Abstract
Women develop chronic inflammatory autoimmune diseases more often than men. The mechanisms causing the increased susceptibility are incompletely understood. Chronic immune stimulation characterizes many autoimmune disorders. We hypothesized that repeated stimulation may cause a different T cell response in women than men. Microarrays were used to compare gene expression in T cells from healthy men and women with and without repeated stimulation. Four days following a single stimulation only 25% of differentially expressed, gender-biased genes were expressed at higher levels in women. In contrast, following restimulation 72% were more highly expressed in women. Immune response genes were significantly over-represented among the genes upregulated in women and among the immune response genes, the inflammatory/cytotoxic effector genes interferon gamma (IFNG), lymphotoxin beta (LTB), granzyme A (GZMA), interleukin-12 receptor beta2 (IL12RB2), and granulysin (GNLY) were among those overexpressed to the greatest degree. In contrast, IL17A was the only effector gene more highly expressed in men. Estrogen response elements were identified in the promoters of half the overexpressed immune genes in women, and in <10% of the male biased genes. The differential expression of inflammatory/cytotoxic effector molecules in restimulated female T cells may contribute to the differences in autoimmune diseases between women and men.
Keywords: Interferon, granzyme A, granulysin, lymphotoxin B, autoimmunity, gender
Introduction
Genetic factors predispose to autoimmunity, and one of the strongest predisposing factors is female sex. Systemic lupus erythematosus (SLE), scleroderma, and rheumatoid arthritis are among the autoimmune diseases more prevalent in women than men1. Why women are more prone to autoimmunity is not completely understood. Estrogen likely contributes to the increased susceptibility and severity of autoimmunity in women 2 and some immune genes have estrogen response elements 3. However, estrogen does not completely explain the female predisposition to autoimmunity. Female predominant diseases like SLE also occur more often in prepubertal girls than boys 4, and cytokine responses are different between boys and girls 1–3 years of age 5, suggesting a role for non-hormonal factors in the female predilection to autoimmunity. Having two X chromosomes may also predispose women to autoimmunity 6, although which X chromosome genes are overexpressed in women to promote autoimmunity is also unclear. Finally, recent studies suggest that sex-specific differences in the regulation of autosomal genes independent of estrogen may also contribute to gender-specific diseases 1. What these genes are is also unknown.
T cells are critical to the development of many forms of autoimmunity. T cells are sufficient to cause lupus-like autoimmunity in animal models 7, 8, and T cell responses are implicated in the pathogenesis of human lupus 9, rheumatoid arthritis 10 and other autoimmune diseases 11. Further, the differentiation of T cells into distinct subsets such as Th1, Th2, Th17, naïve, memory, regulatory (Treg) and others, each with distinct functions, suggests that gender-specific differences in T cell gene expression, perhaps due to gender-specific subset differentiation, could also contribute to the female predisposition to autoimmunity. The repeated T cell stimulation that occurs in chronic autoimmunity could promote differentiation into subsets with distinct repertoires in men and women.
We hypothesized that T cells from men and women express different levels or repertoires of effector molecules under conditions of repeated stimulation such as that occurring in chronic relapsing autoimmune diseases. As an initial test of this hypothesis we used expression microarrays to compare gene expression patterns in T cells from healthy young men and women following two rounds of stimulation. Differentially expressed immune genes were identified, and overexpression of effector molecules was validated by RT-PCR and protein analyses. The overexpressed genes were further analyzed for estrogen response elements as well as chromosomal location.
Results
Gene expression in stimulated and restimulated male and female T cells
To approximate the repeated stimulation characterizing autoimmune responses, PBMC from 3 pairs of healthy men and women were stimulated with PHA, cultured for 4 days, then restimulated or not with PMA and ionomycin. Six hours later T cells were purified and gene expression compared using microarrays. Using the Genomatix ChipInspector program and a false discovery rate (FDR) of 4%, analysis of the unrestimulated T cells revealed a total of 6295 significantly up or down regulated probes, corresponding to 305 distinct transcripts that were differentially expressed between women and men. With a FDR of 4%, approximately 12 of the 305 genes are expected to be false-positive. Interestingly, men overexpressed more genes than women 4 days following the single stimulation. The men overexpressed 228 of the 305 genes (75%), while 77 of the 305 genes (25% of the total) were female biased.
In contrast, when the T cells were restimulated and gene expression similarly compared, a total of 24791 probes were significantly up or down regulated, corresponding to 1953 differentially expressed genes, of which 74 are expected to be false positives. Also in contrast to the unrestimulated cells, 1408 (72%) of the dimorphic genes were female biased while 545 (28%) were male biased, indicating a greater response in the female cells. Lists of the sexually dimorphic genes in unrestimulated and restimulated T cells are shown in Supplemental Table S1 and Supplemental Table S2.
