Abstract
The biarsenical tetracysteine motif is a useful tag for genetic labeling of proteins with small molecules in living cells. The present study concerns the structure of a 12-amino acid peptide FLNCCPGCCMEP bound to the fluorophore ReAsH, based on resorufin. 1H-NMR was used to determine the solution structure of the complex formed between the peptide and the ReAsH moiety. Structure calculations based on the NMR results showed that the backbone structure of the peptide is fairly well defined with a hairpin-like turn, similar to a β turn type II, formed by the central CPGC segment. The most stable complex was formed when As2 was bonded to C4 and C5, and As1 was bonded to C8 and C9. Two clear NOESY crosspeaks between the Phe1 sidechain and ReAsH confirmed the close positioning of the phenyl ring of Phe1 and ReAsH. Phe1 was found to have an edge-face geometry relative to ReAsH. The close interaction between Phe1 and ReAsH may be highly significant for the fluorescence properties of the ReAsH complex.
Genetically targeted labeling of proteins with small molecules in intact living cells has become popular in the last decade because such tagging combines genetic precision of targeting with the versatile properties of small molecules. This strategy requires that the protein of interest be fused genetically to a peptide or protein domain that can bind a small ligand tightly and selectively enough so that the ligand labels the chimeric protein nearly stoichiometrically and exclusively in a living cell. Many such peptide domain–small molecule ligand pairs have been introduced, but the first to be introduced, the tetracysteine–biarsenical system, has been the most widely used1–4. Here the peptide domain is of the form X3C2X2C2X3, where C = cysteine and X = a non-cysteine amino acid, and the small molecule is a tricyclic fluorophore with As(III) substituents at the 4,5-positions. Each As(III) reversibly forms two covalent bonds to cysteine thiols. We have used NMR to determine the solution structure of the 12-mer peptide FLNCCPGCCMEP, in complex with the ReAsH fluorophore. This peptide system was previously selected after several generations of optimization, culminating in high-throughput genetic screens and fluorescence-activated cell sorting3. The present threedimensional structure results show a relatively tight peptide hairpin formed around the fluorophore, interacting closely with the N-terminal phenylalanine sidechain.
The advantages of the investigated tetracysteine–biarsenical label system include the small size of the peptide domain, only 12 residues, the modest size of the biarsenical (≤ 750 Da), the membrane permeability of the latter, and the very tight binding (sub-picomolar dissociation constants). Low micromolar concentrations of 1,2-ethanedithiol (EDT) or 2,3-dimercaptopropanol prevent toxicity by diverting the biarsenical from binding to most endogenous molecules containing pairs of thiol groups. Millimolar concentrations of the same vic-dithiols can competitively remove the biarsenical from the tetracysteine domains, so that the labeling can be reversed if desired. The fluorescence of the biarsenical moiety is strongly quenched until it binds to the peptide domain. However, the biggest limitation remains background labeling of endogenous proteins. The main strategy for reducing such background has been to evolve tetracysteine domains with higher affinity so that good labeling can be maintained with lower concentrations of biarsenical, higher concentrations of vic-dithiol competitor, or both2, 3.
Despite many biological applications of this and previous tetracysteine domains, no structure of a biarsenical complex has been reported. Attempts at crystallization either alone or fused to fluorescent proteins have not yet been successful. Here we turned to NMR, which would reveal the three-dimensional structure in aqueous solution. We had a choice between the two most widely used (and commercially available) biarsenicals, “FlAsH” and “ReAsH”, based on fluorescein and resorufin, respectively. Complexes of FlAsH with tetracysteine peptides usually can be resolved by HPLC into two closely-spaced peaks of identical amplitude and mass spectra, probably atropisomers with opposite orientations of the carboxyphenyl group relative to the xanthene rings and the peptide. ReAsH lacks the bulky carboxyphenyl substituent, gives single peaks on HPLC, and was considered preferable. The complex of ReAsH with FLNCCPGCCMEP (C-terminus amidated) was not soluble enough for natural-abundance multidimensional NMR, so we acetylated the N-terminus with hydrophilic substituents such as sulfo, succinyl, and MeO(CH2CH2O)nCH2CH2CO-. Succinyl, −OOCCH2CH2CO-, was chosen because it increased the solubility the most (to 1 mM).
