Abstract
Because life is often unpredictable, dynamic, and complex, all animals have evolved remarkable abilities to cope with changes in their external environment and internal physiology. This regulatory plasticity leads to shifts in behavior and metabolism, as well as to changes in development, growth, and reproduction, which is thought to improve the chances of survival and reproductive success. In favorable environments, the nematode Caenorhabditis elegans develops rapidly to reproductive maturity, but in adverse environments, animals arrest at the dauer diapause, a long-lived stress resistant stage. A molecular and genetic analysis of dauer formation has revealed key insights into how sensory and dietary cues are coupled to conserved endocrine pathways, including insulin/IGF, TGF-β, serotonergic, and steroid hormone signal transduction, which govern the choice between reproduction and survival. These and other pathways reveal a molecular basis for metazoan plasticity in response to extrinsic and intrinsic signals.
Keywords: Dauer, heterochrony, nuclear receptor, insulin, TFG-β
All living things can sense change in their environment and physiologic milieu and adapt accordingly, revealing remarkable plasticity. Changes in behavior and metabolism are well-recognized forms of plasticity, which are typically rapid and geared to maintain organismal homeostasis. Sustained environmental challenge, such as nutrient limitation, stress, and shifts in photoperiod or temperature, can culminate in long lasting changes in metabolism, behavior, growth, and development, most dramatically resulting in alternate life strategies, such as hibernation, or diapause, a state of developmental arrest. Moreover, restricted dietary intake can delay reproduction and extend organismal life span in diverse species. Collectively, these patterns and their variations comprise an animal's life history. Although determined by genotype, life history traits are largely regulated and plastic, not passively dictated by resource availability. This view is supported by the discovery of myriads of regulatory layers, including neural signaling, hormones, and signal transduction pathways that form logical circuits governing these traits.
Despite its invariant cellular development (Sulston 1988), the worm Caenorhabditis elegans reveals evident plasticity in its life history, which ultimately enhances its chance of survival through environmental hazards, and ensures that somatic growth and reproduction match available resources. Notably, in favorable environments, C. elegans will develop rapidly to reproductive maturity, but in unfavorable environments, animals will arrest at the dauer diapause, a larval stage geared for survival (Fig. 1A). A genetic analysis of C. elegans dauer formation has illuminated how environmental and dietary cues are coupled to evolutionarily conserved molecular pathways, including neurosensory, TGF-β, insulin/IGF, serotonergic, and steroid hormone signal transduction, which impact growth, metabolism, maturation, survival, and aging. These and related studies have led to key insights into similar processes in other metazoans, with implications for understanding disease states of cancer, diabetes, obesity, and other age-related diseases. Here, we review some of the processes, pathways, and molecules involved in the regulatory plasticity of dauer formation and other life history traits.
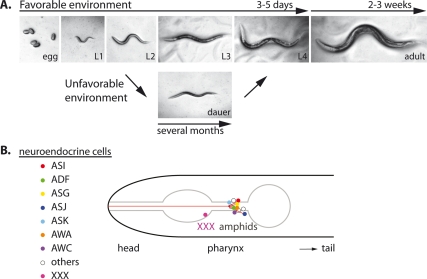
Figure 1.
(A) C. elegans life cycle. In favorable environments, C. elegans undergoes reproductive development and progresses rapidly from embryo through four larval stages (L1–L4) to the adult in 3–5 d (15°C–20°C). Adults then live another 2–3 wk. In unfavorable conditions, including overcrowding, limited food, and high temperature, C. elegans undergoes development to a specialized third larval stage called the dauer diapause (L3d), which can live several months. Upon return to favorable environments, dauer larvae recover to reproductive adults with normal life spans. (B) Schematic of neuroendocrine cells. Integration of environmental cues (dauer pheromone, nutrients, and temperature) are transformed into endocrine signals by amphid neurons (ASI, ADF, ASG, ASJ, ASK, AWA, and AWC). Serotonergic signaling from ADF and cGMP signaling from ASJ and ASK influence production of TGF-β from ASI and the ILPs from ASI, ASJ, and other cells. Insulin/IGF-1 signaling and TGF-β signaling converge on steroid hormone signaling in the XXX cells. Not shown are the other tissues where ILP and steroid hormone production take place. SKN-1 also works in the ASI neuron to regulate DR-induced longevity.
The dauer diapause
C. elegans dauer diapause is a dramatic form of regulatory plasticity that exemplifies the links of environment to life history traits. Although little studied in the wild, C. elegans is thought to feed on bacteria and other microbes as well as micronutrients found in the soil, but must often endure periods without food or extremes of temperature. In the laboratory, it is conveniently cultivated on agar plates containing Escherichia coli, salts, and cholesterol. In favorable laboratory environments, the worm develops rapidly from embryo through four larval stages (L1–L4) to adult in 3.5 d (20°C), called reproductive development (Fig. 1A). Hermaphrodites typically reproduce over a 3- to 5-d period and live another 2–3 wk. When hatched into environments without food, they arrest at the L1 diapause, and fail to initiate larval development (Baugh and Sternberg 2006). If exposed to overcrowded conditions with limiting food during early larval life, animals will divert development to a specialized third larval stage called the dauer diapause (L3d), which can live for months (Cassada and Russell 1975). Other less well understood diapause-like stages have also been described (Ruaud and Bessereau 2006), revealing plasticity throughout the life cycle.
Dauer formation entails vast coordinate changes throughout the body, suggestive of an endocrine mechanism (Riddle and Albert 1997). Prior to the dauer molt, they feed and store fat and carbohydrate. During morphogenesis, they suppress food intake, undergo radial constriction, and remodel various tissues of the body. Once in diapause, they are nonfeeding and spend down their reserves, converting fat to glucose through the glyoxylate cycle (Riddle and Albert 1997). Aerobic respiration is suppressed in favor of glycolysis and fermentative metabolism (Vanfleteren and De Vreese 1996; Holt and Riddle 2003). While often immobile, they can move quickly when prodded, or will rear their bodies onto vertical substrates, behaviors helpful for dispersal. Sealed at all orifices, they have a thickened cuticle resistant to desiccation, with cuticular treads called alae for rapid movement. Generally, they are highly resistant to all forms of stress, including starvation, heat, and oxidative stress (Larsen 1993; Lithgow et al. 1995; Riddle and Albert 1997). Despite their apparent quiescence, several thousand genes continue to be expressed, explicitly important for survival (Wang and Kim 2003). In toto, these defenses enable them to endure literally months under adversity and prevail. When returned to favorable conditions, dauer rapidly recover to post-dauer L3/L4 stages to reproductive adults, with normal life spans. This reveals that animal longevity is plastic and under both environmental and genetic control. Similarly, in response to environmental and nutrient stress, some mammals go through hibernation, torpor, or enter into comparable states of thrifty metabolism, delayed reproduction, and extended survival (Finch 1994).
Genetics of dauer formation
A genetic dissection of dauer diapause has revealed key molecular insights into developmental plasticity and longevity. Over 30 Daf loci inappropriately regulate dauer formation (Riddle et al. 1981; Albert and Riddle 1988). Constitutive mutants (Daf-c) always enter diapause, while defective mutants (Daf-d) bypass diapause, regardless of conditions. Many Daf-c mutants are conditional, forming dauer larvae only at moderately elevated temperatures (25°C). Prior to their molecular characterization, loci were grouped into pathways based on patterns of genetic epistasis and synergy, as well as phenotypic profile (Vowels and Thomas 1992; Thomas et al. 1993; Gottlieb and Ruvkun 1994). Identified pathways include components of sensory neuron structure and function (Shakir et al. 1993), cGMP signaling (Birnby et al. 2000) and serotonergic neurotransmission (Sze et al. 2000), which impinge on TGF-β (Ren et al. 1996) and insulin/IGF-1 signaling (IIS) (Kimura et al. 1997). Ultimately these pathways converge on steroid hormone receptor transcriptional cascades, which are instructive in the dauer decision (Antebi et al. 2000). Molecular genetic studies suggest that when hormones (serotonin, TGF-β, insulin, and steroids) are up, animals select reproductive growth. When down, they select dauer diapause. Parallel processing and molecular cross-talk ensure a robust all or none response when conditions lapse below a critical level. While screens for these major constituents are near saturation, various modulators have been identified through enhancer screens (Ohkura et al. 2003; Yabe et al. 2005; Hu et al. 2006), or by examination of dauer formation at 27°C (Ailion and Thomas 2000, 2003). Below we discuss the cellular and molecular pathways in more detail.
Sensory perception
Animal sensory systems detect and integrate a wide variety of environmental stimuli to evaluate current and predict future events. In C. elegans, plasticity first arises from the detection and processing of graded environmental cues that govern dauer formation—food, temperature, and pheromone—which are integrated by the nervous system to mediate developmental choice (Golden and Riddle 1982, 1984). Unidentified exudates from bacterial food sources and moderate temperatures (15°C–25°C) favor reproductive growth. Scarce food, high temperatures (27°C), and a secreted dauer pheromone (an indicator of population density) bias dauer formation. Dauer pheromone is the most potent inducer of diapause (Golden and Riddle 1982, 1984), and presumably may best predict environmental quality. Recently, its precise chemical structure has been elucidated, revealing three related species of ascarosides: one conjugated to a saturated fatty acid, the second to an unsaturated fatty acid, and the third to a ketone (Jeong et al. 2005; Butcher et al. 2007). The latter two are the most potent, working in the nanomolar range. Pheromones in other species are well known to influence sexual behavior, but are also likely to impact development and reproduction as well. For example, dominant male orangutans not only develop dramatic secondary features that announce their α status, such as facial flaps, large body size, and deep voice, but can suppress the development of subdominant males for years, presumably through release of pheromones (Harrison and Chivers 2007).
Neurons regulating diapause
C. elegans hermaphrodites harbor 302 neurons, including 60 ciliated sensory cells that respond to various modalities. Two major head sensory organs, called amphids, are critical to dauer formation. Specific amphid cells, such as ASI, ADF, and ASG regulate dauer formation (Fig. 1B); Bargmann and Horvitz 1991). Their removal in L1 by laser ablation triggers transient entry to dauer even in the presence of food. Further removal of the amphid ASJ caused constitutive arrest. By inference, these neurons provoke reproductive growth and dauer recovery. Moreover, ASJ, and to a lesser extent ASK, also promote dauer formation in the presence of dauer pheromone (Schackwitz et al. 1996). Aside from their role in chemosensation, specific amphids produce various secreted factors that not only impact dauer (see below), but also influence other traits, including fat storage, foraging, feeding, egg laying, and body size. In fact, hormones that promote reproductive development partly map to some of the neurons named above: DAF-7/TGF-β is expressed exclusively in ASI (Ren et al. 1996), DAF-28/insulin is expressed in ASI and ASJ (Li et al. 2003), and serotonin is synthesized in ADF (Sze et al. 2000). To summarize, the amphids receive sensory input and produce hormones that govern diverse bodily functions, not unlike vertebrate endocrine axes.
G-protein-coupled receptors (GPCRs) and G-protein signaling
GPCRs sense the chemical landscape and represent the first interface between environment and the nervous system (Fig. 2). With >1000 resident GPCRs, an unidentified subset presumably detects dauer pheromone, as well as food cues relevant for dauer formation. A handful of GPCRs show dynamic changes in expression in response to dauer signals, revealing plasticity in the repertoire that likely alters perception (Peckol et al. 2001; Lanjuin and Sengupta 2002). Typically, olfactory and gustatory receptors work in conjunction with heterotrimeric G-proteins. The worm has 21 α, two β, and two γ subunits, including a handful that regulate dauer formation. Notably, mutants of gpa-1, gpa-2, gpa-3, gpc-1, and odr-3 have reduced sensitivity to dauer pheromone, while gpc-11 overexpression leads to greater sensitivity (Zwaal et al. 1997; Lans and Jansen 2007). These G-proteins reside in various neurons, consistent with distributed processing.
