Abstract
Vaccines able to stimulate CD8+T cells are crucial in controlling a broad range of infectious diseases and tumors. To induce effective CD8+T cell responses, exogenous antigen has to be cross-presented onto major histocompatibility complex (MHC) class I molecules by dendritic cells. Although particle size has been recognized as a critical factor of vaccine design, it is unclear how the size of vaccine carriers impacts the intracellular processing of exogenous antigen and cross-presentation onto MHC class I molecules. In this study, by using polystyrene beads with narrowly defined sizes as model antigen carriers, we demonstrate that particle size mediates the efficiency of cross-presentation of exogenous antigens. By examining the intracellular trafficking, kinetics of phagosomal pH and degradation of antigens bounded to beads, we illustrate the possible mechanisms attributed to the profound effect of particle size on the efficiency of cross-presentation. Antigen bounded to 50 nm beads was shuttled rapidly to an acidic environment within half an hour post-exposure to cells, leading to its rapid and unregulated degradation and inefficient cross-presentation. In contrast, antigen bounded to 500 nm and 3 μm beads remained in a more neutral environment, which preserved the majority of antigens, leaving it available for the generation of peptides to be loaded onto MHC class I molecules. We conclude that the size of antigen carriers plays a critical role in directing antigen to the class I antigen presentation pathway. Our results, together with previous in vivo studies on the effect of particle size on CD8+T cell responses, provide insight into the rational design of vaccines for the stimulation of cell-mediated immunity.
Keywords: Nanoparticles, Vaccine, Antigen presentation, Dendritic cells, Phagosomal pH
1. Introduction
Cytotoxic T lymphocyte (CTL) responses are crucial in the immunological control of tumors, viral infections and immune surveillance [1,2]. Vaccines that generate antibody production have been widely used in the past century, but the design of vaccines able to stimulate CTLs remains a challenge. To initiate potent CTL responses, exogenously-delivered vaccines have to be presented by dendritic cells (DCs) to CD8+T cells through a process called “cross-presentation” [3–5].
Particulate antigen delivery systems have been shown to effectively cross-present antigens and elicit potent CTL responses compared to soluble antigens [6–9]. Size has been recognized as a critical factor of particulate vaccine carriers for the stimulation of CTLs. Early studies have observed that antigen delivered by iron oxide or latex microparticles (0.5–1.5 μm) resulted in more effective cross-presentation than small (<0.5 μm) and large (10 μm) particles [6,8]. Recent studies, on the contrary, suggest that nanoparticles (40–50 nm) induce stronger CD8+T cell responses than particles of other sizes [10–12]. These studies attributed the profound effect of particle size on the immunogenecity of vaccines to the ability of particles to access DEC-205+DCs, which were particularly adept at presenting extracellular antigen on MHC class I to CD8+T cells. The discrepancy among previous studies indicates that the enhanced uptake by relevant cell populations of different sizes of particulate antigen may not be the sole factor attributed to the effectiveness of cross-presentation.
Cross-presentation requires the delivery of exogenous antigen through endosomes and phagosomes where it is partially and controlledly degraded through specialized mechanisms found in dendritic cells [13,14]. Antigen fragments are then exported to the cytosol where they are further degraded by proteosomes into 8 or 9 amino acid peptides before being transported into the lumen of the endoplasmic reticulum (ER) or ER–phagosomes and loaded onto MHC class I molecules [15]. During this process, the initial degradation in phagosomes has to be tightly controlled to prevent complete degradation of immunogenic peptides and to maximize the array of antigenic peptides for potential recognition by CD8+T cells [13]. This precision can be achieved by the accurate control of the composition and activities of lysosomal proteases, and hence degradation of antigens. Phagosomal pH is one of the key factors in the regulation of the composition and activities of lysosomal proteases and protein degradation [14,16]. It has been shown that particulate antigens influence the function of phagocytic antigen-presenting cells through effects on the intracellular trafficking of antigen [17,18]. Antigen prepared in large particles (~500 nm) are shuttled to early endosomal compartments with a pH of 6.0, whereas small particles (<200 nm) are delivered to late endosomal/lysosomal compartments with a pH of 4.5–5.0 [18]. Built upon these early observations, we hypothesize that the size of particles would have significant impact on the profiles of phagosomal pH and antigen degradation, and thus the efficiency of cross-presentation of exogenous antigen.
To test this hypothesis, we initially examine the effect of the size of particles on the efficiency of cross-presentation in vitro, and then quantitatively measure the profiles of phagosomal pH and antigen degradation exhibited by beads of varying sizes. We demonstrate that particles of different sizes exhibit distinct kinetics of phagosomal pH, and thus varying kinetics and extent of antigen degradation. These distinct mechanisms of intracellular antigen processing are linked to the more efficient cross-presentation of antigen bound to microparticles than nanoparticles.
2. Materials and methods
2.1. Cells
A dendritic cell line, DC2.4 (a gift from K.L. Rock, University of Massachusetts Medical School) was maintained in RPMI 1640 supplemented with 10% fetal calf serum (FBS), 2 mM L-glutamine, 10 mM HEPES, 50 μM 2-mercaptoethanol, and 100 units/ml penicillin and 100 μg/ml streptomycin. The B3Z T cell hybridoma (a gift from N. Shastri, University of California, Berkeley), engineered to secrete β-galactosidase when its T-cell receptor engages the OVA257–264:Kb complex, was maintained in RPMI 1640 supplemented with 10% fetal calf serum (FBS), 2 mM L-glutamine, 10 mM HEPES, 50 μM 2-mercaptoethanol, 1 mM sodium pyruvate and 100 units/ml penicillin and 100 μg/ml streptomycin. Cells were maintained at 37 °C under an atmosphere with 5% CO2.