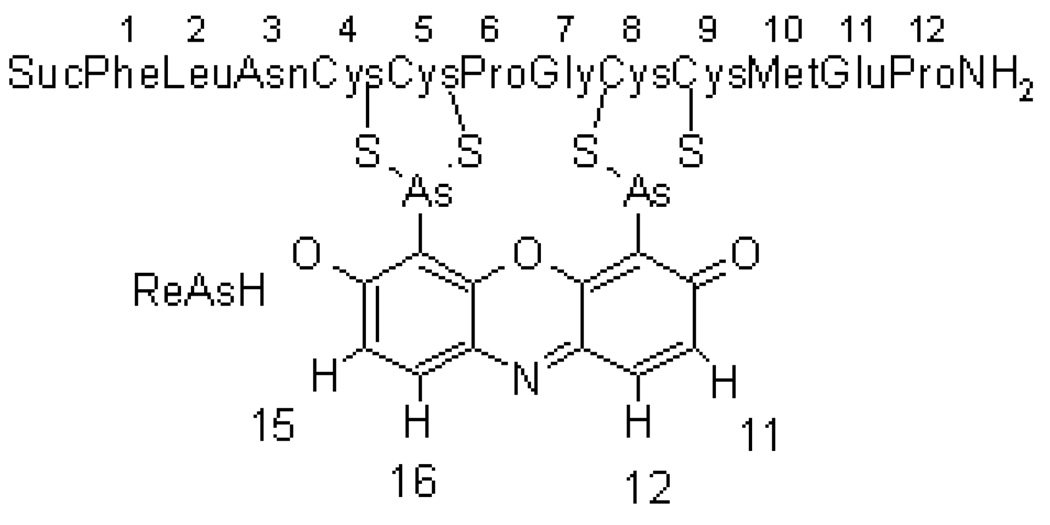
Figure 1 shows the schematic tetracysteine-biarsenical system investigated in this study, with the numbering of the peptide residues and the protons of the resorufin moiety.
Figure 1.
Schematic picture of ReAsH bound to Cys4, Cys5, Cys8 and Cys9 of the peptide. ReAsH protons are numbered 11, 12, 15 and 16. Suc refers to the succinyl group at the N-terminus of the peptide.
CD spectra of the biarsenical complex showed a minimum at 195 nm (Figure S1), suggesting that random coil dominates the secondary structure. A certain contribution from β-structure was suggested by a negative shoulder at approximately 212 nm.
The peptide-ReASH complex in an aqueous solution was investigated by NMR. Using standard homonuclear NMR procedures5, all peptide backbone proton resonances and essentially all sidechain proton resonances were assigned (Table S1). ReAsH by itself is symmetrical and there are two non-equivalent protons in each equivalent ring. A natural abundance 13C-1H HSQC spectrum gave assignments of proton-bonded carbons, in the peptide as well as in resorufin (Table S1), and verified some of the proton assignments. 1H chemical shifts were referenced with respect to the H2O signal at 4.96 ppm6 and 13C chemical shifts were referenced indirectly7.
There are rather large deviations from random coil values of chemical shifts8 for some proton resonances (Figure S2). The presence of the thiol-arsenic bonds causes an approximately 10 ppm down-field shift of the Cβ resonances of the cysteines. The tight structure and potential ring-current shifts are confounded with the presence of the As atoms, making deduction of structural constraints from secondary chemical shifts uncertain.
An NMR translational diffusion experiment9 was performed in order to determine the size of the complex and its oligomeric state in solution. The result showed a translational diffusion coefficient of 12.5×10−11 m2s−1. From this coefficient a hydrodynamic radius and a molecular weight of 1616 Da was determined10 for the diffusing species. This molecular weight is close to that of a monomer (1772.7 Da), showing that the ReAsH bound to the peptide exists as a monomer in the solution at pH 7.4 and 278K.
