Abstract
The PHD finger motif is a signature chromatin-associated motif that is found throughout eukaryotic proteomes. Here we have determined the histone methyl-lysine binding activity of the PHD fingers present within the Saccharomyces cerevisiae proteome. We provide evidence on the genomic scale that PHD fingers constitute a general class of effector modules for histone H3 trimethylated at lysine 4 (H3K4me3) and histone H3 trimethylated at lysine 36 (H3K36me3). Structural modeling of PHD fingers demonstrates a conserved mechanism for recognizing the trimethyl moiety and provides insight into the molecular basis of affinity for the different methyl-histone ligands. Together, our study suggests that a common function for PHD fingers is to transduce methyl-lysine events and sheds light on how a single histone modification can be linked to multiple biological outcomes.
One of the major mechanisms for regulating chromatin structure involves the reversible covalent post-translational modification of histone proteins by chemical moieties such as acetyl, methyl, and phospho groups (1, 2). Different histone modifications are linked to discrete chromatin states and are thought to regulate the extent of accessibility of DNA to transacting factors. In this context, the proteins and domains that recognize histone modification, named “effectors,” are thought to define the functional consequences of specific modifications by transducing molecular events at chromatin to biological outcomes (3). Accumulating evidence suggested that evolutionally conserved domains commonly found within chromatin regulatory proteins, such as the bromodomain and chromodomain, function as such effectors (4, 5).
The PHD finger (plant homeodomain) is a signature chromatin-associated domain that is found throughout eukaryotic proteomes. Accordingly, the PHD fingers from the ING2 and BPTF proteins were recently demonstrated to be highly specific effectors of histone H3 trimethylated at lysine 4 (H3K4me3) (6, 7), suggesting that H3K4me3 recognition, or more generally, histone methyl-lysine binding, might be a common feature of PHD fingers. However, contrary to this hypothesis, a number of other PHD fingers either bind to nucleosomes in a manner independent of post-translational modifications or have no detectable histone binding activity (6, 8, 9). To obtain a genomic scale understanding of PHD finger function, here we have determined the histone methyl-lysine binding activity of the PHD fingers present within the Saccharomyces cerevisiae proteome. Our study identifies either H3K4me3 or H3K36me3 recognition as an activity common to the majority of yeast PHD fingers, arguing that a general function for this domain is to transduce lysine methylation events.
MATERIALS AND METHODS
Peptide Synthesis and Cloning
Biotinylated histone peptides were synthesized as described previously (6). The coding sequences for the PHD domains listed in Table 1 were amplified by PCR from yeast genomic DNA (generous gift of M. Cyert, Stanford Unviersity) and cloned into pGEX-6P vector.
TABLE 1.
Summary of biochemical and genetic functional interactions for S. cerevisiae PHD finger proteins
| Name | PHD domain | Methyl-lysine interactionsa |
HMT interactions |
Chromatin functionsb | ||||
|---|---|---|---|---|---|---|---|---|
| K4me | K36me | K79me | Set1 | Set2 | Dot1 | |||
| Pho23 | 280–329 | √√√ | − | − | + | + | Rpd3/HDAC complex | |
| Yng1 | 222–271 | √√√ | − | − | + | + | NuA3 HAT complex | |
| Yng2 | 155–204 | √√√ | − | − | + | NuA4 HAT complex | ||
| Bye1 | 74–132 | √√√ | − | − | Negative transcription elongation regulator | |||
| Cti6 | 74–121 | √√√ | − | − | + | Rpd3/HDAC complex | ||
| Jhd1 | 6–70 | √√√ | − | − | H3K36 HDM | |||
| Spp1 | 24–70 | √√√ | − | − | + | + | Set1c/HMT complex | |
| Set3 | 119–164 | √√√ | − | − | + | + | Set3c/HDAC complex Candidate HMT | |
| Ecm5 | 1240–1288 | − | √√ | − | Candidate HMD | |||
| Nto1 | 1: 265–311 | # | √√ | − | + | + | NuA3 HAT complex | |
| 2: 375–439 | NS | NS | − | |||||
| Rco1 | 1: 262–307 | NS | NS | − | + | Rpd3/HDAC complex | ||
| 2: 416–470 | NS | NS | NS | |||||
| Snt2 | 1: 319–367 | − | − | − | Unknown | |||
| 2: 1040–95 | − | √ | NS | |||||
| 3: 1178–1249 | − | − | − | |||||
| Set4 | 162–208 | − | − | − | Candidate HMT | |||
| Yj89 | 237–283 | − | − | − | Candidate HDM | |||
√√√, strong interaction;√√, moderate interaction;√, weak interaction; NS, binds to peptide irrespective of lysine methylation;
, Nto1-1 binds weakly to unmethylated H3 peptide amino acids 1–21 (see Fig. 2B).