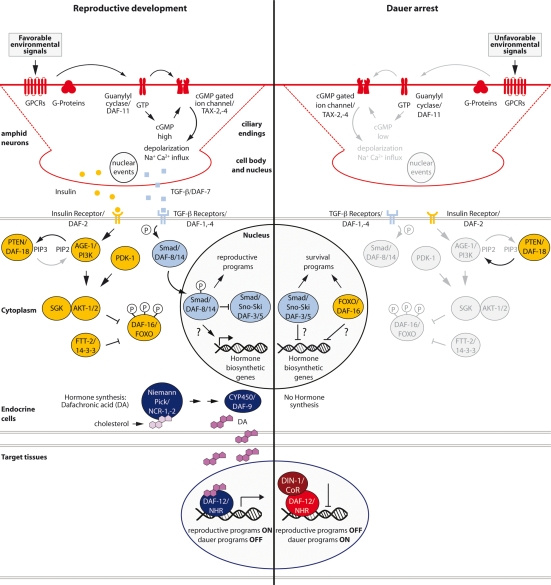
Figure 2.
A model for the regulatory plasticity of dauer formation. Environmental cues (dauer pheromone and nutrients) are detected by GPCRs that reside in the ciliary endings of amphid neurons. These signals may be transduced via G-proteins together with the transmembrane GCY DAF-11, which converts GTP to cGMP. cGMP gated ion channels, TAX-2/TAX-4, transduce cGMP levels into ion influx. (Left) For reproductive development, high cGMP levels, directly or indirectly, lead to production of TGF-β (in ASI) and ILPs in amphids and other tissues. In endocrine cells (e.g., XXX, hypodermal, intestinal cells), binding of ILP agonists to the insulin/IGF-1 receptor DAF-2 results in activation of the AGE-1/PI3Kinase and PIP3 production. Presence of PI3 lipids and activating kinase PDK-1, leads to activation of AKT1,2 and SGK kinases, which phosphorylate DAF-16/FOXO, resulting in cytoplasmic retention by FTT-2/14-3-3 proteins. Binding of DAF-7/TGF-β to the DAF-1/4 receptor kinases results in phosphorylation of DAF-8 and DAF-14/ SMADs, and inhibition of DAF-3/SMAD and DAF-5/SNO-SKI complexes. Nuclear localization of DAF-8 and DAF-14 SMADs directly or indirectly regulates expression of hormone synthesis enzymes (e.g., DAF-9/CYP450), which produce dafachronic acids, the ligands for DAF-12/NHR. Niemann-Pick homologs NCR-1,2 deliver cholesterol and other sterols for use in hormone biosynthesis. In target tissues, liganded DAF-12 promotes reproductive and inhibits dauer programs. (Right) In dauer inducing conditions, low levels of cGMP diminish ILPs and TGF-β production. Down-regulated ILP and TGF-β production results in nuclear translocation of DAF-16/FOXO and DAF-3/DAF-5/SMADs to promote dauer programs and directly or indirectly inhibit expression of hormone biosynthetic genes. In target tissues, unliganded DAF-12 promotes dauer programs and through association with its corepressor DIN-1/SHARP represses reproductive programs. Note that the identified signaling pathways do not necessarily work in a strictly hierarchical fashion, can work in parallel, and have independent outputs.
cGMP signaling
cGMP signaling is critical to neurosensory transduction in animals, and works in close association with G-proteins. Best studied in worms are daf-11, which encodes a transmembrane guanylyl cyclase (GCY) (Birnby et al. 2000), and tax-2 and tax-4, encoding subunits of a cGMP-gated ion channel (Fig. 2; Coburn and Bargmann 1996; Komatsu et al. 1996). daf-11/GCY loss-of-function mutants display potent Daf-c phenotypes, as well as defects in chemosensation and olfaction (Vowels and Thomas 1994). Transmembrane GCYs convert GTP to cGMP, often in response to G-protein signaling or extracellular ligands. Accordingly, daf-11/GCY, but not tax-4 mutants, are rescued by exogenous cGMP analogues (Birnby et al. 2000), consistent with the former producing cGMP and the latter responding to it. Another mutant daf-21 (gain of function) has phenotypes and patterns of epistasis resembling daf-11/GCY. Intriguingly, it encodes an HSP90 homolog but its mode of action is not well understood (Birnby et al. 2000). In contrast to daf-11, tax-2 and tax-4 mutants have weak Daf-c phenotypes, as well as defects in thermotaxis. Epistasis experiments reveal that the strong Daf-c phenotypes of daf-11/GCY depend on functional tax-4, the sensory cilium components (below), as well as the presence of the ASJ neuron (Vowels and Thomas 1992; Schackwitz et al. 1996; Coburn et al. 1998). Taken together, these studies suggest that cGMP levels could be instructive, actively triggering dauer when low, and reproductive growth when high, with signaling mediated through the cilium. Accordingly, daf-11/GCY is expressed in a handful of amphids, including ASI, ASJ, and ASK, and localizes predominantly in ciliary endings (Birnby et al. 2000).
Several lines of evidence support the notion that cGMP signaling works upstream of IIS and TGF-β pathways. First, daf-11/GCY Daf-c mutants are partially suppressed by Daf-d mutants, daf-5/SNO-SKI and daf-16/FOXO (Vowels and Thomas 1992; Thomas et al. 1993), which mediate the transcriptional output of TGF-β and insulin signaling (below). Second, cilium structure mutants suppress the Daf-c phenotypes of daf-11/GCY, but not those of daf-7/TGF-β or daf-2/insulin receptor (InR) mutants. Third, exogenous cGMP rescues daf-11/GCY but not TGF-β or IIS mutants. Finally, daf-11/GCY mutants fail to express daf-7/TGF-β in ASI (Murakami et al. 2001), and daf-28/insulin-like peptide (ILP) in ASI and ASJ (Li et al. 2003), revealing regulatory outputs onto these pathways.
Cilium components
In all animals, ciliated sensory endings of neurons contain components of sensory signal transduction, including receptors, ion channels, and G-proteins. A large class of Daf mutants displays various defects in ciliary structure and biogenesis (Fig. 2; Albert et al. 1981; Perkins et al. 1986), accompanied by deficits in chemosensation (che, osm), an inability of exposed neurons to absorb fluorescent dyes (dyf), and often increased fat accumulation, and longevity (Starich et al. 1995; Apfeld and Kenyon 1999; Ashrafi et al. 2003; Mukhopadhyay et al. 2005). Most cilium structure mutants are Daf-d below 25°C, but weakly Daf-c above it, revealing that the chemosensory organ functions to both prevent or promote dauer formation (Ailion and Thomas 2000), and consistent with the behavior of the ASJ amphid described above. Cilium structure genes encode evolutionarily ancient proteins implicated in intraflagellar transport, including various kinesins, dyneins, and motor regulators, as well as structural components of the cilia themselves (Inglis et al. 2007), many of which are implicated in human conditions such as polycystic kidney disease and Bardet-Biedl syndrome (Barr and Sternberg 1999; Ansley et al. 2003). Among the strongest mutants, daf-19 encodes a highly conserved RFX/Regulatory factor X transcription factor that regulates ciliogenesis in worms, flies, and mammals (Swoboda et al. 2000; Dubruille et al. 2002). Comparable to the worm, mutations in the murine RFX-3 affect ciliogenesis in pancreatic cells, and impairs insulin production and glucose tolerance (Ait-Lounis et al. 2007).
TGF-β signaling
The TGF-β pathway constitutes one of the major endocrine pathways regulating dauer formation (Fig. 2; Savage-Dunn 2005). Genetically grouped together, daf-1, daf-4, daf-7, daf-8, and daf-14 mutants (all Daf-c) are all efficiently suppressed by daf-3 and daf-5 (both Daf-d) (Thomas et al. 1993). These Daf-c loci also show altered social feeding and foraging, increased fat deposition, and egg retention. These loci identify core components of TGF-β signaling: DAF-7 is a novel TGF-β (Ren et al. 1996), and DAF-1 and DAF-4 are type I and II serine/threonine kinase TGF-β receptors, respectively (Georgi et al. 1990; Estevez et al. 1993). DAF-3, DAF-8, and DAF-14 are SMADs, nuclear effectors of the pathway (Patterson et al. 1997; Inoue and Thomas 2000). Finally, DAF-5 is a SNO/SKI transcription factor, which physically interacts with DAF-3/SMAD, similar to the human counterparts (da Graca et al. 2004; Tewari et al. 2004).
DAF-7/TGF-β is expressed solely in the ASI neurosensory cells (Ren et al. 1996; Schackwitz et al. 1996), while downstream receptors and transcription factors are widely expressed (Patterson et al. 1997; Gunther et al. 2000; Inoue and Thomas 2000; da Graca et al. 2004), providing striking evidence for a neuroendocrine signaling mechanism (Fig. 2). As predicted by the genetics, DAF-7/TGF-β expression specifies reproductive growth, with its production positively regulated by food, and inhibited by dauer pheromone and high temperature, showing clear coupling to environmental signals (Ren et al. 1996; Schackwitz et al. 1996). Genetic and molecular data from worms and biochemical data from other systems suggest a simple model whereby graded DAF-7/TGF-β expression from the ASI neuron couples environmental cues to shifts in metabolism and stage alternatives (da Graca et al. 2004). In favorable conditions, high levels of secreted DAF-7/TGF-β bind and activate the DAF-1/4 receptor kinases in target tissues. These receptors are thought to phosphorylate DAF-8 and DAF-14 SMADs, resulting in their nuclear activation, where they antagonize a complex of DAF-3/SMAD and DAF-5/SNO-SKI, and promote energy utilization and reproductive growth (Fig. 2). In unfavorable conditions, reduced or absent DAF-7/TGF-β signaling permits the DAF-3/DAF-5 complex to specify dauer programs, energy storage, and other traits. In addition to these core components, several other loci have been identified as modulators of the pathway (Daniels et al. 2000; Morita et al. 2001; Aoyama et al. 2004; Wang and Levy 2006).
Intriguingly, expression of DAF-4/type II receptor or DAF-5/SNO–SKI primarily in the nervous system rescues phenotypes throughout the body, suggesting that downstream secondary hormones ultimately govern the dauer decision (Inoue and Thomas 2000; da Graca et al. 2004). Consistent with an endocrine mechanism, expression profiles reveal that TGF-β signaling regulates ILPs, as well as cytochrome P450s implicated in steroid hormone production (Liu et al. 2004). Additional evidence also reveals substantial cross-talk with IIS. Notably, TGF-β Daf-c mutants are partly suppressed by daf-16/FOXO (Vowels and Thomas 1992), and cause DAF-16/FOXO to be nuclear localized (Lee et al. 2001). Moreover, DAF-3/SMAD and DAF-16/FOXO share many putative target genes and often have associated binding sites, as revealed by expression profiling (Liu et al. 2004; Shaw et al. 2007). Interestingly, vertebrate SMADs and FOXO also cooperate in cytostatic responses in epidermal, neural, and muscle development (Seoane et al. 2004; Gomis et al. 2006; Allen and Unterman 2007). Until recently, it was thought that cross-talk between TGF-β and IIS in the worm was limited to dauer formation, and that TGF-β signaling had little influence on adult longevity. However, many of the TGF-β Daf-c mutants are substantially long lived in a manner dependent on DAF-16/FOXO, when animals are treated with FuDR to suppress death due to egg retention (Shaw et al. 2007). Thus, TGF-β signaling converges on IIS to promote states of survival, and it will be interesting to see if this holds true in other metazoans.
IIS
The IIS pathway is a pivotal regulator of many life history traits, and exemplifies regulatory plasticity. This pathway has been extensively reviewed, so we highlight only salient features here. IIS specifies alternatives of self-preservation and survival, versus rapid growth and reproduction. IIS plays a critical role in dauer formation, but is best known for its influence on life span. Notably, a modest reduction of the pathway by mutation in the insulin/IGF receptor homolog daf-2 or the PI3-kinase age-1 results in animals that live one- to twofold longer than wild type (Friedman and Johnson 1988; Kenyon et al. 1993). Closely coupled with longevity are associated traits of resistance to heat, oxidative stress, UV, immune infection, and other forms of stress (Lithgow and Walker 2002; Garsin et al. 2003). Remarkably, reduction of IIS in flies and mice also results in longevity and stress resistance, revealing that both the pathway and process are evolutionarily conserved (Clancy et al. 2001; Tatar et al. 2001; Holzenberger et al. 2003). In the worm, strong daf-2/InR mutants also constitutively enter dauer diapause, store fat, are uncoordinated, have extended reproductive periods and reduced broods (Ogg et al. 1997; Gems et al. 1998). However, these traits can be largely uncoupled from adult longevity, revealing that these do not entail obligate tradeoffs (Dillin et al. 2002).
