Abstract
Background
Ventricular dysfunction (VnD) after primary coronary artery bypass grafting is associated with increased hospital stay and mortality. Natriuretic peptides have compensatory vasodilatory, natriuretic and paracrine influences on myocardial failure and ischemia. We hypothesized that natriuretic peptide system gene variants independently predict risk of VnD after primary coronary artery bypass grafting.
Methods
1164 patients undergoing primary coronary artery bypass grafting with cardiopulmonary bypass at two institutions were prospectively enrolled. After prospectively defined exclusions, 697 Caucasian patients (76 with VnD) were analyzed. VnD was defined as need for ≥ 2 new inotropes and/or new mechanical ventricular support after coronary artery bypass grafting. 139 haplotype-tagging SNPs within 7 genes (NPPA; NPPB; NPPC; NPR1; NPR2; NPR3; CORIN) were genotyped. SNPs univariately associated with VnD were entered into logistic regression models adjusting for clinical covariates predictive of VnD. To control for multiple comparisons, permutation analyses were conducted for all SNP associations.
Results
After adjusting for clinical covariates and multiple comparisons within each gene, seven NPPA/NPPB SNPs (rs632793, rs6668352, rs549596, rs198388, rs198389, rs6676300, rs1009592) were associated with decreased risk of postoperative VnD (additive model; odds ratios 0.44–0.55; P = 0.010–0.036), and four NPR3 SNPs (rs700923, rs16890196, rs765199, rs700926) were associated with increased risk of postoperative VnD (recessive model; odds ratios 3.89–4.28; P = 0.007–0.034).
Conclusions
Genetic variation within the NPPA/NPPB and NPR3 genes is associated with risk of VnD after primary coronary artery bypass grafting. Knowledge of such genotypic predictors may result in better understanding of the molecular mechanisms underlying postoperative VnD.
INTRODUCTION
Ventricular dysfunction (VnD) has been reported after cardiac surgery in 10–20%1–3 of patients and has been associated with increased hospital stay and long-term mortality after primary coronary artery bypass graft (CABG) surgery with cardiopulmonary bypass (CPB).2 B-type natriuretic peptide (BNP) is a member of the natriuretic peptide family which includes three structurally homologous peptides: A-type (ANP), BNP, and C-type natriuretic peptides.4 ANP and BNP are primarily released from atrial and ventricular myocytes in response to increased cardiac atrial and ventricular wall stress.4 C-type natriuretic peptide is released from vascular endothelial cells and other tissues.4 Natriuretic peptides bind to two guanylyl cyclase-coupled effector receptors (natriuretic peptide receptors A and B) and a clearance receptor (natriuretic peptide receptor C). In addition to compensatory natriuretic, diuretic, and vasorelaxant properties, natriuretic peptides also have mitigating paracrine influences upon myocardial ischemia-reperfusion injury and cardiovascular remodeling.4–14
Circulating plasma BNP levels are used to assess the severity of heart failure and myocardial ischemia, as well as to predict adverse outcomes in patients with acute coronary syndromes or undergoing cardiovascular surgery.1,2,15–18 Natriuretic peptide system gene variants have been associated with cardiovascular disease states such as hypertension,19–21 stroke22,23 and myocardial infarction.24 However, to date, the association between natriuretic peptide system gene variants and development of VnD after cardiac surgery with CPB has not been examined. Knowledge of such genotypic predictors may enhance our understanding of the molecular mechanisms underlying postoperative VnD. We thus hypothesized that variants within the natriuretic peptide precursor protein (NPPA, NPPB, NPPC), receptor (NPR1, NPR2, NPR3) and precursor converting enzyme (CORIN) genes are independently associated with the risk of postoperative VnD in Caucasians undergoing primary CABG surgery with CPB.
MATERIALS AND METHODS
Study Population
Between August 2001 and June 2005, 1164 men and women aged 20 to 89 years scheduled for isolated primary CABG surgery with CPB at Brigham and Women’s Hospital, Boston, MA and Texas Heart Institute, St. Luke’s Episcopal Hospital, Houston, TX were prospectively enrolled into an ongoing parent study known as the CABG Genomics Study.* Respective Institutional Review Board approval and subject written informed consent were obtained. The overall objective of the CABG Genomics Study is to identify associations between genotypic variants and adverse perioperative outcomes. CABG Genomics Study exclusion criteria include a preoperative hematocrit < 25% or transfusion of leukocyte-rich blood products within 30 days before surgery. Enrolled subjects were additionally prospectively excluded from analysis for this study if they underwent emergency surgery, received a preoperative inotrope, intra-aortic balloon pump or ventricular assist device support, had severe preoperative renal dysfunction (preoperative hemodialysis or preoperative serum creatinine > 3 mg/dL), or were missing preoperative or peak postoperative plasma BNP or cardiac troponin I (cTnI) measurements. To avoid confounding due to population stratification, subjects were further prospectively excluded from analysis if they reported having a non-Caucasian parent or grandparent, or if assessment of genomic control single nucleotide polymorphisms (SNPs) indicated non-Caucasian ethnicity.25
Data and Blood Collection
Data were collected for each enrolled subject during primary hospitalization using a detailed prospectively designed study case report form that includes: 1) preoperative demographic characteristics, comorbidities and medications; 2) surgical characteristics; and 3) postoperative in-hospital events. Data were subject to automated range and logic checking as well as additional manual audit of a proportion of records.
Plasma samples obtained preoperatively and on postoperative days 1–5 were stored in vapor-phase liquid nitrogen until analysis. Plasma BNP and cTnI were measured at all time points using immunoassays conducted at a single reference laboratory (Biosite, San Diego, California). DNA was extracted from white blood cells using standard procedures. Genotyping was performed using Sequenom MassARRAY® iPLEX (Sequenom, San Diego, California) at Helmholtz Zentrum München (Neuherberg, Germany). Raw genotyping signals were analyzed with SpectroTyper 3.4 software (Sequenom, San Diego, California) and were manually curated.
Seven candidate genes related to the natriuretic peptide system were selected a priori: NPPA, NPPB, NPPC, NPR1, NPR2, NPR3, and CORIN. SNPs were selected for genotyping to achieve approximately 2500 base pair spacing across each gene region. SNPs were preferentially selected if they were assessed in prior cardiovascular studies, were non-synonymous coding SNPs, were within exon regions, were within 50 base-pairs of intron-exon boundaries, were in promoter regions, or if they “tagged” haplotype blocks (as per the Caucasian HapMap resource).† Genotyping was performed for 155 SNPs. SNPs were excluded from analysis for one or more of the following reasons: genotyping rate < 90% (3 SNPs), observed minor allele frequency < 1% (13 SNPs) or not being in Hardy Weinberg equilibrium in control subjects (P≤0.001; 1 SNP). No SNPs required exclusion for differential missingness > 10% between case and control groups. 71 additional genomic control SNPs were also genotyped in order to exclude subjects with non-Caucasian ethnicities. In order to account for potential stratification by Northern versus Southern European ancestry, 6 SNPs within the lactase gene (rs182549, rs2322659, rs3754689, rs3769005, rs4954490, rs4988235) were also genotyped.25,26 Subjects were additionally excluded if they were missing > 10% of their genotyping data.
