Abstract
Background
Boosting low HDL levels is a current strategy for preventing clinical events that result from cardiovascular disease (CAD). We previously showed that HDL3 of subjects with CAD is enriched in apolipoprotein (apo) E and that the lipoprotein carries a distinct protein cargo. This observation suggests that altered protein composition might affect the anti-atherogenic and anti-inflammatory properties of HDL. We hypothesized that an intervention that increases HDL levels—combined statin and niacin therapy—might reverse these changes.
Methods and Results
HDL3 isolated from six CAD subjects prior to and one year after combination therapy was analyzed by liquid chromatography—Fourier transform—mass spectrometry. Alterations in protein composition were detected by spectral counting and confirmed using extracted ion chromatograms. We found that combination therapy decreased the abundance of apoE in HDL3 while increasing the abundance of other macrophage proteins implicated in reverse cholesterol transport. Treatment-induced decreases in apoE levels of HDL3 were validated biochemically in a second group of eighteen CAD subjects. Interestingly, the changes in HDL3 proteome with niacin/statin treatment resulted in a protein composition that more closely resembled that of HDL3 in healthy control subjects.
Conclusions
We conclude that combined statin and niacin therapy partially reverses the changes in the protein composition seen in HDL3 in CAD subjects. Our observations raise the possibility that quantifying the HDL proteome could provide insights into the therapeutic efficacy of anti-atherosclerotic interventions.
Keywords: high density lipoprotein, atherosclerosis, inflammation, cardiovascular disease, drugs
Introduction
Epidemiological and clinical studies demonstrate that low levels of high density lipoprotein (HDL) cholesterol are an independent risk factor for premature coronary artery disease (CAD)1, 2. A primary mechanism by which HDL protects against atherosclerosis is by removing cholesterol from artery wall macrophages through reverse cholesterol transport3, 4. However, HDL exhibits other biological activities that may contribute to its anti-atherogenic properties, including the ability to reduce oxidative stress and combat inflammation5, 6. The protein component of HDL plays critical roles in mediating these biological activities.
Apolipoprotein (apo) A-I accounts for ∼70% of HDL protein mass, and apoA-II accounts for ∼20%7-10. However, HDL contains a wide array of other proteins, and therefore exists as a family of distinct particles that vary in protein composition7. Moreover, changes to these proteomes can alter both the functions and cardioprotective effects of HDL. For example, animal studies demonstrate that increasing the apoA-II content of HDL promotes atherosclerosis11, 12. In both humans and animals, acute and chronic inflammation changes HDL protein content13-16, perhaps impairing its cardioprotective effects17. It has been proposed, for example, that alterations in the balance between pro- and anti-oxidative enzymes in HDL play a key role in rendering the lipoprotein atherogenic5, 7, 9, 18-20.
Recently, mass spectrometry has been used to elucidate the proteome of both HDL19-21 and HDL320, its dense subfraction. These studies revealed that HDL contains multiple proteins that regulate the complement system as well as a diverse array serine-type endopeptidases20. Many acute-phase response proteins were also identified, supporting a central role for HDL in inflammation20. The protein composition of HDL also differs in normolipidemic and hyperlipidemic subjects22. Moreover, HDL3 in subjects with established CAD is enriched in several proteins, including apoE20, indicating that these proteins may serve as markers—and perhaps mediators—of vascular disease.
There is intense interest in pharmacological approaches to promoting HDL’s anti-atherogenic effects. Most clinical studies have focused on increasing HDL cholesterol levels, but studies indicate that HDL levels can be dissociated from the lipoprotein’s cardioprotective functions5, 6, 11, 12. Indeed, a recent study was terminated prematurely because the rate of cardiovascular events increased when an agent that elevates HDL cholesterol was added to statin therapy in established CAD subjects23-25. Collectively, these observations indicate that alterations in HDL cholesterol levels may not be the only determinant of the HDL’s cardioprotective effects.
We hypothesized that combination therapy with a statin and niacin, which increases HDL cholesterol levels and reduces CAD risk26, would modify the proteome of HDL3 in CAD subjects and that these modifications might provide insights into the lipoprotein’s anti-atherogenic and anti-inflammatory properties. To test this proposal, we used mass spectrometry to investigate the impact of intensive lipid-lowering therapy with atorvastatin and extended-release niacin on the HDL proteome of CAD subjects. We found that combined treatment altered the protein composition of HDL3 to more closely resemble that of control subjects. Our observations raise the possibility that monitoring the protein composition of HDL could provide a measure of insight into the therapeutic efficacy of lipid interventions.
Methods
Subjects
We investigated HDL’s proteome in two groups of CAD subjects enrolled in the Carotid Plaque Composition Study and in a group of healthy control subjects (Table 1)20, 27. The first group (N=6) was used for proteomic analysis of HDL3, and the second group (N=18) was used to quantify apoE in HDL3 biochemically. All subjects were men recently diagnosed with CAD documented by at least one stenotic lesion (>50%) on coronary. All subjects were treated with extended-release niacin (2g daily) and statin atorvastatin (10-20mg daily) for 12 months. Blood was drawn from each patient at baseline and after 12 months of treatment. All subjects were concurrently taking aspirin and anti-hypertensive medication (5/6 beta-blockers and 4/6 on ACE inhibitors), and 4 of 6 subjects were also using clopidogrel. Concurrent medications did not change over the course of the study.
Table 1.
Clinical characteristics of study subjects.
| N | Status | Age (years) |
Cholesterol (mg/dL) |
TG (mg/dL) |
LDL-C (mg/dL) |
HDL-C (mg/dL) |
HDL2-C (mg/dL) |
HDL3-C (mg/dL) |
|
|---|---|---|---|---|---|---|---|---|---|
|
Group 1: Proteomics Study |
6 | CAD | 58 (5) | 230 (93) | 165 (103) |
168 (27) | 42 (12) | 6.4 (3.8) | 36 (10) |
| 6 | CAD + Niacin/Statin |
59 (5) | 142 (29)* | 68 (39)* | 79 (16)* | 56 (16)* | 11 (6.9)* | 45 (11)* | |
| 6 | Healthy Control |
54 (14) | 183 (29)** | 117 (61)** | 121(22)** | 45 (12) | 8.2 (4.3) | 37 (10) | |
|
Group 2: ApoE Validation Study |
18 | CAD | 54 (7) | 227 (33) | 159 (75) | 160 (29) | 41 (9) | 6.6 (3) | 36 (7) |
| 18 | CAD + Niacin/Statin |
55 (7) | 145 (29)* | 78 (41)* | 83 (25)* | 52 (15)* | 10.6 (5.6)* |
41 (10)* | |
Results represent means (SD).