The forkhead transcription factor DAF-16/FOXO has emerged as the key nuclear mediator of longevity and stress resistance, integrating IIS signals to promote survival (Lin et al. 1997; Ogg et al. 1997). Nearly all daf-2/InR phenotypes are strictly dependent on daf-16/FOXO, revealing that IIS antagonizes DAF-16/FOXO (Kenyon et al. 1993; Gottlieb and Ruvkun 1994). A now classic model for IIS action, based on molecular genetic work in worms and biochemical studies in mammals, links the DAF-2/InR receptor to the nucleus (Fig. 2). It is thought that in favorable environments, secreted ILP agonists bind to the receptor and cause recruitment of the IST-1/IRS adaptor protein (Wolkow et al. 2002), resulting in the activation of the AGE-1/PI3Kinase and production of PIP3 (Morris et al. 1996). In the presence of PIP3 lipids and the activating kinase PDK (Paradis et al. 1999), the AKT1,2 and SGK kinases phosphorylate DAF-16/FOXO (Paradis and Ruvkun 1998; Hertweck et al. 2004). This potentiates DAF-16/FOXO nuclear export (Henderson and Johnson 2001; Lee et al. 2001; Lin et al. 2001) and cytoplasmic retention by FTT-2/14–3–3 proteins (Wang et al. 2006; Li et al. 2007). Consequently, animals undergo reproductive growth and have a normal life span. In unfavorable environments, IIS is inactivated, DAF-16/FOXO enters the nucleus, where it turns on genes for stress resistance, dauer formation, and longevity. In addition, the DAF-18/PTEN phosphatase opposes the activity of AGE-1, down-regulating the pathway (Ogg and Ruvkun 1998; Gil et al. 1999; Mihaylova et al. 1999; Rouault et al. 1999).
Nuclear localization of DAF-16/FOXO is not sufficient to recapitulate its physiology, suggesting other activities are involved (Henderson and Johnson 2001; Lee et al. 2001; Lin et al. 2001). In addition to the core pathway, multiple inputs converge onto DAF-16/FOXO including several kinases such as JNK (Oh et al. 2005), MAPK (Nanji et al. 2005; Troemel et al. 2006), and STE20-like (Lehtinen et al. 2006) and AMP-dependent kinases (Apfeld et al. 2004; Greer et al. 2007). Similarly, various nuclear complexes, including heat-shock factor (Hsu et al. 2003), β-catenin (Essers et al. 2005), sirtuin protein deacetylase (Berdichevsky et al. 2006; Wang et al. 2006), SMK-1 nuclear coregulator (Wolff et al. 2006), and DAF-12 nuclear receptor (Berman and Kenyon 2006), modulate distinct aspects of DAF-16/FOXO function.
Further signaling diversity is achieved through ILPs, which number 38 in the worm (Pierce et al. 2001; Li et al. 2003). Interestingly, some function as receptor agonists (e.g., INS-7) and others as antagonists (e.g., INS-1) (Pierce et al. 2001; Murphy et al. 2003). Expressed primarily in neurons, but also in gut, muscle, and other tissues, the ILPs presumably work combinatorially to carry out multiple aspects of IIS biology. Importantly, daf-28/ILP is transcriptionally inhibited by dauer pheromone and low food, showing clear regulation by sensory cues (Li et al. 2003). Moreover, proteins involved in synaptic transmission such as UNC-64/syntaxin and UNC-31/CAPS (Ailion et al. 1999; Munoz and Riddle 2003), as well the conserved ASNA-1/ATPase, govern ILP release (Kao et al. 2007).
In wild-type adults, DAF-16/FOXO nuclear localization is regulated in response to environmental stress. Oxidative challenge, heat, and food deprivation send FOXO into the nucleus, giving visible molecular evidence of plasticity (Henderson and Johnson 2001; Lee et al. 2001; Lin et al. 2001). The transcriptional output of FOXO includes genes that mitigate stress and promote repair, including those involved in ROS detoxification, heat shock, innate immunity, heavy metal stress, and xenobiotic detoxification (McElwee et al. 2003; Murphy et al. 2003). Moreover, expression changes suggest increased levels of gluconeogenesis, glycolysis, and β oxidation. Targets that are repressed include genes involved in growth (e.g., TOR pathway components, Jia et al. 2004) and reproduction (e.g., vitellogenins). Interestingly, daf-2/InR and daf-16/FOXO regulate dauer formation and longevity cell-nonautonomously, principally from neurons and intestine, suggesting that downstream targets include hormones (Apfeld and Kenyon 1998; Wolkow et al. 2000; Libina et al. 2003). Accordingly, target genes include ILPs themselves, such as INS-7, as well as numerous cytochrome P450s and short chain dehydrogenases that could be involved in steroid hormone metabolism (McElwee et al. 2003; Murphy et al. 2003). Hormonal communication between tissues reflects a high level of systemic decision-making critical to plasticity.
Neurosensory control of longevity
One of the most provocative findings is that sensory perception also influences adult life span, primarily through IIS. Many of the mutants involved in sensory signal transduction, including cilium structure mutants, tax-4/cGMP channel subunit, various G-proteins, and the str-2/GPCR extend adult life span to various extents (Apfeld and Kenyon 1999; Alcedo and Kenyon 2004; Lans and Jansen 2007). Similarly, ablation of the amphid sheath cell or specific gustatory (ASI and ASG) and olfactory (AWA and AWC) neurons modestly prolong life (Fig. 1B). Interestingly, ablation of ASK and ASJ suppresses longevity due to ASI loss, suggesting a neuronal relay that influences life span (Alcedo and Kenyon 2004). These manipulations are thought to down-regulate ILP agonists or up-regulate ILP antagonists, thereby activating transcription factor DAF-16/FOXO. Accordingly, sensory mutants result in FOXO nuclear translocation and longevity is often largely DAF-16/FOXO-dependent (Apfeld and Kenyon 1999; Lin et al. 2001). Interestingly, food derived odors can curtail the life extension due to dietary restriction in Drosophila, and olfactory mutants such as or83b are long lived (Libert et al. 2007). Moreover, insulin is released in response to the smell and sight of food in humans (Sjostrom et al. 1980). Altogether these observations suggest that perception—one of the clearest examples of neural plasticity—can deeply impact organismal metabolism, development, and life span.
Serotonin
Serotonergic transmission is an evolutionarily ancient signaling pathway that couples food and stress signals to various parts of the body. Serotonin can work in classical synaptic transmission or at a distance as a neurohumoral modulator of peripheral tissues. Moreover, it regulates synthesis and release of a wide variety of neuropeptides, hormones, and cytokines to influence physiology and behavior related to satiety, energy expenditure, immune function, and thermoregulation (Lucki 1998; Mossner and Lesch 1998). Similarly, serotonergic signaling in C. elegans plays a role in adaptive behavior, physiology, development, and pathogen avoidance, coupling sensory information to IIS and TGF-β signaling pathways (Fig. 3; Sze et al. 2000; Zhang et al. 2005; Liang et al. 2006; Chase and Koelle 2007).
Figure 3.
Heterochronic loci and control of developmental timing. The heterochronic pathway controls temporal development specifying stage appropriate programs for each larval stage. Upon hatching, food signals lead to up-regulation of lin-4/miRNA, which down-regulates its targets LIN-14/NUCLEAR PROTEIN and LIN-28/RNA BINDING PROTEIN, resulting in L1-to-L2 transitions. Up-regulation of let-7 microRNA family members (mir-48, mir-84, and mir-241) down-regulate hbl-1/HUNCHBACK to trigger the L2/L3 transitions. DAF-12/NHR integrates environmental signals from the dauer pathways (cGMP, serotonin, insulin, and TGF-β). In unfavorable environmental conditions, unliganded DAF-12 together with its corepressor DIN-1/SHARP shut down the heterochronic circuit and specify dauer development. In favorable environmental conditions, liganded DAF-12 advances the heterochronic circuit to the L3 stage, and thus promotes reproductive development and maturation (for details, see Fig. 2 and the text). Several gene products control late larval development by preventing expression of LIN-29/ZnF transcription factor, which is instructive for adult development. The larval-to-adult transition is triggered through expression of let-7/miRNA, which leads to down-regulation of LIN-41/RBCC protein and up-regulation of LIN-29/ZnF transcription factor. Not all heterochronic activites are shown.
TPH-1 tryptophan hydroxylase is the rate-limiting enzyme for serotonin biosynthesis.
In C. elegans, tph-1 mutants exhibit depressed feeding and egg retention—behaviors seen in wild type under starvation conditions (Sze et al. 2000). In addition, mutants display increased fat deposition and weak Daf-c phenotypes at all temperatures. Notably, the latter phenotypes are suppressed by both daf-3/SMAD and daf-16/FOXO, suggesting inputs onto both TGF-β and IIS pathways. Accordingly, tph-1 mutants synergize with daf-7/TGF-β mutants for Daf-c phenotypes and depress daf-7/TGF-β expression in ASI. Moreover, tph-1 mutants induce DAF-16 nuclear accumulation, evoke resistance to heat stress and immune challenge, and up-regulate SOD-3 expression—all in a daf-16/FOXO-dependent manner (Liang et al. 2006). Conceivably, serotonin impacts IIS by regulating production of ILPs, but this has not yet been shown. Consistent with reduced IIS signaling, tph-1 mutants also exhibit an extended reproductive period, but surprisingly, demonstrate normal longevity (Sze et al. 2000; Murakami and Murakami 2007). By contrast, the activity of two different serotonin receptors, ser-1 and ser-4, reduce and extend adult longevity, respectively, perhaps neutralizing one another's effect (Murakami and Murakami 2007). Importantly then, serotonergic signaling converges on IIS, coupling sensory information to the stress response. Similarly, in humans, drugs that augment serotonergic signaling may improve insulin sensitivity (Lam and Heisler 2007).
TPH-1 is expressed in a handful of serotonergic neurons, including ADF. In accord with cell ablation experiments described above, TPH-1 activity in ADF regulates dauer, DAF-16 nuclear localization, and stress responses (Liang et al. 2006). TPH-1 expresssion in ADF itself is down-regulated in response to starvation and high temperature. Sensory transduction mutants, ocr-2 and osm-9, which encode TRPV ion channel subunits, reduce TPH-1 expression, while CaM-kinase gain-of-function mutants suppress this phenotype (Zhang et al. 2004). Consistent with a unified pathway, ocr-2, and osm-9, like tph-1, enhance the Daf-c phenotype of daf-7/TGF-β mutants. This has led to a model whereby food cues result in opening of TRPV channel subunits. Ca2+ influx activates CaM-kinase, which in turn stimulates nuclear transcription of the tph-1 gene. Intriguingly, TPH-1 expression itself is also regulated by IIS and TGF-β pathways: tph-1∷gfp is down-regulated in daf-2/InR mutants but up-regulated in daf-7/TGF-β mutants, revealing substantial cross-talk or feedback among the three neuroendocrine pathways (Estevez et al. 2006).
Steroid hormone signaling
Nuclear receptors are transcription factors that often respond to lipid hormones to regulate gene transcription, and are well poised to coordinate life history traits. Typically, they contain an N-terminal DNA-binding domain (DBD), consisting of paired C4 Zn fingers, and a C-terminal ligand-binding domain (LBD), which also docks coactivator and corepressor complexes. The worm genome harbors 284 nuclear receptors, fivefold more than vertebrates, with roughly 15 conserved across taxa (Robinson-Rechavi et al. 2005). Among them, DAF-12/NHR is perhaps best understood for its various roles in dauer formation, fat metabolism, developmental timing, gonadal maturation, and longevity. DAF-12/NHR is most closely related to the vertebrate vitamin D and LXR receptors (Antebi et al. 2000; Snow and Larsen 2000), but regulates reproductive maturation analogous to the estrogen receptor. Genetic epistasis experiments reveal that it works at the end of the dauer pathways, downstream from cGMP, TGF-β, and IIS pathways (Fig. 2; Riddle et al. 1981; Vowels and Thomas 1992; Thomas et al. 1993). Null mutations, which typically reside in the DBD, are Daf-d, and suppress all known Daf-c mutants in upstream pathways. By contrast, lesions in the LBD are often Daf-c, suggesting that loss of hormone binding specifies diapause.