Definitions
Postoperative VnD was defined as a new requirement for two or more inotropes or new placement of an intra-aortic balloon pump or ventricular assist device either during the intraoperative period after the patient separated from CPB or postoperatively in the intensive care unit. Inotrope support was defined as continuous infusion of amrinone, milrinone, dobutamine, dopamine (> 5µg/kg/min), epinephrine, isoproterenol, norepinephrine or vasopressin. Dyspnea score was derived using the New York Heart Association classification, with score 1 = no dyspnea, score 2 = mild impairment of daily functioning, score 3 = substantial functional impairment when not at rest, and score 4 = functional impairment at rest.27 Urgent CABG surgery was defined as occurring within the same hospitalization as the diagnosis of an acute coronary event or coronary artery disease. Stenosis of > 50% of the left anterior descending, left circumflex or right coronary arteries or their major branches were quantified based on cardiac catheterization data and scored as regions of coronary arterial disease (1, 2 or 3 regions total). Stenosis of > 50% of the left main coronary artery was counted as 2 regions of significant disease. Peak postoperative plasma BNP and cTnI was assessed if a subject had at least 3 postoperative day 1–5 measures, or, for peak postoperative cTnI, at least postoperative day 1 and 2 measures.
Statistical Analysis
Categorical and continuous patient and clinical characteristics were compared between case and control groups with chi-squared, Fisher’s Exact or Wilcoxon rank sum tests as indicated using SAS (version 9.1.3, SAS Institute, Cary, NC). Continuous plasma BNP and cTnI data was log transformed to normalize data distribution prior to analysis. Multivariable logistic regression models of non-genetic clinical and biomarker predictors of VnD were also created using SAS. One model was created using preoperative and intraoperative covariates, while the other model also included peak postoperative cTnI in order to adjust additionally for the effects of postoperative myocardial injury. The two multivariable clinical prediction models were created using combined forwards and backwards regression modeling with covariate entry into the models determined by univariate association with VnD (P<0.15; Table 1) and exit from the models determined by P value of 0.05. Preoperative cTnI >0.1µg/L was kept in the model that includes preoperative and intraoperative characteristics because it has been associated with VnD previously in a larger study group.2 CPB time >120 minutes was maintained in the model that includes postoperative peak cTnI concentration, as this covariate independently predicts VnD in the multivariable model that includes only preoperative and intraoperative covariates. Demographic characteristics of age, gender, institution and body mass index >30 kg/m2 were locked into both multivariable models regardless of significance. Simultaneous inclusion of preoperative myocardial infarction in the model with peak postoperative cTnI did not improve the predictive ability of the model.
Table 1.
Patient demographics, medications and perioperative risk factors in 697 Caucasians with and without VnD after primary coronary artery bypass graft surgery.
| Preoperative Demographics and Risk Factors | No VnD (n =621) |
VnD (n = 76) |
P Value |
|---|---|---|---|
| Age (years) | 64 ± 10 | 64 ± 11 | 0.82 |
| Female gender | 116 (18.7%) | 12 (15.8%) | 0.64 |
| Institution | |||
| Brigham and Women’s Hospital | 486 (78.3%) | 65 (85.5%) | 0.18 |
| Texas Heart Institute | 135 (21.7%) | 11 (14.5%) | |
| Diabetes mellitus | 168 (27.1%) | 23 (30.3%) | 0.59 |
| Hypertension | 466 (75.3%) | 59 (77.6%) | 0.78 |
| Hypercholesterolemia | 469 (75.8%) | 59 (78.7%) | 0.67 |
| Obesity (BMI > 30 kg/m2) | 240 (38.7%) | 27 (35.5%) | 0.62 |
| Smoking, > 30 pack year history | 167 (27.8%) | 29 (42.0%) | 0.02 |
| Renal insufficiency (creatinine = 1.6–3.0 mg/dL) | 26 (4.2%) | 9 (11.8%) | 0.009 |
| Myocardial infarction ≤ 2 weeks preoperatively | 91 (14.7%) | 27 (35.5%) | <0.0001 |
| LVEF % | 54 ± 12 | 42 ± 15 | <0.0001 |
| Dyspnea score (based on NYHA classification) | |||
| 0.06 | |||
| None or mild | 307 (53.7%) | 32 (44.4%) | |
| Moderate | 205 (35.8%) | 26 (36.1%) | |
| Severe | 60 (10.5%) | 14 (19.4%) | |
| Coronary artery regions with > 50% stenosis | |||
| 0.07 | |||
| 0–1 region | 33 (5.3%) | 1 (1.3%) | |
| 2 regions | 100 (16.2%) | 7 (9.2%) | |
| 3 regions | 485 (78.5%) | 68 (89.5%) | |
| Mitral insufficiency (moderate or severe) | 12 (2.0%) | 5 (6.9%) | 0.03 |
| Past arrhythmia | 60 (9.7%) | 8 (10.5%) | 0.84 |
| Median preoperative BNP (pg/mL) [10th, 90th percentile] | 15.5 [1.6, 102.8] |
61.0 [3.4, 369.4] |
<0.0001 |
| Preoperative cTnI > 0.1µg/L | 76 (12.2%) | 21 (27.6%) | <0.0001 |
| Preoperative Medications | |||
| ACE-inhibitor | 290 (46.8%) | 48 (63.2%) | 0.008 |
| Diuretic | 118 (19.0%) | 25 (32.9%) | 0.007 |
| Statin | 468 (75.4%) | 55 (72.4%) | 0.58 |
| Digoxin | 19 (3.1%) | 4 (5.3%) | 0.30 |
| Beta blocker | 469 (75.5%) | 61 (80.3%) | 0.40 |
| Aspirin | 462 (74.4%) | 54 (71.1%) | 0.58 |
| Non-aspirin platelet inhibitor | 106 (17.1%) | 17 (22.4%) | 0.26 |
| Perioperative Risk Factors | |||
| Urgent surgery | 326 (52.6%) | 44 (57.9%) | 0.40 |
| Cardiopulmonary bypass time, minutes | 93.2 ± 36.8 | 111.4 ± 34.4 | <0.0001 |
| Aortic cross clamp time, minutes | 69.5 ± 31.3 | 79.0 ± 29.3 | 0.007 |
| Peak postoperative cTnI [median, 10th, 90th percentile], µg/L |
1.57 [0.50, 3.37] |
3.14 [0.74, 8.60] |
<0.0001 |
Data are shown as n (%) for dichotomous variables and mean ± standard deviation or median [10th, 90th percentiles] for continuous variables. ACE = angiotensin converting enzyme; BMI = body mass index; BNP = B-type natriuretic peptide; LVEF = left ventricular ejection fraction; NYHA = New York Heart Association; VnD = ventricular dysfunc
Genetic statistical analyses were conducted with PLINK (version 1.01).28 Hardy Weinberg equilibrium was evaluated using Fisher’s Exact tests. Potential confounding of associations between natriuretic peptide SNP frequencies and VnD by subjects’ Northern versus Southern European ancestries was determined by investigating association between lactase gene SNPs and VnD. Individual SNP associations with VnD were estimated using logistic regression for additive (each copy of the minor allele has an equivalent additional additive predictive value, i.e., 0, 1, 2), dominant (1 or 2 copies of the minor allele versus 0 copies of the minor allele) and recessive (2 copies of the minor allele versus 0 or 1 copies of the minor allele) genetic models, and corresponding odds ratios (OR) and 95% confidence intervals (CI) calculated for each SNP according to the role of the SNP’s minor allele in each genetic model. SNPs associated with VnD in single covariate logistic assessment were then individually entered into the multivariable clinical logistic regression models for predicting VnD, and multivariate adjusted ORs and 95% CIs were determined for each SNP with regards to developing VnD.