P<0.01 by a paired two-tailed Student’s t-test
P<0.01 compared to untreated CAD subjects by two-tailed Student’s t-test.
TG, triglycerides; C, cholesterol
A healthy control group of age-matched men (N=6) had no known history of CAD, were not hyperlipidemic or diabetic, had no family history of premature CAD, and were not receiving lipid-lowering therapy. All studies involving human material were approved by the Human Studies Committee at the University of Washington.
Preparation of HDL3
Blood anticoagulated with EDTA was collected from subjects who had fasted overnight. HDL3 (d=1.110-1.210 g/mL) was isolated from EDTA plasma stored at -80°C by sequential density ultracentrifugation using KBr28.
Protein digestion
HDL3 digested with trypsin20 (1:20, w/w) was reconstituted with 0.1% acetic acid and desalted using an Agilent HP1100 HPLC system interfaced with a peptide macrotrap (Michrom BioSciences, Inc., CA). Desalted samples were freeze-dried and stored at -80°C.
Liquid chromatography—Fourier transform ion cyclotron resonance—mass spectrometry (LC-FT-MS)
Interleaved, randomly ordered samples were analyzed on a hybrid LTQ-FT mass spectrometer (Thermo Electron, Germany) with a nanoelectrospray source29 (Molecular Profiling Proteomics Group, Merck). Each sample was resuspended in 10μL 0.1% acetic acid, and 1μL injected onto the C18 reverse-phase capillary column (New Objective, MA). Chromatographic separations were performed with an Agilent HP1100 HPLC (Palo Alto, CA) using a 50 min linear gradient of 0.1M acetic acid and 0.1M acetic acid in 90% acetonitrile at 1μl/min. One FT-MS scan, one linear ion trap MS scan, and three data-dependant MS/MS scans were acquired. For data-dependent MS/MS acquisition, the dynamic exclusion settings were: repeat count 2; repeat time 30s; exclusion list set at 50; and exclusion time 180s.
MS/MS data were converted to dta format using Extract_msn in Bioworks 3.0 (Thermo Inc.)(15 ions minimum, each MS/MS spectrum exported individually, charge state not determined) and searched against the complete human IPI database30 (version 3.01) using SEQUEST31 (version 2.7). Tryptic peptides (up to 2 missed cleavages), fixed carbamidomethyl on cysteine residues, and variable oxidation on methionine residues were allowed in the search. Precursor ion tolerance was ±2.8m/z, and fragment ion tolerance was 0.8m/z. SEQUEST results were validated using PeptideProphet32 and ProteinProphet33 with peptide probability of ≥0.90 and protein probability ≥0.95 resulting in maximum 0.4 false identifications. Each charge state of a peptide was considered a unique identification. Only protein identifications with at least 2 unique peptides detected in at least 2 samples were considered valid for further evaluation.
Identification of differentially expressed HDL3 proteins by spectral counting
Spectral counting was performed as previously described20, 34-36. Student’s paired t-test was used to compare the total number of peptides identified in CAD patients before and during niacin/statin treatment. For proteins found in only one group of subjects, a one-sample t-test was used to compare the number of total peptides to a theoretical mean of 0.
Quantifying HDL3 proteins by extracted ion chromatograms
HDL3 proteins identified as potentially different in relative abundance by spectral counting were quantified by extracted ion chromatograms, using between two and seven peptides for each protein35. Extracted ion chromatograms were constructed from the FT-MS data and peak area was determined using LCQuan software (Thermo Electron). Ion chromatograms were reconstructed for the peptide charge state identified by SEQUEST using accurate monoisotopic mass with 0.05Da tolerance. Peptides selected for further analysis exhibited no evidence of interference from closely eluting isomeric or isobaric peptides. For apoC-II, apoE, apoJ, PLTP, and apoF, we quantified all peptides that met the criteria and were identified in at least 50% of the samples. Differences in protein levels were determined by analyzing obtained peak areas for multiple peptides from each protein, using a repeated measure two-way analysis-of-variance with treatment and peptide as within-subjects factors based on natural log transformed values. We used this analytical strategy as spectral counting offers a rapid, simple mechanism of detecting differences in protein levels whereas spectral peak intensity provides more quantitative estimates of such ratios35, 37, 38.
Biochemical quantification of apoE levels
ApoE levels in HDL3 were determined by nephelometry, using Dade-Behring reagents and normalized to protein concentration determined by the BCA method.
Statistical analysis
Results are reported as means and standard deviations (SD). Differences in lipoprotein values were determined by a paired two-tail t-test and differences in apoE values were determined using Wilcoxon matched pairs test. Pearson product moment correlation coefficients were used to assess linear relationships between variables. P<0.05 was considered significant.
The authors had full access to and take full responsibility for the integrity of the data. All authors have read and agree to the manuscript as written.
Results
HDL3 was isolated from six recently diagnosed CAD patients at baseline and after one year of treatment with extended-release niacin and atorvastatin. The lipoprotein was digested with trypsin, analyzed by LC-MS/MS, and the resulting spectra searched against the IPI database. We studied the HDL dense subfraction, HDL3, because total HDL and HDL2 from CAD subjects is contaminated with apoB10020, an LDL protein, likely reflecting the presence of small dense LDL. Further, we previously established that changes in the HDL3 proteome associate with CAD20.