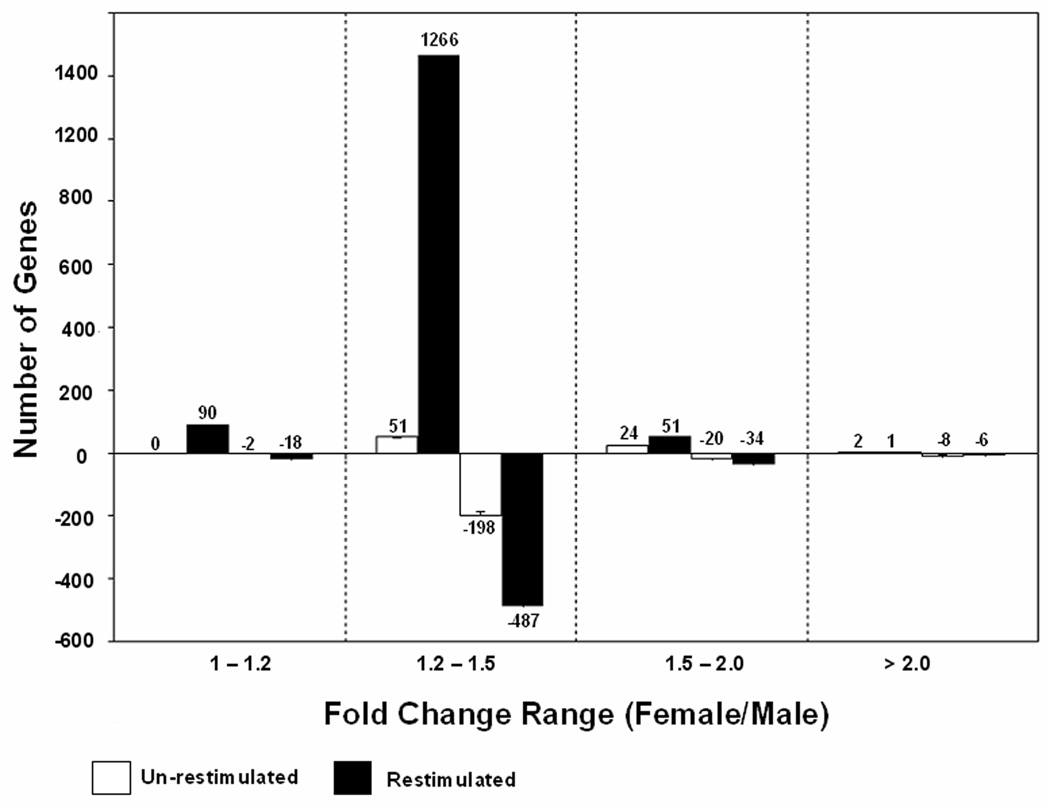
Figure 1 illustrates the distribution of the sexually dimorphic genes, categorized by the relative magnitude of the difference in expression (fold change 1–1.2, 1.21–1.5, 1.51–2.0, and >2.0) in unrestimulated and restimulated T cells. In this figure a positive number is used to indicate a greater increase in T cells from women, while a negative number indicates a greater increase in T cells from men. Regardless of the sex of the donor, the majority of the sexually dimorphic genes had a fold change difference in the range of 1.2–1.5. The functional significance of the relatively few genes with a fold-change difference of only 1.0–1.2 is uncertain, and thus were not further analyzed.
FIGURE 1. Expression profile of T cell genes differentially expressed between men and women.
Bars shown greater than zero represent the numbers of genes expressed at higher levels in women than men, while the negative values represent the numbers of genes expressed more highly in men. Error bars represent the FDR of 4%.
Quality controls included comparison of gender-specific sex chromosome gene transcript levels. XIST (X (inactive)-specific transcript) is expressed exclusively from the inactive X in women, and was among the most strongly female biased genes (7.9 fold increase relative to men). Similarly, the Y chromosome genes EIF1AY (fold increase 6.7 men/women), and RPS4Y (fold increase 15.5 men/women) were among the male biased genes with greatest differences (Supplementary Table S1&Supplementary Table S2). The majority of the remaining differentially expressed genes were localized to autosomal chromosomes.
The functional significance of the differentially expressed genes was estimated using Gene Ontology (GO) terms. Since the magnitude of the biologic effect is likely to be proportional to the magnitude of the difference in immune effector molecule levels, those genes with a ≥ 1.4-fold difference in the level of expression between the sexes were analyzed as likely to be most reflective of functional differences. Using this criterion, 109 of the 305 unrestimulated transcripts, and 289 of the 1953 restimulated transcripts, were classified by GO terms. The broad Biological Process filter identified 95 differentially expressed genes in the unrestimulated cells, and 249 in the restimulated cells. These genes were then analyzed for GO subcategories. The GO categories were only considered significant if they contained at least 4 genes and their Z-score was ≥1.65, corresponding to p ≤ 0.05, or inclusion in the top 5% of overrepresented genes.
Gender specific differences in restimulated T cell gene expression were found in multiple GO categories. The categories are shown in Table 1, ranked by the number of genes included in each relative to restimulated female T cells, which contained the greatest number of genes affected. The 150 female-biased biological process gene transcripts were most significantly (Z ≥ 4) overrepresented in the following GO classifications: response to stimulus, immune response, immune system process, and signal transduction (Table 1). In contrast, only cell communication and immune system process were enriched to this degree among the male biased genes in the restimulated cells. In unrestimulated cells, the categories of response to stimulus, cell differentiation, immune system process, immune response, cell death and apoptosis were highly (Z >4) enriched in the women, while only immune system process and immune response were enriched to this degree in the men.
Table 1.