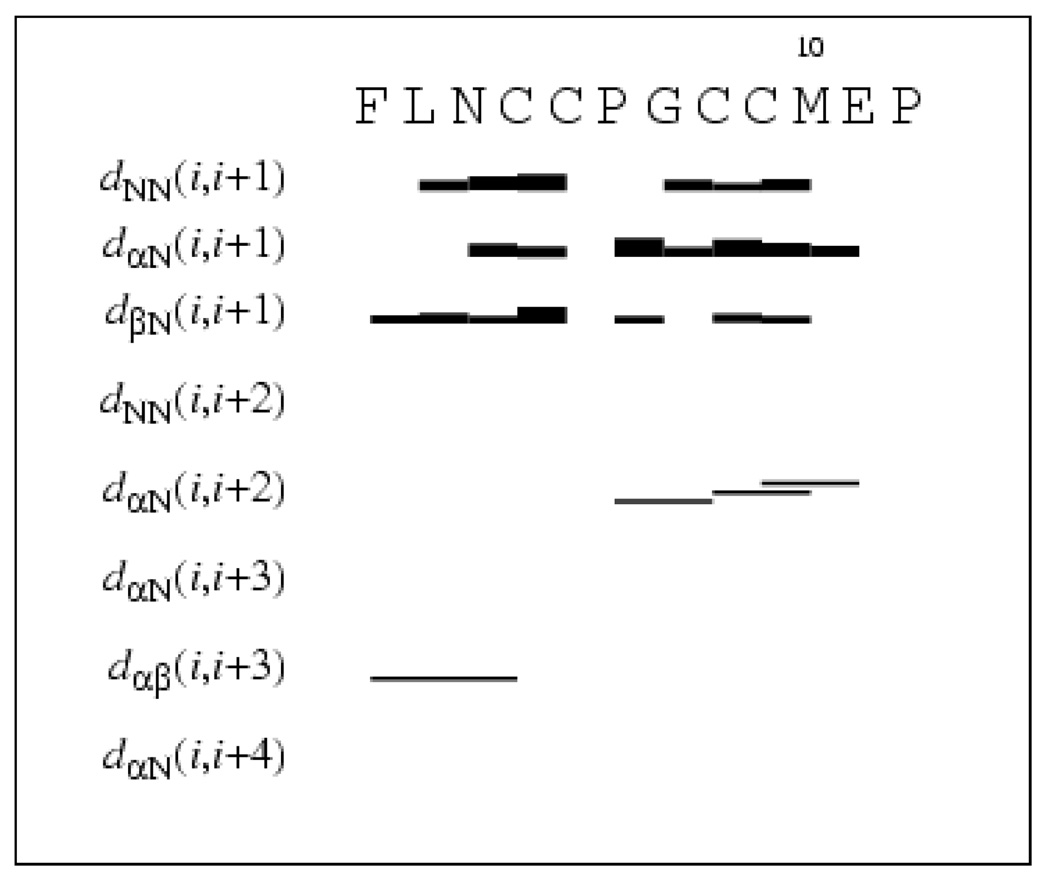
Crosspeaks in a 1H-1H NOESY spectrum recorded with the mixing time of 300 ms were used to calculate distance constraints for structure calculation using the program CYANA11,12. The intensities (volumes) of the crosspeaks were converted into upper distance constraints. No long-range NOEs were observed, but almost all sequential inter-residue NOEs and some non-sequential medium range NOEs were found (Figure 2). We observed NOEs between the β protons of Cys4 and theα, β, and aromatic protons of Phe1 (Table 1). Two NOEs were found between ReAsH and the peptide: between the δ and ε (ζ) protons of the sidechain of Phe1 and the 16 (12) proton of ReAsH (Figure S3).
Figure 2.
Inter-residue connectivities used for the structure calculation for the peptide bound to ReAsH. The NOE constraints were obtained from a 500 MHz NOESY spectrum recorded with a mixing time of the 300 ms at 278K. Thick and thin bars represent strong and weak intensities of sequential NOEs, while lines between non-sequential residues indicate observed medium range NOEs 5. Data are from CYANA (version 2.0).
Table 1.