Empirical P values were determined for all SNP associations based on permuting case/control status (15,000 repetitions) and adjusting for family-wise type 1 error within each gene. Point-wise permutation analyses were conducted for each SNP to adjust for potentially random occurrence of the SNP’s minor allele in VnD cases versus controls. Permutation analyses adjusting for family-wise type 1 error were conducted to adjust for the probability of making false positive discoveries when assessing multiple SNPs within a gene region for association with the VnD outcome. NPPA and NPPB were assessed as one genetic region, as these two genes lie in close proximity on chromosome 1.
Linkage disequilibrium blocks and associated haplotypes for NPPA/NPPB (Figure 1) and NPR3 (Figure 2) gene regions were derived in Haploview (version 4.1; The Broad Institute, Cambridge, MA) using default criteria.29,30 Omnibus tests were conducted to assess if variance in haplotype frequencies within each linkage disequilibrium block had an overall significant association with the VnD phenotype. Haplotypes within linkage disequilibrium blocks that had omnibus test P values < 0.05 were then assessed with Pearson chi-squared tests for association with VnD. Since these univariate haplotype results were consistent with observed SNP associations, additional permutation analyses and multivariable adjustments for clinical covariates were not conducted.
Figure 1.
Block structure of linkage disequilibrium for the 28 successfully genotyped single nucleotide polymorphisms (SNPs) in the natriuretic peptide precursor protein A and B gene regions (NPPA/NPPB) on chromosome 1p36.22. SNPs surrounded by boxes are the SNPs that were significantly associated with the ventricular dysfunction phenotype after adjustment for clinical covariates and multiple comparisons (P<0.05). Stronger correlations between SNPs are noted by darker color in the intersecting squares linking each pair of SNPs. Stronger correlation indicates a tendency for the minor allele of one SNP to be inherited with the minor allele of the other SNP that it is being linked with in this figure. Correlations and block structures were estimated using Haploview (version 4.1; The Broad Institute, Cambridge, MA). 30
Figure 2.
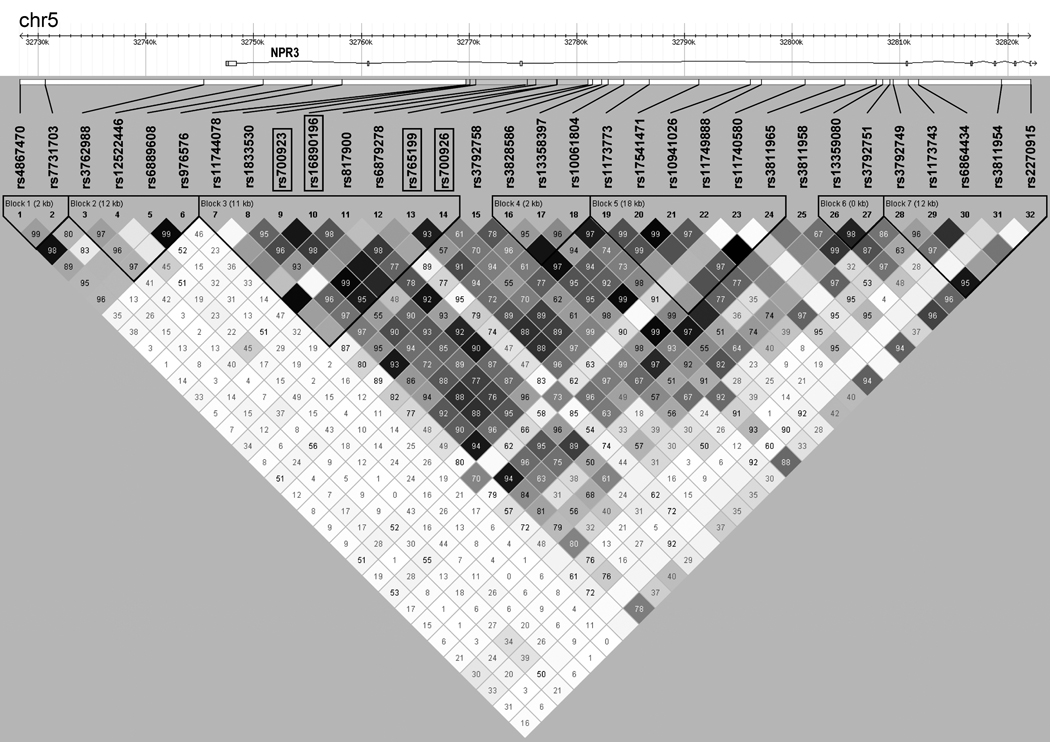
Block structure of linkage disequilibrium for the 32 successfully genotyped single nucleotide polymorphisms (SNPs) in the natriuretic peptide receptor C gene region (NPR3) on chromosome 5p13.3. SNPs surrounded by boxes are the SNPs that were significantly associated with the ventricular dysfunction phenotype after adjustment for clinical covariates and multiple comparisons (P<0.05). Stronger correlations between SNPs are noted by darker color in the intersecting squares linking each pair of SNPs. Stronger correlation indicates a tendency for the minor allele of one SNP to be inherited with the minor allele of the other SNP that it is being linked with in this figure. Correlations and block structures were estimated using Haploview (version 4.1; The Broad Institute, Cambridge, MA). 30
RESULTS
Of the 1164 subjects enrolled into the source cohort during the study period, 407 were prospectively excluded from analysis for one or more of the following exclusion criteria: non-Caucasian ethnicity (n=176), no CABG surgery performed (n=6), prior cardiac surgery (n=4), concurrent valve surgery performed (n=51), emergency surgery (n=4), CPB not used (n=39), aortic cross-clamp not used (n=68), preoperative ventricular assist device (n=1), preoperative intra-aortic balloon pump (n=27), preoperative inotropes (n=4), no preoperative or peak postoperative BNP or cTnI measurements (n=72), preoperative hemodialysis (n=1), preoperative serum creatinine > 3 mg/dL (n=4), or no genotyping performed (n=83). In addition to these exclusions 60 subjects were excluded for having less than 90% success for their individual genotyping.
Patient Characteristics
Perioperative patient characteristics for 697 subjects included in the study analysis are shown in Table 1, stratified by occurrence of postoperative VnD (n=76). None of the subjects who developed the VnD phenotype underwent ventricular assist device placement. Twelve VnD subjects received an intra-aortic balloon pump intraoperatively after separating from CPB. An additional 7 VnD subjects underwent intra-aortic balloon pump insertion postoperatively in the intensive care unit. Forty-seven VnD subjects (6.2%) required ≥ 2 inotropes intraoperatively, and 43 subjects (6.7%) required ≥ 2 inotropes postoperatively.
Patients who developed postoperative VnD were significantly more likely to have a > 30 pack year history of smoking, renal insufficiency, recent myocardial infarction, lower preoperative left ventricular ejection fraction, higher preoperative plasma BNP concentration, moderate to severe mitral regurgitation, or be taking an angiotensin converting enzyme inhibitor or diuretic. Subjects who developed VnD had significantly longer CPB and aortic cross-clamp times and higher peak postoperative cTnI concentrations. Multivariable logistic regression models of patient and clinical predictors of postoperative VnD in the 697 study subjects are shown in Table 2 and are consistent with our previous report from a larger, multiethnic cohort, that preoperative left ventricular ejection fraction, BNP, and CPB time >120 minutes are independent clinical predictors of VnD after CABG.2
Table 2.