Subject characteristics
CAD patients had symptoms consistent with angina and were recently diagnosed, as documented by the presence of at least one stenotic lesion (>50%) on coronary angiography. These subjects were clinically stable, and at least 3 months had elapsed since their acute coronary syndrome. None smoked cigarettes, had liver or renal disease, or had received lipid-lowering medication for at least 4 weeks before their blood was collected.
The subjects’ clinical characteristics and lipid values before and during aggressive lipid-lowering therapy are shown in Table 1. Niacin/statin treatment significantly decreased both total plasma cholesterol and LDL cholesterol, with an average reduction of 53% (SD 7.0%, P<0.001) and 40% (SD 9.5%, P<0.001), respectively. HDL cholesterol increased by an average of 34% (SD 12.3%, P=0.001) after one year of combination therapy, with a 26% increase in the HDL3 fraction (SD 17.3%, P=0.003). A control group with no known history of CAD and not receiving lipid-lowering therapy was also analyzed (Table 1; Group 1). These healthy subjects had lower levels of plasma total cholesterol, LDL, and triglycerides than CAD subjects at baseline. Importantly, subjects’ weight remained stable over the study (212 lbs, SD 30 at baseline and 207 lbs, SD 30 at 1 yr). There was no significant difference between the healthy subjects’ and CAD subjects’ body weights (205 lbs, SD 30 and 212 lbs, SD 30, respectively, p=0.67) or BMIs (30.2, SD 3.3 and 27.0, SD 4.7, respectively, p=0.17).
Mass spectrometric analysis of HDL3
Our LC-FT-MSMS analysis of the HDL3 proteome detected 27 proteins in HDL3 (Supplemental Table 1), including 8 of the 12 known HDL apolipoproteins20. We identified proteins implicated in lipid metabolism (lecithin:cholesterol acyl transferase [LCAT], phospholipid transfer protein [PLTP], paraoxonase-1 [PON1]), serum amyloid proteins (SAA1 and SAA4), complement pathway regulatory proteins (complement C3, complement C4B, vitronectin), and endopeptidase inhibitors (α-1-antitrypsin, kininogen-1). This analysis detected 25 of 32 proteins previously identified in HDL320. The lower number of proteins identified in this analysis as compared to a previous analysis20 likely reflects differences in chromatography, data acquisition, and mass spectrometers used for the studies. In addition, we identified kininogen and vitamin D-binding protein, previously found in HDL20 but not specifically in the HDL3 subfraction.
Impact of combination therapy on the proteome of HDL3 isolated from CAD subjects
Spectral counting identified apoE, apoF, and PLTP as differentially expressed (Table 2) in HDL3 isolated from six recently diagnosed CAD patients at baseline and after one year of treatment (Table 1, Group 1). ApoE levels decreased while apoF and PLTP levels increased. Two additional proteins had borderline significant differences: apoC-II and apoJ (P=0.13 and P=0.12, respectively). Spectral counting did not detect a significant change in apoA-I levels (P=0.49). It is important to note that proteins with low spectral counts exhibit greater relative variability39. Consistent with this proposal, we find an average %CV of 3% for apoA-I, 4%–6% for apoC-II and apoE, and 9%–12% for apoF, apoJ, and PLTP in our analyses.
Table 2.
Differentially expressed HDL3 proteins detected by spectral counting.
| Protein Identification Number1 |
Protein Name | CAD2 | CAD + Niacin/Statin2 |
Average Ratio (on therapy to off therapy) 3 |
P value |
|---|---|---|---|---|---|
| IPI00022733 | PLTP | 0 | 3.0 (2.7) | >3 | 0.02 |
| IPI00480119 | Apolipoprotein F | 3.8 (1.1) | 5.2 (0.75) | 1.4 | 0.005 |
| IPI00291262 | Clusterin (Apolipoprotein J) |
0.3 (0.8) | 1.8 (2.2) | 6.2 | 0.12 |
| IPI00021842 | Apolipoprotein E | 15 (3.5) | 11 (2.1) | 0.74 | 0.002 |
| IPI00021856 | Apolipoprotein C- II |
15 (5.5) | 11 (4.2) | 0.80 | 0.13 |
| IPI00021841 | Apolipoprotein A-I | 242 (48) | 257 (45) | 1.08 | 0.43 |
Protein identification numbers are from the International Protein Index database, v3.01.
Numbers shown are total number of peptides identified (SD) for spectral counting.
Average ratios were calculated from the individual ratio for each protein for each subject at baseline and at 1 yr niacin/statin therapy.
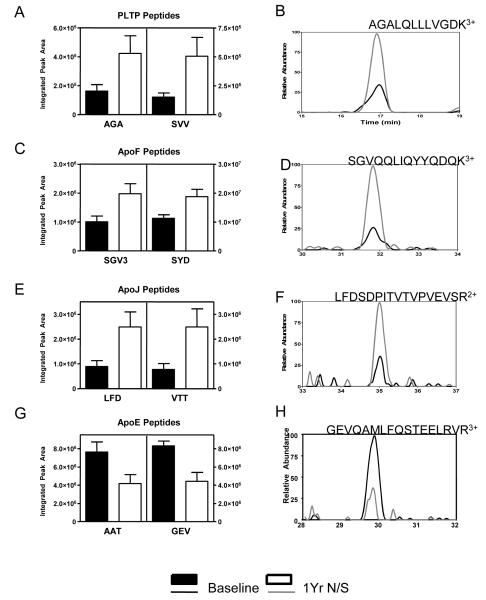
We used extracted ion chromatograms to confirm changes in relative abundance of HDL3 proteins in CAD subjects on combination therapy. ApoF and PLTP as well as apoJ levels in HDL3 increased by 85%, 247%, and 198%, respectively, as a result of therapy (Fig. 1A-F; Supplementary Table 2; F=42.6, 7.2, 17.2, respectively; P=0.001, 0.02, 0.009, respectively). In contrast, levels of apoE in HDL3 were significantly reduced an average of 41% following niacin/statin treatment (Fig. 1G,H; Supplementary Table 2, F=11.35 and P=0.02) for all seven apoE peptides. Although levels of three peptides from apoC-II decreased by an average of 38%, the change was of borderline significance (F=5.96, P=0.059; Supplementary Table 2).