2.2. Antigen adsorption/conjugation to beads
Ovalbumin (OVA) was adsorbed to polystyrene beads (Polysciences) with diameters ranging from 50 nm to 3 μm to initially explore a broad size range of particles. Beads were re-suspended at 1 wt% in citrate buffer at pH 4.2. OVA was added for a final concentration of 1 mg/ml and incubated at 4 °C for 24 h. Beads were extensively washed and re-suspended in phosphate-buffered saline (PBS). The amount of protein adsorbed was determined by measuring the concentration in the supernatant after adsorption. A control sample containing 1 mg/ml OVA without beads was included to account for protein adsorption on the container walls. Alternatively, OVA was covalently coupled to 3 μm, 500 nm or 50 nm carboxylated polystyrene beads. Carboxylated beads allowed coupling of fluorescent proteins used for different intracellular assays. Briefly, 1% w/v carboxylated beads were activated with 5 mM N-hydroxysulfosuccinimide (sulfo-NHS) and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDAC) for 2 h at room temperature. OVA was then added to the mixture to give a final concentration of 1 mg/ml. The mixture was then incubated for an additional 1 h at room temperature. The reaction was quenched for 30 min by adding glycine to a final concentration of 100 mM. Particles were then washed by centrifugation and stored in a 1 mg/ml bovine serum albumin (BSA) solution at 4 °C.
2.3. Cross-presentation assays
DC2.4 cells were seeded in triplicates at a density of 5 × 104 per well in 96-well round-bottomed plates and incubated overnight. DC2.4 cells were loaded with beads to deliver an equivalent of 100 μg/ml OVA and incubated with cells for 4 h. Cells were then washed three times with PBS and co-incubated with 1 × 105 B3Z T cell hybridomas for 20–24 h in 200 μl of culture media. For cross-presentation kinetics experiments, DC2.4 were fixed with 1% paraformaldehyde (PFA) for 20 min at room temperature after incubation with beads for the indicated time points, and washed with PBS before incubation with B3Z T cells. B3Z stimulation was determined through detecting the reporter enzyme β-galactosidase by replacing the medium with 150 μl lysis buffer containing 0.1 mM 2-mercaptoethanol, 9 mM MgCl2, 0.1% Triton X-100, 0.15 mM chlorophenol red-β-D-galactopyranoside per well. Following 90 min incubation, the absorbance at 570 nm (OD570) was measured using a microplate reader (Molecular Devices).
For enzyme inhibition experiments, cells were incubated with antigens in medium containing 100 μM leupeptin and 10 μM pepstatin or 1 mM NH4Cl for 4 h. Cells were thenwashed with PBS, fixed with 1% PFA for 20 min at room temperature, washed with PBS, and incubated with B3Z T cells. T cell stimulation was then assayed as above.
2.4. Confocal microscopy analysis of intracellular distribution of particles
DC2.4 cells were cultured on round glass coverslips in a 24-well tissue culture dish at a density of 5 × 105 cells/well. Cells were pulsed with 50 nm or 3 μm Fluoresbrite YG polystyrene beads (Polysciences) for 30 min, washed three times with PBS, and incubated for the indicated time. During the last 10 min of each incubation period, LysoTracker Red (Molecular Probes) was added to a final concentration of 3 μM. Cells were then washed three times with PBS, fixed with 4% PFA for 20 min at room temperature, washed three times with PBS, and mounted with Vectashield Hardset Mounting Medium with 4′,6-diamidino-2-phenylindole (DAPI) to label the cell nuclei (Vector Laboratories). Images were acquired with a Leica multiphoton confocal microscope (Keck Microscopy Facility, University of Washington) using a 63× objective.
2.5. Phagocytosis measurement by flow cytometry
Phagocytosis was determined using polystyrene beads of various diameters adsorbed with OVA–fluorescein isothiocyanate (OVA–FITC) conjugates (Molecular Probes). Cells were incubated with increasing dilutions of beads for 4 h at 37 °C in complete medium. The cells were washed three times with PBS and analyzed by flow cytometry. Phagocytosis was quantified by determining the geometric mean of fluorescence of FITC using Flowjo (Treestar, Inc.).
2.6. Measurement of phagosomal pH
Carboxylated polystyrene beads with diameters of 3 μm, 500 nm, and 50 nm (Polysciences) were activated with sulfo-NHS and EDAC for 2 h and coupled with OVA–FITC (pH-sensitive) and OVA–AlexaFluor647 (pH-insensitive) conjugates (Molecular Probes) for 1 h at room temperature. The reaction was quenched with 100 mM glycine and extensively washed with PBS and re-suspended in a 1 mg/ml BSA solution. Coupled beads were incubated with cells at 37 °C for 30 min at a concentration of 100 μg/ml of OVA–FITC. Particles were removed and cells were washed three times with PBS. The cells were then further incubated at 37 °C in media for the indicated times and immediately analyzed by flow cytometry. The ratio of the mean fluorescence intensity of the OVA–FITC and OVA–AlexaFluor647 probes for each sample was determined (FL1/FL3 ratio). Values were calibrated against a standard curve obtained by resuspending cells that were exposed to coupled beads for 2 h. Cells used for obtaining calibration curves were fixed with 4% PFA, permeabilized with 0.1% Triton X-100, incubated in citrate buffers with defined pH and immediately analyzed by flow cytometry. To confirm that the decrease in OVA–FITC fluorescence observed at low pH was not due to the loss of FITC from beads, low pH samples were neutralized after measurement with sodium hydroxide and re-measured.