Numerous lines of evidence reveal that DAF-12 is regulated hormonally. Notably, DAF-9 is a cytochrome P450 related to steroidogenic hydroxylases that works proximal to DAF-12/NHR. daf-9/CYP450-null mutants are unconditionally Daf-c, but will slowly recover to sterile adults that are stress resistant and long-lived (Gerisch et al. 2001; Jia et al. 2002). These phenotypes are potently suppressed by daf-12-null mutants, suggesting that the unliganded receptor specifies states of diapause and survival. DAF-9 is expressed in a pair of neuroendocrine cells called XXX, the epidermis, and the hermaphrodite spermatheca (Fig. 1). The XXX cells are a critical point of signal integration, where many dauer components reside (Ohkura et al. 2003; Li et al. 2004; Hu et al. 2006). However, the most visible DAF-9/CYP450 regulation is seen in the epidermis in response to food, pheromone, temperature, cholesterol, as well as TGF-β, cGMP, and insulin signaling. Interestingly, much of this regulation ultimately depends on DAF-12/NHR, suggesting feedback control (Gerisch and Antebi 2004; Mak and Ruvkun 2004). Consistent with an endocrine mechanism, DAF-9/CYP450 works cell-nonautonomously, while DAF-12/NHR is nuclear and widely expressed (Gerisch and Antebi 2004; Mak and Ruvkun 2004). Another hormone biosynthetic enzyme, DAF-36, is a Rieske-like oxygenase (Rottiers et al. 2006), whose Drosophila homolog has also been implicated in ecdysone production (Yoshiyama et al. 2006). DAF-36 resides predominately in the intestine, revealing that the hormone biosynthetic pathway is distributed among tissues. A distributed biosynthetic pathway may be one way in which different tissues can register their physiologic state. Transport of sterols is likely mediated by Niemann-Pick Type C proteins, NCR-1,2, and possibly the multidrug resistance transporter, MRP-1, whose human homologs are implicated in cholesterol/steroid transport (Sym et al. 2000; Li et al. 2004; Yabe et al. 2005). Indeed, cholesterol deprivation triggers Daf-c phenotypes, thus linking dietary sterols directly to dauer signal transduction (Gerisch et al. 2001).
In accord with the hormone hypothesis, endogenous ligands for DAF-12/NHR have been chemically identified as bile acid-like steroids called Δ-4 and Δ-7 dafachronic acid (DA) (Motola et al. 2006). These molecules transcriptionally activate DAF-12/NHR in the nanomolar range. A related bile acid, 25S-cholestenoic acid also serves as a low affinity ligand to DAF-12 as well as for its mammalian homolog LXR (Song and Liao 2000; Held et al. 2006). As predicted from the genetics, DA supplementation efficiently rescues the dauer and adult longevity phenotypes of daf-9 mutants. Additionally, DA rescues Daf-c phenotypes of upstream IIS and TGF-β mutants, but not those of downstream daf-12 LBD mutants. As expected for endogenous ligands, DAs are present in wild type but absent from daf-9 mutants (Motola et al. 2006). Like its vertebrate relatives, DAF-12 assembles with coregulators in a ligand regulated fashion. Notably, DIN-1/CoR, a homolog of mammalian SHARP corepressor, forms a complex with DAF-12 to specify dauer diapause and associated traits (Ludewig et al. 2004). This complex associates in the absence of ligand but dissociates in its presence. Moreover, in the presence of DA, DAF-12 can assemble with mammalian coactivator SRC-1, although endogenous coactivators have yet to be found (Motola et al. 2006). Altogether, these studies show that from biology to mechanism, steroid control of reproduction is evolutionarily ancient. Furthermore, they provide pioneering evidence that bile acids influence metazoan longevity (Gerisch et al. 2007).
The outline of a branched biosynthetic pathway for DAs has been deduced from the activities of biosynthetic enzymes and known sterol intermediates. Namely, DAF-9/CYP450 carries out the last steps, oxidizing the sterol sidechain to a carboxylic acid (Motola et al. 2006) a reaction that is biochemically orthologous to mammalian CYP27A1, a key enzyme in bile acid synthesis (Russell 2003). DAF-36/Rieske is involved in the first step, converting cholesterol to 7-dehydrocholesterol (Rottiers et al. 2006). Both enzymes are controlled by various upstream inputs of the dauer pathways, providing important links in the endocrine network.
Altogether, this has led to a unified model for dauer formation whereby graded signals from the environment regulate a hormone-dependent switch (Fig. 2). In favorable conditions, cues integrated by the nervous system and perhaps other tissues result in the production of TGF-β and ILPs. Activation of their respective signaling pathways stimulates synthesis of DA. When ligand bound, DAF-12/NHR and presumptive coactivators specify reproductive growth. In unfavorable conditions, failure to produce TGF-β and ILPs results in down-regulation of DA synthesis, whereby unliganded DAF-12/NHR together with corepressor DIN-1/CoR specify dauer diapause and extended survival. Despite the simplicity of this model, many of the identified pathways are unlikely to work in a strictly hierarchical manner and are likely to show feedback. Moreover, many of the mechanistic connections and cellular communication between the various signaling pathways have yet to be determined.
In other animals, regulation of reproduction by similar endocrine pathways suggests these comprise critical components of an evolutionarily ancient mechanism. In Drosophila, InR mutants have impaired ecdysteroid biosynthesis and are infertile (Tu et al. 2002). In mice, genetic ablation of insulin signaling in the brain reveals a role in fertility (Bruning et al. 2000; Burks et al. 2000), and IGF-1 and TGF-β signaling in the hypothalamus influence the expression and release of gonadotropes that promote maturation (Bouret et al. 2004; Daftary and Gore 2005). Moreover, TGF-β, IIS, and steroid hormone signaling converge in the ovary to orchestrate folliculogenesis (Castrillon et al. 2003; Park et al. 2005). Further study may reveal intimate connections between neural and dietary cues, and hormones to other aspects of animal life history.
Dietary restriction
Dietary restriction, a reduction of dietary intake without malnutrition, has health benefits and extends the life span of a range of animals, suggesting an evolutionarily conserved process. DR is thought to invoke a regulated mechanism that couples nutrient availability to metabolism, growth, and reproduction. Indeed, worms undergoing DR utilize fat, reduce growth, and diminish reproductive output (Houthoofd and Vanfleteren 2006). Extended life span likely arises as a secondary consequence of increased protection, hormesis, or thrifty metabolism. Until recently, the molecular basis of DR-induced longevity remained obscure. The protein deacetylase SIR-2 is proposed to mediate DR in yeast, worms, flies, and mice (Lin et al. 2000; Rogina and Helfand 2004; Wang and Tissenbaum 2006; Bordone et al. 2007), but this issue remains somewhat unresolved (Kaeberlein et al. 2004; Hansen et al. 2007). IIS is an obvious candidate given the intimate connection between diet, metabolism, and longevity. Indeed, in some DR regimens a role for daf-16/FOXO is implicated (Greer et al. 2007), but other regimens suggest otherwise (Lakowski and Hekimi 1998; Houthoofd et al. 2003; Bishop and Guarente 2007; Panowski et al. 2007). Recently, two transcription factors, the FOXA homolog, PHA-4, and the NRF homolog SKN-1 appear to specifically mediate DR-induced longevity in worms (Bishop and Guarente 2007; Panowski et al. 2007). Intriguingly, SKN-1 does so from the ASI neurons, suggesting a hormonal mechanism coordinates the DR response. It remains to be seen whether these functions are evolutionarily conserved. Finally, genetic manipulations that down-regulate TOR kinase signaling, and protein synthesis itself, extend life span, but the relationship to DR and IIS appears complex (Vellai et al. 2003; Jia et al. 2004; Hansen et al. 2007; Pan et al. 2007; Syntichaki et al. 2007).
Body size
Body size is a phenotypically plastic trait that is genetically programmed, but also dynamically regulated. In particular, it would be beneficial to couple environmental cues, like nutrient availability, to appropriate somatic growth. In mammals, body size is principally governed by growth hormone and IGF-1 endocrine axes. In C. elegans, the SMA pathway (for small/male tail) regulates body size, male tail development, and immune function (Savage-Dunn 2005). It is represented by a canonical TGF-β pathway distinct from that used for dauer, although the daf-4/Type II receptor is common to both. The accumulated evidence suggests that body size is determined by cell volume, rather than number, and is governed by the degree of endoreplication in the epidermis (Morita et al. 2002; Lozano et al. 2006).
Like other traits, body size may be regulated by neurosensory input and diet. Expression of DBL-1/TGF-β in the nervous system is sufficient to rescue body size defects for the whole organism, implying that neural or sensory inputs could regulate growth. Accordingly, mutants in cilium structure (e.g., che-2, che-3, and osm-6) or neurosensory signal tranduction (e.g., tax-2, tax-4, and kin-29) result in smaller than normal worms. Conversely, mutants of the egl-4/cGMP-dependent kinase result in larger animals, as well as defects in olfactory plasticity (Fujiwara et al. 2002; Lanjuin and Sengupta 2002; Hirose et al. 2003). Many of these mutations also affect dauer formation and longevity. Size differences are not due to altered nutrient input, since animals often have normal dietary intake and broods. Wild-type animals with reduced food intake, or genetic mimetics of dietary restriction such as eat-2 mutants, are smaller. Interestingly, DR may reduce growth in part by limiting epidermal polyploidization (Tain et al. 2008). However, dietary intake may work independently of the SMA pathway, since eat-2 sma double mutants are even smaller (Morck and Pilon 2006). Conceivably, at the molecular level, growth is regulated through TOR signaling, ribosomal biogenesis, or remodeling by autophagy (Morck and Pilon 2007).
Developmental timing
Temporal patterns of post-embryonic development are determined by the genome, but are highly responsive to environmental conditions and show great diversity throughout the animal kingdom. In C. elegans, the heterochronic loci regulate temporal patterning, specifying stage appropriate programs during each of the larval stages in a variety of tissues, including stem cell divisions of epidermal seam cells and migration programs of the gonadal distal tip cells. Mutations in the heterochronic genes transform the temporal identity of life stage programs, such that larval programs are expressed in the adult (delayed) or adult programs are expressed in the larva (precocious). Notably, most identified loci are evolutionarily conserved, including the first discovered microRNAs (lin-4 and let-7), 22-nucleotide-long RNAs that down-regulate gene expression by base-pairing with the 3′ untranslated region (UTR) of mRNA targets (Lee et al. 1993; Wightman et al. 1993; Pasquinelli et al. 2000; Reinhart et al. 2000; Lagos-Quintana et al. 2002). These loci have been placed into a regulatory hierarchy by performing genetic epistasis and molecular experiments; essentially the components work as switches to transition from one stage program to the next (Moss 2007). Their study has not only led to important insights into developmental timing, but also into stem cell biology, differentiation, and cancer.
An important question is whether the heterochronic circuit is hard wired, or shows regulatory plasticity. Intriguingly, plasticity is already revealed at the initiation of larval development: Only upon a food signal is lin-4/miRNA up-regulated, which in turn down-regulates LIN-14/nuclear protein, enabling the heterochronic circuit to proceed from the L1 to the L2 stage programs (Fig. 3; Lee et al. 1993; Wightman et al. 1993). In the absence of food, animals enter the L1 diapause, sustaining LIN-14 expression (Ambros and Moss 1994). In L2, LIN-14 and LIN-28/RNA-binding protein are further down-regulated by lin-4 (Moss et al. 1997). In addition, members of the let-7 microRNA family (mir-48, mir-84, and mir-241) specify the L2/L3 transition, in part through down-regulation of hbl-1/HUNCHBACK (Abbott et al. 2005; Li et al. 2005). Perhaps most dramatically, DAF-12/NHR reveals plasticity within the heterochronic circuit itself. It works at the convergence of the dauer and heterochronic pathways, coupling environmental information into the circuit (Antebi et al. 1998). DAF-12 mutants repeat L2 programs inappropriately at the L3 stage, in epidermis and gonad. By inference, DAF-12 is required for L2/L3 transitions as well as L2/L3d (dauer) transitions. Importantly, in unfavorable environments, DAF-12 shuts down the heterochronic circuit, while in favorable environments, DAF-12 drives forward development toward sexual maturation (Fig. 3). This may be analogous to the way in which nutrient levels regulate the onset of puberty (Frisch 1987).