Figure 1. Niacin/statin treatment remodels the HDL3 proteome in six subjects with CAD.
HDL3 was isolated from 6 CAD subjects before and after 1 year of therapy with sustained-release niacin and atorvastatin and tryptic digests of HDL3 analyzed by LC-MS/MS. Proteins identified as differentially expressed before (black bars) and during treatment (empty bars) by spectral counting were quantified by extracted ion chromatograms, using the area under the curve (AUC) of the chromatograms. Average AUCs for two representative peptides from each protein are shown (left column) along with a sample chromatograph for a peptide from each protein (right column). The peptides analyzed were: (A,B) PLTP, AGA (AGALQLLLVGDK3+) and SVV (SSVDELVGIDYSLM3+) (C,D) apoF, SGV (SGVQQLIQYYQDQK3+) and SYD (SYDLDPGAGSLEI); (E,F) apoJ, LFD (LFDSDPITVTVPVEVSR2+) and VTT (VTTVASHTSDSDVPSGVTEVVVK3+); (G,H) apoE, AAT (AATVGSLAGQPLQER2+) and GEV (GEVQAMLGQSTEELRVR3+). The effect of treatment on protein levels was determined by using all peptides quantified for each protein by repeated measures two-way analysis of variance. P<0.05 for all proteins other than apoA-I (P=0.44) and apoC-II (P=0.06). Shown are mean±SEM.
Since the combined statin/niacin therapy increases HDL levels, we were interested whether this change involves also changes in apoA-I levels of HDL3. Five peptides from apoA-I were monitored (Supplementary Table 2) and increased with niacin/statin therapy by an average of 26%, however this change was not statistically significant (F=0.71, P=0.44). As an additional control, we analyzed peptides from three additional proteins whose abundance did not change by spectral counting: apoA-II, apoC-III, and albumin. None of these proteins showed significant changes in peak area (F=0.37, 0.48, and 0.073, respectively; P= 0.57, 0.52, and 0.80, respectively, Supplementary Table 2).
The protein composition of HDL from subjects on combination therapy resembles that of control subjects
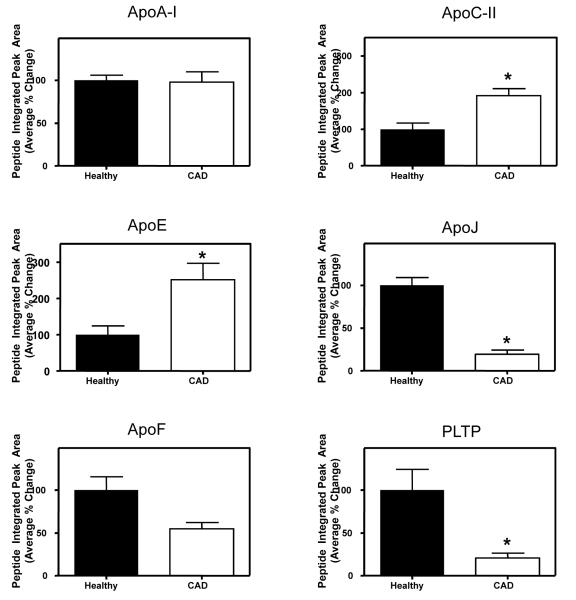
To determine whether combination therapy might reverse CAD-associated changes in HDL3 protein composition, we analyzed HDL3 from apparently healthy male subjects (N=6) who were age-matched with the CAD subjects. As assessed by extracted ion chromatograms, mean levels of apoE and apoC-II were 60% and 51% lower, respectively, in HDL3 of control subjects than in CAD subjects (Fig. 2). In contrast, levels of apoJ, apoF, and PLTP were significantly higher in healthy control HDL3 (by 81%, 45%, and 76%, respectively). The two groups had comparable levels of apoA-I. Taken together, these observations indicate that combination therapy with a statin and niacin remodels the HDL3 proteome, resulting in protein composition that resembles that of apparently healthy subjects.
Figure 2. The HDL3 proteomes of healthy subjects and subjects with CAD differ.
The peptides listed in Supplementary Table 2 were quantified by extracted ion chromatogram, using AUCs of the chromatograms from HDL3 isolated from six healthy subjects and six CAD subjects. The average normalized change for all peptides examined for each protein is shown. *P<0.01 for all proteins other than apoF (P=0.08) and apoA-I (P=0.39). Shown are mean±SEM
Biochemical and mass spectrometric analyses of apoE levels in HDL
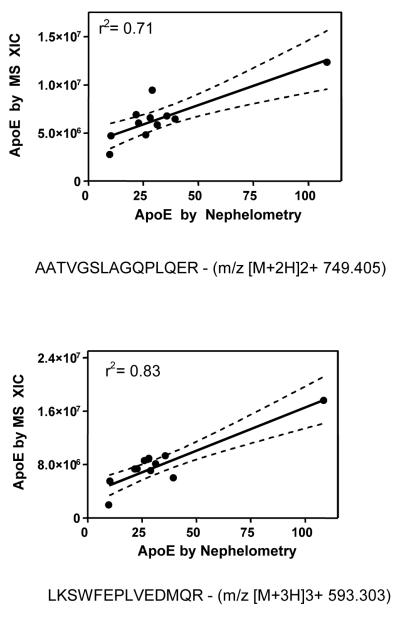
Model system studies indicate that mass spectral peak intensities correlate well with independent measures of protein concentration in biological materials34, 36, 38. To validate the utility of our approach, we compared the extracted ion chromatogram peak area for seven different apoE peptides in HDL3 to apoE as quantified by nephelometry (Fig. 3). The MS and immunochemical analyses were performed on two independent HDL3 preparations. The seven peptides showed significant correlations with nephelometric values (P< 0.01 for 6 of the 7 peptides), with an overall correlation coefficient of r2=0.62 (SD 0.17). Moreover, the levels of apoC-II in HDL3 determined biochemically correlated with those of two apoC-II peptides as quantified by MS (r2=0.48 and 0.55). These observations provide strong evidence that extracted ion chromatograms afford quantitative assessments of protein abundance in HDL3.