2.7. Measurement of phagosomal protein degradation
DQ OVA (Molecular Probes) is a self-quenched OVA conjugate that exhibits fluorescence after proteolytic degradation. DQ OVA was coupled to activated polystyrene beads with diameters of 50 nm, 500 nm, and 3 μm. Cells were incubated with beads for 30 min at 37 °C and washed three times with PBS. They were further incubated for the indicated times and analyzed by flow cytometry to quantify the fluorescence intensity of the DQ OVA. The cumulative degradation was calculated as (MFIt – MFI0)/MFI0, where MFI denotes the mean fluorescent intensity and the subscripts represent the time points, with MFI0 representing the initial MFI after a 30 min incubation with beads.
2.8. Statistical analysis
The data shown are the average of at least two independent experiments. For antigen presentation experiments, triplicate samples were performed and the averages were shown. The error bars represent the standard deviation of the samples.
3. Results
3.1. The efficiency of cross-presentation is size-dependent
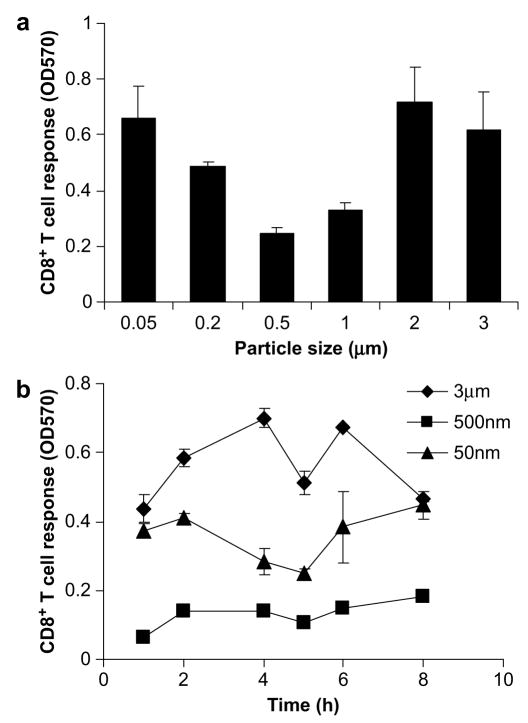
To examine the effect of particle size on the efficiency of cross-presentation, we initially exposed a DC cell line, DC2.4, to OVA (a model antigen), adsorbed to polystyrene beads of varying sizes (50 nm–3 μm). Antigen was initially adsorbed to beads because of the limited availability of particles of varying sizes containing surface carboxylate groups for conjugation. For all particle sizes, 100 μg/ml OVA in total was delivered. The magnitude of K.K. Tran, H. Shen/Biomaterials 30 (2009) 1356–1362 cross-presentation differed depending on the size of particles; both large (1–3 μm) and small (50 nm) particles exhibited higher levels of cross-presentation than intermediate particles (Fig. 1a).
Fig. 1.
The effect of particle size on cross-presentation. (a) OVA was adsorbed on polystyrene beads ranging from 50 nm to 3 μm and incubated with DC2.4 at a concentration of 100 μg/ml OVA. The CD8+T cell responses were then assessed. Soluble OVA controls of 100 μg/ml were included and induced negligible levels of T cell stimulation. (b) The kinetics of cross-presentation of antigen adsorbed to 3 μm, 500 nm, and 50 nm was examined. Cells were exposed to beads for the indicated time, fixed with 1% PFA, and incubated with T cells. The amount of T cell stimulation was expressed as OD570.
To determine whether the difference in the efficiency of cross-presentation by beads of different sizes was time-dependent, we examined the kinetics of cross-presentation by 3 μm, 500 nm, and 50 nm beads. DC2.4 cells were exposed to beads for various time points from 1 h to 8 h. Antigen coupled to 3 μm and 50 nm beads were consistently cross-presented at a higher efficiency compared to 500 nm beads at all time points (Fig. 1b), demonstrating that the kinetics of uptake or antigen presentation that may be associated with different particle sizes do not lead to the observed size-dependency of cross-presentation.
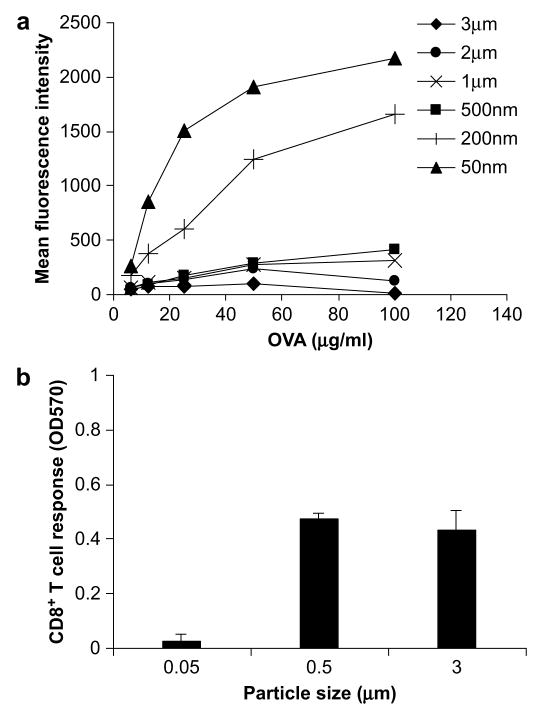
Since particle size also affects the amount of antigen uptake by cells [19], the enhancement observed can be attributed to both the efficiency of intracellular antigen processing and presentation and the total amount of antigen taken up by cells. We titrated the amount of antigen delivered for each particle size and examined their uptake by flow cytometry (Fig. 2a). When 100 μg/ml of antigen was delivered, antigen coupled to 50 nm and 200 nm beads were taken up with far greater efficiency compared to larger beads (>200 nm); when 10 μg/ml of antigen was delivered, equal levels of antigen were taken up by cells for the different particle sizes (Fig. 2a). Therefore, to eliminate the effect of antigen uptake on the cross-presentation efficiency, we re-examined the cross-presentation efficiency of 3 μm, 500 nm, and 50 nm beads conjugated to OVA when 10 μg/ml of antigen was delivered. As illustrated in Fig. 2b, the cross-presentation efficiencies of 3 μm and 500 nm beads are greater than those of 50 nm beads. Experiments with beads adsorbed with OVA at this concentration also showed a size-dependent response, with 3 μm beads eliciting the highest level of cross-presentation, followed by 500 nm and 50 nm beads (data not shown). These results suggest that intracellular antigen processing may attribute to the difference in the efficiency of cross-presentation of particulate antigen exhibited by beads of different sizes.