See supplemental material for references. HDAC, histone deacetylase; HAT, histone acetyltransferase; HDM, histone demethylase.
Peptide Pulldown Assays and Affinity Measurements
0.5 μg of biotinylated histone peptides with different modifications were incubated with 1 μg of GST4-fused PHD fingers in binding buffer as previously described (6), using either 150 or 300 mM NaCl as indicated. Tryptophan fluorescence spectroscopy experiments to determine the dissociation constants (Kd values) of PHD finger interactions were performed as described previously (10).
Peptide Microarray
Biotinylated histone peptides were printed in triplicates onto a streptavidin-coated slide (ArrayIt) using a VersArray Compact Microarrayer (Bio-Rad). After a short blocking with biotin (Sigma), the slides were incubated with the GST-fused PHD fingers in binding buffer (50 mM Tris-HCl, pH 7.5, 300 mM NaCl, 0.1% Nonidet P-40, 1mM phenylmethylsulfonyl fluoride, 20% fetal bovine serum) for overnight at 4 °C with gentle agitation. After washing with the same buffer, slides were probed with anti-GST antibody and then fluorescein-conjugated secondary antibody and visualized with GenePix 4000 scanner (Molecular Devices).
Protein Microarray
Protein microarrays were generated essentially as described previously (11). Briefly, 0.25 μg of GST fusion proteins with PHD fingers from S. cerevisiae were spotted in duplicates onto a glass slide precoated with nitrocellulose polymer (Schleicher & Schuell) using a FLEXYS ® robot (Genomic Solutions). After blocking, the arrayed slides were incubated with Cy3-labeled histone peptides and the fluorescent signal detected using a GeneTAC™ LSIV scanner (Genomic Solutions). The arrays were also probed with an anti-GST antibody to control for loading.
In Silico Analysis of Functions and Interactions of PHD Finger-containing Proteins in S. cerevisiae
PHD finger proteins were assigned functions and scored as interactors with Set1, Set2, and Dot1 based on 1) BioGRID data base searches, 2) Saccharomyces Genome Data base searches, and 3) the presence of domains with characterized enzymatic activity.
Molecular Modeling of PHD Fingers
Homology models of PHD fingers with and without bound peptides were built with MODELER (12) (Ecm5(PHD), Cti6(PHD), and Nto1(PHD)) or with SWISSMODEL (13) (Yng1(PHD), Yng2(PHD), and Pho23(PHD) and Spp1(PHD), Set3(PHD), and Set4(PHD)) using structures of ING2 and BPTF PHD domains (Protein Data Bank IDs: 2G6Q and 2F6N, respectively) as templates. The conformation of the side chains were predicted by SCWRL3 algorithm (14) except for residues that coordinate metal ions, where the original conformers were kept. Glide docking algorithm (15) was used to establish the binding modes of H3(33–38)K36me peptide to Emc5(PHD). The models were analyzed using PyMOL visualization software (16).
