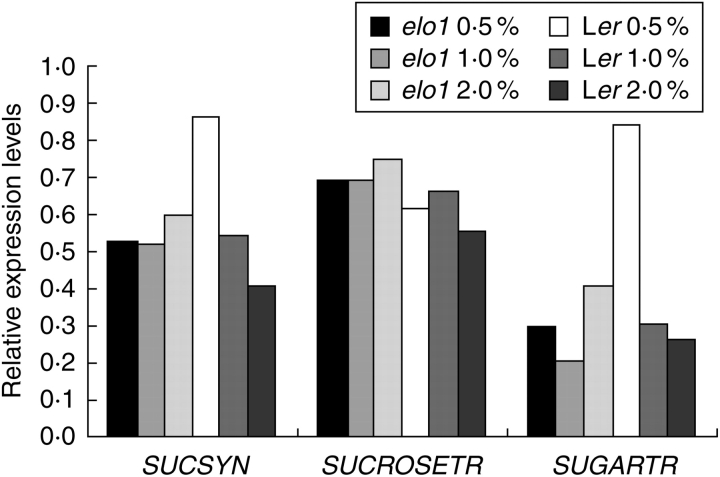
The expression of three genes, sucrose synthase (At4g02280), sucrose transporter (At1g71890) and sugar transporter (At5g18840), with a function in sucrose metabolism was analysed by qPCR in Ler and elo1 seedlings grown for 15 d on medium containing 0·5, 1 and 2 % sucrose concentration. A three-factor ANOVA analysis was performed (Table 2) and showed that the expression of the sucrose transporter and sugar transporter were dependent on the elo1 genotype (P < 0·032 and P < 0·001, respectively), implying that the mutation alters their gene expression significantly. The sucrose transporter gene expression was upregulated in elo1 at all sucrose concentrations (Fig. 7). The expression of the sugar transporter and sucrose synthase was significantly affected by the medium (P < 0·002) and was genotype-dependent (genotype × medium interaction). Indeed, their gene expression was slightly up-regulated in elo1 when the sucrose concentration was increased, although their expression was strongly down-regulated by sucrose addition in the medium (Fig. 7).
Table 2.
Statistical analysis of gene expression in elo1 and Ler seedlings grown for 15 d on medium with different sucrose concentrations (0·5, 1 and 2 %)
| Gene | Name | Genotype | Medium | Genotype × Medium | Run | Levene |
|---|---|---|---|---|---|---|
| At4g02280 | Sucrose synthase | <0·002 | <0·001 | <0·001 | >0·05 | |
| At1g71890 | Sucrose transporter | <0·032 | <0·001 | >0·05 | ||
| At5g18840 | Sugar transporter | <0·001 | <0·001 | <0·001 | <0·041 | >0·05 |
The P values were calculated by using a three-factor ANOVA (run as a random factor, genotype and medium as fixed factor and genotype × medium interaction). Actin was used as housekeeping gene.
Fig. 7.
qPCR analysis of three sucrose metabolism/transport-related genes, sucrose synthase (SUCSYN), sucrose transport (SUCROSETR) and sugar transport (SUGARTR). Relative expression levels were calculated and the actin gene, whose expression was not affected by the media with different sucrose concentrations, was used as reference gene.
DISCUSSION
In the present work, a clear relationship between leaf development, growth conditions and Elongator function has been detected by investigating growth responses and specific gene expression, under different nutrient conditions, in both wild-type and an arabidopsis ‘narrow leaves’ mutant (elo1), belonging to the elongata class (Berná et al., 1999), which has a mutation in one of the components of the histone acetyl transferase Elongator complex.
Leaf morphology is the result of endogenous and exogenous factors (Tsukaya, 2005), which act in a co-ordinated manner to optimize leaf functions (Van Volkenburgh, 1999). Favourable environmental conditions, ranging from nutrient availability to physical and biotic factors, and active sensing mechanisms working in the plants can co-operatively enhance leaf growth, which is achieved through cell division initially, but prominently through cell expansion thereafter (Dale, 1988; Beemster et al., 2005).