Logistic regression models of clinical predictors of ventricular dysfunction after primary coronary artery bypass graft surgery with cardiopulmonary bypass.
| Model Including Preoperative cTnI > 0.1 µg/L (n=672*; AIC = 416.30) |
Model Including Log10 Peak Postoperative cTnI (n=672*; AIC = 403.08) |
|||||
|---|---|---|---|---|---|---|
| Predictor | Odds Ratio | 95% CI | P value | Odds Ratio | 95% CI | P value |
| Age (ten year increment) | 0.91 | (0.70, 1.18) | 0.46 | 0.91 | (0.70, 1.18) | 0.47 |
| Gender (female) | 0.81 | (0.40, 1.64) | 0.55 | 0.79 | (0.38, 1.61) | 0.51 |
| Institution | 0.75 | (0.35, 1.60) | 0.45 | 0.73 | (0.33, 1.58) | 0.42 |
| BMI > 30 kg/m2 | 0.95 | (0.55, 1.65) | 0.85 | 0.91 | (0.52, 1.60) | 0.74 |
| Preoperative LVEF (1% increment) |
0.94 | (0.92, 0.96) | <0.0001 | 0.94 | (0.92, 0.96) | <0.0001 |
| Log10 preoperative BNP | 1.56 | (1.04, 2.36) | 0.03 | 1.61 | (1.07, 2.42) | 0.02 |
| Preoperative cTnI > 0.1µg/L | 1.76 | (0.94, 3.30) | 0.08 | - | - | - |
| Log10 peak postoperative cTnI | - | - | - | 2.80 | (1.70, 4.60) | <0.0001 |
| CPB time >120 min | 2.06 | (1.14, 3.70) | 0.02 | 1.55 | (0.85, 2.86) | 0.16 |
AIC =Akaike information criterion; BMI = body mass index; BNP = B-type natriuretic peptide; CI = confidence interval; CPB = cardiopulmonary bypass; cTnI = cardiac troponin I; LVEF = left ventricular ejection fraction
25 subjects were missing one or more of the models’ predictor variables and are not included in this analysis.
Natriuretic Peptide System SNP Associations with Ventricular Dysfunction
The 139 SNPs that passed quality control criteria are listed in Supplemental Digital Content along with corresponding P values for univariate associations with VnD as assessed with additive, dominant and recessive genetic models (see Appendix, Supplemental Digital Content 1: 139 single nucleotide polymorphisms genotyped within 7 natriuretic peptide system gene regions in 697 Caucasians with and without ventricular dysfunction after primary coronary artery bypass graft surgery). The genotyping rate for these 139 SNPs was 98.8%. The minor alleles of 13 NPPA/NPPB gene region SNPs were univariately associated with a decreased incidence of VnD after CABG surgery, and the minor alleles of 7 NPR3 gene SNPs were univariately associated with an increased incidence of VnD after CABG surgery (asymptotic P<0.05; Table 3). SNPs in the NPPC, NPR1, and CORIN gene regions were not associated with VnD. One NPR3 SNP rs11740580 and one NPR2 region SNP rs3808864 were univariately associated with decreased VnD, but these associations did not survive statistical adjustments for multiple comparisons.
Table 3.
SNPs univariately associated with VnD after primary coronary artery bypass graft surgery in 697 Caucasians.
| SNP | Chromosomal Position NCBI hg36 |
Gene Region and SNP Role |
Minor/ Major Allele |
No VnD (n=621) (aa/aA/AA) |
VnD (n=76) (aa/aA/AA) |
Genetic Model § |
Odds Ratio |
95% CI | Asymptotic P value* |
Point-wise Empirical P value¶ |
Family-wise Empirical P value± |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs198358 | chr1:11826663 | NPPA; Downstream | C/T | 46/235/340 | 2/24/49 | Additive | 0.647 | 0.422, 0.992 | 0.046 | 0.039 | 0.382 |
| rs632793 | chr1:11833264 | NPPA; Promoter NPPB; Downstream |
C/T | 104/302/212 | 7/32/37 | Additive | 0.616 | 0.427, 0.889 | 0.010 | 0.009 | 0.108 |
| rs6668352 | chr1:11837416 | NPPA; Promoter NPPB; Downstream |
T/C | 43/234/332 | 1/21/53 | Additive | 0.509 | 0.320, 0.809 | 0.004 | 0.004 | 0.0503 |
| rs549596 | chr1:11838682 |
NPPA; Promoter NPPB; Downstream |
C/T | 124/306/191 | 9/30/37 | Additive | 0.577 | 0.402, 0.827 | 0.003 | 0.002 | 0.037 |
| rs198388 | chr1:11839927 | NPPA; Promoter NPPB; Downstream |
T/C | 118/291/186 | 8/27/33 | Additive | 0.586 | 0.402, 0.856 | 0.006 | 0.004 | 0.060 |
| rs198389 | chr1:11841858 | NPPB; Promoter | C/T | 118/304/195 | 9/32/35 | Additive | 0.631 | 0.441, 0.902 | 0.012 | 0.011 | 0.121 |
| rs3753580 | chr1:11843635 | NPPB; Promoter | C/T | 74/277/265 | 4/28/43 | Additive | 0.600 | 0.405, 0.890 | 0.011 | 0.009 | 0.118 |
| rs12406089 | chr1:11843768 | NPPB; Promoter | G/C | 75/281/258 | 4/28/42 | Additive | 0.593 | 0.399, 0.881 | 0.010 | 0.008 | 0.108 |
| rs12406383 | chr1:11844580 | NPPB; Promoter | T/G | 48/241/330 | 1/25/49 | Additive | 0.588 | 0.381, 0.909 | 0.017 | 0.014 | 0.187 |
| rs6668659 | chr1:11844885 | NPPB; Promoter | G/T | 75/284/256 | 4/26/41 | Additive | 0.574 | 0.383, 0.862 | 0.007 | 0.006 | 0.084 |
| rs6676300 | chr1:11847887 | NPPB; Promoter | C/T | 78/281/236 | 4/26/41 | Additive | 0.538 | 0.358, 0.809 | 0.003 | 0.002 | 0.038 |
| rs12562952 | chr1:11849643 | NPPB; Promoter | C/T | 9/122/450 | 0/8/64 | Additive | 0.431 | 0.207, 0.899 | 0.025 | 0.020 | 0.254 |
| rs1009592 | chr1:11851301 | NPPB; Promoter | C/G | 91/282/248 | 5/29/42 | Additive | 0.587 | 0.402, 0.857 | 0.006 | 0.005 | 0.061 |
| rs700923 | chr5:32770625 | NPR3; Intron | G/A | 34/226/352 | 13/19/40 | Recessive | 3.746 | 1.873, 7.490 | 0.0002 | <0.0001 | 0.004 |
| rs16890196 | chr5:32775418 | NPR3; Intron | G/A | 25/205/384 | 10/18/47 | Recessive | 3.630 | 1.667, 7.882 | 0.001 | 0.001 | 0.020 |
| rs765199 | chr5:32778222 | NPR3; Intron | A/G | 32/235/351 | 13/23/39 | Recessive | 3.840 | 1.915, 7.699 | 0.0002 | <0.0001 | 0.003 |
| rs700926 | chr5:32781040 | NPR3; Intron | C/A | 37/257/322 | 13/24/39 | Recessive | 3.229 | 1.630, 6.395 | 0.0008 | 0.001 | 0.015 |
| rs17541471 | chr5:32791346 | NPR3; Intron | G/A | 29/207/380 | 9/21/45 | Recessive | 2.760 | 1.253, 6.082 | 0.012 | 0.007 | 0.131 |
| rs10941026 | chr5:32796132 | NPR3 | G/A | 30/207/383 | 9/20/47 | Recessive | 2.642 | 1.203, 5.801 | 0.015 | 0.014 | 0.183 |
| rs11749888 | chr5:32797186 | NPR3; Intron | A/G | 6/132/475 | 3/19/54 | Recessive | 4.156 | 1.018, 16.98 | 0.047 | 0.062 | 0.464 |
aa/aA/AA = subjects with 2 minor alleles/subjects with 1 minor allele and 1 major allele/ subjects with 2 major alleles A=adenine; C=cytosine; G=Guanine; T=Thymine
CI = confidence interval; NPPA = natriuretic peptide precursor A; NPPB = natriuretic peptide precursor B; NPR3 = natriuretic peptide receptor 3; VnD = ventricular dysfunction; SNP = single nucleotide polymorphism;
Genetic model with most significant association shown, although other models may have resulted in asymptotic P≤0.05
asymptotic P value* = P value before permutation adjustment for cohort case-control status (point-wise¶ empirical P value) or adjustment for testing multiple SNP hypotheses within a gene region (family-wise± empirical P value)
Bold type signifies SNPs that remain significantly associated with the VnD phenotype after family-wise permutation adjustment for multiple comparisons within a gene region.