RESULTS AND DISCUSSION
In contrast to the human epigenome where abundant potential methylation sites exist, the S. cerevisiae epigenome is suitable for a genomic scale analysis, as it is thought to contain only three methylated lysine residues: H3K4, H3K36, and H3K79. SMART and Pfam data base searches of the S. cerevisiae genome revealed a total of 18 canonical and non-canonical PHD fingers, present within 14 proteins (Table 1). Virtually all these proteins are nuclear proteins with known or candidate chromatin-regulatory functions, including histone demethylases (Jhd1, Ecm5, and Yj89), histone methyltransferases, and associated proteins (Set3, Set4, and Spp1) modulators of histone acetylation (Cti6, Nto1, Pho23, Rco1, Yng1, and Yng2), and transcriptional regulators (Bye1) (see Table 1). To screen these PHD fingers for histone methyl-lysine binding activity, histone peptide microarrays containing methylated histone peptides or the corresponding unmodified control peptide were probed with PHD fingers fused to GST (Fig. 1A). In agreement with previous work, this analysis revealed strong binding to H3K4me2/3 peptides for the PHD fingers from the three yeast ING2 family members, Yng1, Yng2, and Pho23 (Fig. 1A) (6, 10, 17). In addition, novel interactions were detected between methylated H3K4 and the PHD fingers from Bye1, Cti6, Jhd1, Set3, and Spp1 (Fig. 1A). A second screening strategy, in which protein microarrays containing the recombinant GST PHD domains were probed with the histone peptides, detected the three yeast ING proteins but weakly or not at all for the other PHD fingers (Fig. 1B) (11). This difference argues that the peptide microarray is more sensitive assay than the protein microarray for screening purposes, possibly due to the difficulty of recombinant PHD fingers properly folding during the printing process (Fig. 1; data not shown). Using both assays, no interactions were detected with methylated Lys36 or Lys79 peptides (Fig. 1A; data not shown).
FIGURE 1. Identification of histone methyl-lysine binding activity for the PHD fingers present within the S. cerevisiae proteome.
A, peptide microarrays identifies the S. cerevisiae PHD fingers with H3K4me binding activity. The indicated biotinylated histone peptides were arrayed in triplicate onto streptavidin-coated slides as shown in the schematic and probed with the indicated GST-PHD finger fusion proteins. ING2(PHD) and GST are shown as positive and negative controls, respectively. B, H3K4me3 peptides bind to PHD fingers printed on protein microarray chips. The indicated GST-PHD finger fusion proteins were arrayed in duplicate onto nitrocellulose slides as shown in the schematic and probed with the indicated histone peptide. Anti-GST antibody probe is shown as a control for protein loading.
To validate the binding data by a third method that in our experience has proven more sensitive,5 in vitro binding assays with biotinylated unmodified and methylated histone peptides were employed (Fig. 2A). Consistent with the data obtained with the protein microarray studies, the PHD fingers of Yng1, Yng2, Pho23, Bye1, Cti6, Jhd1, Set3, and Spp1 were all found to bind specifically to H3K4me, with no binding to K36me or K79me peptides observed (Fig. 2a; for disassociation constants see Fig. 2c). Moreover, a preference for binding to the trimethylated species versus lower methylation species was observed for all PHD fingers apart from Spp1 (Fig. 2A). No further PHD finger in the yeast proteome bound to H3K4me. Based on these data, we conclude that 8 of the 14 proteins that contain PHD fingers in the S. cerevisiae proteome (8 of 18 PHD fingers) possess H3K4me recognition activity and are thus novel candidate effector domains for this modification (Table 1).
FIGURE 2. H3K4me3 or H3K36me3 recognition is a common property of S. cerevisiae PHD fingers.
A, preferential binding to higher states of H3K4 methylation by S. cerevisiae PHD fingers. Western analysis of histone peptide pulldowns under stringent conditions (300 mM NaCl) with the indicated GST fusion proteins and biotinylated peptides (aa: amino acids). B, a number of S. cerevisiae PHD fingers bind to H3K36me3. The indicated proteins were tested in pulldown assays as described for A, except 150 mM NaCl binding buffer was utilized. C, disassociation constants (Kd) of the indicated PHD fingers with the indicated peptide were determined by Trp fluorescence.
The PHD fingers that failed to bind H3K4me were also tested for H3K36me and H3K79me recognition activity. Notably, a number of PHD fingers bound weakly to H3K36me, with the PHD finger from the putative histone demethylase Ecm5, and a PHD finger present within the NuA3-associated protein Nto1, binding best to H3K36me3 (Fig. 2B). These interactions were confirmed by tryptophan fluorescence experiments and disassociation constants determined (Fig. 2C). The inability to detect H3K36me-binding by the Ecm5 and Nto1 PHD fingers using microarray screening (see Fig. 1) indicates that the threshold for detection using these methodologies requires Kd values < 100 μM (Fig. 2C). A number of additional PHD fingers present within different proteins bound to methylated and unmethylated histone peptides but without significant affinity for a specific ligand (Fig. 2b). Thus, 10 of 18 yeast PHD fingers show methyllysine binding activity (Table 1). Based on these data we conclude that histone methyl-lysine recognition activity is a common feature of PHD fingers present within the S. cerevisiae proteome.