Fig. 2.
Cross-presentation with equivalent antigen uptake. (a) To account for antigen uptake in the evaluation of CD8+T cell responses, OVA–FITC was conjugated onto polystyrene beads of varying sizes and incubated with DC2.4 at varying concentrations. Uptake was assessed by flow cytometry and quantified as the geometric mean fluorescence intensity of FITC. (b) OVA conjugated to 3 μm, 500 nm, and 50 nm beads was delivered at a concentration of 10 μg/ml and the CD8+T cell response was assessed.
3.2. Phagosomal pH profiles are size-dependent
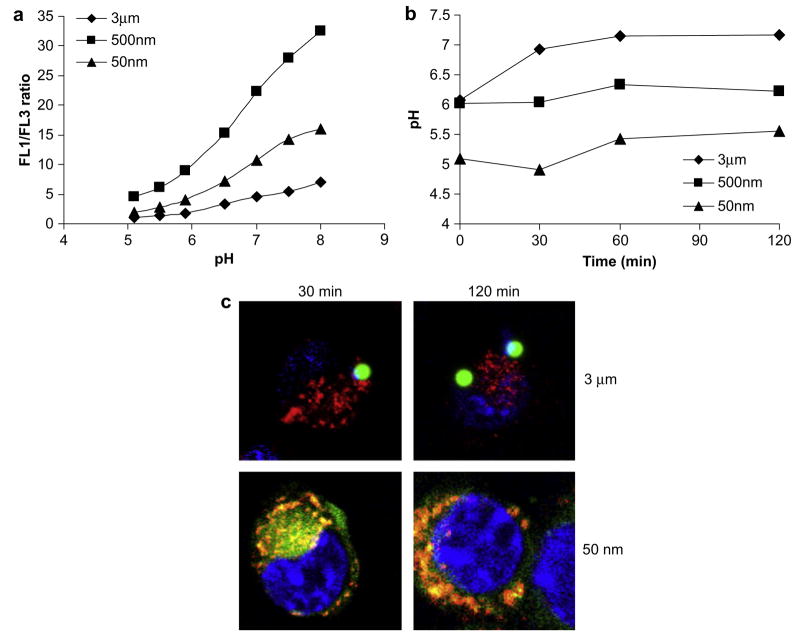
To quantitatively measure the pH experienced by varying sizes of beads, we adapted a fluorescence ratiometric method to monitor phagosomal pH [13,20]. Polystyrene beads of varying sizes were coupled to OVA–FITC (pH-sensitive) and OVA–AlexaFluor647 (pH-insensitive) and incubated with cells. At given time points post-30 min pulse, the fluorescence intensities of the two dyes were quantified using flowcytometry. The ratio of fluorescence intensities between the two dyes (expressed as FL1/FL3 ratio) correlated with the pH in the phagosomes. The absolute value of phagosomal pH was calibrated with a standard curve established with permeabilized cellsimmersed in buffers of fixed pH(Fig. 3a). As shown in Fig. 3b, the profiles of phagosomal pH are size-dependent. The pH of phagosomes containing 50 nm beads rapidly decreased to pH 5.0 at which it remained. The pH of phagosomes containing 500 nm beads, however, remained at 6.0. The pH of phagosomes with 3 μm beads initially was 6.0 as that of phagosomes containing 500 nm beads. Interestingly, the phagosomal pH then gradually increased to 7.2 during the first 60 min at which it remained.
Fig. 3.
The effect of particle size on kinetics of phagosomal pH and intracellular localization. (a) Phagosomal pH calibration curves determined by the ratio of OVA–FITC to OVA– AlexaFluor647 (FL1/FL3) conjugated to 50 nm, 500 nm, and 3 μm polystyrene beads. (b) Beads were delivered to cells at a concentration of 100 μg/ml OVA–FITC. The phagosomal pH profiles of 50 nm, 500 nm, and 3 μm polystyrene beads were determined at varying time points. (c) 3 μm and 50 nm fluorescent polystyrene beads (green) were incubated with DCs for 30 min, washed, further incubated for either 30 or 120 min and examined by confocal microscopy to determine the intracellular localization of beads. DCs were labeled with LysoTracker Red (red) to label lysosomes and DAPI (blue) to label nuclei.
To confirm the pH profiles obtained above, we examined the intracellular localization of particles of two size ranges. DCs were pulsed with either 3 μm or 50 nm fluorescent polystyrene beads. After a 30 min incubation, 3 μm particles did not co-localize with lysosomes, which exhibit an environment pH of 5.0 ± 0.5 [21], whereas a majority of 50 nm particles did (Fig. 3c). Even after 120 min incubation, 3 μm particles did not co-localize with lysosomes. In contrast, 50 nm particles completely merged with lysosomes, indicating that all particles were routed into lysosomes. In addition, 50 nm particles and lysosomes were concentrated on the periphery of the cell nucleus (labeled blue by DAPI), a characteristic feature of late endosomal and lysosomal compartments. These results are consistent with the phagosomal pH profiles obtained for different particle sizes.