Late larval development and the larval-to-adult transition are tightly controlled by several loci that prevent expression of LIN-29/C2H2 ZnF transcription factor, which specifies terminal differentiation and adult development (Fig. 3; Rougvie and Ambros 1995). These loci include HBL-1/HUNCHBACK (Abrahante et al. 2003; Lin et al. 2003), LIN-41/RBCC (Slack et al. 2000), and DRE-1/F-box protein (Fielenbach et al. 2007). Up-regulation of let-7/miRNA in L4 results in down-regulation of lin-41 and hbl-1 allowing LIN-29 expression and the larval-to-adult transition (Reinhart et al. 2000; Slack et al. 2000; Vella et al. 2004). Interestingly, circadian rhythm homologs LIN-42/PERIOD, TIM-1/TIMELESS, and KIN-20/DOUBLETIME may also play a role in this transition (Jeon et al. 1999; Banerjee et al. 2005).
Although mammalian circadian rhythm proteins integrate environmental signals, such as daylight cycle and temperature, no such connection has been found in the worm. However, the heterochronic circuit, like the circadian circuit, resets in response to nutrients. For example, many heterochronic mutants are suppressed by transition through dauer or starvation (Liu and Ambros 1991; Euling and Ambros 1996). How this reprogramming is achieved is not understood. Intriguingly, lin-4 and lin-14 also impact adult longevity, working through IIS and DAF-16/FOXO (Boehm and Slack 2005). In particular, the lin-4 mutant is short-lived, while lin-14 mutants are long-lived in a daf-16/FOXO-dependent manner. It is therefore reasonable to speculate that timing circuits might retard or accelerate organismal maturation, with secondary consequences on aging.
Reproduction
Many aspects of reproduction show plasticity, and here we mention only a few. First, brood size is tightly coupled to nutrient availability, and animals with restricted dietary intake have reduced broods (Klass 1977; Lakowski and Hekimi 1998; Houthoofd and Vanfleteren 2006). Similarly, mutants in many of the pathways described above including IIS, steroid, TGF-β, and SMA, have reduced fecundity, but it is unknown whether this reflects regulation or pleiotropy. Secondly, the reproductive schedule can be extended by dietary restriction as well as by reduction in IIS and serotonergic signaling (Gems et al. 1998; Sze et al. 2000; Crawford et al. 2007). Egg laying is an important, often overlooked aspect of reproductive plasticity; wild-type animals retain eggs in response to starvation, which can lead to a process called bagging (Chen and Caswell-Chen 2004). Bagging may be beneficial, since the progeny that hatch inside feed off the mother's carcass often make it to the dauer stage. Serotonin and TGF-β signaling influence this behavior as well (Schafer 2006). The heterochronic gene lin-29/ZnF regulates maturation of the serotonergic egg-laying neuron HSN (Bettinger et al. 1997; Schafer 2006), again suggesting that some plasticity could be exerted through developmental timing pathways. Finally, gonadal development itself is somehow tied to nutrients; animals starved for food or sterols will often have unreflexed gonads, not unlike certain DAF-12/NHR LBD mutants (Antebi et al. 1998; Gerisch et al. 2001).
Evolutionary theory suggests a tradeoff between growth and reproduction and somatic maintenance (Partridge et al. 2005). Given limited resources, energy channeled into one process comes at the expense of another. If so, clearly this cannot be passive since many mutants uncouple food availability, which is unlimited in the laboratory, from reproductive output. Most discussed is the tradeoff between reproduction and longevity (Partridge et al. 2005). Although many long-lived mutants diminish reproduction, some do not, notably weak daf-2/InR mutants (Gems et al. 1998; Tissenbaum and Ruvkun 1998). Another example is the germline longevity pathway. Ablation of the germline stem cells through laser microsurgery or mutation extends hermaphrodite life span 50%–60%, suggesting a tradeoff with reproduction (Hsin and Kenyon 1999; Arantes-Oliveira et al. 2002). However, further removal of somatic gonadal support cells results in animals that are still infertile, but yet not long lived, instead suggesting that germline and somatic gonad produce antagonistic signals that reduce or extend life span, respectively. Molecules required for germline longevity include DAF-16/FOXO, KRI-1 ankyrin repeat protein, DAF-12/NHR and hormone biosynthetic enzymes DAF-9/CYP450 and DAF-36/Rieske oxygenase, as well as dafachronic acid and the steroid pregnenolone (Hsin and Kenyon 1999; Gerisch et al. 2001, 2007; Berman and Kenyon 2006; Rottiers et al. 2006; Broue et al. 2007). Genetic analysis suggests a model where in the absence of germline derived signals, KRI-1 and dafachronic acid promote translocation of DAF-16/FOXO into the nucleus of intestinal cells, where it works in conjunction with DAF-12/NHR to extend life. Interestingly, the germline longevity pathway appears to be evolutionarily conserved, as ablation of germline stem cells in Drosophila also results in extension of life in a manner that implicates IIS (Flatt et al. 2008). In nature, germline longevity may represent a type of plasticity that links the state of the germline stem cells to the soma. For example, a delay in germline development could delay aging so that reproduction is coordinated throughout the organism.
Perspective
Clearly, many aspects of C. elegans post-embryonic development, reproductive maturation, and longevity are plastic and subject to regulation. It begins with perception of the environment through the nervous system and dietary intake. By coupling environmental information to conserved endocrine signaling pathways, close coordination of states of survival, growth, and maturation can be achieved. Feedback and communication among tissues as well as molecular cross-talk ensure robust decision making. In addition, stochastic factors provide an important source of variation, which impacts these processes.
Despite the wealth of knowledge gleaned from the worm, many questions remain. What specific nutrient cues impact these processes and how are they sensed? How are the environmental cues integrated in the nervous system and transformed into endocrine signals? How is cross-talk achieved between the endocrine pathways? What molecular pathways mediate dietary restriction? Do nutrient sensing pathways impinge directly on timing mechanisms to retard or advance development, growth, and aging? What are the important effectors of these pathways? Although there are many unknowns, these signaling pathways have measurable molecular and physiological outputs, with the potential to yield detailed, systematic, and multidimensional understanding of the relationship between environment, genome, and physiology. It seems plausible that, in the future, systems approaches that combine empirical observations with models of these signaling networks will yield important insights into the plasticity of life history regulation. Because much of the machinery, pathways, and physiology are evolutionarily conserved, studies in these simpler organisms are bound to illuminate similar regulatory plasticity in other animals.
Acknowledgments
We thank members of the Antebi laboratory for comments on the manuscript and sources of funding (NIH, NIA, Ellison Foundation, and AFAR).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1701508.
References
- Abbott A.L., Alvarez-Saavedra E., Miska E.A., Lau N.C., Bartel D.P., Horvitz H.R., Ambros V. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahante J.E., Daul A.L., Li M., Volk M.L., Tennessen J.M., Miller E.A., Rougvie A.E. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- Ailion M., Thomas J.H. Dauer formation induced by high temperatures in C. elegans. Genetics. 2000;156:1047–1067. doi: 10.1093/genetics/156.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ailion M., Thomas J.H. Isolation and characterization of high-temperature-induced dauer formation mutants in Caenorhabditis elegans. Genetics. 2003;165:127–144. doi: 10.1093/genetics/165.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ailion M., Inoue T., Weaver C.I., Holdcraft R.W., Thomas J.H. Neurosecretory control of aging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 1999;96:7394–7397. doi: 10.1073/pnas.96.13.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-Lounis A., Baas D., Barras E., Benadiba C., Charollais A., Nlend Nlend R., Liegeois D., Meda P., Durand B., Reith W. Novel function of the ciliogenic transcription factor RFX3 in development of the endocrine pancreas. Diabetes. 2007;56:950–959. doi: 10.2337/db06-1187. [DOI] [PubMed] [Google Scholar]
- Albert P.S., Riddle D.L. Mutants of Caenorhabditis elegans that form dauer-like larvae. Dev. Biol. 1988;126:270–293. doi: 10.1016/0012-1606(88)90138-8. [DOI] [PubMed] [Google Scholar]
- Albert P.S., Brown S.J., Riddle D.L. Sensory control of dauer larva formation in Caenorhabditis elegans. J. Comp. Neurol. 1981;198:435–451. doi: 10.1002/cne.901980305. [DOI] [PubMed] [Google Scholar]
- Alcedo J., Kenyon C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron. 2004;41:45–55. doi: 10.1016/s0896-6273(03)00816-x. [DOI] [PubMed] [Google Scholar]
- Allen D.L., Unterman T.G. Regulation of myostatin expression and myoblast differentiation by FoxO and SMAD transcription factors. Am. J. Physiol. Cell Physiol. 2007;292:C188–C199. doi: 10.1152/ajpcell.00542.2005. [DOI] [PubMed] [Google Scholar]
- Ambros V., Moss E.G. Heterochronic genes and the temporal control of C. elegans development. Trends Genet. 1994;10:123–127. doi: 10.1016/0168-9525(94)90213-5. [DOI] [PubMed] [Google Scholar]
- Ansley S.J., Badano J.L., Blacque O.E., Hill J., Hoskins B.E., Leitch C.C., Kim J.C., Ross A.J., Eichers E.R., Teslovich T.M., et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- Antebi A., Culotti J.G., Hedgecock E.M. daf-12 regulates developmental age and the dauer alternative in C. elegans. Development. 1998;125:1191–1205. doi: 10.1242/dev.125.7.1191. [DOI] [PubMed] [Google Scholar]
- Antebi A., Yeh W.H., Tait D., Hedgecock E.M., Riddle D.L. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes & Dev. 2000;14:1512–1527. [PMC free article] [PubMed] [Google Scholar]
- Aoyama Y., Urushiyama S., Yamada M., Kato C., Ide H., Higuchi S., Akiyama T., Shibuya H. MFB-1, an F-box-type ubiquitin ligase, regulates TGF-β signalling. Genes Cells. 2004;9:1093–1101. doi: 10.1111/j.1365-2443.2004.00792.x. [DOI] [PubMed] [Google Scholar]
- Apfeld J., Kenyon C. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell. 1998;95:199–210. doi: 10.1016/s0092-8674(00)81751-1. [DOI] [PubMed] [Google Scholar]
- Apfeld J., Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 1999;402:804–809. doi: 10.1038/45544. [DOI] [PubMed] [Google Scholar]
- Apfeld J., O'Connor G., McDonagh T., DiStefano P.S., Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes & Dev. 2004;18:3004–3009. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arantes-Oliveira N., Apfeld J., Dillin A., Kenyon C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295:502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- Ashrafi K., Chang F.Y., Watts J.L., Fraser A.G., Kamath R.S., Ahringer J., Ruvkun G. Genome-wide RNAi analysis of C. elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- Banerjee D., Kwok A., Lin S.Y., Slack F.J. Developmental timing in C. elegans is regulated by kin-20 and tim-1, homologs of core circadian clock genes. Dev. Cell. 2005;8:287–295. doi: 10.1016/j.devcel.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Bargmann C.I., Horvitz H.R. Control of larval development by chemosensory neurons in C. elegans. Science. 1991;251:1243–1246. doi: 10.1126/science.2006412. [DOI] [PubMed] [Google Scholar]
- Barr M.M., Sternberg P.W. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 1999;401:386–389. doi: 10.1038/43913. [DOI] [PubMed] [Google Scholar]
- Baugh L.R., Sternberg P.W. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr. Biol. 2006;16:780–785. doi: 10.1016/j.cub.2006.03.021. [DOI] [PubMed] [Google Scholar]
- Berdichevsky A., Viswanathan M., Horvitz H.R., Guarente L. C. elegans SIR-2.1 interacts with 14–3–3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- Berman J.R., Kenyon C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell. 2006;124:1055–1068. doi: 10.1016/j.cell.2006.01.039. [DOI] [PubMed] [Google Scholar]