Figure 3. ApoE levels measured by nephelometry correlate with apoE levels quantified by extracted ion chromatogram (XIC).
The peptides used for the extracted ion chromatograms were (A) AAT (AATVGSLAGQPLQER2+, m/z [M+2H]2+ 749.405) and (B) LKS (LKSWFEPLVEDMQR3+, m/z [M+3H]3+ 593.303). Nephelometry and mass spectrometry were performed on two different preparations of HDL3 from the same plasma sample. The solid line represents the fit, and the dashed lines show the 95% confidence intervals in linear regression analysis.
Biochemical validation that statin/niacin treatment alters apoE levels of HDL3 in an independent group of subjects
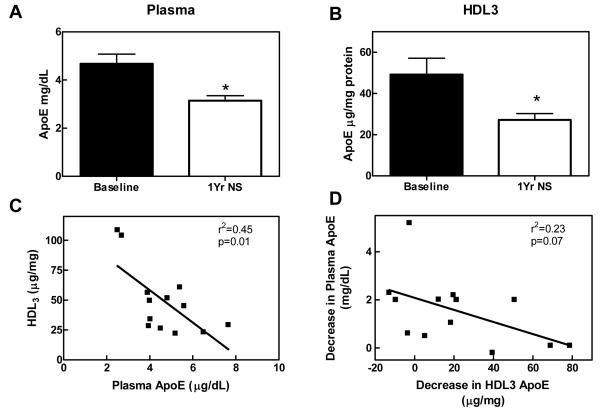
To extend and confirm our observations biochemically, we used an antibody-based approach to quantify apoE in HDL3 isolated from a second set of subjects: 18 men with established CAD (age=54 years, SD 7) prior to and during niacin/statin therapy. As expected, combination therapy significantly lowered LDL cholesterol and triglyceride levels and elevated HDL cholesterol levels (Table 1, Group 2). Niacin/statin treatment lowered HDL3-associated apoE in 15 of 19 subjects (P=0.02) with an average reduction of 17μg apoE per mg HDL protein (Fig. 4A). Importantly, changes in the protein composition of HDL3 isolated from subjects before and during therapy, as assessed by mass spectrometry and biochemical assays, were similar, supporting the proposal that extracted ion chromatograms quantitatively assess relative protein abundance in biological material. Thus, niacin/statin therapy reduced apoE levels in HDL3 of male subjects with established CAD using three independent methods.
Figure 4. Niacin/statin treatment reduces apoE levels in HDL3.
ApoE levels from 18 subjects (Table 1, Group 2) were measured by nephelometry in HDL3 (A) and plasma (B) before and after 1 year of combined niacin/statin (N/S) therapy. *P=0.02. Shown is the correlation between baseline (C) and changes with N/S therapy (D) in apoE between plasma and HDL3. Shown are mean±SEM
We next examined the relationship between the levels of apoE in HDL3 and plasma. Combination therapy reduced apoE levels in plasma (Fig. 4B) as well as HDL3 (Fig. 4A). Plasma levels of apoE before therapy correlated negatively with HDL3 levels (Fig. 4C; N=13, r2=0.45, P=0.01) and treatment-related decreases in plasma apoE did not correlate significantly with decreases in HDL3-associated apoE (Fig. 4D; r2=0.23, P=0.07). These data suggest that the alterations of HDL3 apoE induced by lipid-lowering therapy were not directly linked to changes of plasma levels of apoE.
Discussion
We used mass spectrometry to test the hypothesis that aggressive lipid therapy with atorvastatin and niacin modifies the HDL proteome in humans with established CAD. To quantify changes in protein abundance we employed two complementary label-free quantification methods, spectral counting and extracted ion chromatograms, which are well-suited for analyzing HDL obtained in clinical studies. Spectral counting is based on the observation that tryptic peptides derived from proteins that are more abundant in a sample have a higher probability of being identified during data-dependent MS/MS scanning34-36, 38. For extracted ion chromatograms, the ion current for a given peptide and charge state are extracted from the full scan mass spectrum and used to construct a chromatogram35. The area under the curve then provides a quantitative measure of relative peptide abundance.
We used spectral counting as an initial screen to identify proteins that appeared to be differentially expressed and then used extracted ion chromatograms to quantify the relative abundance of these proteins in HDL3 isolated from CAD subjects before and during treatment. This approach offers two important advantages. First, extracted ion chromatograms estimate protein ratios more accurately than spectral counting35. Second, it is possible to compare the extracted ion chromatogram ratios of multiple peptides detected from the same protein, which should increase confidence in the results.
Spectral counting identified three HDL3 proteins whose relative abundance appeared to change as a result of treatment: apoE, apoF, and PLTP. Levels of apoE fell, whereas levels of apoF and PLTP rose. Spectral counting also detected trends toward lower apoC-II and higher apoJ levels with borderline statistical significance. The extracted ion chromatogram results confirmed that therapy with atorvastatin and niacin significantly lowered apoE and increased apoJ, apoF, and PLTP levels in HDL3 isolated from CAD subjects. We previously showed biochemically and by mass spectrometry that apoE levels are elevated in HDL3 isolated from CAD subjects20 indicate that therapy reverses this change in the HDL3 proteome. Although elevation of plasma apoA-I levels is a well-established effect of niacin, there was no significant change in the apoA-I content of HDL3, which is consistent with the proposal that niacin increases the number of HDL particles but not the amount of apoA-I per particle. It is also possible that niacin increases apoA-I levels in HDL species distinct from HDL3.
We used two approaches to confirm that our mass spectrometric techniques quantify changes in the HDL proteome. First, we observed a strong linear correlation between apoE levels (r2=0.62) assessed by extracted ion chromatograms and nephelometry when we analyzed HDL3 isolated from a different set of 13 subjects. Second, we used a biochemical approach to confirm that combination therapy with niacin and statin reduced levels of apoE in HDL3 in an independent group of subjects. The validity of our approach to assessing global changes in HDL’s protein composition is further supported by reports of decreased apoE levels in both plasma and HDL during atorvastatin treatment of hypertriglyceridemic subjects40 or hypertriglyceridemic subjects with type 2 diabetes mellitus41.