The ‘narrow leaves’ phenotype of the Arabidopsis elo1 mutant is mainly the result of, at the cytological level, a reduced number of palisade cells (Nelissen et al., 2003, 2005) that also show an altered pattern of cell expansion. During leaf growth cell division and cell expansion may spatially and temporally overlap and, normally, both events are tightly related to carbohydrate availability. Of relevance here is the role played by sucrose (Wobus and Weber, 1999; Paul and Pellny, 2003; Gibson, 2004; Koch, 2004). It has been widely demonstrated that sucrose depletion or abundance triggers enormous metabolic changes in order to reach a new equilibrium in nutrient balance and that sucrose plays important roles in gene expression, metabolic pathways and, ultimately, plant growth and differentiation (Koch, 1996, 2004; Li et al., 2001; Paul and Pellny, 2003; Nielsen et al., 2004; Loreti et al., 2005). For instance, glucose and fructose, which originate from sucrose cleavage, are able to modulate gene expression by inducing/repressing so-called ‘famine’ and ‘feast’ genes (Koch, 1996). In this context, it has been shown that under hexose depletion, genes involved in photosynthesis and nutrient mobilization are up-regulated in ‘source’ cells, while genes involved in storage and utilization are inhibited. By contrast, under hexose abundance, storage and utilization genes are up-regulated, to keep apart nutrients as much as possible and release energy for optimum metabolic processes (Van Volkenburgh, 1999; Roitsch and González, 2004).
Using several cytological approaches, it was shown here that changes in the sucrose concentrations of the growth medium differentially affected both germination and growth in elo1 seeds and plantlets as compared with the wild-type. In addition, growth stage-based analysis revealed that no sucrose effects were evident during the first week after vernalization, thus suggesting a late stage-specific action mechanism of sucrose. Furthermore, sucrose-dependent differences between the two samples also characterized leaf morphology. Indeed, even if strong sucrose abundance (>3 %) or sucrose depletion (0 %) greatly inhibited or limited general plant growth, under different sucrose concentrations lamina growth showed opposite patterns in the two phenotypes: increasing the sucrose concentration from 0·5 to 2 % narrowed leaf lamina in elo1 mutants as compared with under standard conditions; by contrast, in wild-type lamina, growth was enhanced by increasing sucrose concentrations, mainly as a consequence of an increased cell area. Thus, it was assessed that in elo1 mutants increasing sucrose concentrations inhibited faster general growth and especially leaf expansion than in wild-type plants.
A second peculiar aspect of the results dealt with the inhibitory action of high sucrose concentrations on cell expansion in elo1 mutants. According to literature data (Nelissen et al., 2003, 2005), under standard conditions the reduction of palisade cell number in elo1 first leaf was accompanied by numerous cell gaps. However, owing to a partial compensatory mechanism, the paradermal area of these cells was larger than in the wild-type. High sucrose concentrations strongly affected cell division, resulting in an even more reduced cell number in both phenotypes, but a compensatory mechanism, leading to an enhancement of cell expansion, was active only in the wild-type. Namely, elo1 cells reduced and Ler cells increased their paradermal cell area, respectively. In addition, whatever sucrose concentrations were tested, dorsal–ventral expansion of palisade cells was reduced, thus showing that in elo1 mutants cell expansion is polarly affected.
Together, these different sucrose-related effects on elo1 and wild-type plants clearly indicate that sucrose and Elongator activity are mutually related to seed germination and plant growth. In this context, it is worth noting that the qPCR analysis for three sucrose metabolism/transport- related genes indicated that the elo1 mutation has an effect on their gene expression. In particular, these results suggest that mechanisms that import sucrose and produce carbon assimilates are differentially regulated in elo1 plants as compared with the wild-type.
On the basis of these results it is suggested that the higher germination rates and stress tolerance mechanisms to NaClO that were detected in the elo1 mutant are tightly related to the enhancement of specific metabolic pathways leading to sucrose accumulation and sugar reallocation, both necessary processes leading to radicle emergence (Gupta and Kaur, 2005; Aoki et al., 2006).
The different expression of sucrose metabolism/transport- related genes in elo1 mutants could also shed light on the cytological and ultrastructural differences observed. Indeed, elo1 cells exhibited a hypotonic vacuole filled with a dense matrix, thus indicating an impairment of water influx, which in turn could account for the lack of cell expansion. In this context, it is suggested that, owing to the mutation in the At3g11220 gene, the elo1 mutant plants no longer sense sucrose as an active metabolite or osmotic product, even if cellular concentrations are elevated. On the other hand, other ultrastructural differences, relating to the size of grana and starch grains in the chloroplasts and the massive presence of Golgi vesicles in the cytoplasm, which have been detected between Ler and elo1 plants, are all related to sucrose accumulation, catabolism or export.
Conversely, in wild-types, a functional Elongator is able to ‘sense’ intracellular sucrose levels and adjust transcription of genes involved in sucrose cleavage and hexose formation, which are known to favour cell division and are important for ATP formation, thus avoiding the establishment of inhibitory levels that may affect general growth. Further speculation may suggest a stage-specific action of Elongator at the critical germination–seedling growth switch, when photosynthesis becomes the main metabolic way that leads to sucrose formation. Elongator could ‘sense’ the intracellular energetic status and control important pathways, leading to sucrose synthesis or sucrose cleavage, thus adjusting intracellular respiratory ratios.