After adjusting for clinical predictors of VnD using either multivariable model shown in Table 2, the minor alleles of eleven NPPA/NPPB gene SNPs and the minor alleles of six NPR3 gene SNPs remained associated with the occurrence of postoperative VnD (asymptotic P<0.05). After additional point-wise permutation adjustment for subject case-control status and family-wise permutation adjustment for total number of SNPs assessed in the gene region, the minor alleles of seven NPPA/NPPB SNPs and four NPR3 SNPs remained associated with occurrence of postoperative VnD using either multivariable model shown in Table 2 (Family-wise empirical P<0.05). In Table 4 we present the gene association results adjusted for the clinical model containing preoperative cTnI>0.1µg/L, but the asymptotic, point-wise, and family-wise significant associations (P<0.05) were unchanged when adjustments were made for the second clinical model.
Table 4.
SNPs associated with VnD after primary coronary artery bypass graft surgery in 697 Caucasians after adjusting for clinical predictors of VnD#
| SNP | Gene Region | Genetic Model§ |
Odds Ratio |
95% CI | Asymptotic P value* |
Point-wise Empirical P value¶ |
Family-wise Empirical P value± |
|---|---|---|---|---|---|---|---|
| rs632793 | NPPA_NPPB | Additive | 0.520 | 0.347, 0.780 | 0.002 | 0.002 | 0.026 |
| rs6668352 | NPPA_NPPB | Additive | 0.437 | 0.264, 0.722 | 0.001 | 0.002 | 0.022 |
| rs549596 | NPPA_NPPB | Additive | 0.488 | 0.330, 0.722 | 0.0003 | 0.0004 | 0.010 |
| rs198388 | NPPA_NPPB | Additive | 0.505 | 0.335, 0.760 | 0.001 | 0.001 | 0.019 |
| rs198389 | NPPA_NPPB | Additive | 0.545 | 0.370, 0.804 | 0.002 | 0.002 | 0.036 |
| rs3753580 | NPPA_NPPB | Additive | 0.525 | 0.342, 0.807 | 0.003 | 0.003 | 0.053 |
| rs12406089 | NPPA_NPPB | Additive | 0.527 | 0.343, 0.812 | 0.004 | 0.003 | 0.057 |
| rs12406383 | NPPA_NPPB | Additive | 0.506 | 0.315, 0.814 | 0.005 | 0.005 | 0.073 |
| rs6668659 | NPPA_NPPB | Additive | 0.521 | 0.335, 0.810 | 0.004 | 0.004 | 0.058 |
| rs6676300 | NPPA_NPPB | Additive | 0.481 | 0.308, 0.751 | 0.001 | 0.001 | 0.023 |
| rs1009592 | NPPA_NPPB | Additive | 0.495 | 0.325, 0.752 | 0.001 | 0.0008 | 0.019 |
| rs700923 | NPR3 | Recessive | 4.282 | 1.937, 9.466 | 0.0003 | 0.0006 | 0.007 |
| rs16890196 | NPR3 | Recessive | 4.090 | 1.680, 9.955 | 0.002 | 0.003 | 0.034 |
| rs765199 | NPR3 | Recessive | 4.270 | 1.934, 9.430 | 0.0003 | 0.0008 | 0.007 |
| rs700926 | NPR3 | Recessive | 3.892 | 1.802, 8.409 | 0.0005 | 0.001 | 0.009 |
| rs17541471 | NPR3 | Recessive | 3.204 | 1.334, 7.696 | 0.009 | 0.007 | 0.114 |
| rs10941026 | NPR3 | Recessive | 3.140 | 1.309, 7.530 | 0.010 | 0.009 | 0.129 |
CI = confidence interval; NPPA = natriuretic peptide precursor A; NPPB = natriuretic peptide precursor B; NPR3 = natriuretic peptide receptor 3; SNP = single nucleotide polymorphism; VnD = ventricular dysfunction;
SNP associations adjusted for multivariable clinical logistic regression model in Table 2 that includes preoperative cTnI > 0.1 µg/L
Genetic model with most significant association shown, although other models may also have also resulted in asymptotic P≤0.05
asymptotic P value* = P value before permutation adjustment for cohort case-control status (point-wise¶ empirical P value) or adjustment for testing multiple SNP hypotheses within a gene region (family-wise± empirical P value)
Bold type signifies SNPs that remain significantly associated with the VnD henotype after family-wise permutation adjustment for multiple comparisons within a gene region
The logistic regression model including demographic, clinical, and non-genetic preoperative biomarker variables (Table 2) predicted 15.9% of the variability (r2) in occurrence of postoperative VnD, and the area under the receiver operating characteristic curve related to this model was 0.781. When the SNPs most significantly associated with VnD from the NPPA/NPPB region (rs549596; additive model) and the NPR3 region (rs700923; recessive model) were both added to the clinical model, the proportion of explained variability (r2) for the model increased to 21.9% (P<0.0001) and the area under the receiver operating characteristic curve for the model increased to 0.828.
Natriuretic Peptide System Haplotype Associations with Ventricular Dysfunction
The seven NPPA/NPPB SNPs that remained associated with a decreased risk of VnD after statistical adjustment for clinical predictors and multiple comparisons are contained within the linkage disequilibrium blocks 2, 3 and 4 (chr1:11,832,122-11,851,301) of the NPPA/NPPB gene region (Figure 1). Omnibus tests for global association between haplotype variation within each NPPA/NPPB linkage disequilibrium block and the VnD phenotype were significant for blocks 1, 3, and 4 (P<0.05; block 2 P=0.055). Within these blocks, analyses of individual haplotype associations with occurrence of VnD were consistent with the NPPA/NPPB region SNP associations.
The four NPR3 SNPs that remained associated with an increased risk of VnD after statistical adjustment for clinical predictors and multiple comparisons are contained within block 3 of the NPR3 gene (chr5:32,769,725–32,781,040; Figure 2). Omnibus tests for global association between haplotype variability within NPR3 linkage disequilibrium blocks and occurrence of the VnD phenotype showed significant associations for blocks 3 and 5 (P<0.05). Within these blocks, analyses of individual haplotype associations were consistent with the associations observed for the NPR3 SNPs.