Our analysis detected 27 HDL3-associated proteins. We previously identified 25 of these in HDL3 as well as seven additional proteins20. The increased protein coverage of our earlier study likely reflects significant differences in the conditions used to separate peptides, which centered on 2-D liquid chromatography. This approach increases peptide separation and improves detection of low abundance peptides by minimizing interference from more abundant peptides. Consistent with this proposal, the proteins we did not detect in the current studies were of low abundance20. Gel electrophoresis with proteomic analysis has also been used to elucidate the HDL proteome. This approach identified 12 proteins associated with HDL319 and 14 proteins with total HDL 21, all of which we have identified in both our previous 20 and current (Supplementary Table 2) analyses of HDL3. Collectively, these results suggest that LC-MS/MS methods are more sensitive than gel-based proteomic methods for detecting HDL-associated proteins.
Changes in lipid metabolism or in the lipid composition of HDL are likely to contribute to the alterations in HDL’s proteome induced by statin/niacin therapy. For example, both apoE and apoC-II are exchangeable apolipoproteins. It is therefore possible that therapy with niacin and atorvastatin might lower their levels in HDL3 by redistributing them to other lipoproteins. It is of interest that the reduction of apoE in HDL previously reported with atorvastatin treatment in humans associated with an increased level of apoE in VLDL particles40, 41. Interestingly, apoE-rich HDL binds with high affinity to the LDL receptor142. Combined statin/niacin therapy may help determine the protein composition of total HDL by upregulating LDL receptors in the liver, thereby promoting the removal of apoE-rich HDL.
Our observations also indicate that combined statin/niacin therapy increases levels of apoJ, apoF and PLTP in HDL3. It is noteworthy that those levels were lower in CAD subjects than in control subjects. Interestingly, both apoJ and PLTP can contribute to reverse cholesterol transport by macrophages43, 44. ApoF, also known as lipid transfer inhibitor protein since it inhibits cholesterol ester transfer protein (CETP), has been proposed to cause redistribution of cholesterol between HDL and LDL45. Increased apoF levels could plausibly contribute to treatment associated increases in plasma HDL-C. A limitation of our study is the lack of statin-only and niacin-only groups. Taken together, these observations indicate that it will be important to extend our observations to monotherapy and suggest that alterations produced in the HDL proteome with combined therapy may have functional significance.
In summary, we also found that combined niacin and statin treatment remodeled the HDL3 proteome in subjects with established CAD, yielding a protein composition that resembled that in apparently healthy age- and sex-matched control subjects. Our observations indicate that mass spectrometry is a useful tool for assessing global changes in HDL protein composition with pharmacotherapy. This demonstration raises the possibility that quantifying the HDL proteome could provide insights into the efficacy of lipid therapy and help identify agents with cardioprotective actions.
Supplementary Material
Supplementary Table 1. HDL3 proteins identified by mass spectrometric analysis.
Supplementary Table 2: Peptides quantified by peak areas of extracted ion chromatograms from HDL3 of CAD subjects before and during niacin/statin (N/S) therapy.
Acknowledgments
Funding Sources This research was supported by grants from the National Institutes of Health (HL086798, P30ES07033, P30DK017047, and PO1HL030086). PSG and TV were supported by Pilot and Feasibility Awards from the Center for Nutrition and Clinical Research Unit and the Diabetes and Endocrinology Research Center, respectively. SP was supported by the Juvenile Diabetes Research Foundation, NIH HL092237, and a Doris Duke Clinical Scientist Development Award. Mass spectrometry experiments were supported by the Mass Spectrometry Resource, Department of Medicine, and the Mass Spectrometry Core, Diabetes and Endocrinology Research Center, University of Washington.
Appendix
There is intense interest in HDL as a target for therapies designed to prevent coronary artery disease (CAD). Although most studies of HDL have focused on HDL-cholesterol levels, recent work has indicated that HDL carries a wide range of previously unsuspected components. These include families of proteins implicated in inhibiting proteolysis or activating the complement system, and both factors are thought to be involved in the pathogenesis of CAD. Moreover, control and CAD subjects appear to carry different protein cargos on their HDL. In the current studies, we used mass spectrometry to examine the protein composition of HDL isolated from subjects with established CAD before and during treatment with extended-release niacin and atorvastatin, which elevate HDL cholesterol and reduce the risk of CAD. Our observations indicate that this combination therapy remodels the HDL proteome so that it resembles the proteome of apparently healthy age- and sex-matched control subjects. This finding raises the possibility that quantifying the HDL proteome of CAD patients could provide insights into the therapeutic efficacy of anti-atherosclerotic interventions.
Footnotes
Author Conflict of Interest Disclosures PSG, TV, JJK, ABM, SM, JB and QZ have nothing to disclose. SP (<10K) and JWH (>10K) are on the Speaker’s Bureau at Merck. JWH consults for Merck, Novartis and Insilicos. RK has grant support from Merck and AstraZeneca, is on the Speaker’s Bureau at Abbott and AstraZeneca, and consults for Abbott. RK has grant support from Merck and AstraZeneca, is on the Speaker’s Bureau at Abbott and AstraZeneca, and consults for Abbott.