Structure calculation data of the peptide bound to ReAsH (30 structures).
| Total number of upper distance constraints | 233 |
| Intraresidual | 152 |
| Sequential (i, i+1) | 64 |
| Medium-range (i, i+2; i, i+3) | 11 |
| ReAsH and Phe1 | 2 |
| ReAsH and Cys(S) | 4 |
| CYANA target function | 0.19+/−0.13 |
| Atom rmsd (Å): | |
| Backbone | 1.00+/−0.48 |
| Heavy atoms | 1.28+/−0.42 |
| Residues 4–8 | 0.24+/−0.16 |
The β-protons of Pro6 were stereospecifically assigned. No stereospecific assignment was meaningful for the other β-protons, which were treated in the pseudo atom representation. The four distances between the As and S atoms (Table 1) were given the same upper and lower limits of 2.25 Å as reported from a crystal structure13. In total 233 upper distance and 6 lower distance constraints were used to calculate the structures (Table 1). The upper limits were adjusted iteratively during the structure calculation. Different combinations of binding between the As atoms and the four S atoms in the cysteines were studied by calculating the corresponding structures and comparing the energies and potential constraint violations. Only the isomer where As2 is connected to C4 and C5, and As1 is connected to C8 and C9 results in low energy structures. Other isomers are incompatible with the experimental distance constraints and result in van der Waals conflicts and high energies. Hence, it is concluded that sequential cysteine residues bind to the same As atom, as previously suggested2, based on data from mass spectrometry and fluorescence experiments on chemically modified compounds. Altogether 100 structures were calculated in CYANA. The 30 best structures, regarding violation energies and CYANA target function, were selected for further investigation of the structure.
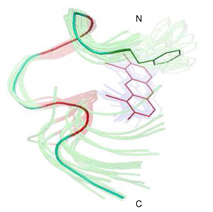
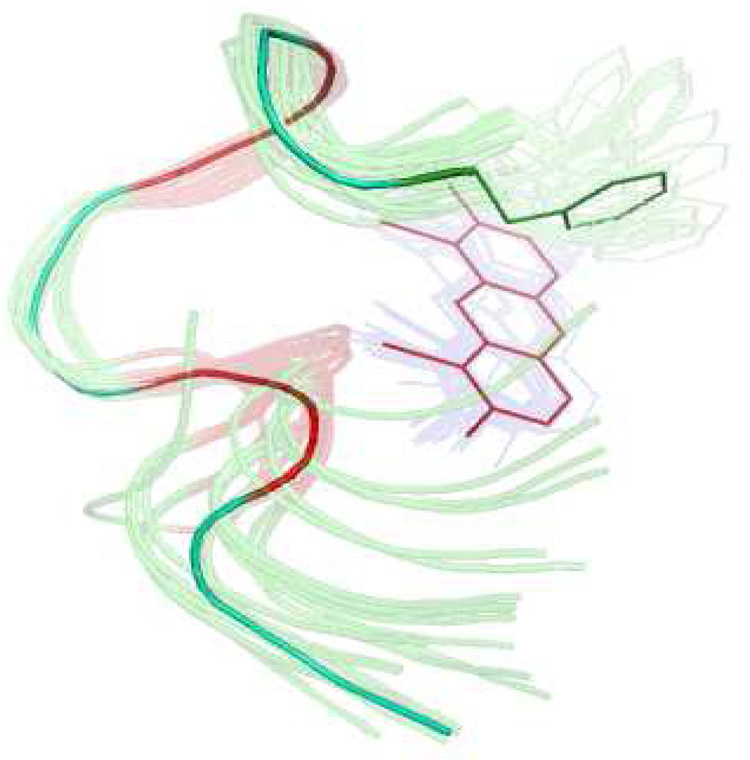
Figure 3 shows an ensemble of the 30 best structures of the peptide backbone bound to the ReAsH based on the violation energies and CYANA target function. The structure calculation data is summarized in Table 1.
Figure 3.
Structure calculation of peptide bound to ReAsH based on the NOESY derived distance constraints. Arsenic atoms are fixed to the cysteine atoms (colored red) at a distance of 2.25 Å. Peptide backbone and ReAsH represented by an ensemble of the 30 best structures appear as semi-transparent, while the average peptide backbone structure from the ensemble, including the Phe1 sidechain and the ReAsH moiety, are shown as a solid line. The overlay was made on the backbone atoms from residue 4 to 8 and also including ReAsH.