- Bettinger J.C., Euling S., Rougvie A.E. The terminal differentiation factor LIN-29 is required for proper vulval morphogenesis and egg laying in Caenorhabditis elegans. Development. 1997;124:4333–4342. doi: 10.1242/dev.124.21.4333. [DOI] [PubMed] [Google Scholar]
- Birnby D.A., Link E.M., Vowels J.J., Tian H., Colacurcio P.L., Thomas J.H. A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in C. elegans. Genetics. 2000;155:85–104. doi: 10.1093/genetics/155.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop N.A., Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Boehm M., Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310:1954–1957. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- Bordone L., Cohen D., Robinson A., Motta M.C., van Veen E., Czopik A., Steele A.D., Crowe H., Marmor S., Luo J., et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:735–737. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- Bouret S., De Seranno S., Beauvillain J.C., Prevot V. Transforming growth factor β1 may directly influence gonadotropin-releasing hormone gene expression in the rat hypothalamus. Endocrinology. 2004;145:1794–1801. doi: 10.1210/en.2003-1468. [DOI] [PubMed] [Google Scholar]
- Broue F., Liere P., Kenyon C., Baulieu E.E. A steroid hormone that extends the lifespan of Caenorhabditis elegans. Aging Cell. 2007;6:87–94. doi: 10.1111/j.1474-9726.2006.00268.x. [DOI] [PubMed] [Google Scholar]
- Bruning J.C., Gautam D., Burks D.J., Gillette J., Schubert M., Orban P.C., Klein R., Krone W., Muller-Wieland D., Kahn C.R. Role of brain insulin receptor in control of body weight and reproduction. Science. 2000;289:2122–2125. doi: 10.1126/science.289.5487.2122. [DOI] [PubMed] [Google Scholar]
- Burks D.J., Font de Mora J., Schubert M., Withers D.J., Myers M.G., Towery H.H., Altamuro S.L., Flint C.L., White M.F. IRS-2 pathways integrate female reproduction and energy homeostasis. Nature. 2000;407:377–382. doi: 10.1038/35030105. [DOI] [PubMed] [Google Scholar]
- Butcher R.A., Fujita M., Schroeder F.C., Clardy J. Small-molecule pheromones that control dauer development in Caenorhabditis elegans. Nat. Chem. Biol. 2007;3:420–422. doi: 10.1038/nchembio.2007.3. [DOI] [PubMed] [Google Scholar]
- Cassada R., Russell R. The Dauer-larva: A post-embryonic developmental variant of the nematode C. elegans. Dev. Biol. 1975;46:326–342. doi: 10.1016/0012-1606(75)90109-8. [DOI] [PubMed] [Google Scholar]
- Castrillon D.H., Miao L., Kollipara R., Horner J.W., DePinho R.A. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science. 2003;301:215–218. doi: 10.1126/science.1086336. [DOI] [PubMed] [Google Scholar]
- Chase D.L., Koelle M.R. 2007. Biogenic amine neurotransmitters in C. elegans (February 20, 2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.J., Caswell-Chen E.P. Facultative vivipary is a life-history trait in Caenorhabditis elegans. J. Nematol. 2004;36:107–113. [PMC free article] [PubMed] [Google Scholar]
- Clancy D.J., Gems D., Harshman L.G., Oldham S., Stocker H., Hafen E., Leevers S.J., Partridge L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Coburn C.M., Bargmann C.I. A putative cyclic nucleotide-gated channel is required for sensory development and function in C. elegans. Neuron. 1996;17:695–706. doi: 10.1016/s0896-6273(00)80201-9. [DOI] [PubMed] [Google Scholar]
- Coburn C.M., Mori I., Ohshima Y., Bargmann C.I. A cyclic nucleotide-gated channel inhibits sensory axon outgrowth in larval and adult C. elegans: A distinct pathway for maintenance of sensory axon structure. Development. 1998;125:249–258. doi: 10.1242/dev.125.2.249. [DOI] [PubMed] [Google Scholar]
- Crawford D., Libina N., Kenyon C. Caenorhabditis elegans integrates food and reproductive signals in lifespan determination. Aging Cell. 2007;6:715–721. doi: 10.1111/j.1474-9726.2007.00327.x. [DOI] [PubMed] [Google Scholar]
- Daftary S.S., Gore A.C. IGF-1 in the brain as a regulator of reproductive neuroendocrine function. Exp. Biol. Med. 2005;230:292–306. doi: 10.1177/153537020523000503. [DOI] [PubMed] [Google Scholar]
- da Graca L.S., Zimmerman K.K., Mitchell M.C., Kozhan-Gorodetska M., Sekiewicz K., Morales Y., Patterson G.I. DAF-5 is a Ski oncoprotein homolog that functions in a neuronal TGF β pathway to regulate C. elegans dauer development. Development. 2004;131:435–446. doi: 10.1242/dev.00922. [DOI] [PubMed] [Google Scholar]
- Daniels S.A., Ailion M., Thomas J.H., Sengupta P. egl-4 acts through a transforming growth factor-β/SMAD pathway in Caenorhabditis elegans to regulate multiple neuronal circuits in response to sensory cues. Genetics. 2000;156:123–141. doi: 10.1093/genetics/156.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A., Crawford D.K., Kenyon C. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science. 2002;298:830–834. doi: 10.1126/science.1074240. [DOI] [PubMed] [Google Scholar]
- Dubruille R., Laurencon A., Vandaele C., Shishido E., Coulon-Bublex M., Swoboda P., Couble P., Kernan M., Durand B. Drosophila regulatory factor X is necessary for ciliated sensory neuron differentiation. Development. 2002;129:5487–5498. doi: 10.1242/dev.00148. [DOI] [PubMed] [Google Scholar]
- Essers M.A., de Vries-Smits L.M., Barker N., Polderman P.E., Burgering B.M., Korswagen H.C. Functional interaction between β-catenin and FOXO in oxidative stress signaling. Science. 2005;308:1181–1184. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
- Estevez M., Attisano L., Wrana J.L., Albert P.S., Massague J., Riddle D.L. The daf-4 gene encodes a bone morphogenetic protein receptor controlling C. elegans dauer larva development. Nature. 1993;365:644–649. doi: 10.1038/365644a0. [DOI] [PubMed] [Google Scholar]
- Estevez A.O., Cowie R.H., Gardner K.L., Estevez M. Both insulin and calcium channel signaling are required for developmental regulation of serotonin synthesis in the chemosensory ADF neurons of Caenorhabditis elegans. Dev. Biol. 2006;298:32–44. doi: 10.1016/j.ydbio.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Euling S., Ambros V. Reversal of cell fate determination in Caenorhabditis elegans vulval development. Development. 1996;122:2507–2515. doi: 10.1242/dev.122.8.2507. [DOI] [PubMed] [Google Scholar]
- Fielenbach N., Guardavaccaro D., Neubert K., Chan T., Li D., Feng Q., Hutter H., Pagano M., Antebi A. DRE-1: An evolutionarily conserved F box protein that regulates C. elegans developmental age. Dev. Cell. 2007;12:443–455. doi: 10.1016/j.devcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Finch C.E. Longevity, senescence and the genome. University of Chicago Press; Chicago: 1994. [Google Scholar]
- Flatt T., Min K.J., D'Alterio C., Villa-Cuesta E., Cumbers J., Lehmann R., Jones D.L., Tatar M. Drosophila germ-line modulation of insulin signaling and lifespan. Proc. Natl. Acad. Sci. 2008;105:6368–6373. doi: 10.1073/pnas.0709128105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D.B., Johnson T.E. A mutation in the age-1 gene in C. elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch R.E. Body fat, menarche, fitness and fertility. Hum. Reprod. 1987;2:521–533. doi: 10.1093/oxfordjournals.humrep.a136582. [DOI] [PubMed] [Google Scholar]
- Fujiwara M., Sengupta P., McIntire S.L. Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron. 2002;36:1091–1102. doi: 10.1016/s0896-6273(02)01093-0. [DOI] [PubMed] [Google Scholar]
- Garsin D.A., Villanueva J.M., Begun J., Kim D.H., Sifri C.D., Calderwood S.B., Ruvkun G., Ausubel F.M. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- Gems D., Sutton A.J., Sundermeyer M.L., Albert P.S., King K.V., Edgley M.L., Larsen P.L., Riddle D.L. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in C. elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgi L.L., Albert P.S., Riddle D.L. daf-1, a C. elegans gene controlling dauer larva development, encodes a novel receptor protein kinase. Cell. 1990;61:635–645. doi: 10.1016/0092-8674(90)90475-t. [DOI] [PubMed] [Google Scholar]
- Gerisch B., Antebi A. Hormonal signals produced by DAF-9/cytochrome P450 regulate C. elegans dauer diapause in response to environmental cues. Development. 2004;131:1765–1776. doi: 10.1242/dev.01068. [DOI] [PubMed] [Google Scholar]
- Gerisch B., Weitzel C., Kober-Eisermann C., Rottiers V., Antebi A. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev. Cell. 2001;1:841–851. doi: 10.1016/s1534-5807(01)00085-5. [DOI] [PubMed] [Google Scholar]
- Gerisch B., Rottiers V., Li D., Motola D.L., Cummins C.L., Lehrach H., Mangelsdorf D.J., Antebi A. A bile acid-like steroid modulates C. elegans lifespan through nuclear receptor signaling. Proc. Natl. Acad. Sci. 2007;104:5014–5019. doi: 10.1073/pnas.0700847104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil E.B., Malone Link E., Liu L.X., Johnson C.D., Lees J.A. Regulation of the insulin-like developmental pathway of C. elegans by a homolog of the PTEN tumor suppressor gene. Proc. Natl. Acad. Sci. 1999;96:2925–2930. doi: 10.1073/pnas.96.6.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden J.W., Riddle D.L. A pheromone influences larval development in the nematode C. elegans. Science. 1982;218:578–580. doi: 10.1126/science.6896933. [DOI] [PubMed] [Google Scholar]
- Golden J.W., Riddle D.L. A pheromone-induced developmental switch in C. elegans: Temperature-sensitive mutants reveal a wild-type temperature-dependent process. Proc. Natl. Acad. Sci. 1984;81:819–823. doi: 10.1073/pnas.81.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis R.R., Alarcon C., He W., Wang Q., Seoane J., Lash A., Massague J. A FoxO-Smad synexpression group in human keratinocytes. Proc. Natl. Acad. Sci. 2006;103:12747–12752. doi: 10.1073/pnas.0605333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S., Ruvkun G. daf-2, daf-16 and daf-23: Genetically interacting genes controlling Dauer formation in C. elegans. Genetics. 1994;137:107–120. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E.L., Dowlatshahi D., Banko M.R., Villen J., Hoang K., Blanchard D., Gygi S.P., Brunet A. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr. Biol. 2007;17:1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther C.V., Georgi L.L., Riddle D.L. A Caenorhabditis elegans type I TGF β receptor can function in the absence of type II kinase to promote larval development. Development. 2000;127:3337–3347. doi: 10.1242/dev.127.15.3337. [DOI] [PubMed] [Google Scholar]
- Hansen M., Taubert S., Crawford D., Libina N., Lee S.J., Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Harrison M.E., Chivers D.J. The orang-utan mating system and the unflanged male: A product of increased food stress during the late Miocene and Pliocene? J. Hum. Evol. 2007;52:275–293. doi: 10.1016/j.jhevol.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Held J.M., White M.P., Fisher A.L., Gibson B.W., Lithgow G.J., Gill M.S. DAF-12-dependent rescue of dauer formation in Caenorhabditis elegans by (25S)-cholestenoic acid. Aging Cell. 2006;5:283–291. doi: 10.1111/j.1474-9726.2006.00218.x. [DOI] [PubMed] [Google Scholar]
- Henderson S.T., Johnson T.E. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode C. elegans. Curr. Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- Hertweck M., Gobel C., Baumeister R. C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev. Cell. 2004;6:577–588. doi: 10.1016/s1534-5807(04)00095-4. [DOI] [PubMed] [Google Scholar]
- Hirose T., Nakano Y., Nagamatsu Y., Misumi T., Ohta H., Ohshima Y. Cyclic GMP-dependent protein kinase EGL-4 controls body size and lifespan in C elegans. Development. 2003;130:1089–1099. doi: 10.1242/dev.00330. [DOI] [PubMed] [Google Scholar]
- Holt S.J., Riddle D.L. SAGE surveys C. elegans carbohydrate metabolism: Evidence for an anaerobic shift in the long-lived dauer larva. Mech. Ageing Dev. 2003;124:779–800. doi: 10.1016/s0047-6374(03)00132-5. [DOI] [PubMed] [Google Scholar]
- Holzenberger M., Dupont J., Ducos B., Leneuve P., Geloen A., Even P.C., Cervera P., Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- Houthoofd K., Vanfleteren J.R. The longevity effect of dietary restriction in Caenorhabditis elegans. Exp. Gerontol. 2006;41:1026–1031. doi: 10.1016/j.exger.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Houthoofd K., Braeckman B.P., Johnson T.E., Vanfleteren J.R. Life extension via dietary restriction is independent of the Ins/IGF-1 signalling pathway in Caenorhabditis elegans. Exp. Gerontol. 2003;38:947–954. doi: 10.1016/s0531-5565(03)00161-x. [DOI] [PubMed] [Google Scholar]