In conclusion, the sucrose-sensing mechanism of elo1 plants has been shown to be not comparable with that of the wild-type. Based on the data presented, it is suggested that in elo1 mutants high sucrose concentration, due to enhanced import and reallocation, could account for the stimulation of germination and inhibition of leaf growth.
ACKNOWLEDGMENTS
We thank Drs Enrico Perrotta and Enza Tenzi for technical support. This project was funded by the European Union in the frame of the PREDEC project (HPMT-CT-2000 00088) and MIUR, Italy (ex 60 % grant).
LITERATURE CITED
- Aoki N, Scofield GN, Wang X-D, Offler CE, Patrick JW, Furbank RT. Pathway of sugar transport in germinating wheat seeds. Plant Physiology. 2006;141:1255–1263. doi: 10.1104/pp.106.082719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GTS, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, et al. Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiology. 2005;138:734–743. doi: 10.1104/pp.104.053884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berná G, Robles P, Micol JL. A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics. 1999;152:729–742. doi: 10.1093/genetics/152.2.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. The Plant Cell. 2001;13:1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnops G, Jover-Gil S, Peters JL, Neyt P, De Block S, Robles P, et al. The rotunda2 mutants identify a role for the LEUNIG gene in vegetative leaf morphogenesis. Journal of Experimental Botany. 2004;55:1529–1539. doi: 10.1093/jxb/erh165. [DOI] [PubMed] [Google Scholar]
- Dale JE. The control of leaf expansion. Annual Review of Plant Plant Physiology and Plant Molecular Biology. 1988;39:267–295. [Google Scholar]
- Fellows J, Erdjument-Bromage H, Tempst P, Svejstrup JQ. The Elp2 subunit of Elongator and elongating RNA polymerase II holoenzyme is a WD40 repeat protein. Journal of Biological Chemistry. 2000;275:12896–12899. doi: 10.1074/jbc.275.17.12896. [DOI] [PubMed] [Google Scholar]
- Fichtner L, Jablonowski D, Schierhorn A, Kitamoto HKK, Stark MJR, Schaffrath R. Elongator's toxin-target (TOT) function is nuclear localization sequence dependent and suppressed by post-translational modification. Molecular Microbiology. 2003;49:1297–1307. doi: 10.1046/j.1365-2958.2003.03632.x. [DOI] [PubMed] [Google Scholar]
- Fleming AJ. The mechanism of leaf morphogenesis. Planta. 2002;216:17–22. doi: 10.1007/s00425-002-0864-8. [DOI] [PubMed] [Google Scholar]
- Fleming AJ. Formation of primordia and phyllotaxy. Current Opinion in Plant Biology. 2005;8:53–58. doi: 10.1016/j.pbi.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Frohloff F, Fichtner L, Jablonowski D, Breunig KD, Schaffrath R. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. The EMBO Journal. 2001;20:1993–2003. doi: 10.1093/emboj/20.8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. Qbase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SI. Sugar and phytohormone response pathways: navigating a signalling network. Journal of Experimental Botany. 2004;55:253–264. doi: 10.1093/jxb/erh048. [DOI] [PubMed] [Google Scholar]
- Gilbert C, Kristjuhan A, Winkler GS, Svejstrup JQ. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Molecular Cell. 2004;14:457–464. doi: 10.1016/s1097-2765(04)00239-4. [DOI] [PubMed] [Google Scholar]
- Gupta AK, Kaur N. Sugar signalling and gene expression in relation to carbohydrate metabolism under abiotic stresses in plants. Journal of Biosciences. 2005;30:761–776. doi: 10.1007/BF02703574. [DOI] [PubMed] [Google Scholar]
- Jablonowski D, Schaffrath R. Saccharomyces cerevisiae RNA polymerase II is affected by Kluyveromyces lactis zymocin. Journal of Biological Chemistry. 2002;277:26276–26280. doi: 10.1074/jbc.M203354200. [DOI] [PubMed] [Google Scholar]
- Jablonowski D, Frohloff F, Fichtner L, Stark MJR, Schaffrath R. Kluyveromyces lactis zymocin mode of action is linked to RNA polymerase II function via Elongator. Molecular Microbiology. 2001;42:1095–1105. doi: 10.1046/j.1365-2958.2001.02705.x. [DOI] [PubMed] [Google Scholar]
- Kessler S, Sinha N. Shaping up: the genetic control of leaf shape. Current Opinion in Plant Biology. 2004;7:65–72. doi: 10.1016/j.pbi.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Kim J-H, Lane WS, Reinberg D. Human Elongator facilitates RNA polymerase II transcription through chromatin. Proceedings of the National Academy of Sciences of the USA. 2002;99:1241–1246. doi: 10.1073/pnas.251672198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch K. Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Current Opinion in Plant Biology. 2004;7:235–246. doi: 10.1016/j.pbi.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Koch KE. Carbohydrate-modulated gene expression in plants. Annual Review in Plant Physiology and Plant Molecular Biology. 1996;47:509–540. doi: 10.1146/annurev.arplant.47.1.509. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Greenblatt JF. Characterization of a six-subunit holo-Elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Molecular and Cellular Biology. 2001;21:8203–8212. doi: 10.1128/MCB.21.23.8203-8212.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine R. Sucrose transporters in plants: update on function and structure. Biochimica et Biophysica Acta. 2000;1465:246–262. doi: 10.1016/s0005-2736(00)00142-5. [DOI] [PubMed] [Google Scholar]