References
- 1.Gordon DJ, Rifkind BM. High-density lipoprotein--clinical implications of recent studies. N Engl J Med. 1989;321:1311–1316. doi: 10.1056/NEJM198911093211907. [DOI] [PubMed] [Google Scholar]
- 2.Barzilai N, Atzmon G, Schechter C, Schaefer EJ, Cupples AL, Lipton R, Cheng S, Shuldiner AR. Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA. 2003;290:2030–2040. doi: 10.1001/jama.290.15.2030. [DOI] [PubMed] [Google Scholar]
- 3.Oram JF, Heinecke JW. ATP-binding cassette transporter A1: a cell cholesterol exporter protects against cardiovascular disease. Physiol Rev. 2005;85:1343–1372. doi: 10.1152/physrev.00005.2005. [DOI] [PubMed] [Google Scholar]
- 4.Tall AR, Costet P, Wang N. Regulation and mechanisms of macrophage cholesterol efflux. J Clin Invest. 2002;110:899–904. doi: 10.1172/JCI16391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barter PJ, Nicholls S, Rye KA, Anantharamaiah GM, Navab M, Fogelman AM. Antiinflammatory properties of HDL. Circ Res. 2004;95:764–772. doi: 10.1161/01.RES.0000146094.59640.13. [DOI] [PubMed] [Google Scholar]
- 6.Vaisar T, Shao B, Green PS, Oda MN, Oram JF, Heinecke JW. Myeloperoxidase and inflammatory proteins: pathways for generating dysfunctional high-density lipoprotein in humans. Curr Atheroscler Rep. 2007;9:417–424. doi: 10.1007/s11883-007-0054-z. [DOI] [PubMed] [Google Scholar]
- 7.Davidson WS, Thompson TB. The structure of apolipoprotein A-I in high density lipoproteins. J Biol Chem. 2007;282:22249–22253. doi: 10.1074/jbc.R700014200. [DOI] [PubMed] [Google Scholar]
- 8.Alaupovic P. Significance of apolipoproteins for structure, function, and classification of plasma lipoproteins. Methods Enzymol. 1996;263:32–60. doi: 10.1016/s0076-6879(96)63004-3. [DOI] [PubMed] [Google Scholar]
- 9.Cheung MC, Albers JJ. Characterization of lipoprotein particles isolated by immunoaffinity chromatography. Particles containing A-I and A-II and particles containing A-I but no A-II. J Biol Chem. 1984;259:12201–12209. [PubMed] [Google Scholar]
- 10.Shiflett AM, Bishop JR, Pahwa A, Hajduk SL. Human high density lipoproteins are platforms for assembly of multi-component innate immune complexes. J Biol Chem. 2005;280:32578–32585. doi: 10.1074/jbc.M503510200. [DOI] [PubMed] [Google Scholar]
- 11.Warden CH, Hedrick CC, Qiao JH, Castellani LW, Lusis AJ. Atherosclerosis in transgenic mice overexpressing apolipoprotein A-II. Science. 1993;261:469–472. doi: 10.1126/science.8332912. [DOI] [PubMed] [Google Scholar]
- 12.Schultz JR, Verstuyft JG, Gong EL, Nichols AV, Rubin EM. Protein composition determines the anti-atherogenic properties of HDL in transgenic mice. Nature. 1993;365:762–764. doi: 10.1038/365762a0. [DOI] [PubMed] [Google Scholar]
- 13.Cabana VG, Reardon CA, Feng N, Neath S, Lukens J, Getz GS. Serum paraoxonase: effect of the apolipoprotein composition of HDL and the acute phase response. J Lipid Res. 2003;44:780–792. doi: 10.1194/jlr.M200432-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Getz GS. Thematic review series: the immune system and atherogenesis. Immune function in atherogenesis. J Lipid Res. 2005;46:1–10. doi: 10.1194/jlr.R400013-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Khovidhunkit W, Duchateau PN, Medzihradszky KF, Moser AH, Naya-Vigne J, Shigenaga JK, Kane JP, Grunfeld C, Feingold KR. Apolipoproteins A-IV and A-V are acute-phase proteins in mouse HDL. Atherosclerosis. 2004;176:37–44. doi: 10.1016/j.atherosclerosis.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 16.Cabana VG, Siegel JN, Sabesin SM. Effects of the acute phase response on concentration and density distribution of plasma lipids and apolipoproteins. J Lipid Res. 1989;30:39–49. [PubMed] [Google Scholar]
- 17.Van Lenten BJ, Hama SY, de Beer FC, Stafforini DM, McIntyre TM, Prescott SM, La Du BN, Fogelman AM, Navab M. Anti-inflammatory HDL becomes pro-inflammatory during acute phase response. J Clin Invest. 1995;96:2758–2767. doi: 10.1172/JCI118345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergt C, Pennathur S, Fu X, Byun J, O’Brien K, McDonald TO, Singh P, Anantharamaiah GM, Chait A, Brunzell J, Geary RL, Oram JF, Heinecke JW. The myeloperoxidase product hypochlorous acid oxidizes HDL in the human artery wall and impairs ABCA1-dependent cholesterol transport. Proc Natl Acad Sci U S A. 2004;101:13032–13037. doi: 10.1073/pnas.0405292101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlsson H, Leanderson P, Tagesson C, Lindahl M. Lipoproteomics II: mapping of proteins in high-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 2005;5:1431–1445. doi: 10.1002/pmic.200401010. [DOI] [PubMed] [Google Scholar]
- 20.Vaisar T, Pennathur S, Green PS, Gharib SA, Hoofnagle AN, Cheung MC, Byun J, Vuletic S, Kassim S, Singh P, Chea H, Knopp RH, Brunzell J, Geary R, Chait A, Zhao XQ, Elkon K, Marcovina S, Ridker P, Oram JF, Heinecke JW. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest. 2007;117:746–756. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rezaee F, Casetta B, Levels JH, Speijer D, Meijers JC. Proteomic analysis of high-density lipoprotein. Proteomics. 2006;6:721–730. doi: 10.1002/pmic.200500191. [DOI] [PubMed] [Google Scholar]
- 22.Heller M, Schlappritzi E, Stalder D, Nuoffer JM, Haeberli A. Compositional protein analysis of high density lipoproteins in hypercholesterolemia by shotgun LC-MS/MS and probabilistic peptide scoring. Mol Cell Proteomics. 2007;6:1059–1072. doi: 10.1074/mcp.M600326-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Barter PJ, Caulfield M, Eriksson M, Grundy SM, Kastelein JJ, Komajda M, Lopez-Sendon J, Mosca L, Tardif JC, Waters DD, Shear CL, Revkin JH, Buhr KA, Fisher MR, Tall AR, Brewer B. Effects of torcetrapib in patients at high risk for coronary events. N Engl J Med. 2007;357:2109–2122. doi: 10.1056/NEJMoa0706628. [DOI] [PubMed] [Google Scholar]