Major GO-rankings for genes differentially expressed between women and men
| Term | GoID | Unrestimulated | Stimulated | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Female Higher |
Male Higher | Female Higher |
Male Higher | ||||||
| No of Genes |
Z score |
No of Genes |
Z score |
No of Genes |
Z score |
No of Genes |
Z score |
||
| biological_process | GO:0008150 | 55 | 2.71 | 40 | 2.26 | 150 | 5.5 | 99 | 3.21 |
| cellular metabolic process | GO:0044237 | 35 | 2.85 | 17 | −0.17 | 78 | 2.28 | 56 | 1.49 |
| signal transduction | GO:0007165 | 18 | 2.16 | 10 | 0.64 | 53 | 4.59 | 31 | 2.41 |
| response to stimulus | GO:0050896 | 21 | 4.81 | 11 | 2.21 | 49 | 6.32 | 22 | 1.92 |
| cell communication | GO:0007154 | 21 | 2.65 | 10 | 0.29 | 36 | 2.97 | 60 | 4.4 |
| transcription | GO:0006350 | 6 | −0.71 | 7 | 0.56 | 30 | 2.15 | 15 | 0.15 |
| regulation of gene expression | GO:0010468 | 6 | −0.83 | 7 | 0.44 | 28 | 1.45 | 17 | 0.53 |
| cell differentiation | GO:0030154 | 17 | 4.46 | 7 | 1.16 | 25 | 2.11 | 19 | 2.26 |
| intracellular signaling cascade | GO:0007242 | 9 | 1.87 | 22 | 3.35 | 15 | 2 | ||
| immune system process | GO:0002376 | 10 | 4.1 | 9 | 4.67 | 21 | 4.7 | 15 | 4.09 |
| immune response | GO:0006955 | 9 | 4.45 | 8 | 4.91 | 20 | 5.68 | 11 | 3.79 |
| cell surface receptor linked signal transduction | GO:0007166 | 6 | 0.48 | 4 | 0.2 | 19 | 1.67 | 14 | 1.67 |
| cell death | GO:0008219 | 11 | 4.68 | 5 | 1.91 | 15 | 2.52 | 14 | 3.65 |
| apoptosis | GO:0006915 | 10 | 4.34 | 4 | 1.36 | 13 | 2.04 | 12 | 3.03 |
Gene sets with fold change ≥1.4 were analyzed using Genomatix BiblioSphere software to identify overrepresented GO groups.
The 3 categories with the greatest enrichment in restimulated T cells were all in women. In descending order these were “response to stimulus”, “immune response”, and “immune system process”. Genes in the category “immune response” were enriched in restimulated T cells from women relative to men (Z scores 5.68 in women vs 3.79, 20 vs 11 genes in men), and “immune system process” was also enriched in women (Z scores 4.7 vs 4.09, 21 vs 15 genes, women vs men).
The GO functional subcategories of genes involved in immune function are shown in Table 2, again ranked by number of genes identified. The top 4 categories for both men and women were “immune system process”, “immune response”, “response to external stimulus” and “defense response”, and all contained more genes and were more significantly enriched in restimulated T cells from women than the men. The categories “regulation of immune system process”, “immune system development” and “innate immune response” were approximately equal in men and women, but contained relatively few genes (4–5) in both sexes. Due to low numbers of genes, further sub-grouping of immune genes was not meaningful for the list of genes from unstimulated cells. The immune genes in the category “response to stimulus” were largely overlapping with these groups, and are shown in Supplementary Table S3 for the interested reader.
Table 2.
GO analysis of immune genes
| Term | GoID | Female Higher |
Male Higher | ||
|---|---|---|---|---|---|
| No of Genes |
Z score |
No of Genes |
Z score |
||
| immune system process | GO:0002376 | 21 | 4.70 | 15 | 4.09 |
| immune response | GO:0006955 | 20 | 5.68 | 11 | 3.79 |
| response to external stimulus | GO:0009605 | 18 | 4.90 | 10 | 2.84 |
| defense response | GO:0006952 | 14 | 3.84 | 10 | 3.34 |
| regulation of immune system process | GO:0002682 | 5 | 2.71 | 4 | 2.76 |
| immune system development | GO:0002520 | 4 | 1.58 | 4 | 2.40 |
| innate immune response | GO:0045087 | 4 | 2.83 | - | - |
Sexually dimorphic genes from the “immune response” category with a ≥ 1.4 fold difference in expression between T cells from women and men are listed in Table 3 and Table 4 respectively. Twenty genes exhibited female-biased expression (Table 3) while 11 genes exhibited male-biased expression (Table 4). Interestingly the women expressed higher levels of the pro-inflammatory/cytotoxic effector molecules granulysin, IFN-γ, granzyme A, and lymphotoxin-β, as well as IL-12 receptor β2 involved in Th1 responses (Table 3). In contrast, men only overexpressed the pro-inflammatory cytokine IL17A, and had higher levels of the Th2 cytokines IL5 and IL10 (Table 4). The women also had higher levels of the chemokines CX3CL1, CX3CL2 and the cytokines IL1F5 and IL16, while men had higher levels of the chemokines XCL1, CXCL9 and CXCL10.
Table 3.
Female-biased immune genes with fold change ≥1.4
| Gene Symbol | Gene ID |
Locus | Gene Name | Fold | EREF |
|---|---|---|---|---|---|
| IFNG | 3458 | 12q14 | interferon, gamma | 1.8 | N |
| GZMA | 3001 | 5q11 | granzyme A(cytotoxic T-lymphocyte-associated serine esterase 3) | 1.7 | N |
| IL12RB2 | 3595 | 1p31 | interleukin 12 receptor, beta 2 | 1.7 | N |
| GNLY | 10578 | 2p11 | granulysin | 1.7 | Y |
| IL1F5 | 26525 | 2q14 | interleukin 1 family, member 5 (delta) | 1.5 | Y |
| CX3CL1 | 6376 | 16q13 | chemokine (C-X3-C motif) ligand 1 | 1.5 | N |
| MIST | 116449 | 4p16 | mast cell immunoreceptor signal transducer | 1.5 | N |
| LTB | 4050 | 6p21 | lymphotoxin beta (TNF superfamily, member 3) | 1.4 | Y |
| TRAT1 | 50852 | 3q13 | T cell receptor associated transmembrane adaptor 1 | 1.4 | N |
| CXCL2 | 2920 | 4q21 | chemokine (C-X-C motif) ligand 2 | 1.4 | N |
| IL16 | 3603 | 15q26 | interleukin 16 (lymphocyte chemoattractant factor) | 1.4 | Y |
| IFITM2 | 10581 | 11p15 | interferon induced transmembrane protein 2 (1–8D) | 1.4 | N |
| TLR10 | 81793 | 4p14 | toll-like receptor 10 | 1.4 | Y |
| DDX58 | 23586 | 9p12 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | 1.4 | Y |
| SPINK5 | 11005 | 5q32 | serine peptidase inhibitor, Kazal type 5 | 1.4 | Y |
| OAS3 | 4940 | 12q24 | 2'-5'-oligoadenylate synthetase 3, 100kDa | 1.4 | Y |
| OAS1 | 4938 | 12q24 | 2',5'-oligoadenylate synthetase 1, 40/46kDa | 1.4 | Y |
| IFI6 | 2537 | 1p35 | interferon, alpha-inducible protein 6 | 1.4 | N |
| TNFRSF11A | 8792 | 18q22 | tumor necrosis factor receptor superfamily, member 11a, | 1.4 | N |
| MBP | 4155 | 18q23 | myelin basic protein | 1.4 | Y |
Table 4.