3.3. Antigen degradation profiles in phagosomes are size-dependent
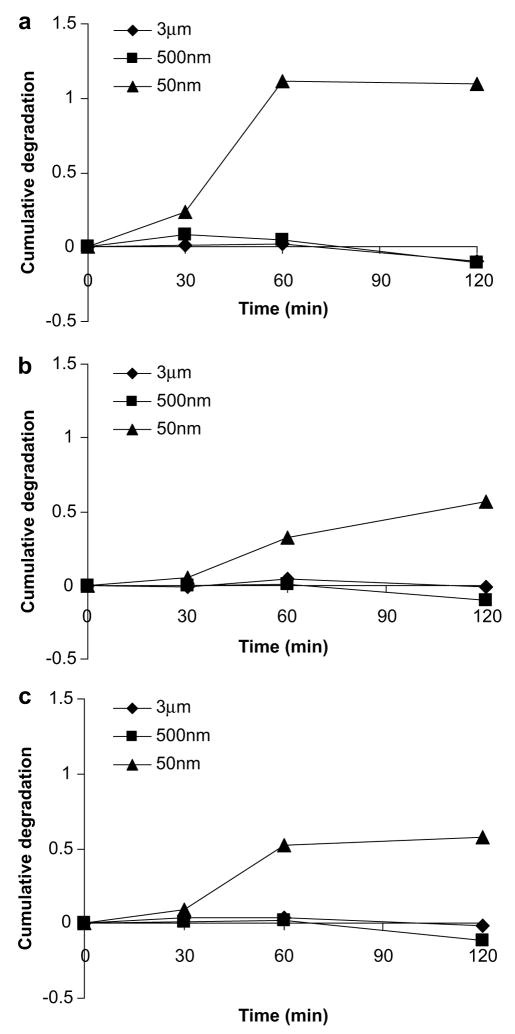
Our results strongly suggest that phagosomes containing particles of different sizes exhibit significantly different pH profiles, which could potentially lead to variations in the degree of protein degradation, and thus the magnitude of antigen cross-presentation. We next examined the functional consequences of different phagosomal pH profiles by particles of various sizes. We monitored the rate of antigen degradation by DQ OVA (Molecular Probes), a self-quenched OVA conjugate that exhibits fluorescence after proteolytic degradation. As expected, antigen bound to 50 nm beads showed the greatest degradation, while degradation was less evident for antigen bound to both 500 nm and 3 μm beads (Fig. 4a).
Fig. 4.
The effect of particle size on the degradation of antigen. (a) DQ OVA was conjugated to 50 nm, 500 nm, and 3 μm polystyrene beads. DCs were pulsed with beads for 30 min and chased for the indicated times. Cells were analyzed by flow cytometry to quantify the fluorescence of the probe and plotted as the normalized, cumulative degradation of antigen. The experiment was repeated in the presence of (b) 100 μM leupeptin/10 μM pepstatin mixture and (c) 1 mM NH4Cl.
To confirm that the observed degradation was largely due to the action of enzyme proteases and not other non-specific mechanisms, the protease inhibitors, pepstatin and leupeptin, were used to inhibit enzymatic activities [13]. As illustrated in Fig. 4b, the degradation of antigen bound to 50 nm beads was not appreciable at the first 30-min pulse and a significant lower level degradation was observed after 30-min pulse compared to untreated samples in Fig. 4a, demonstrating that the degradation of antigen is mainly due to the activity of proteases. In addition, NH4Cl, which prevents acidification of phagosomes, also reduced the degree of degradation of antigen bound to 50 nm beads, as shown in Fig. 4c. Therefore, antigen bound to 50 nm beads was routed to more acidic environments where majority of protein was degraded, whereas antigen bound to 500 nm and 3 μm beads was exposed to more alkaline environments where protein degradation was much less severe.
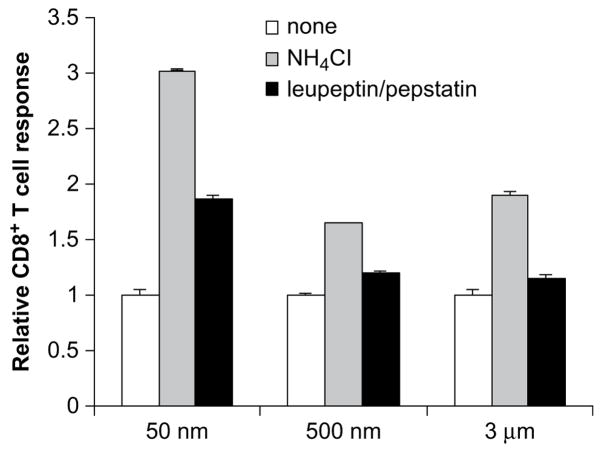
3.4. Efficiency of cross-presentation in the presence of protease inhibitors and NH4Cl
Lastly, we examined whether we could increase the efficiency of cross-presentation of antigen bound to 50 nm beads by inhibiting the activities of proteases and increasing the phagosomal pH by NH4Cl. As shown in Fig. 5, cross-presentation by 50 nm beads increased two- and three-fold in the presence of leupeptin/pepstatin and NH4Cl, respectively. As for 3 μm and 500 nm beads, the presence of protease inhibitors had little effect on the magnitude of cross-presentation; however, the presence of NH4Cl enhanced cross-presentation of antigen on both 3 μm and 500 nm beads. Taken together, our results demonstrated that antigen bound to particles of different sizes was exposed to different pH environment and thus subjected to varying degrees of degradation by proteases, which resulted in size-dependent efficiencies of cross-presentation.