- Li H, Shen J-J, Zheng Z-L, Lin Y, Yang Z. The Rop GTPase switch controls multiple developmental processes in Arabidopsis. Plant Physiology. 2001;126:670–684. doi: 10.1104/pp.126.2.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Poggi A, Novi G, Alpi A, Perata P. A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiology. 2005;137:1130–1138. doi: 10.1104/pp.104.057299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micol JL, Hake S. The development of plant leaves. Plant Physiology. 2003;131:389–394. doi: 10.1104/pp.015347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, Clarke JH, De Block M, De Block S, Vanderhaeghen R, Zielinski RE, et al. DRL1, a homolog of the yeast TOT4/KTI12 protein, has a function in meristem activity and organ growth in plants. The Plant Cell. 2003;15:639–654. doi: 10.1105/tpc.007062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, Fleury D, Bruno L, Robles P, De Veylder L, Traas J, et al. The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proceedings of the National Academy of Sciences of the USA. 2005;102:7754–7759. doi: 10.1073/pnas.0502600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen TH, Rung JH, Villadsen D. Fructose-2,6-bisphosphate: a traffic signal in plant metabolism. Trends in Plant Science. 2004;9:556–563. doi: 10.1016/j.tplants.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Otero G, Fellows J, Li Y, de Bizemont T, Dirac AMG, Gustafsson CM, et al. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Molecular Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- Paul MJ, Pellny TK. Carbon metabolite feedback regulation of leaf photosynthesis and development. Journal of Experimental Botany. 2003;54:539–547. doi: 10.1093/jxb/erg052. [DOI] [PubMed] [Google Scholar]
- Pozzi C, Rossini L, Agosti F. Patterns and symmetries in leaf development. Seminars in Cell & Developmental Biology. 2001;12:363–372. doi: 10.1006/scdb.2001.0265. [DOI] [PubMed] [Google Scholar]
- Roitsch T, González M-C. Function and regulation of plant invertases: sweet sensations. Trends in Plant Science. 2004;9:606–613. doi: 10.1016/j.tplants.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Slaugenhaupt SA, Gusella JF. Familial dysautonomia. Current Opinion in Genetics and Development. 2002;12:307–311. doi: 10.1016/s0959-437x(02)00303-9. [DOI] [PubMed] [Google Scholar]
- Sturm A. Invertases. Primary structures, functions and roles in plant development and sucrose partitioning. Plant Physiology. 1999;121:1–7. doi: 10.1104/pp.121.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H. Leaf development. In: Somerville CR, Meyerowitz EM, editors. The Arabidopsis book. Rockville, MD: American Society of Plant Biologists; 2002. (doi/10·1199/tab.0072; http://www.aspb.org/publications/arabidopsis. ) [Google Scholar]
- Tsukaya H. Leaf shape: genetic controls and environmental factors. International Journal of Developmental Biology. 2005;49:547–555. doi: 10.1387/ijdb.041921ht. [DOI] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proceedings of the National Academy of Sciences of the USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Ray N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3:R34. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Volkenburgh E. Leaf expansion – an integrating plant behaviour. Plant, Cell and Environment. 1999;22:1463–1473. [Google Scholar]
- Weber H, Borisjuk L, Heim U, Sauer N, Wobus U. A role for sugar transporters during seed development: molecular characterization of a hexose and a sucrose carrier in fava bean seeds. The Plant Cell. 1997;9:895–908. doi: 10.1105/tpc.9.6.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weschke W, Panitz R, Gubatz S, Wang Q, Radchuk R, Weber H, Wobus U. The role of invertases and hexose transporters in controlling sugar ratios in maternal and filial tissues of barley caryopses during early development. The Plant Journal. 2003;33:395–411. doi: 10.1046/j.1365-313x.2003.01633.x. [DOI] [PubMed] [Google Scholar]
- Winkler GS, Kristjuhan A, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proceedings of the National Academy of Sciences of the USA. 2002;99:3517–3522. doi: 10.1073/pnas.022042899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus U, Weber H. Sugars as signal molecules in plant seed development. Biological Chemistry. 1999;380:937–944. doi: 10.1515/BC.1999.116. [DOI] [PubMed] [Google Scholar]