Male-biased immune genes with fold change ≥1.4
| Gene Symbol | Gene ID |
Locus | Gene Name | Fold | EREF |
|---|---|---|---|---|---|
| AIM2 | 9447 | 1q22 | absent in melanoma 2 | −1.4 | N |
| FYN | 2534 | 6q21 | FYN oncogene related to SRC, FGR, YES | −1.4 | Y |
| IRF8 | 3394 | 16q24 | interferon regulatory factor 8 | −1.4 | N |
| SWAP70 | 23075 | 11p15 | SWAP-70 protein | −1.4 | N |
| IL17A | 3605 | 6p12 | interleukin 17A | −1.4 | N |
| IL10 | 3586 | 1q31 | interleukin 10 | −1.5 | N |
| IL5 | 3567 | 5q31 | interleukin 5 (colony-stimulating factor, eosinophil) | −1.5 | N |
| XCL1 | 6375 | 1q23 | chemokine (C motif) ligand 1 | −1.7 | N |
| CXCL10 | 3627 | 4q21 | chemokine (C-X-C motif) ligand 10 | −1.7 | N |
| CXCL9 | 4283 | 4q21 | chemokine (C-X-C motif) ligand 9 | −1.8 | N |
| HLA-DQA1 | 3117 | 6p21 | major histocompatibility complex, class II, DQ alpha 1 | −2 | N |
Sex hormones regulate gene expression and may play an important role in predisposing women to autoimmunity 12. We therefore analyzed the gender-specific genes for estrogen response elements. Ten of the 20 female biased immune gene promoters exhibited estrogen response element family (EREF) binding sites in the promoter region (Table 3), suggesting that some, but not all genes differentially expressed in women may be directly influenced by hormone levels. In contrast, only 1 of the 11 male biased genes had an EREF binding site (Table 4).
We also considered the possibility that X chromosome genes may be overexpressed in women due to processes such as X chromosome demethylation or skewing, as has been reported in some female-predominant autoimmune diseases 6, 13. However, none of the immune genes overexpressed in restimulated T cells were encoded on the X chromosome (Table 3).
Validation of gender-biased genes
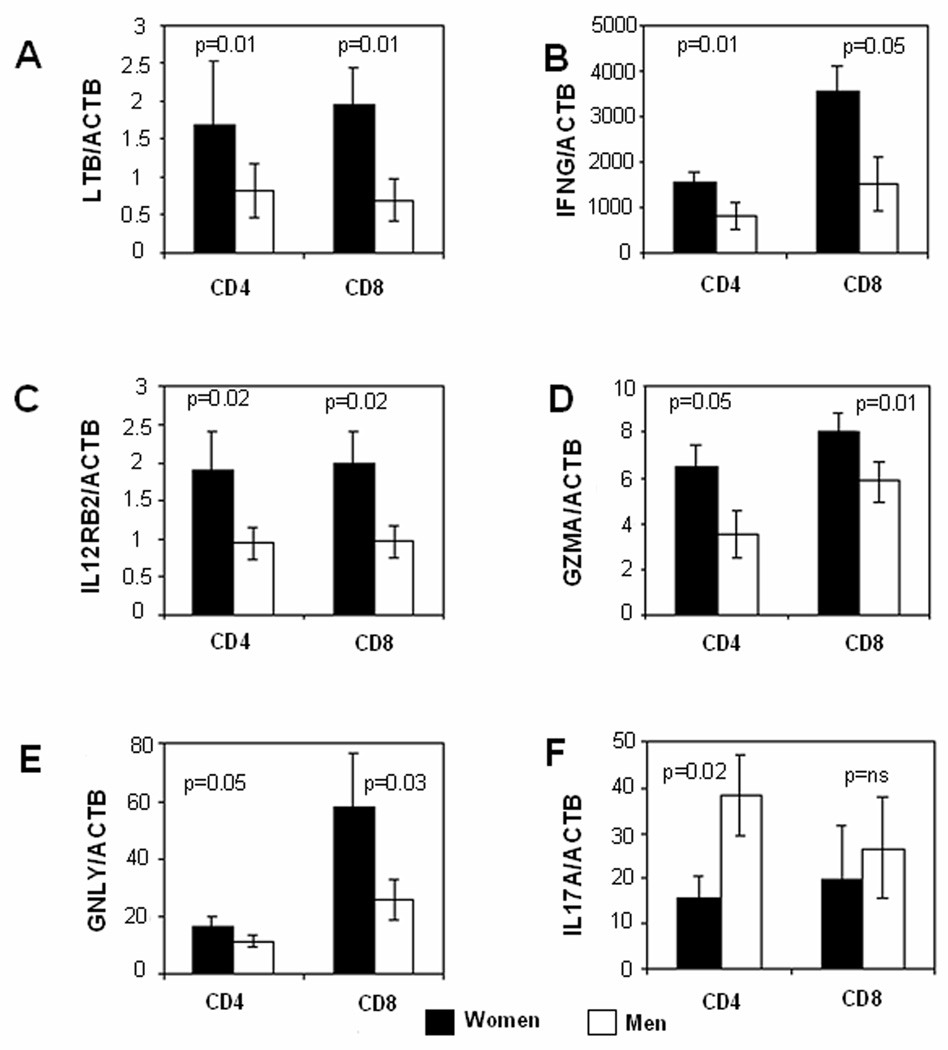
As noted above, the pro-inflammatory/cytotoxic molecules IFNG, GNLY, IL12RB2, GZMA and LTB were among the female-biased immune genes detected by the microarrays. The microarray results were validated by comparing levels of these transcripts in restimulated CD4+ and CD8+ cells from 10 pairs of healthy men and women. PBMC were similarly isolated from the men and women, stimulated with PHA, then restimulated for 6 hours with PMA + ionomycin on day 4 as before. CD4+ and CD8+ T cells were then purified, and LTB, IFNG, IL12RB2, GZMA