The peptide backbone makes a rather well-defined threedimensional structure where P6 and G7 contribute to a relatively tight turn. The conformation of the central CPGC fragment resembles a β-turn type II, where the i+1 and i+2 residues should adopt the dihedral angles ϕ ψ = (−60°,131°) and ((84°,1°)14, respectively. For the PG fragment we find ϕ ψ ≈ (−70°, 163°) for the P residue and (52°, −32°) for the G residue and that the cysteines in the i (C5) and i+3 (C8) positions adopt β conformation. The resemblance to a type II turn explains why the peptide with the spacer residues PG displayed much better binding to ReAsH than peptides with other spacer sequences2.
The preference of the peptide sequence for a proline in position i+1 in type II turns is very strong15 with proline preferred over all other residue types. The preference for glycine in position i+2 is even higher15, Interestingly, cysteine has the highest preference of all residues in position i+3 and the second highest preference for position i15. Hence, the CPGC sequence should fit almost perfectly into the observed structure when bound to ReAsH, which explains why the apparent dissociation constant Kd is very favorable2. However, the ReAsH peptide complex is not a classical β-hairpin. There is no evidence for hydrogen bonds between Cys5 and Cys8, neither do Cys4 and Cys9 adopt β-conformation or display hydrogen bonds. The cysteine in position 9 adopts an αL conformation to make the interaction between its sulfur atom and the arsenic atom possible. The cysteine in position 4 has the dihedral angles ϕ ψ ≈ (−78°,−72°). The constraints imposed by the covalent S-As bonds to the rigid fluorophore may be the cause of this unusual conformation. If Cys4 and Cys9 would adopt β-conformation, as in a classical β-hairpin, their sidechains would point away from the As atoms.
The four (Cys)S-As-C angles are relatively well defined: from Cys4, 122 +/− 2 deg standard deviation in the 30 best structures; Cys5, 126 +/− 15 deg; Cys8, 112 +/− 15 deg; Cys9, 113 +/− 9 deg. The position of ReAsH relative to the peptide was investigated by calculating the angle between the Phe1 sidechain plane and the resorufin plane in the calculated structures. The Phe1 sidechain is located almost perpendicular to the resorufin plane (angle 92 +/− 21 deg). This edge-face geometry is not unusual for interacting aromatic systems16 and allows the partially positive hydrogens of the phenyl ring to contact the highest electron density at the center of the anionic dye. The close positioning of the phenyl ring of Phe1 and ReAsH explains why Phe is strongly preferred at position 1, as already implied by alanine scanning3. However, the structure provides no explanation for the slight preference that alanine scanning had revealed3 for Asn at position 3.
The average distance between the N-terminus (amide N atom) and the C-terminus (carboxy C atom) of the peptide in the 30 best structures was determined to be 12.5 Å +/− 2.2 Å. This distance is relatively small compared to e.g. an α-helix (17.8 Å) with the same number of residues, confirming the hairpin nature of the construct. The hairpin geometry suggests that this tetracysteine-biarsenical label system may also be internally inserted within a protein structure without too much structure disturbance. Previous insertions were limited to minimal CCXXCC sequences17, The present dodecapeptide sequence should enable insertions with increased affinity and fluorescence brightness.
To test whether Phe1 is uniquely favorable for the fluorescence properties and complex stability, complexes were prepared with Phe1 replaced by His, Tyr or Trp. The results (Table 2) showed that the fluorescence of all three variants was quenched compared to the Phe1 variant, probably by electron transfer from the electron-rich sidechains to the fluorophore. The complex stability as assessed by time constants for ReAsH displacement by EDT was decreased by factors of about 2 or more (Table 2). The NMR structure indicated one surprisingly accessible face of the resorufin that could be available for additional interaction with the C-terminal flanking residues. Interchanging this sequence with the corresponding 3 residues from another optimized peptide, HRWCCPGCCKTF3, which also contains a potentially interacting F, to give FLNCCPGCCKTF did not change the fluorescence properties but decreased the stability to EDT 4-fold.
Table 2.