The three HMTs that catalyze methylation of H3K4, H3K36, and H3K79 are Set1, Set2, and Dot1, respectively. We thus determined if proteins with PHD fingers that have methyl-histone binding activity are reported to be genetically or physically linked to one of these HMTs. Notably, all of the H3K4me binders with the exception of Bye1 and Jhd1 are identified as Set1 interactors, and Nto1, which binds to H3K36me3, is linked to Set2 (Table 1; see “Discussion”). No links to HMTs were found for the four proteins that fail to bind methyled histones (Table 1). These data suggest a link between the methyl-histone recognition property of PHD fingers and the function of their cognate protein.
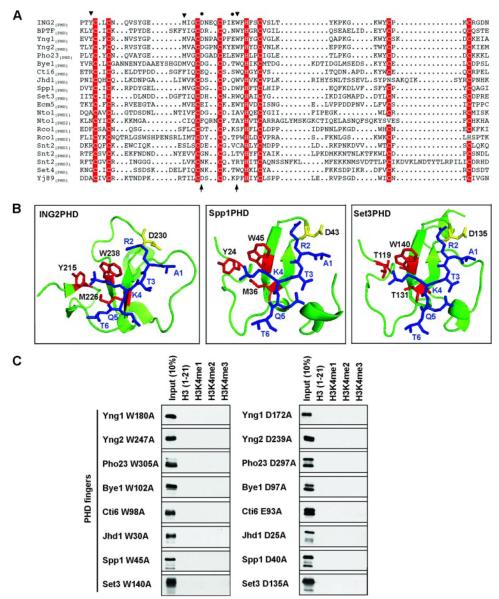
The molecular basis of specificity by the PHD fingers of ING2 and BPTF for H3K4me3 versus other methylated lysines is achieved by a coordinated fitting of the trimethyl moiety of H3K4 and the long side chain of H3R2 into adjacent surface pockets bracketed by an invariant tryptophan (Fig. 3b) (10, 18). In this context, alignment of the sequences from the S. cerevisiae PHD fingers and those of ING2 and BPTF revealed that many of the yeast PHD fingers include residues predicted critical for the interaction in the appropriate linear positions (Fig. 3A). For example, the ING2 homologues and Spp1(PHD) have the full complement of conserved residues and indeed bind to H3K4me3. Other PHD fingers, including Bye1(PHD), Cti6(PHD), Jhd1(PHD), and Set3(PHD) are only partially conserved yet still bind to H3K4me3, whereas Ecm5(PHD), despite having virtually all the known conserved residues required for H3K4me3 binding, recognizes H3K36me3 rather than H3K4me3 (Figs. 2 and 3A). Finally, Set4(PHD), which contains the invariant tryptophan residue, has no detectable histone methyl-lysine binding activity, even after attempts to engineer gain-of-function Set4(PHD) protein via introduction of the full set of residues essential for the activity of ING2(PHD) and BPTF(PHD) (supplemental Fig. 1). Based on these data, we argue that the conserved sequence information provides an important but limited element of predictive value with respect to potential histone methyl-lysine binding activity for PHD fingers.
FIGURE 3. Molecular features of H3K4me3 binding by S. cerevisiae PHD fingers.
A, sequence alignment of PHD fingers from S. cerevisiae with those from human ING2 and BPTF. Zinc-coordinating residues are shaded in red; residues in the ING2 PHD finger essential for interaction of methyl-H3K4 (triangles) and H3R2 (circles) are indicated. Note that BPTF(PHD) utilizes an extra tyrosine to form a K4me-binding cage (18). Residues substituted for the mutagenesis studies are indicated with arrows at the bottom of the alignment. Sequence alignment was produced with the ESPript web server. B, structural models of Spp1(PHD) and Set3(PHD) complexed with H3(1–6)K4me3 peptide. Blue, histone peptide; red, residues that form methyl-K4 binding cage; yellow, residues essential for H3R2 interaction. Structure of the ING2(PHD)-H3K4me3 complex is shown for comparison. C, peptide pulldown assay with the indicated PHD finger mutants (left, W>A; right, D>A) performed as described for Fig. 2A.