and GNLY transcripts measured by quantitative RT-PCR. mRNA levels of all 5 genes were significantly higher in women compared to men. LTB expression was significantly higher (p = 0.01) in women in both CD4+ and CD8+ cells (Fig. 2A). Restimulation similarly caused a female-specific increase in INFG expression in both CD4+ (p=0.01) and CD8+ (p =0.05) cells (Fig 2B). Significant (p=0.02) female-biased expression of IL12RB2 was also seen in both CD4+ and CD8+ cells (Fig 2C), and GZMA similarly exhibited higher expression levels in women for both CD4+ (p= 0.05) and CD8+ (p = 0.01) T cells (Fig 2D). GNLY was expressed at higher levels in CD8+ than CD4+ cells, although statistically female-biased expression was observed in both CD4+ (p =0.02) and CD8+ (p=0.03) T cells (Fig 2E). Figure 2F confirms that men overexpress IL17A in CD4+ (p=0.02) but not CD8+ T cells relative to women.
FIGURE 2. Expression levels of pro-inflammatory transcripts in restimulated T cells from men and women.
PBMC from 10 racially matched male-female pairs were stimulated with PHA, cultured 4 days, then restimulated for 6 hours with PMA + ionomycin. CD4+ and CD8+ T cells were isolated and expression levels of (A) LTB, (B) IFNG, (C) IL12RB2, (D) GZMA, (E) GNLY and (F) IL17A were measured by qRT- PCR relative to β-actin. Results are presented as the mean±SEM of the 10 determinations per group. Dark bars represent transcript levels in the women, and light bars transcript levels in the men.
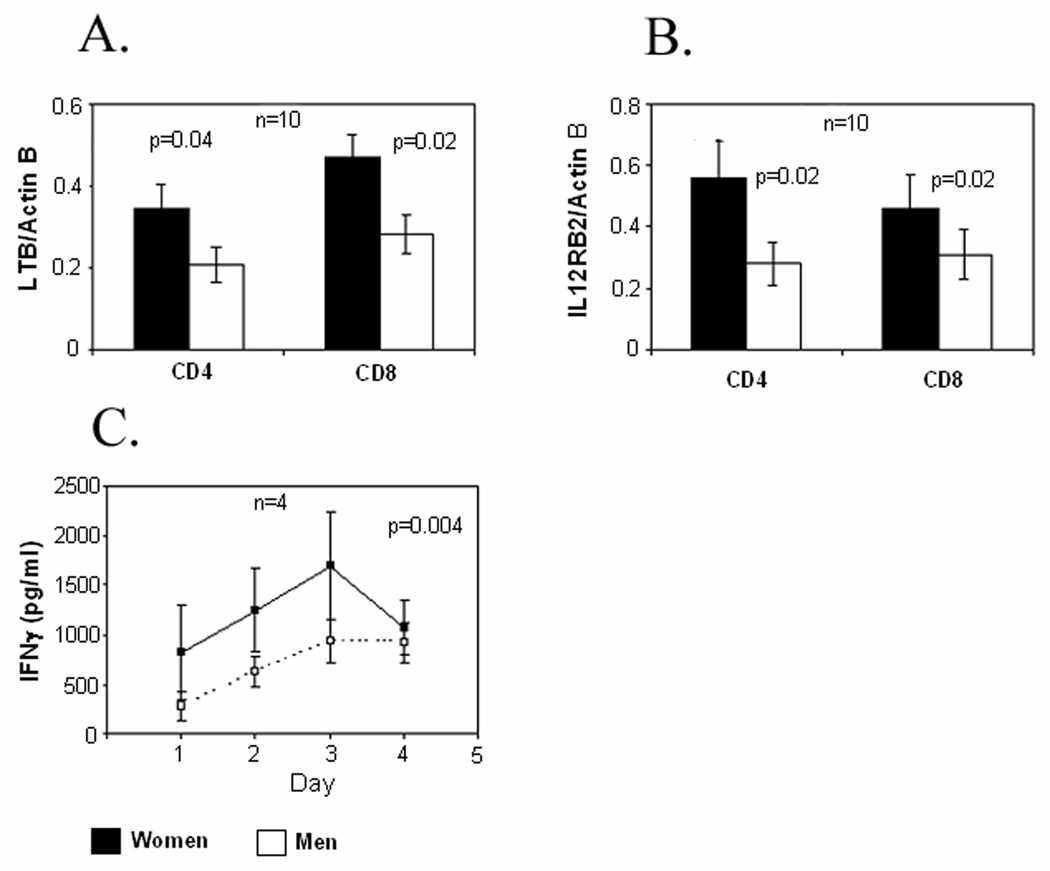
The same restimulation protocol was used to compare LTB, IL12RB2, and IFN-γ protein levels in T cells from men and women. LTB and IL12RB2 were quantitated by immunoblotting using protein isolated from the same restimulated CD4+ and CD8+ T cells from 10 men and 10 women used for the experiments shown in Fig 2, while IFN-γ was measured by ELISA in the supernatants of serially restimulated T cell aliquots from 4 additional men and women. Figure 3A shows that women have greater levels of LTB in restimulated CD4+ and CD8+ T cells relative to the men, and Figure 3B confirms a similar increase in IL12RB2 in CD4+ and CD8+ T cells in women relative to men. Figure 3C shows that female T cells secreted greater amounts of IFN-γ relative to the men, with the greatest secretion on day 3 following the initial stimulation (p = 0.004 by ANOVA).