DISCUSSION
We have identified novel regions of genetic variation within the NPPA, NPPB, and NPR3 genes that are associated with VnD after primary CABG surgery. After adjusting for environmental covariates and multiple comparisons the minor alleles of the NPPA/NPPB SNPs identified in this study associate with a decreased risk of postoperative VnD, and the minor alleles of the NPR3 SNPs identified in this study associate with an increased risk of postoperative VnD. Furthermore, simultaneous addition of the NPPA/NPPB SNP rs549596 and the NPR3 rs700923 into the clinical multivariable logistic regression model for predicting postoperative VnD improved the predictive ability of the model.
While NPPA and NPPB SNPs have not been assessed previously in relation to adverse perioperative cardiovascular outcomes, several NPPA/NPPB gene variants have been examined for association with ambulatory cardiovascular diseases.19,22–24,31–35 In the only study examining NPPA/NPPB variants in ambulatory heart failure patients, the NPPA nonsynonymous coding polymorphism rs5065 was found to be associated with elevated plasma BNP and amino-terminal BNP (NT-proBNP) levels in patients who were New York Heart Association class III-IV, but not in patients who were New York Heart Association class I–II.31 rs5065 has also been associated with nonfatal myocardial infarction,24 coronary artery disease,24 and along with the rare nonsynonymous coding polymorphism rs5063, has been associated with stroke,22 hypertension,19 and left ventricular mass in hypertensive patients.34 We did not identify any association between rs5065 or rs5063 and VnD after CABG surgery.
In addition to the above studies of NPPA/NPPB gene variants in relation to cardiovascular disease, the NPPA/NPPB promoter SNPs rs198388 and rs198389 have been associated with elevated circulating NT-proBNP concentrations in ambulatory diabetics with nephropathy,33 and these same two SNPs along with NPPA/NPPB SNPs rs632793, rs6668352, 6676300 and rs198375 have been associated with elevated plasma BNP levels in a large ambulatory Japanese cohort.32 The strong overlap between these NPPA/NPPB SNPs and the NPPA/NPPB SNPs associated with VnD in the present study supports the need for further studies regarding how genetic variation in the region containing these SNPs may interact with the natriuretic peptide system and influence postoperative VnD. The NPPB promoter region has multiple cis regulatory elements known to be gene regulators, and multiple physiologic stimuli including mechanical stretch, ischemic injury and hypoxia, and inflammatory mediators are known to stimulate regulation of the NPPB gene.36 Furthermore, processing of natriuretic peptide preprohormones to more active fragments is complex and at present incompletely understood.13 Autocrine and paracrine effects of natriuretic peptides on myocardial ischemia-reperfusion injury and remodeling are also being identified.13
In comparison with the NPPA/NPPB gene region, less is known about the association between SNPs within the natriuretic peptide receptor C (NPR3) gene and adverse cardiovascular outcomes. While natriuretic peptide receptor C was initially identified as a natriuretic peptide clearance receptor, animal studies indicate that this receptor may locally modulate the physiologic effects of natriuretic peptides and may influence microvascular permeability and cardiac sarcolemmal Na+-K+ pump activity.12,37,38 The major allele of the NPR3 promoter SNP rs9716700 has been associated with increased family history of hypertension, as well as lower ANP levels and higher systolic blood pressure in obese hypertensives,20,21 while a different study found a univariate association between the major allele of rs9716700 and higher plasma ANP and BNP levels in ambulatory patients without heart failure but not in patients with heart failure.31 While rs9716700 was not assessed in the present study, we did not find associations between other SNPs in the NPR3 promoter region and VnD after CABG. Furthermore, the NPR3 SNPs that we did find to be associated with VnD are within a linkage disequilibrium block that does not include the NPR3 promoter region and is demarcated from the promoter region by a strong recombination point. Therefore, the NPR3 SNPs associated with VnD in this study are unlikely to be related to rs9716700.
While the NPR3 SNPs (rs700923, rs16890196, rs765199, rs700926) that we identified as significantly associated with decreased postoperative VnD lie within currently assumed non-coding intronic sequences, there are several potential mechanisms that could explain their association with the study’s clinical VnD outcome and which warrant further study. These SNPs may be in linkage disequilibrium with promoter SNPs that have not yet been identified or that were not genotyped in this study. Furthermore these intronic SNPs may have promoter functions that have not yet been identified. Finally, intronic variants may potentially affect receptor function through alternative splicing mechanisms. For example, recent reports have demonstrated associations between intronic SNPs unrelated to the NPR3 gene and alternatively spliced mRNA transcripts that alter function of ultimately translated protein.39
There are several potential limitations to this study. First, within the present cardiac surgical literature there is no standardized outcome definition for ventricular dysfunction after cardiac surgery. Many primary CABG patients at both study institutions do not undergo perioperative monitoring with transesophageal echocardiography or pulmonary artery catheters. We elected to define VnD after CABG surgery as a need for two or more inotropes or new IABP or VAD support in order to best ensure that we were not including patients with normal ventricular function. It is not standard organizational or surgeon based practice at either study institution to separate from CPB on prophylactic inotropes. Furthermore, we have previously reported that patients with our definition of VnD experience significantly prolonged hospital stays and increased up to 5 year all-cause mortality after primary CABG surgery.2 Second, we limited analysis to Caucasian subjects only, as we had insufficient non-Caucasian subjects to date to allow for statistical accommodation of genetic admixture. Thus, further studies are warranted to replicate our findings in non-Caucasian cardiac surgical patient populations. Third, while our study’s sample and effect sizes allowed adjustment for multiple comparisons within each gene, our findings should be viewed as exploratory until further validation studies are conducted in other Caucasian cardiac surgical populations. Fourth, while we have identified regions in NPPA/NPPB and NPR3 that contain genetic variants independently associated with VnD after CABG surgery, there is considerable linkage disequilibrium within these regions. Consequently we cannot conclude that the SNPs identified in this study are directly related to occurrence of VnD after CABG surgery. Finally, our findings do not identify mechanistic pathways that link identified SNP associations to occurrence of postoperative VnD. The natriuretic peptide system and its regulation is complex.36 Further investigation is required to determine how the NPPA/NPPB and NPR3 SNPs associations identified in this study relate to development of postoperative VnD.
CONCLUSIONS
VnD after CABG surgery with CPB is associated with increased postoperative hospital stay and mortality. Genetic variation within defined regions of the NPPA/NPPB and NPR3 natriuretic peptide system genes is associated with VnD after CABG surgery and improves ability to predict the VnD outcome beyond what can be predicted using clinical covariates alone. Further investigation of these regions is warranted, as knowledge of such genotypic predictors may enhance our understanding of the molecular mechanisms underlying postoperative VnD and heart failure.