We turned to structural modeling to obtain a better insight into the molecular determinants of methyl-lysine affinity and specificity for the various PHD fingers. This analysis revealed that the analogous structural surface entities formed by the residues critical for determining H3K4me3 specificity for ING2(PHD) and BPTF(PHD), comprised of an aromatic cage and an adjacent groove bracketed by a tryptophan residue, are predicted to be present in all of the H3K4me-binding yeast PHD fingers (Fig. 3b and supplemental Fig. 2) (10, 18). Notably, the PHD fingers from Bye1, Cti6, Jhd1, and Set3 lack a conserved tyrosine residue that forms a part of the aromatic cage for ING2 and BPTF and in lieu have a nearby similarly positioned bulky residue (Fig. 3b and supplemental Fig. 2). These data argue that S. cerevisiae PHD fingers use a molecular mechanism similar to that of mammalian ING2(PHD) and BPTF(PHD) to distinguish H3K4me3 versus other methylated lysines. We directly tested this notion by generating alanine mutants of all the H3K4me3-binding PHD fingers, targeting the equivalent residues as Asp230 (which forms critical electrostatic interactions with H3R2) and Trp238 of ING2(PHD) (Fig. 3A, arrows). As shown in Fig. 3C, each mutant abolished the ability of the cognate PHD finger to bind H3K4me3, providing experimental evidence to support the structural modeling.
Next, we asked if modeling Ecm5(PHD) and Nto1(PHD-1) might provide insight into the molecular basis of H3K36me3 recognition. Notwithstanding the considerable differences in primary amino acid sequence, modeling predicts that both PHD fingers use the same surface to form the K36me3-binding pocket as that used by the known K4me3-binders (Fig. 4, A and B). In this regard, mutagenesis within the methyl-Lys binding pocket largely eliminated the interaction with H3K36me3 peptides for both PHD fingers (Fig. 4C). The specificity for methylated Lys36 versus Lys4 is likely due to the surrounding amino acid sequence. Specifically, the large side chains of Glu1254 in Ecm5(PHD) and Phe285 in Nto1(PHD-1) are in a position to interfere sterically with the long side chain of H3R2 (Fig. 4, A and B). In contrast, the surrounding region of H3K36 is more flexible and thus steric hindrance is less of a factor. Together, these data suggest that PHD fingers may use the same mechanism for binding the trimethyl group, with additional contacts between the sequence surrounding the methylated lysine and the PHD finger surface determining the ligand specificity.
FIGURE 4. Molecular features of H3K36me3-binding by S. cerevisiae PHD fingers.
A, structural models of Ecm5(PHD) in complex with H3(1–6)K4me3 (right panel) and H3(33–38)K36me3 (left panel) peptides as in Fig. 3B, except that the yellow residue indicates Glu1254 (see “Results and Discussion”). B, structural model of Nto1(PHD), residues that form the aromatic cage are indicated. Residue Phe285 is shown in yellow (see “Results and Discussion”). C, peptide pulldown assay with the indicated GST-fused PHD mutants as described for Fig. 2B.
Methylation events at H3K4 and H3K36 have been linked to multiple diverse activities (5, 19). Our genomic scale analysis indicates that PHD fingers might be critical molecular mediators that transduce these methylation events into different epigenetic programs (20). In this context, within the yeast proteome, PHD fingers with methyl-lysine binding activity are present within proteins of diverse functions, including modulators of methylation and acetylation (Table 1). These and other data raise the issue of how the same modification is recognized by effectors present within different proteins to potentially manifest distinct biological outcomes. There is considerable evidence that an isolated recognition event by an effector domain is necessary, but not sufficient, for determining epigenomic positioning of chromatin-regulatory proteins. Sufficiency is most likely determined by multiple concurrent interactions functioning in a combinatorial fashion to generate a specific localization (21). In this regard, many chromatin-regulatory proteins and complexes contain multiple effector domains. For example, the NuA3 complex has two PHD finger proteins, Yng1 and Nto1 that bind to H3K4me3 and H3K36me3, respectively, and the enzymes that generate H3K4me3 (SET1) and H3K36me3 (SET2) are both required for NuA3 association with chromatin (17, 22). Numerous additional molecular interactions, such as protein-protein interaction networks seeded by locus-specific transcription factors, are expected to contribute to epigenomic localization of chromatin-regulatory activities. Thus, recognition of a specific methyl-lysine event will manifest differently depending on the surrounding chromatin context, and as such, offers substantial signaling flexibility.