- Hsin H., Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- Hsu A.L., Murphy C.T., Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Hu P.J., Xu J., Ruvkun G. Two membrane-associated tyrosine phosphatase homologs potentiate C. elegans AKT-1/PKB signaling. PLoS Genet. 2006;2:e99. doi: 10.1371/journal.pgen.0020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis P.N., Ou G., Leroux M.R., Scholey J.M. 2007. The sensory cilia of Caenorhabditis elegans (November 27, 2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Thomas J.H. Targets of TGF-β signaling in C. elegans dauer formation. Dev. Biol. 2000;217:192–204. doi: 10.1006/dbio.1999.9545. [DOI] [PubMed] [Google Scholar]
- Jeon M., Gardner H.F., Miller E.A., Deshler J., Rougvie A.E. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science. 1999;286:1141–1146. doi: 10.1126/science.286.5442.1141. [DOI] [PubMed] [Google Scholar]
- Jeong P.Y., Jung M., Yim Y.H., Kim H., Park M., Hong E., Lee W., Kim Y.H., Kim K., Paik Y.K. Chemical structure and biological activity of the Caenorhabditis elegans dauer-inducing pheromone. Nature. 2005;433:541–545. doi: 10.1038/nature03201. [DOI] [PubMed] [Google Scholar]
- Jia K., Albert P.S., Riddle D.L. DAF-9, a cytochrome P450 regulating C. elegans larval development and adult longevity. Development. 2002;129:221–231. doi: 10.1242/dev.129.1.221. [DOI] [PubMed] [Google Scholar]
- Jia K., Chen D., Riddle D.L. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M., Kirkland K.T., Fields S., Kennedy B.K. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal. pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao G., Nordenson C., Still M., Ronnlund A., Tuck S., Naredi P. ASNA-1 positively regulates insulin secretion in C. elegans and mammalian cells. Cell. 2007;128:577–587. doi: 10.1016/j.cell.2006.12.031. [DOI] [PubMed] [Google Scholar]
- Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kimura K.D., Tissenbaum H.A., Liu Y., Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in C. elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Klass M.R. Aging in the nematode C. elegans: Major biological and environmental factors influencing life span. Mech. Ageing Dev. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Komatsu H., Mori I., Rhee J.S., Akaike N., Ohshima Y. Mutations in a cyclic nucleotide-gated channel lead to abnormal thermosensation and chemosensation in C. elegans. Neuron. 1996;17:707–718. doi: 10.1016/s0896-6273(00)80202-0. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lakowski B., Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam D.D., Heisler L.K. Serotonin and energy balance: Molecular mechanisms and implications for type 2 diabetes. Expert Rev. Mol. Med. 2007;9:1–24. doi: 10.1017/S1462399407000245. [DOI] [PubMed] [Google Scholar]
- Lanjuin A., Sengupta P. Regulation of chemosensory receptor expression and sensory signaling by the KIN-29 Ser/Thr kinase. Neuron. 2002;33:369–381. doi: 10.1016/s0896-6273(02)00572-x. [DOI] [PubMed] [Google Scholar]
- Lans H., Jansen G. Multiple sensory G proteins in the olfactory, gustatory and nociceptive neurons modulate longevity in Caenorhabditis elegans. Dev. Biol. 2007;303:474–482. doi: 10.1016/j.ydbio.2006.11.028. [DOI] [PubMed] [Google Scholar]
- Larsen P.L. Aging and resistance to oxidative damage in C. elegans. Proc. Natl. Acad. Sci. 1993;90:8905–8909. doi: 10.1073/pnas.90.19.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee R.Y., Hench J., Ruvkun G. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr. Biol. 2001;11:1950–1957. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- Lehtinen M.K., Yuan Z., Boag P.R., Yang Y., Villen J., Becker E.B., DiBacco S., de la Iglesia N., Gygi S., Blackwell T.K., et al. A conserved MST–FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- Li W., Kennedy S.G., Ruvkun G. daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes & Dev. 2003;17:844–858. doi: 10.1101/gad.1066503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Brown G., Ailion M., Lee S., Thomas J.H. NCR-1 and NCR-2, the C. elegans homologs of the human Niemann-Pick type C1 disease protein, function upstream of DAF-9 in the dauer formation pathways. Development. 2004;131:5741–5752. doi: 10.1242/dev.01408. [DOI] [PubMed] [Google Scholar]
- Li M., Jones-Rhoades M.W., Lau N.C., Bartel D.P., Rougvie A.E. Regulatory mutations of mir-48, a C. elegans let-7 family MicroRNA, cause developmental timing defects. Dev. Cell. 2005;9:415–422. doi: 10.1016/j.devcel.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Li J., Tewari M., Vidal M., Lee S.S. The 14–3–3 protein FTT-2 regulates DAF-16 in Caenorhabditis elegans. Dev. Biol. 2007;301:82–91. doi: 10.1016/j.ydbio.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B., Moussaif M., Kuan C.J., Gargus J.J., Sze J.Y. Serotonin targets the DAF-16/FOXO signaling pathway to modulate stress responses. Cell Metab. 2006;4:429–440. doi: 10.1016/j.cmet.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Libert S., Zwiener J., Chu X., Vanvoorhies W., Roman G., Pletcher S.D. Regulation of Drosophila life span by olfaction and food-derived odors. Science. 2007;315:1133–1137. doi: 10.1126/science.1136610. [DOI] [PubMed] [Google Scholar]
- Libina N., Berman J.R., Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- Lin K., Dorman J.B., Rodan A., Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of C. elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Lin S.J., Defossez P.A., Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Lin K., Hsin H., Libina N., Kenyon C. Regulation of the C. elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat. Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- Lin S.Y., Johnson S.M., Abraham M., Vella M.C., Pasquinelli A., Gamberi C., Gottlieb E., Slack F.J. The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- Lithgow G.J., Walker G.A. Stress resistance as a determinate of C. elegans lifespan. Mech. Ageing Dev. 2002;123:765–771. doi: 10.1016/s0047-6374(01)00422-5. [DOI] [PubMed] [Google Scholar]
- Lithgow G.J., White T.M., Melov S., Johnson T.E. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc. Natl. Acad. Sci. 1995;92:7540–7544. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Ambros V. Alternative temporal control systems for hypodermal cell differentiation in C. elegans. Nature. 1991;350:162–165. doi: 10.1038/350162a0. [DOI] [PubMed] [Google Scholar]
- Liu T., Zimmerman K.K., Patterson G.I. Regulation of signaling genes by TGFβ during entry into dauer diapause in C. elegans. BMC Dev. Biol. 2004;4:11. doi: 10.1186/1471-213X-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano E., Saez A.G., Flemming A.J., Cunha A., Leroi A.M. Regulation of growth by ploidy in Caenorhabditis elegans. Curr. Biol. 2006;16:493–498. doi: 10.1016/j.cub.2006.01.048. [DOI] [PubMed] [Google Scholar]
- Lucki I. The spectrum of behaviors influenced by serotonin. Biol. Psychiatry. 1998;44:151–162. doi: 10.1016/s0006-3223(98)00139-5. [DOI] [PubMed] [Google Scholar]
- Ludewig A.H., Kober-Eisermann C., Weitzel C., Bethke A., Neubert K., Gerisch B., Hutter H., Antebi A. A novel nuclear receptor/coregulator complex controls C. elegans lipid metabolism, larval development, and aging. Genes & Dev. 2004;18:2120–2133. doi: 10.1101/gad.312604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak H.Y., Ruvkun G. Intercellular signaling of reproductive development by the C. elegans DAF-9 cytochrome P450. Development. 2004;131:1777–1786. doi: 10.1242/dev.01069. [DOI] [PubMed] [Google Scholar]
- McElwee J., Bubb K., Thomas J.H. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- Mihaylova V.T., Borland C.Z., Manjarrez L., Stern M.J., Sun H. The PTEN tumor suppressor homolog in C. elegans regulates longevity and dauer formation in an insulin receptor-like signaling pathway. Proc. Natl. Acad. Sci. 1999;96:7427–7432. doi: 10.1073/pnas.96.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morck C., Pilon M. C. elegans feeding defective mutants have shorter body lengths and increased autophagy. BMC Dev. Biol. 2006;6:39. doi: 10.1186/1471-213X-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morck C., Pilon M. Caloric restriction and autophagy in Caenorhabditis elegans. Autophagy. 2007;3:51–53. doi: 10.4161/auto.3418. [DOI] [PubMed] [Google Scholar]
- Morita K., Shimizu M., Shibuya H., Ueno N. A DAF-1-binding protein BRA-1 is a negative regulator of DAF-7 TGF-β signaling. Proc. Natl. Acad. Sci. 2001;98:6284–6288. doi: 10.1073/pnas.111409798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Flemming A.J., Sugihara Y., Mochii M., Suzuki Y., Yoshida S., Wood W.B., Kohara Y., Leroi A.M., Ueno N. A Caenorhabditis elegans TGF-β, DBL-1, controls the expression of LON-1, a PR-related protein, that regulates polyploidization and body length. EMBO J. 2002;21:1063–1073. doi: 10.1093/emboj/21.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J.Z., Tissenbaum H.A., Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in C. elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Moss E.G. Heterochronic genes and the nature of developmental time. Curr. Biol. 2007;17:R425–R434. doi: 10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- Moss E.G., Lee R.C., Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Mossner R., Lesch K.P. Role of serotonin in the immune system and in neuroimmune interactions. Brain Behav. Immun. 1998;12:249–271. doi: 10.1006/brbi.1998.0532. [DOI] [PubMed] [Google Scholar]
- Motola D.L., Cummins C.L., Rottiers V., Sharma K., Sunino K., Xu E., Auchus R., Antebi A., Mangelsdorf M. Identification of DAF-12 ligands that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A., Deplancke B., Walhout A.J., Tissenbaum H.A. C. elegans tubby regulates life span and fat storage by two independent mechanisms. Cell Metab. 2005;2:35–42. doi: 10.1016/j.cmet.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Munoz M.J., Riddle D.L. Positive selection of C. elegans mutants with increased stress resistance and longevity. Genetics. 2003;163:171–180. doi: 10.1093/genetics/163.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H., Murakami S. Serotonin receptors antagonistically modulate C. elegans longevity. Aging Cell. 2007;6:483–488. doi: 10.1111/j.1474-9726.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- Murakami M., Koga M., Ohshima Y. DAF-7/TGF-β expression required for the normal larval development in C. elegans is controlled by a presumed guanylyl cyclase DAF-11. Mech. Dev. 2001;109:27–35. doi: 10.1016/s0925-4773(01)00507-x. [DOI] [PubMed] [Google Scholar]
- Murphy C.T., McCarroll S.A., Bargmann C.I., Fraser A., Kamath R.S., Ahringer J., Li H., Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Nanji M., Hopper N.A., Gems D. LET-60 RAS modulates effects of insulin/IGF-1 signaling on development and aging in Caenorhabditis elegans. Aging Cell. 2005;4:235–245. doi: 10.1111/j.1474-9726.2005.00166.x. [DOI] [PubMed] [Google Scholar]
- Ogg S., Ruvkun G. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol. Cell. 1998;2:887–893. doi: 10.1016/s1097-2765(00)80303-2. [DOI] [PubMed] [Google Scholar]
- Ogg S., Paradis S., Gottlieb S., Patterson G.I., Lee L., Tissenbaum H.A., Ruvkun G. The fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Oh S.W., Mukhopadhyay A., Svrzikapa N., Jiang F., Davis R.J., Tissenbaum H.A. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc. Natl. Acad. Sci. 2005;102:4494–4499. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura K., Suzuki N., Ishihara T., Katsura I. SDF-9, a protein tyrosine phosphatase-like molecule, regulates the L3/dauer developmental decision through hormonal signaling in C. elegans. Development. 2003;130:3237–3248. doi: 10.1242/dev.00540. [DOI] [PubMed] [Google Scholar]