FIGURE 3. LTB, IL12RB2 and IFN-γ protein levels in restimulated T cells from men and women.
(A) LTB and (B) IL12RB2 were measured by immunoblotting in the same restimulated CD4+ and CD8+ cells shown in figure 2. Results again are presented as the mean±SEM of the 10 determinations per group. (C) IFN-γ release by similarly restimulated T cells from 4 additional racially matched male-female pairs was measured by ELISA on the days indicated. Results again represent the mean±SEM of the 4 determinations per group.
Discussion
Microarray approaches have demonstrated sexually dimorphic gene expression in mouse liver, adipose tissue, brain and muscle 14. To the best of our knowledge though, the present studies represent the first use of microarrays to compare T cell gene expression between men and women, particularly under conditions of repeated stimulation. Following restimulation, T cells from women overexpressed a greater number of genes than T cells from men, and the inflammatory/cytotoxic effector molecules IFNG, GNLY, GZMA, IL12RB2 and LTB were the among the most highly overexpressed (1.4–1.8 times higher than men). Since the number of male-female pairs studied by array was limited, overexpression of these genes was confirmed in a separate cohort of men and women using RT-PCR as well as immunoblotting and ELISAs.
In contrast, the only pro-inflammatory gene overexpressed to the same degree in restimulated T cells from men was IL17A (1.4 times higher than in women). IL17 defines a pro-inflammatory T cell subset found in autoimmune diseases like rheumatoid arthritis and multiple sclerosis 15 and could contribute to the pathogenesis of these conditions in men, although these diseases still have a female predominance 1. Interestingly, the arrays also indicted overexpression of the anti-inflammatory Th2 gene IL10 and the Th2 cytokine IL5 in men (1.5-fold relative to women), suggesting a more mixed T cell effector response in men. In support of this, UV-induced immune suppression is greater in men, and mediated by IL-10 16. Overall, these results suggest a greater inflammatory/cytotoxic T cell response in restimulated T cells from women than men, at least within the age range and time frame tested.
Sex-specific differences in gene expression are not unique to humans or the immune system. Sex-specific differences in autosomal gene regulation have also been described in worms, flies, fish, rodents and primates. Others have observed that sex-biased genes tend to evolve rapidly at the DNA sequence level 17, and differences in gene regulation between sexes tend to be evolutionarily conserved 1, 18, 19 implying functional importance. Such differences have been proposed to contribute to other gender-specific diseases such as hypertension and schizophrenia1. Women may also be predisposed to some diseases due to gender-related differences in exposure to environmental agents, differences in metabolism of exogenous substances, and to differences in behavior 20. Sex specific differences in T cell gene regulation such as those reported here and by others 1 likely also contribute to the development of autoimmunity in women. Why women might have evolved a more inflammatory T cell response is unclear. However, a stronger inflammatory response during the years of peak estrogen levels may help women resist infections during their peak childbearing years as proposed by others 21.
The mechanisms causing differential T cell gene expression between men and women are not completely understood, but estrogen likely plays a role. In the present studies half the highly expressed gender-biased genes in restimulated female T cells had an EREF, suggesting that estrogen contributes directly to the overexpression in some of the genes detected. Further, estrogen receptor-α (ER-α) can also exert its effects indirectly by binding transcription factors such as AP-1 or SP1, or by forming signaling complexes at the plasma membrane with proteins such as Src 22, and so could contribute to overexpression of the female biased genes lacking an EREF. Future studies using estrogen inhibition/stimulation and similar gene surveys would help address this question.
T cells also differentiate throughout life into subsets such as Th1, Th2, Th17, Treg, naïve, memory and others, and gender-biased differentiation could potentially contribute differences in gene expression in women. In support of this, others have reported that healthy women have greater numbers of CD4+CD45RO+ “memory” T cells 23, which are functionally diverse and can develop within 72 hours 24, thus potentially contributing to a difference in gene expression between women and men such as that observed in the present study. However, to date no study as systematically tested whether estrogen alters T cell repertoire or alters thymic deletion of autoreactive T cells 22.
Other mechanisms proposed for female biased autoimmunity include X chromosome skewing, which characterizes some forms of female-biased autoimmunity such as thyroiditis 6, and reactivation of immune genes on the silenced X chromosome 13. However, none of the highly expressed gender biased genes detected in this study were encoded on the X chromosome, arguing against these mechanisms occurring in normal women.
In summary, these studies indicate that T cell responses can differ between men and women particularly when restimulated. The overexpression of inflammatory/cytotoxic effector molecules in women could contribute to the development and severity of autoimmune diseases such as lupus in women.
Materials and Methods
Subjects
Healthy men and women ages 25–35 years were recruited by advertising from the general population at the University of Michigan (Ann Arbor, MI). All procedures involving human subjects were reviewed and approved by the University of Michigan Institutional Review Board for Human Subject Research.
Cell culture
PBMC were isolated from venous blood of pairs of age and racially matched healthy donors using Ficoll-Paque (GE Healthcare, Uppsula, Sweden) density gradient centrifugation, stimulated with 1 µg/ml phytohemaglutinin (PHA) (Remel, Lenexa, KS) and cultured at 37°C in a 5% CO2 balanced air atmosphere in RPMI 1640 media (Hyclone, Logan, UT) supplemented with 10% FBS (Gibco, Grand Island NY), 100 U/ml penicillin (Gibco) and 100 µg/ml streptomycin. At the times indicated, the cells were washed and restimulated or not with 10 ng/ml PMA (Sigma Chemical Co, St Louis, MO) and 1 µg/ml ionomycin (Sigma). Six hours later the CD4+ and CD8+ cells were isolated using magnetic bead selection (Miltenyi Biotech, Auburn, CA) for gene expression assays.