Supplementary Material
Acknowledgments
Funding: Biosite Incorporated, San Diego, California; Society of Cardiovascular Anesthesiologists Research Starter Grant, Richmond, Virginia (Principle Investigator: A. A. Fox); Foundation for Anesthesia Education and Research Research Starter Grant, Rochester, Minnesota (Principle Investigator: A. A. Fox); Eleanor and Miles Shore Fellowship from Harvard Medical School via the Department of Anesthesiology, Perioperative and Pain Medicine, Brigham and Women’s Hospital, Boston, Massachusetts (Principle Investigator: A. A. Fox); National Institute of Health grant, Bethesda, Maryland (K23-HL068774; Principle Investigator: S. C. Body); Funding from the Department of Anesthesiology, Perioperative and Pain Medicine, Brigham and Women’s Hospital, Boston, Massachusetts and Baylor College of Medicine Division of Cardiovascular Anesthesia at the Texas Heart Institute, Saint Luke’s Episcopal Hospital, Houston, Texas
Footnotes
Presented at: The American Society of Anesthesiologists’ 2008 Annual Meeting, Orlando, Florida, Oral Discussion, October 20th, 2008
Summary Statement: Genetic variation within the NPPA/NPPB and NPR3 natriuretic peptide genes is associated with postoperative ventricular dysfunction in Caucasians undergoing primary coronary artery bypass graft surgery with cardiopulmonary bypass.
http://clinicaltrials.gov/show/NCT00281164, posted December 10th, 2008
www.hapmap.org, last accessed March 10th, 2007
REFERENCES
- 1.Hutfless R, Kazanegra R, Madani M, Bhalla MA, Tulua-Tata A, Chen A, Clopton P, James C, Chiu A, Maisel AS. Utility of B-type natriuretic peptide in predicting postoperative complications and outcomes in patients undergoing heart surgery. J Am Coll Cardiol. 2004;43:1873–1879. doi: 10.1016/j.jacc.2003.12.048. [DOI] [PubMed] [Google Scholar]
- 2.Fox AA, Shernan SK, Collard CD, Liu KY, Aranki SF, DeSantis SM, Jarolim P, Body SC. Preoperative B-type natriuretic peptide is as independent predictor of ventricular dysfunction and mortality after primary coronary artery bypass grafting. J Thorac Cardiovasc Surg. 2008;136:452–461. doi: 10.1016/j.jtcvs.2007.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Provenchere S, Berroeta C, Reynaud C, Baron G, Poirier I, Desmonts JM, Iung B, Dehoux M, Philip I, Benessiano J. Plasma brain natriuretic peptide and cardiac troponin I concentrations after adult cardiac surgery: association with postoperative cardiac dysfunction and 1-year mortality. Crit Care Med. 2006;34:995–1000. doi: 10.1097/01.CCM.0000206110.94385.C4. [DOI] [PubMed] [Google Scholar]
- 4.Cea LB. Natriuretic peptide family: new aspects. Curr Med Chem Cardiovasc Hematol Agents. 2005;3:87–98. doi: 10.2174/1568016053544309. [DOI] [PubMed] [Google Scholar]
- 5.Itoh H, Suga S, Ogawa Y, Komatsu Y, Tamura N, Igaki T, Yamashita J, Ikeda T, Doi K, Chun TH, Inoue M, Matsuda K, Yoshimasa T, Ueda M, Ban T, Nakao K. Significance of vascular natriuretic peptide system in vascular remodeling in humans and its application to gene therapy. Ann N Y Acad Sci. 1997;811:533–541. doi: 10.1111/j.1749-6632.1997.tb52037.x. [DOI] [PubMed] [Google Scholar]
- 6.Kawakami R, Saito Y, Kishimoto I, Harada M, Kuwahara K, Takahashi N, Nakagawa Y, Nakanishi M, Tanimoto K, Usami S, Yasuno S, Kinoshita H, Chusho H, Tamura N, Ogawa Y, Nakao K. Overexpression of brain natriuretic peptide facilitates neutrophil infiltration and cardiac matrix metalloproteinase-9 expression after acute myocardial infarction. Circulation. 2004;110:3306–3312. doi: 10.1161/01.CIR.0000147829.78357.C5. [DOI] [PubMed] [Google Scholar]
- 7.Cerisano G, Pucci PD, Sulla A, Tommasi M, Raspanti S, Santoro GM, Antoniucci D. Relation between plasma brain natriuretic peptide, serum indexes of collagen type I turnover, and left ventricular remodeling after reperfused acute myocardial infarction. Am J Cardiol. 2007;99:651–656. doi: 10.1016/j.amjcard.2006.09.114. [DOI] [PubMed] [Google Scholar]
- 8.Soeki T, Kishimoto I, Okumura H, Tokudome T, Horio T, Mori K, Kangawa K. C-type natriuretic peptide, a novel antifibrotic and antihypertrophic agent, prevents cardiac remodeling after myocardial infarction. J Am Coll Cardiol. 2005;45:608–616. doi: 10.1016/j.jacc.2004.10.067. [DOI] [PubMed] [Google Scholar]
- 9.Tamura N, Ogawa Y, Chusho H, Nakamura K, Nakao K, Suda M, Kasahara M, Hashimoto R, Katsuura G, Mukoyama M, Itoh H, Saito Y, Tanaka I, Otani H, Katsuki M. Cardiac fibrosis in mice lacking brain natriuretic peptide. Proc Natl Acad Sci U S A. 2000;97:4239–4244. doi: 10.1073/pnas.070371497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Souza SP, Yellon DM, Martin C, Schulz R, Heusch G, Onody A, Ferdinandy P, Baxter GF. B-type natriuretic peptide limits infarct size in rat isolated hearts via KATP channel opening. Am J Physiol Heart Circ Physiol. 2003;284:H1592–H1600. doi: 10.1152/ajpheart.00902.2002. [DOI] [PubMed] [Google Scholar]
- 11.Ren B, Shen Y, Shao H, Qian J, Wu H, Jing H. Brain natriuretic peptide limits myocardial infarct size dependent of nitric oxide synthase in rats. Clin Chim Acta. 2007;377:83–87. doi: 10.1016/j.cca.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 12.Hobbs A, Foster P, Prescott C, Scotland R, Ahluwalia A. Natriuretic peptide receptor-C regulates coronary blood flow and prevents myocardial ischemia/reperfusion injury: novel cardioprotective role for endothelium-derived C-type natriuretic peptide. Circulation. 2004;110:1231–1235. doi: 10.1161/01.CIR.0000141802.29945.34. [DOI] [PubMed] [Google Scholar]
- 13.Burley DS, Hamid SA, Baxter GF. Cardioprotective actions of peptide hormones in myocardial ischemia. Heart Fail Rev. 2007;12:279–291. doi: 10.1007/s10741-007-9029-y. [DOI] [PubMed] [Google Scholar]
- 14.D'Souza SP, Davis M, Baxter GF. Autocrine and paracrine actions of natriuretic peptides in the heart. Pharmacol Ther. 2004;101:113–129. doi: 10.1016/j.pharmthera.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Maisel AS, Krishnaswamy P, Nowak RM, McCord J, Hollander JE, Duc P, Omland T, Storrow AB, Abraham WT, Wu AH, Clopton P, Steg PG, Westheim A, Knudsen CW, Perez A, Kazanegra R, Herrmann HC, McCullough PA. Rapid measurement of B-type natriuretic peptide in the emergency diagnosis of heart failure. N Engl J Med. 2002;347:161–167. doi: 10.1056/NEJMoa020233. [DOI] [PubMed] [Google Scholar]