Fig. 5.
Cross-presentation of particles of varying sizes in the presence of NH4Cl and enzyme inhibitors. OVA was conjugated to 50 nm, 500 nm, and 3 μm polystyrene beads and incubated with DCs at a concentration of 10 μg/ml in the presence of a mixture of 100 μM leupeptin/10 μM pepstatin, 1 mM NH4Cl, or media for 4 h. Cells were fixed with 1% PFA before incubation with 1 × 105 B3Z T cells. CD8+T cell responses were assessed after 20–24 h incubation with B3Z T cells. The CD8+T cell responses were normalized with the response in the absence of NH4Cl and enzyme inhibitors.
4. Discussion
The current study has provided a mechanistic understanding of the size of particulate antigen carriers on the efficiency of cross-presentation of exogenous antigens. Microparticles (>500 nm) demonstrated a higher efficiency in shuttling exogenous antigen to the cross-presentation pathway than nanoparticles (<200 nm). This variation was attributed to the phagosomal pH environment that particles of different sizes were exposed to. Nanoparticles were quickly transported to an acidic environment with a pH of 5 while microparticles remained in a more neutral environment, with a pH of 6.0 for 500 nm particles and a pH of 7.2 for 3 μm particles. The different phagosomal pH profiles resulted in varying levels of antigen degradation, and thus efficiency of cross-presentation.
The induction of CD8+T cell responses is critical for the immunological control of intracellular pathogens and tumors. The discovery of the cross-presentation pathway in a critical population of antigen presentation cells, DCs, offers a strategy for eliciting effective CD8+T cell responses through exogenously-delivered antigen. However, the efficiency of cross-presentation is very low for soluble antigens. Several studies have demonstrated that particulate antigens can be more efficiently cross-presented onto MHC class I molecules, though the detailed mechanisms are still debated [6,8]. Currently, two major mechanisms are generally accepted. One is that particulate antigen is phagocytosed, escape into cytosol and then utilize the classical antigen presentation pathway to load onto MHC class I molecules. Another theory is that particulate antigen is phagocytosed into ER–phagosomal compartments, exported into the cytosol, selectively degraded by proteasomes into 8–9 amino acids and translocated back into the ER–phagosomal compartment through the transporter associated with antigen processing (TAP), and then loaded onto MHC class I molecules [22–24]. However, the existence of ER–phagosomal compartments is being debated [25,26]. Nevertheless, the residence time and degradative environment of exogenously-delivered antigen in phagosomes are critical for preserving peptides available for loading onto MHC class I molecules [13,27].
Upon closure, phagosomes are not static but quickly fuse with other intracellular vesicles. In general, they first fuse with early endosomes and late endosomes, and then lysosomes in a process called phagosome maturation [28,29]. During this process, they acquire progressively both the acidification machinery and the proteases characteristic of endocytic compartments, which become adept in the degradation of proteins and pathogens [30]. It has been demonstrated that DCs possess specialized endocytic mechanisms to control protein degradation for efficient antigen presentation through the regulation of the pH environment in phagosomes [13,14,31]. For example, lysosome acidification in immature DCs has been shown to be inefficient due to limited recruitment of vacuolar-ATPase (v-ATPase) subunits [14]. Upon maturation, recruitment of the subunits is enhanced, resulting in increased acidification and thus degradation of proteins into peptides for antigen presentation. In this study, we demonstrate that an important physical feature of the particulate vaccine itself, particle size, can alter antigen trafficking, processing, and cross-presentation efficiencies. Antigen bound to 50 nm beads was quickly routed to an environment with a pH of 5.0. In contrast, antigen bound to 500 nm beads remained in phagosomes with a pH of 6.0, which corresponds to early endosomes. Interestingly, phagosomes containing 3 μm beads initially exhibited a pH of 6.0 and increased to a pH of 7.2 within 30 min. The increase in pH may be due to the production of reactive oxygen species which consume protons generated by v-ATPase, a unique mechanism in DCs for controlled degradation of antigens as suggested by other studies [13].
Previously, several groups have reported that exacerbated antigen degradation by proteases in phagosomes or lysosomes is detrimental to the cross-presentation of exogenous antigen, and that elevated pH in phagosomes can protect antigen from complete degradation [13,32]. In addition, Delamarre et al. have demonstrated that antigens more resistant to lysosomal proteases result in greater immunogenicity in vivo [33]. Weak bases such as chloroquine and NH4Cl, which inhibit phagosomal acidification, have been shown to enhance cross-presentation in vivo, leading to effective CTL responses against viral infections [34]. The results from our studies are consistent with the studies of other groups in that antigen bound to microparticles experience more alkaline pH environments, reducing protein degradation, and thus increasing the efficiency of cross-presentation. Antigen bound to 50 nm beads was subjected to an acidic environment, where many lysosomal proteases were most active and potentially destroyed peptides for loading onto MHC class I molecules. As a result, the magnitude of cross-presentation was reduced. However, in a more neutral environment that antigen bound to 3 μm beads experienced, a majority of lysosomal proteases were in general not active. Consequently, more antigens were available for controlled degradation to generate peptides with 8–9 amino acids for loading onto MHC class I molecules. Our studies provide the first evidence that phagosomal pH regulates the variation of efficiency of cross-presentation associated with particles of different sizes.
Based on our and previous studies regarding the effect of particle size on the induction of CTLs, there exists a paradox for the design of vaccines to achieve optimal CTLs. The current study suggests that microparticles can potentially generate higher levels of CTLs than nanoparticles in terms of intracellular antigen processing and presentation, whereas other studies have demonstrated that nanoparticles may be superior to microparticles in terms of their ability to efficiently access DC populations in vivo [10,11].