RNA and protein isolation
T cell RNA was extracted with the Qiagen RNAeasy Mini kit (Valencia CA) and the RNase-free DNase set following the manufacturer’s instructions. For simultaneous RT-PCR and the protein analysis, DNA, RNA, and protein were purified from CD4+ and CD8+ T cells using the Qiagen All Prep DNA/RNA/Protein Mini Kit according to manufacturer's instructions.
Microarray analyses
RNA samples were analyzed on Affymetrix (Santa Clara, CA) GeneChip Human Genome Plus 2.0 (HG-133 Plus 2.0) microarrays by the University of Michigan Comprehensive Cancer Center (UMCCC) Affymetrix and Microarray Core Facility http://www.umich.edu/~caparray/). The resulting data were then analyzed using the Genomatix (http://www.genomatix.de) ChipInspector program. Differential gene expression was detected using men as the control, and a false discovery rate (FDR) ≤ 4% was applied to identify differentially expressed genes. Microarray data mining was performed using BiblioSphere, Gene2Promoter and GEMS-Launcher applications of the Genomatix software suite, Gene Ontology (GO) classifications were performed using the BiblioSphere Biological Process filter. To identify potential Estrogen Response Elements Family (EREF) binding sites, promoter sequences flanking the transcriptional start sites (−500 kbp to + 100 bp) were extracted using Gene2Promoter and queried for potential ER sites.
Real Time quantitative PCR
The RNA from the isolated CD4+ and CD8+ cells converted to cDNA synthesis with the Invitrogen (Carlsbad, CA) SuperScript first strand synthesis kit then the cDNA was subjected to real-time PCR. Roche (Basle) FastStart universal SYBR green master mix containing 1 µl template cDNA and 0.5 µM forward and reverse primers in a total volume of 20 µl was annealed at 56 °C for a total of 40 cycles. The fold change of expression was calculated using β-actin as an internal reference gene. The gene-specific primers used to are shown in Table 5.
Table 5.
Gene specific primer sequences
| Primer | Sequence |
|---|---|
| ACTB | f- GGA CTT CGA GCA AGA GAT GG |
| r- AGC ACT GTG TTG GCG TAC AG | |
| GZMA | f- ATTCTGGAAGCCCTTTGTTGTGCG |
| r- AGAATATAGACACCAGGCCCACGA | |
| IFNG | f- TGAACTGTCGCCAGCAGCTAAA |
| r- AGGCAGGACAACCATTACTGGGAT | |
| LTB | f- ATTACCTCTACTGTCTCGTCGGCT |
| r- TCCAGCACTGGAGTCACCGTCT | |
| GNLY | f- AAACACAGGAGCTGGGCCGTGACTA |
| r- GGTCGCAGCATTGGAAACACTTCT | |
| IL12RB2 | f- CTGTGAACGCTGAGCACACGATTT |
| r- TGCTGTGTCGCCTTGAGCAAGATA | |
| IL17A | f- AATCTCCACCGCAATGAGGA |
| r- ACGTTCCCATCAGCGTTGA |
Immunoblotting
Protein was isolated using an Qiagen All-Prep kit, resuspended in 5% SDS/water and quantitated by absorbance at 280 nm using a Nanodrop spectrophotometer (Nanodrop Technologies Willington, DE). Ten to 20 µg of protein/sample was loaded onto a discontinuous 5–15% gradient SDS-PAGE gel (Bio-Rad, Los Angeles, CA). Following electrophoretic separation, the proteins were transferred to polyvinylidene difluoride (PVDF) filter membranes (Millipore, Bedford, MA) and blocked with 5% non-fat milk (Carnation, Glendale, CA) in 20 mM Tris-buffered saline containing 0.05% Tween-20 (TBST) buffer overnight at 4°C. The membrane was washed thrice with TBST then incubated with primary antibody overnight at 4°C. The following antibodies from Santa Cruz Biotechnology (Santa Cruz, CA) were used at a 1:200 dilution in 5% non-fat dry milk in TBST: Goat anti-actin (SC1616), goat anti-LTB (SC23561), goat anti-IL12Rb2 (SC18648), and goat anti-granulysin (SC16968). Binding of the primary antibodies was detected using a 1:2,000 horseradish peroxidase-conjugated donkey anti-goat (Santa Cruz). Signal detection was performed with SuperSignal chemiluminescent substrate (Pierce, Rockford, IL) and X-Omatic X-ray film following the manufacturer’s instructions. For densitometry analysis, protein profiles were digitized using Canoscan (Canon, Lake Success, NY) software followed by quantification of the bands using UN-SCAN-IT gel 6.1 software (Silk Scientific, Orem, UT). The protein bands were then normalized to the β-actin band.
ELISAs
Secreted IFN-γ was measured using a BD Pharmingen (San Jose CA) OPTI-EIA Human Gamma Interferon ELISA kit and reference recombinant human IFN-γ according to the manufacturer’s instructions.
Statistical analysis
Apart from microarray analyses, Student's t-or and ANOVA as appropriate were used to determine the significance of differences between groups. Results are presented as the mean ± SEM.
Supplementary Material
Acknowledgements
The authors thank Ms. Cindy Bourke for her expert secretarial assistance. This work was supported by PHS grants AR42525, AG25877 and ES015214, and a Merit grant from the Dept. of Veterans Affairs.