- 16.Feringa HH, Schouten O, Dunkelgrun M, Bax JJ, Boersma E, Elhendy A, de Jonge R, Karagiannis SE, Vidakovic R, Poldermans D. Plasma N-terminal pro-B-type natriuretic peptide as long-term prognostic marker after major vascular surgery. Heart. 2007;93:226–231. doi: 10.1136/hrt.2006.093716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahla E, Baumann A, Rehak P, Watzinger N, Vicenzi MN, Maier R, Tiesenhausen K, Metzler H, Toller W. N-Terminal Pro-brain Natriuretic Peptide Identifies Patients at High Risk for Adverse Cardiac Outcome after Vascular Surgery. Anesthesiology. 2007;106:1088–1095. doi: 10.1097/01.anes.0000267591.34626.b0. [DOI] [PubMed] [Google Scholar]
- 18.Morrow DA, de Lemos JA, Sabatine MS, Murphy SA, Demopoulos LA, DiBattiste PM, McCabe CH, Gibson CM, Cannon CP, Braunwald E. Evaluation of B-type natriuretic peptide for risk assessment in unstable angina/non-ST-elevation myocardial infarction: B-type natriuretic peptide and prognosis in TACTICS-TIMI 18. J Am Coll Cardiol. 2003;41:1264–1272. doi: 10.1016/s0735-1097(03)00168-2. [DOI] [PubMed] [Google Scholar]
- 19.Conen D, Glynn RJ, Buring JE, Ridker PM, Zee RY. Natriuretic peptide precursor a gene polymorphisms and risk of blood pressure progression and incident hypertension. Hypertension. 2007;50:1114–1119. doi: 10.1161/HYPERTENSIONAHA.107.097634. [DOI] [PubMed] [Google Scholar]
- 20.Sarzani R, Dessi-Fulgheri P, Salvi F, Serenelli M, Spagnolo D, Cola G, Pupita M, Giantomassi L, Rappelli A. A novel promoter variant of the natriuretic peptide clearance receptor gene is associated with lower atrial natriuretic peptide and higher blood pressure in obese hypertensives. J Hypertens. 1999;17:1301–1305. doi: 10.1097/00004872-199917090-00010. [DOI] [PubMed] [Google Scholar]
- 21.Pitzalis MV, Sarzani R, Dessi-Fulgheri P, Iacoviello M, Forleo C, Lucarelli K, Pietrucci F, Salvi F, Sorrentino S, Romito R, Guida P, Rappelli A, Rizzon P. Allelic variants of natriuretic peptide receptor genes are associated with family history of hypertension and cardiovascular phenotype. J Hypertens. 2003;21:1491–1496. doi: 10.1097/00004872-200308000-00012. [DOI] [PubMed] [Google Scholar]
- 22.Rubattu S, Ridker P, Stampfer MJ, Volpe M, Hennekens CH, Lindpaintner K. The gene encoding atrial natriuretic peptide and the risk of human stroke. Circulation. 1999;100:1722–1726. doi: 10.1161/01.cir.100.16.1722. [DOI] [PubMed] [Google Scholar]
- 23.Rubattu S, Stanzione R, Di Angelantonio E, Zanda B, Evangelista A, Tarasi D, Gigante B, Pirisi A, Brunetti E, Volpe M. Atrial natriuretic peptide gene polymorphisms and risk of ischemic stroke in humans. Stroke. 2004;35:814–818. doi: 10.1161/01.STR.0000119381.52589.AB. [DOI] [PubMed] [Google Scholar]
- 24.Gruchala M, Ciecwierz D, Wasag B, Targonski R, Dubaniewicz W, Nowak A, Sobiczewski W, Ochman K, Romanowski P, Limon J, Rynkiewicz A. Association of the ScaI atrial natriuretic peptide gene polymorphism with nonfatal myocardial infarction and extent of coronary artery disease. Am Heart J. 2003;145:125–131. doi: 10.1067/mhj.2003.52. [DOI] [PubMed] [Google Scholar]
- 25.Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, Kessing BD, Malasky MJ, Scafe C, Le E, De Jager PL, Mignault AA, Yi Z, De The G, Essex M, Sankale JL, Moore JH, Poku K, Phair JP, Goedert JJ, Vlahov D, Williams SM, Tishkoff SA, Winkler CA, De La Vega FM, Woodage T, Sninsky JJ, Hafler DA, Altshuler D, Gilbert DA, O'Brien SJ, Reich D. A high-density admixture map for disease gene discovery in african americans. Am J Hum Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bersaglieri T, Sabeti PC, Patterson N, Vanderploeg T, Schaffner SF, Drake JA, Rhodes M, Reich DE, Hirschhorn JN. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisher JD. New York Heart Association Classification. Arch Intern Med. 1972;129:836. [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 30.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 31.Vassalle C, Andreassi MG, Prontera C, Fontana M, Zyw L, Passino C, Emdin M. Influence of ScaI and natriuretic peptide (NP) clearance receptor polymorphisms of the NP System on NP concentration in chronic heart failure. Clin Chem. 2007;53:1886–1890. doi: 10.1373/clinchem.2007.088302. [DOI] [PubMed] [Google Scholar]
- 32.Takeishi Y, Toriyama S, Takabatake N, Shibata Y, Konta T, Emi M, Kato T, Kawata S, Kubota I. Linkage disequilibrium analyses of natriuretic peptide precursor B locus reveal risk haplotype conferring high plasma BNP levels. Biochem Biophys Res Commun. 2007;362:480–484. doi: 10.1016/j.bbrc.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 33.Lajer M, Tarnow L, Jorsal A, Parving HH. Polymorphisms in the B-type natriuretic peptide (BNP) gene are associated with NT-proBNP levels but not with diabetic nephropathy or mortality in type 1 diabetic patients. Nephrol Dial Transplant. 2007;22:3235–3239. doi: 10.1093/ndt/gfm360. [DOI] [PubMed] [Google Scholar]
- 34.Rubattu S, Bigatti G, Evangelista A, Lanzani C, Stanzione R, Zagato L, Manunta P, Marchitti S, Venturelli V, Bianchi G, Volpe M, Stella P. Association of atrial natriuretic peptide and type a natriuretic peptide receptor gene polymorphisms with left ventricular mass in human essential hypertension. J Am Coll Cardiol. 2006;48:499–505. doi: 10.1016/j.jacc.2005.12.081. [DOI] [PubMed] [Google Scholar]
- 35.Dedoussis GV, Maumus S, Skoumas J, Choumerianou DM, Pitsavos C, Stefanadis C, Visvikis-Siest S. Natriuretic peptide Val7Met substitution and risk of coronary artery disease in Greek patients with familial hypercholesterolemia. J Clin Lab Anal. 2006;20:98–104. doi: 10.1002/jcla.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Rumayor A, Richards AM, Burnett JC, Januzzi JL., Jr Biology of the natriuretic peptides. Am J Cardiol. 2008;101:3–8. doi: 10.1016/j.amjcard.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 37.Matsukawa N, Grzesik WJ, Takahashi N, Pandey KN, Pang S, Yamauchi M, Smithies O. The natriuretic peptide clearance receptor locally modulates the physiological effects of the natriuretic peptide system. Proc Natl Acad Sci U S A. 1999;96:7403–7408. doi: 10.1073/pnas.96.13.7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.William M, Hamilton EJ, Garcia A, Bundgaard H, Chia KK, Figtree GA, Rasmussen HH. Natriuretic peptides stimulate the cardiac sodium pump via NPR-C-coupled NOS activation. Am J Physiol Cell Physiol. 2008;294:C1067–C1073. doi: 10.1152/ajpcell.00243.2007. [DOI] [PubMed] [Google Scholar]
- 39.Medina MW, Gao F, Ruan W, Rotter JI, Krauss RM. Alternative splicing of 3-hydroxy-3-methylglutaryl coenzyme A reductase is associated with plasma low-density lipoprotein cholesterol response to simvastatin. Circulation. 2008;118:355–362. doi: 10.1161/CIRCULATIONAHA.108.773267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.