A possible approach to address this paradox is to carry out a detailed study on the accessibility of varying sizes of particles to the DC population of interest through various routes of administration, and then tune the chemistry of particles to direct their intracellular trafficking for optimal antigen processing and presentation. Some studies have attempted this approach by modifying nanoparticles to facilitate antigen escape from phagosomal compartments into the cytosol for more efficient cross-presentation [7,35,36]. However, it is unclear whether the facilitation of particles into the cytosol leads to more efficient cross-presentation than retaining particles in a more neutral ER–phagosomal environment. Further studies are required to address this issue.
5. Conclusions
In summary, the current study provides a mechanistic understanding on how the size of particulate antigen carriers affects the efficiency of intracellular antigen processing and cross-presentation. Microparticles (>500 nm) demonstrated a higher efficiency in shuttling exogenous antigen to the cross-presentation pathway than nanoparticles (<200 nm). Microparticles were shuttled to a more basic environment where antigen degradation was limited. In contrast, nanoparticles were quickly transported to acidic, lysosomal compartments, resulting in higher levels of antigen degradation. Thus, the phagosomal pH profiles exhibited by different particle sizes contribute to the size-dependent cross-presentation efficiency. Together with other previous studies, these findings will aid in more rational design of particulate vaccine carriers and delivery strategies to stimulate effective CTLs for the control of tumors and viral infections.
Acknowledgments
This work was supported by the grant R21EB007494-01 from the National Institutes of Health and by the National Science Foundation CAREER Award awarded to H. Shen.
Appendix
Figures with essential color discrimination. Fig. 3C in this article is difficult to interpret in black and white. The full colour images can be found in the online version, at doi:10.1016/j.biomaterials. 2008.11.034.
References
- 1.Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YT, et al. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 2.Heath WR, Carbone FR. Cross-presentation in viral immunity and self-tolerance. Nat Rev Immunol. 2001;1:126–34. doi: 10.1038/35100512. [DOI] [PubMed] [Google Scholar]
- 3.Rock KL, Shen L. Cross-presentation: underlying mechanisms and role in immune surveillance. Immunol Rev. 2005;207:166–83. doi: 10.1111/j.0105-2896.2005.00301.x. [DOI] [PubMed] [Google Scholar]
- 4.den Haan JMM, Bevan MJ. Antigen presentation to CD8+T cells: cross-priming in infectious diseases. Curr Opin Immunol. 2001;13:437–41. doi: 10.1016/s0952-7915(00)00238-7. [DOI] [PubMed] [Google Scholar]
- 5.Heath WR, Carbone FR. Cytotoxic T lymphocyte activation by cross-priming. Curr Opin Immunol. 1999;11:314–8. doi: 10.1016/s0952-7915(99)80050-8. [DOI] [PubMed] [Google Scholar]
- 6.Kovacsovicsbankowski M, Clark K, Benacerraf B, Rock KL. Efficient major histocompatibility complex class-I presentation of exogenous antigen upon phagocytosis by macrophages. Proc Natl Acad Sci U S A. 1993;90:4942–6. doi: 10.1073/pnas.90.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen H, Ackerman AL, Cody V, Giodini A, Hinson ER, Cresswell P, et al. Enhanced and prolonged cross-presentation following endosomal escape of exogenous antigens encapsulated in biodegradable nanoparticles. Immunology. 2006;117:78–88. doi: 10.1111/j.1365-2567.2005.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falo LD, Kovacsovicsbankowski M, Thompson K, Rock KL. Targeting antigen into the phagocytic pathway in-vivo induces protective tumor immunity. Nat Med. 1995;1:649–53. doi: 10.1038/nm0795-649. [DOI] [PubMed] [Google Scholar]
- 9.Rejman J, Oberle V, Zuhorn IS, Hoekstra D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem J. 2004;377:159–69. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reddy ST, Rehor A, Schmoekel HG, Hubbell JA, Swartz MA. In vivo targeting of dendritic cells in lymph nodes with poly(propylene sulfide) nanoparticles. J Controlled Release. 2006;112:26–34. doi: 10.1016/j.jconrel.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Fifis T, Gamvrellis A, Crimeen-Irwin B, Pietersz GA, Li J, Mottram PL, et al. Size-dependent immunogenicity: therapeutic and protective properties of nanovaccines against tumors. J Immunol. 2004;173:3148–54. doi: 10.4049/jimmunol.173.5.3148. [DOI] [PubMed] [Google Scholar]
- 12.Reddy ST, van der Vlies AJ, Simeoni E, Angeli V, Randolph GJ, O’Neill CP, et al. Exploiting lymphatic transport and complement activation in nanoparticle vaccines. Nat Biotechnol. 2007;25:1159–64. doi: 10.1038/nbt1332. [DOI] [PubMed] [Google Scholar]
- 13.Savina A, Jancic C, Hugues S, Guermonprez P, Vargas P, Moura IC, et al. Nox2 controls phagosomal pH to regulate antigen processing during cross-presentation by dendritic cells. Cell. 2006;126:205–18. doi: 10.1016/j.cell.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 14.Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I. Activation of lysosomal function during dendritic cell maturation. Science. 2003;299:1400–3. doi: 10.1126/science.1080106. [DOI] [PubMed] [Google Scholar]