Absorbance, fluorescence and stability to EDT of ReAsH complexes with tetracysteine peptides. Emax, fluorescence emission maximum; ϕ, fluorescence quantum yield; τ, time constant of displacement of ReAsH from complex by 0.5 mM EDT in the presence of 5 mM 2-mercaptoethanesulfonate. Spectra and stability to EDT were measured in 100 mM K•MOPS pH 7.2
| Tetracysteine Peptide | λmax nm |
Emax nm |
εmax M−1cm−1 |
ϕ | τ, sec |
|---|---|---|---|---|---|
| FLNCCPGCCMEP | 597 | 608 | 69000 | 0.48 | 476 |
| YLNCCPGCCMEP | 594 | 607 | 68000 | 0.094 | 256a, 1667 |
| WLNCCPGCCMEP | 601 | 607 | 56000 | 0.034 | 133b, 909 |
| HLNCCPGCCMEP | 596 | 607 | 67000 | 0.20 | 138c, 5000 |
| FLNCCPGCCKTF | 597 | 608 | 65000 | 0.47 | 115 |
comprising 60% of total decrease
comprising 55% of total decrease
comprising 77% of total decrease of resorufin fluorescence.
The NMR results and solution structure calculations of the optimized ReAsH peptide complex suggest that the N-terminus of the 12-mer peptide is relatively tightly constrained. The residues on the N-terminal side of the binding site interact strongly with the resorufin. A hairpin structure with some resemblance to a type II β-turn is formed. The C-terminus is not as well constrained by the NMR results. This could mean that the C-terminus is more dynamic and not strongly interacting with the fluorophore. The N-and C-terminal tripeptides may interact with each other while binding the fluorophore. Further optimization of the tetracysteine motif will require screening of the full 12-mer or even longer sequences.
Supplementary Material
Acknowledgment
This study was supported by the Swedish Research Council (to A.G.), the Knut and Alice Wallenberg Foundation (to A.G.), NIH GM072033 (to R.Y.T.), and HHMI (to R.Y.T.).
Footnotes
Supporting Information Available: Experimental methods, Figures S1–S3, Table S1. The structure coordinates of the FLNCCPGCCMEP_ReAsH_ motif and the NMR chemical shift list have been deposited (BioMagResBank ID code 16041).
REFERENCES
- 1.Griffin BA, Adams SR, Tsien RY. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 2.Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY. J. Am. Chem. Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 3.Martin BR, Giepmans BN, Adams SA, Tsien RY. Nat..Biotechnol. 2005;23:1308–1314. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- 4.Luedtke NW, Dexter RJ, Fried DB, Schepartz A. Nat. Chem. Biol. 2007;3:779–784. doi: 10.1038/nchembio.2007.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wüthrich K. NMR of proteins and nucleic acids. New York: Wiley & Sons; 1986. [Google Scholar]
- 6.Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skelton NJ. Protein NMR Spectroscopy. 2nd ed. USA: Elsevier Inc.; 2007. pp. 236–237. [Google Scholar]
- 7.Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wüthrich K. J. Mol. Biol. 1998;280:933–952. doi: 10.1006/jmbi.1998.1852. [DOI] [PubMed] [Google Scholar]
- 8.Wishart DS, Sykes BD. Meth. Enzymol. 1994;239:363–392. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- 9.Damberg P, Jarvet J, Gräslund A. J. Magn. Reson. 2001;148:343–348. doi: 10.1006/jmre.2000.2260. [DOI] [PubMed] [Google Scholar]
- 10.Danielsson J, Jarvet J, Damberg P, Gräslund A. Magn. Reson. Chem. 2002;40:89–97. [Google Scholar]
- 11.Güntert P, Mumenthaler C, Wüthrich K. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 12.Güntert P. Methods Mol. Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 13.Cruse WBT, James MNG. Acta Crystallogr B. 1972;28:1325–1331. [Google Scholar]
- 14.Hutchinson E, Thornton J. Protein Science. 1994;3:2207–2221. doi: 10.1002/pro.5560031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guruprasad K, Rajkumar S. J. Biosci. 2000;25:143–156. [PubMed] [Google Scholar]
- 16.Waters ML. Curr. Opin Chem. Biol. 2002;6:736–741. doi: 10.1016/s1367-5931(02)00359-9. [DOI] [PubMed] [Google Scholar]
- 17.See for example Ignatova Z, Gierasch LM. Proc. Natl. Acad. Sci. USA. 2004;101:523–528. doi: 10.1073/pnas.0304533101.; Hoffmann C, Gaietta G, Bünemann M, Adams SR, Oberdorff-Maass S, Behr B, Vilardaga JP, Tsien RY, Ellisman MH, Lohse MJ. Nat. Meth. 2005;2:171–176. doi: 10.1038/nmeth742..
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.