Footnotes
Supplementary information is available at the Genes & Immunity’s website (http://www.nature.com/gene/index.html)
References
- 1.Ober C, Loisel DA, Gilad Y. Sex-specific genetic architecture of human disease. Nat Rev Genet. 2008;9(12):911–922. doi: 10.1038/nrg2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roubinian JR, Talal N, Greenspan JS, Goodman JR, Siiteri PK. Effect of castration and sex hormone treatment on survival, anti-nucleic acid antibodies, and glomerulonephritis in NZB/NZW F1 mice. J Exp Med. 1978;147(6):1568–1583. doi: 10.1084/jem.147.6.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, McMurray RW. Effects of estrogen receptor subtype-selective agonists on autoimmune disease in lupus-prone NZB/NZW F1 mouse model. Clin Immunol. 2007;123(2):219–226. doi: 10.1016/j.clim.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Huang JL, Yao TC, See LC. Prevalence of pediatric systemic lupus erythematosus and juvenile chronic arthritis in a Chinese population: a nation-wide prospective population-based study in Taiwan. Clin Exp Rheumatol. 2004;22(6):776–780. [PubMed] [Google Scholar]
- 5.Uekert SJ, Akan G, Evans MD, Li Z, Roberg K, Tisler C, et al. Sex-related differences in immune development and the expression of atopy in early childhood. J Allergy Clin Immunol. 2006;118(6):1375–1381. doi: 10.1016/j.jaci.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 6.Invernizzi P, Pasini S, Selmi C, Miozzo M, Podda M. Skewing of X chromosome inactivation in autoimmunity. Autoimmunity. 2008;41(4):272–277. doi: 10.1080/08916930802024574. [DOI] [PubMed] [Google Scholar]
- 7.Sawalha AH, Jeffries M, Webb R, Lu Q, Gorelik G, Ray D, et al. Defective T-cell ERK signaling induces interferon-regulated gene expression and overexpression of methylation-sensitive genes similar to lupus patients. Genes Immun. 2008;9(4):368–378. doi: 10.1038/gene.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yung RL, Quddus J, Chrisp CE, Johnson KJ, Richardson BC. Mechanism of drug-induced lupus. I. Cloned Th2 cells modified with DNA methylation inhibitors in vitro cause autoimmunity in vivo. J Immunol. 1995;154(6):3025–3035. [PubMed] [Google Scholar]
- 9.Richardson B. Primer: epigenetics of autoimmunity. Nat Clin Pract Rheumatol. 2007;3(9):521–527. doi: 10.1038/ncprheum0573. [DOI] [PubMed] [Google Scholar]
- 10.Fox DA. The role of T cells in the immunopathogenesis of rheumatoid arthritis: new perspectives. Arthritis Rheum. 1997;40(4):598–609. doi: 10.1002/art.1780400403. [DOI] [PubMed] [Google Scholar]
- 11.Korman BD, Kastner DL, Gregersen PK, Remmers EF. STAT4: genetics, mechanisms, and implications for autoimmunity. Curr Allergy Asthma Rep. 2008;8(5):398–403. doi: 10.1007/s11882-008-0077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy HS, Sun Q, Murphy BA, Mo R, Huo J, Chen J, et al. Tissue-specific effect of estradiol on endothelial cell-dependent lymphocyte recruitment. Microvasc Res. 2004;68(3):273–285. doi: 10.1016/j.mvr.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J Immunol. 2007;179(9):6352–6358. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- 14.Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, et al. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16(8):995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korn T, Oukka M, Kuchroo V, Bettelli E. Th17 cells: effector T cells with inflammatory properties. Semin Immunol. 2007;19(6):362–371. doi: 10.1016/j.smim.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Damian DL, Patterson CR, Stapelberg M, Park J, Barnetson RS, Halliday GM. UV radiation-induced immunosuppression is greater in men and prevented by topical nicotinamide. The Journal of investigative dermatology. 2008;128(2):447–454. doi: 10.1038/sj.jid.5701058. [DOI] [PubMed] [Google Scholar]
- 17.Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 2007;8(9):689–698. doi: 10.1038/nrg2167. [DOI] [PubMed] [Google Scholar]
- 18.Reinius B, Saetre P, Leonard JA, Blekhman R, Merino-Martinez R, Gilad Y, et al. An evolutionarily conserved sexual signature in the primate brain. PLoS Genet. 2008;4(6) doi: 10.1371/journal.pgen.1000100. e1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Sturgill D, Parisi M, Kumar S, Oliver B. Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature. 2007;450(7167):233–237. doi: 10.1038/nature06323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lockshin MD. Sex ratio and rheumatic disease. Autoimmun Rev. 2002;1(3):162–167. doi: 10.1016/s1568-9972(02)00034-4. [DOI] [PubMed] [Google Scholar]
- 21.Ansar Ahmed S, Penhale WJ, Talal N. Sex hormones, immune responses, and autoimmune diseases. Mechanisms of sex hormone action. Am J Pathol. 1985;121(3):531–551. [PMC free article] [PubMed] [Google Scholar]
- 22.Pernis AB. Estrogen and CD4+ T cells. Curr Opin Rheumatol. 2007;19(5):414–420. doi: 10.1097/BOR.0b013e328277ef2a. [DOI] [PubMed] [Google Scholar]
- 23.Bisset LR, Lung TL, Kaelin M, Ludwig E, Dubs RW. Reference values for peripheral blood lymphocyte phenotypes applicable to the healthy adult population in Switzerland. Eur J Haematol. 2004;72(3):203–212. doi: 10.1046/j.0902-4441.2003.00199.x. [DOI] [PubMed] [Google Scholar]
- 24.McKinstry KK, Strutt TM, Swain SL. The effector to memory transition of CD4 T cells. Immunol Res. 2008;40(2):114–127. doi: 10.1007/s12026-007-8004-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.