- 15.Cresswell P, Ackerman AL, Giodini A, Peaper DR, Wearsch PA. Mechanisms of MHC class I-restricted antigen processing and cross-presentation. Immunol Rev. 2005;207:145–57. doi: 10.1111/j.0105-2896.2005.00316.x. [DOI] [PubMed] [Google Scholar]
- 16.Mallya SK, Partin JS, Valdizan MC, Lennarz WJ. Proteolysis of the major yolk glycoproteins is regulated by acidification of the yolk platelets in sea-urchin embryos. J Cell Biol. 1992;117:1211–21. doi: 10.1083/jcb.117.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harding CV, Collins DS, Slot JW, Geuze HJ, Unanue ER. Liposome-encapsulated antigens are processed in lysosomes, recycled, and presented to T-cells. Cell. 1991;64:393–401. doi: 10.1016/0092-8674(91)90647-h. [DOI] [PubMed] [Google Scholar]
- 18.Brewer JM, Pollock KGJ, Tetley L, Russell DG. Vesicle size influences the trafficking, processing, and presentation of antigens in lipid vesicles. J Immunol. 2004;173:6143–50. doi: 10.4049/jimmunol.173.10.6143. [DOI] [PubMed] [Google Scholar]
- 19.Desai MP, Labhasetwar V, Walter E, Levy RJ, Amidon GL. The mechanism of uptake of biodegradable microparticles in caco-2 cells is size dependent. Pharm Res. 1997;14:1568–73. doi: 10.1023/a:1012126301290. [DOI] [PubMed] [Google Scholar]
- 20.Jankowski A, Scott CC, Grinstein S. Determinants of the phagosomal pH in neutrophils. J Biol Chem. 2002;277:6059–66. doi: 10.1074/jbc.M110059200. [DOI] [PubMed] [Google Scholar]
- 21.Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B. Rab7: a key to lysosome biogenesis. Mol Biol Cell. 2000;11:467–80. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guermonprez P, Saveanu L, Kleijmeer M, Davoust J, van Endert P, Amigorena S. ER–phagosome fusion defines an MHC class I cross-presentation compartment in dendritic cells. Nature. 2003;425:397–402. doi: 10.1038/nature01911. [DOI] [PubMed] [Google Scholar]
- 23.Houde M, Bertholet S, Gagnon E, Brunet S, Goyette G, Laplante A, et al. Phagosomes are competent organelles for antigen cross-presentation. Nature. 2003;425:402–6. doi: 10.1038/nature01912. [DOI] [PubMed] [Google Scholar]
- 24.Ackerman AL, Kyritsis C, Tampe R, Cresswell P. Early phagosomes in dendritic cells form a cellular compartment sufficient for cross presentation of exogenous antigens. Proc Natl Acad Sci U S A. 2003;100:12889–94. doi: 10.1073/pnas.1735556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Touret N, Paroutis P, Terebiznik M, Harrison RE, Trombetta S, Pypaert M, et al. Quantitative and dynamic assessment of the contribution of the ER to phagosome formation. Cell. 2005;123:157–70. doi: 10.1016/j.cell.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 26.Gagnon E, Duclos S, Rondeau C, Chevet E, Cameron PH, Steele-Mortimer O, et al. Endoplasmic reticulum-mediated phagocytosis is a mechanism of entry into macrophages. Cell. 2002;110:119–31. doi: 10.1016/s0092-8674(02)00797-3. [DOI] [PubMed] [Google Scholar]
- 27.Howland SW, Wittrup KD. Antigen release kinetics in the phagosome are critical to cross-presentation efficiency. J Immunol. 2008;180:1576–83. doi: 10.4049/jimmunol.180.3.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desjardins M, Nzala NN, Corsini R, Rondeau C. Maturation of phagosomes is accompanied by changes in their fusion properties and size-selective acquisition of solute materials from endosomes. J Cell Sci. 1997;110:2303–14. doi: 10.1242/jcs.110.18.2303. [DOI] [PubMed] [Google Scholar]
- 29.Desjardins M. Biogenesis of phagolysosomes – the kiss and run hypothesis. Trends Cell Biol. 1995;5:183–6. doi: 10.1016/s0962-8924(00)88989-8. [DOI] [PubMed] [Google Scholar]
- 30.Jutras I, Desjardins M. Phagocytosis: at the crossroads of innate and adaptive immunity. Annu Rev Cell Dev Biol. 2005;21:511–27. doi: 10.1146/annurev.cellbio.20.010403.102755. [DOI] [PubMed] [Google Scholar]
- 31.Savina A, Amigorena S. Phagocytosis and antigen presentation in dendritic cells. Immunol Rev. 2007;219:143–56. doi: 10.1111/j.1600-065X.2007.00552.x. [DOI] [PubMed] [Google Scholar]
- 32.McCoy KL, Miller J, Jenkins M, Ronchese F, Germain RN, Schwartz RH. Diminished antigen processing by endosomal acidification mutant antigenpresenting cells. J Immunol. 1989;143:29–38. [PubMed] [Google Scholar]
- 33.Delamarre L, Couture R, Mellman I, Trombetta ES. Enhancingimmunogenicity by limiting susceptibility to lysosomal proteolysis. J Exp Med. 2006;203:2049–55. doi: 10.1084/jem.20052442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Accapezzato D, Visco V, Francavilla V, Molette C, Donato T, Paroli M, et al. Chloroquine enhances human CD8+T cell responses against soluble antigens in vivo. J Exp Med. 2005;202:817–28. doi: 10.1084/jem.20051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwon YJ, Standley SM, Goh SL, Frechet JMJ. Enhanced antigen presentation and immunostimulation of dendritic cells using acid-degradable cationic nanoparticles. J Controlled Release. 2005;105:199–212. doi: 10.1016/j.jconrel.2005.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Y, Litwin T, Nagaraja AR, Kwong B, Katz J, Watson N, et al. Cytosolic delivery of membrane-impermeable molecules in dendritic cells using pH-responsive core–shell nanoparticles. Nano Lett. 2007;7:3056–64. doi: 10.1021/nl071542i. [DOI] [PubMed] [Google Scholar]