- 24.Bots ML, Visseren FL, Evans GW, Riley WA, Revkin JH, Tegeler CH, Shear CL, Duggan WT, Vicari RM, Grobbee DE, Kastelein JJ. Torcetrapib and carotid intima-media thickness in mixed dyslipidaemia (RADIANCE 2 study): a randomised, double-blind trial. Lancet. 2007;370:153–160. doi: 10.1016/S0140-6736(07)61088-5. [DOI] [PubMed] [Google Scholar]
- 25.Rader DJ. Illuminating HDL--is it still a viable therapeutic target? N Engl J Med. 2007;357:2180–2183. doi: 10.1056/NEJMe0707210. [DOI] [PubMed] [Google Scholar]
- 26.Brown BG, Zhao XQ, Chait A, Fisher LD, Cheung MC, Morse JS, Dowdy AA, Marino EK, Bolson EL, Alaupovic P, Frohlich J, Albers JJ. Simvastatin and niacin, antioxidant vitamins, or the combination for the prevention of coronary disease. N Engl J Med. 2001;345:1583–1592. doi: 10.1056/NEJMoa011090. [DOI] [PubMed] [Google Scholar]
- 27.Zhao XQ, Phan BA, Chu B, Bray F, Moore AB, Polissar NL, Dodge JT, Jr., Lee CD, Hatsukami TS, Yuan C. Testing the hypothesis of atherosclerotic plaque lipid depletion during lipid therapy by magnetic resonance imaging: study design of Carotid Plaque Composition Study. Am Heart J. 2007;154:239–246. doi: 10.1016/j.ahj.2007.04.035. [DOI] [PubMed] [Google Scholar]
- 28.Mendez AJ, Oram JF, Bierman EL. Protein kinase C as a mediator of high density lipoprotein receptor-dependent efflux of intracellular cholesterol. J Biol Chem. 1991;266:10104–10111. [PubMed] [Google Scholar]
- 29.Meng F, Wiener MC, Sachs JR, Burns C, Verma P, Paweletz CP, Mazur MT, Deyanova EG, Yates NA, Hendrickson RC. Quantitative analysis of complex peptide mixtures using FTMS and differential mass spectrometry. J Am Soc Mass Spectrom. 2007;18:226–233. doi: 10.1016/j.jasms.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 30.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 31.Yates JR, 3rd, Morgan SF, Gatlin CL, Griffin PR, Eng JK. Method to compare collision-induced dissociation spectra of peptides: potential for library searching and subtractive analysis. Anal Chem. 1998;70:3557–3565. doi: 10.1021/ac980122y. [DOI] [PubMed] [Google Scholar]
- 32.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 33.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The International Protein Index: An integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 34.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 35.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Wiener MC, Sachs JR, Deyanova EG, Yates NA. Differential mass spectrometry: a label-free LC-MS method for finding significant differences in complex peptide and protein mixtures. Anal Chem. 2004;76:6085–6096. doi: 10.1021/ac0493875. [DOI] [PubMed] [Google Scholar]
- 37.Fu X, Gharib SA, Green PS, Aitken ML, Frazer DA, Park DR, Vaisar T, Heinecke JW. Spectral Index for Assessment of Differential Protein Expression in Shotgun Proteomics. J Proteome Res. 2008 doi: 10.1021/pr070271+. [DOI] [PubMed] [Google Scholar]
- 38.Chelius D, Bondarenko PV. Quantitative profiling of proteins in complex mixtures using liquid chromatography and mass spectrometry. J Proteome Res. 2002;1:317–323. doi: 10.1021/pr025517j. [DOI] [PubMed] [Google Scholar]
- 39.Fu X, Gharib SA, Green PS, Aitken ML, Frazer DA, Park DR, Vaisar T, Heinecke JW. Spectral index for assessment of differential protein expression in shotgun proteomics. J Proteome Res. 2008;7:845–854. doi: 10.1021/pr070271+. [DOI] [PubMed] [Google Scholar]
- 40.Le NA, Innis-Whitehouse W, Li X, Bakker-Arkema R, Black D, Brown WV. Lipid and apolipoprotein levels and distribution in patients with hypertriglyceridemia: effect of triglyceride reductions with atorvastatin. Metabolism. 2000;49:167–177. doi: 10.1016/s0026-0495(00)91169-7. [DOI] [PubMed] [Google Scholar]
- 41.Bach-Ngohou K, Ouguerram K, Frenais R, Maugere P, Ripolles-Piquer B, Zair Y, Krempf M, Bard JM. Influence of atorvastatin on apolipoprotein E and AI kinetics in patients with type 2 diabetes. J Pharmacol Exp Ther. 2005;315:363–369. doi: 10.1124/jpet.105.085522. [DOI] [PubMed] [Google Scholar]
- 42.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 43.Lee-Rueckert M, Vikstedt R, Metso J, Ehnholm C, Kovanen PT, Jauhiainen M. Absence of endogenous phospholipid transfer protein impairs ABCA1-dependent efflux of cholesterol from macrophage foam cells. J Lipid Res. 2006;47:1725–1732. doi: 10.1194/jlr.M600051-JLR200. [DOI] [PubMed] [Google Scholar]
- 44.Gelissen IC, Hochgrebe T, Wilson MR, Easterbrook-Smith SB, Jessup W, Dean RT, Brown AJ. Apolipoprotein J (clusterin) induces cholesterol export from macrophage-foam cells: a potential anti-atherogenic function? Biochem J. 1998;331:231–237. doi: 10.1042/bj3310231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paromov VM, Morton RE. Lipid transfer inhibitor protein defines participation of high density lipoprotein subfractions in lipid transfer reactions mediated by cholesterol ester transfer protein (CETP) J Biol Chem. 2003;278:40859–40866. doi: 10.1074/jbc.M306580200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. HDL3 proteins identified by mass spectrometric analysis.
Supplementary Table 2: Peptides quantified by peak areas of extracted ion chromatograms from HDL3 of CAD subjects before and during niacin/statin (N/S) therapy.