- Pan K.Z., Palter J.E., Rogers A.N., Olsen A., Chen D., Lithgow G.J., Kapahi P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski S.H., Wolff S., Aguilaniu H., Durieux J., Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- Paradis S., Ruvkun G. C. elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes & Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S., Ailion M., Toker A., Thomas J.H., Ruvkun G. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in C. elegans. Genes & Dev. 1999;13:1438–1452. doi: 10.1101/gad.13.11.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y., Maizels E.T., Feiger Z.J., Alam H., Peters C.A., Woodruff T.K., Unterman T.G., Lee E.J., Jameson J.L., Hunzicker-Dunn M. Induction of cyclin D2 in rat granulosa cells requires FSH-dependent relief from FOXO1 repression coupled with positive signals from Smad. J. Biol. Chem. 2005;280:9135–9148. doi: 10.1074/jbc.M409486200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L., Gems D., Withers D.J. Sex and death: What is the connection? Cell. 2005;120:461–472. doi: 10.1016/j.cell.2005.01.026. [DOI] [PubMed] [Google Scholar]
- Pasquinelli A.E., Reinhart B.J., Slack F., Martindale M.Q., Kuroda M.I., Maller B., Hayward D.C., Ball E.E., Degnan B., Muller P., et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Patterson G.I., Koweek A., Wong A., Liu Y., Ruvkun G. The DAF-3 Smad protein antagonizes TGF-β-related receptor signaling in the C. elegans dauer pathway. Genes & Dev. 1997;11:2679–2690. doi: 10.1101/gad.11.20.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckol E.L., Troemel E.R., Bargmann C.I. Sensory experience and sensory activity regulate chemosensory receptor gene expression in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 2001;98:11032–11038. doi: 10.1073/pnas.191352498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins L.A., Hedgecock E.M., Thomson J.N., Culotti J.G. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Pierce S.B., Costa M., Wisotzkey R., Devadhar S., Homburger S.A., Buchman A.R., Ferguson K.C., Heller J., Platt D.M., Pasquinelli A.A., et al. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes & Dev. 2001;15:672–686. doi: 10.1101/gad.867301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in C. elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Ren P., Lim C.S., Johnsen R., Albert P.S., Pilgrim D., Riddle D.L. Control of C. elegans larval development by neuronal expression of a TGF-β homolog. Science. 1996;274:1389–1391. doi: 10.1126/science.274.5291.1389. [DOI] [PubMed] [Google Scholar]
- Riddle D.L., Albert P.S. Genetic and environmental regulation of Dauer larva development. In: Riddle D.L., et al., editors. C elegans II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 739–768. [PubMed] [Google Scholar]
- Riddle D.L., Swanson M.M., Albert P.S. Interacting genes in nematode dauer larva formation. Nature. 1981;290:668–671. doi: 10.1038/290668a0. [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M., Maina C.V., Gissendanner C.R., Laudet V., Sluder A. Explosive lineage-specific expansion of the orphan nuclear receptor HNF4 in nematodes. J. Mol. Evol. 2005;60:577–586. doi: 10.1007/s00239-004-0175-8. [DOI] [PubMed] [Google Scholar]
- Rogina B., Helfand S.L. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottiers V., Motola D.L., Gerisch B., Cummins C.L., Nishiwaki K., Mangelsdorf D.J., Antebi A. Hormonal control of C. elegans dauer formation and life span by a Rieske-like oxygenase. Dev. Cell. 2006;10:473–482. doi: 10.1016/j.devcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Rouault J.P., Kuwabara P.E., Sinilnikova O.M., Duret L., Thierry-Mieg D., Billaud M. Regulation of dauer larva development in C. elegans by daf- 18, a homologue of the tumour suppressor PTEN. Curr. Biol. 1999;9:329–332. doi: 10.1016/s0960-9822(99)80143-2. [DOI] [PubMed] [Google Scholar]
- Rougvie A.E., Ambros V. The heterochronic gene lin-29 encodes a zinc finger protein that controls a terminal differentiation event in Caenorhabditis elegans. Development. 1995;121:2491–2500. doi: 10.1242/dev.121.8.2491. [DOI] [PubMed] [Google Scholar]
- Ruaud A.F., Bessereau J.L. Activation of nicotinic receptors uncouples a developmental timer from the molting timer in C. elegans. Development. 2006;133:2211–2222. doi: 10.1242/dev.02392. [DOI] [PubMed] [Google Scholar]
- Russell D.W. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- Savage-Dunn C. 2005. TGF-β signaling (September 9, 2005) [Google Scholar]
- Schackwitz W.S., Inoue T., Thomas J.H. Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron. 1996;17:719–728. doi: 10.1016/s0896-6273(00)80203-2. [DOI] [PubMed] [Google Scholar]
- Schafer W.F. Genetics of egg-laying in worms. Annu. Rev. Genet. 2006;40:487–509. doi: 10.1146/annurev.genet.40.110405.090527. [DOI] [PubMed] [Google Scholar]
- Seoane J., Le H.V., Shen L., Anderson S.A., Massague J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- Shakir M.A., Fukushige T., Yasuda H., Miwa J., Siddiqui S.S. C. elegans osm-3 gene mediating osmotic avoidance behaviour encodes a kinesin-like protein. Neuroreport. 1993;4:891–894. doi: 10.1097/00001756-199307000-00013. [DOI] [PubMed] [Google Scholar]
- Shaw W.M., Luo S., Landis J., Ashraf J., Murphy C.T. The C. elegans TGF-β dauer pathway regulates longevity via insulin signaling. Curr. Biol. 2007;17:1635–1645. doi: 10.1016/j.cub.2007.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjostrom L., Garellick G., Krotkiewski M., Luyckx A. Peripheral insulin in response to the sight and smell of food. Metabolism. 1980;29:901–909. doi: 10.1016/0026-0495(80)90031-1. [DOI] [PubMed] [Google Scholar]
- Slack F.J., Basson M., Liu Z., Ambros V., Horvitz H.R., Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- Snow M.I., Larsen P.L. Structure and expression of daf-12: A nuclear hormone receptor with three isoforms that are involved in development and aging in Caenorhabditis elegans. Biochim. Biophys. Acta. 2000;1494:104–116. doi: 10.1016/s0167-4781(00)00224-4. [DOI] [PubMed] [Google Scholar]
- Song C., Liao S. Cholestenoic acid is a naturally occurring ligand for liver X receptor α. Endocrinology. 2000;141:4180–4184. doi: 10.1210/endo.141.11.7772. [DOI] [PubMed] [Google Scholar]
- Starich T.A., Herman R.K., Kari C.K., Yeh W.H., Schackwitz W.S., Schuyler M.W., Collet J., Thomas J.H., Riddle D.L. Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics. 1995;139:171–188. doi: 10.1093/genetics/139.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. Cell lineage. The Nematode Caenorhabditis elegans. In: Wood W.B., editor. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1988. pp. 123–155. [Google Scholar]
- Swoboda P., Adler H.T., Thomas J.H. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol. Cell. 2000;5:411–421. doi: 10.1016/s1097-2765(00)80436-0. [DOI] [PubMed] [Google Scholar]
- Sym M., Basson M., Johnson C. A model for Niemann-Pick type C disease in the nematode C. elegans. Curr. Biol. 2000;10:527–530. doi: 10.1016/s0960-9822(00)00468-1. [DOI] [PubMed] [Google Scholar]
- Syntichaki P., Troulinaki K., Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature. 2007;445:922–926. doi: 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- Sze J.Y., Victor M., Loer C., Shi Y., Ruvkun G. Food and metabolic signalling defects in a C. elegans serotonin-synthesis mutant. Nature. 2000;403:560–564. doi: 10.1038/35000609. [DOI] [PubMed] [Google Scholar]
- Tain L.S., Lozano E., Saez A.G., Leroi A.M. Dietary regulation of hypodermal polyploidization in C. elegans. BMC Dev. Biol. 2008;8:28. doi: 10.1186/1471-213X-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M., Kopelman A., Epstein D., Tu M.P., Yin C.M., Garofalo R.S. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- Tewari M., Hu P.J., Ahn J.S., Ayivi-Guedehoussou N., Vidalain P.O., Li S., Milstein S., Armstrong C.M., Boxem M., Butler M.D., et al. Systematic interactome mapping and genetic perturbation analysis of a C. elegans TGF-β signaling network. Mol. Cell. 2004;13:469–482. doi: 10.1016/s1097-2765(04)00033-4. [DOI] [PubMed] [Google Scholar]
- Thomas J.H., Birnby D.A., Vowels J.J. Evidence for parallel processing of sensory information controlling dauer formation in C. elegans. Genetics. 1993;134:1105–1117. doi: 10.1093/genetics/134.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum H.A., Ruvkun G. An insulin-like signaling pathway affects both longevity and reproduction in C. elegans. Genetics. 1998;148:703–717. doi: 10.1093/genetics/148.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel E.R., Chu S.W., Reinke V., Lee S.S., Ausubel F.M., Kim D.H. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.131/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu M.P., Yin C.M., Tatar M. Impaired ovarian ecdysone synthesis of Drosophila melanogaster insulin receptor mutants. Aging Cell. 2002;1:158–160. doi: 10.1046/j.1474-9728.2002.00016.x. [DOI] [PubMed] [Google Scholar]
- Vanfleteren J.R., De Vreese A. Rate of aerobic metabolism and superoxide production rate potential in the nematode Caenorhabditis elegans. J. Exp. Zool. 1996;274:93–100. doi: 10.1002/(SICI)1097-010X(19960201)274:2<93::AID-JEZ2>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Vella M.C., Choi E.Y., Lin S.Y., Reinert K., Slack F.J. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes & Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T., Takacs-Vellai K., Zhang Y., Kovacs A.L., Orosz L., Muller F. Genetics: Influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Vowels J.J., Thomas J.H. Genetic analysis of chemosensory control of dauer formation in C. elegans. Genetics. 1992;130:105–123. doi: 10.1093/genetics/130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels J.J., Thomas J.H. Multiple chemosensory defects in daf-11 and daf-21 mutants of C. elegans. Genetics. 1994;138:303–316. doi: 10.1093/genetics/138.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Kim S.K. Global analysis of dauer gene expression in C. elegans. Development. 2003;130:1621–1634. doi: 10.1242/dev.00363. [DOI] [PubMed] [Google Scholar]
- Wang Y., Levy D.E. C. elegans STAT cooperates with DAF-7/TGF-β signaling to repress dauer formation. Curr. Biol. 2006;16:89–94. doi: 10.1016/j.cub.2005.11.061. [DOI] [PubMed] [Google Scholar]
- Wang Y., Tissenbaum H.A. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech. Ageing Dev. 2006;127:48–56. doi: 10.1016/j.mad.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Wang Y., Oh S.W., Deplancke B., Luo J., Walhout A.J., Tissenbaum H.A. C. elegans 14-3-3 proteins regulate life span and interact with SIR-2.1 and DAF-16/FOXO. Mech. Ageing Dev. 2006;127:741–747. doi: 10.1016/j.mad.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wolff S., Ma H., Burch D., Maciel G.A., Hunter T., Dillin A. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell. 2006;124:1039–1053. doi: 10.1016/j.cell.2005.12.042. [DOI] [PubMed] [Google Scholar]
- Wolkow C.A., Kimura K.D., Lee M.S., Ruvkun G. Regulation of C. elegans life-span by insulin-like signaling in the nervous system. Science. 2000;290:147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- Wolkow C.A., Munoz M.J., Riddle D.L., Ruvkun G. Insulin receptor substrate and p55 orthologous adaptor proteins function in the Caenorhabditis elegans daf-2/insulin-like signaling pathway. J. Biol. Chem. 2002;277:49591–49597. doi: 10.1074/jbc.M207866200. [DOI] [PubMed] [Google Scholar]
- Yabe T., Suzuki N., Furukawa T., Ishihara T., Katsura I. Multidrug resistance-associated protein MRP-1 regulates dauer diapause by its export activity in Caenorhabditis elegans. Development. 2005;132:3197–3207. doi: 10.1242/dev.01909. [DOI] [PubMed] [Google Scholar]
- Yoshiyama T., Namiki T., Mita K., Kataoka H., Niwa R. Neverland is an evolutionally conserved Rieske-domain protein that is essential for ecdysone synthesis and insect growth. Development. 2006;133:2565–2574. doi: 10.1242/dev.02428. [DOI] [PubMed] [Google Scholar]
- Zhang S., Sokolchik I., Blanco G., Sze J.Y. Caenorhabditis elegans TRPV ion channel regulates 5HT biosynthesis in chemosensory neurons. Development. 2004;131:1629–1638. doi: 10.1242/dev.01047. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Lu H., Bargmann C.I. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438:179–184. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]
- Zwaal R.R., Mendel J.E., Sternberg P.W., Plasterk R.H. Two neuronal G proteins are involved in chemosensation of the Caenorhabditis elegans dauer-inducing pheromone. Genetics. 1997;145:715–727. doi: 10.1093/genetics/145.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]