The observation that more than half of S. cerevisiae PHD fingers have methyl-lysine binding activity in conjunction with the considerable diversity in sequences of these PHD fingers argues that the number of methyl-lysine binders in the human proteome is certain to be far greater than predicted based solely on homology to the PHD fingers of ING2 and BPTF. Finally, the fact that a number of PHD fingers fail to bind to methylated histones raises the intriguing possibility that these PHD fingers recognize methylated lysines present on non-histone proteins (23).
Supplementary Material
Footnotes
This work was supported by grants from the National Institutes of Health (to S. D. B., P. J. U., and T. G. K.), the Canadian Institutes of Health and Michael Smith Foundation (to L. H.), the Baxter Foundation and Dana Foundation (to P. J. U.), the Welch Foundation (to M. T. B.), and the Kimmel Foundation (to O. G.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- GST
- glutathione S-transferase
- HMT
- histone methyltransferase.
X. Shi and O. Gozani, unpublished observations.
REFERENCES
- 1.Turner BM. BioEssays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 2.Strahl BD, Allis CD. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 3.Jenuwein T, Allis CD. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 4.de la Cruz X, Lois S, Sanchez-Molina S, Martinez-Balbas MA. BioEssays. 2005;27:164–175. doi: 10.1002/bies.20176. [DOI] [PubMed] [Google Scholar]
- 5.Daniel JA, Pray-Grant MG, Grant PA. Cell Cycle. 2005;4:919–926. doi: 10.4161/cc.4.7.1824. [DOI] [PubMed] [Google Scholar]
- 6.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 8.Eberharter A, Vetter I, Ferreira R, Becker PB. EMBO J. 2004;23:4029–4039. doi: 10.1038/sj.emboj.7600382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ragvin A, Valvatne H, Erdal S, Arskog V, Tufteland KR, Breen K, AM OY, Eberharter A, Gibson TJ, Becker PB, Aasland R. J. Mol. Biol. 2004;337:773–788. doi: 10.1016/j.jmb.2004.01.051. [DOI] [PubMed] [Google Scholar]
- 10.Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sali A, Blundell TL. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 13.Schwede T, Kopp J, Guex N, Peitsch MC. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canutescu AA, Shelenkov AA, Dunbrack RL., Jr. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 16.DeLano WL. PyMOL Molecular Graphics System. DeLano Scientific LLC; Palo Alto, CA: 2002. [Google Scholar]
- 17.Martin DG, Baetz K, Shi X, Walter KL, Macdonald VE, Wlodarski MJ, Gozani O, Hieter P, Howe L. Mol. Cell. Biol. 2006;26:7871–7879. doi: 10.1128/MCB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bannister AJ, Kouzarides T. Methods Enzymol. 2004;376:269–288. doi: 10.1016/S0076-6879(03)76018-2. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y. Nat. Struct. Mol. Biol. 2006;13:572–574. doi: 10.1038/nsmb0706-572. [DOI] [PubMed] [Google Scholar]
- 21.Becker PB. Nature. 2006;442:31–32. doi: 10.1038/442031a. [DOI] [PubMed] [Google Scholar]
- 22.Martin DG, Grimes DE, Baetz K, Howe L. Mol. Cell. Biol. 2006;26:3018–3028. doi: 10.1128/MCB.26.8.3018-3028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang K, Lin W, Latham JA, Riefler GM, Schumacher JM, Chan C, Tatchell K, Hawke DH, Kobayashi R, Dent SY. Cell. 2005;122:723–734. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.