Abstract
Mutations in the voltage-gated sodium channel SCN1A are responsible for a number of seizure disorders including Generalized Epilepsy with Febrile Seizures Plus (GEFS+) and Severe Myoclonic Epilepsy of Infancy (SMEI). To determine the effects of SCN1A mutations on channel function in vivo, we generated a bacterial artificial chromosome (BAC) transgenic mouse model that expresses the human SCN1A GEFS+ mutation, R1648H. Mice with the R1648H mutation exhibit a more severe response to the proconvulsant kainic acid compared with mice expressing a control Scn1a transgene. Electrophysiological analysis of dissociated neurons from mice with the R1648H mutation reveal delayed recovery from inactivation and increased use-dependent inactivation only in inhibitory bipolar neurons, as well as a hyperpolarizing shift in the voltage dependence of inactivation only in excitatory pyramidal neurons. These results demonstrate that the effects of SCN1A mutations are cell type-dependent and that the R1648H mutation specifically leads to a reduction in interneuron excitability.
Keywords: Sodium channel, SCN1A, GEFS+, SMEI, epilepsy, mutation
Introduction
Voltage-gated sodium channels (VGSCs) play a critical role in the regulation of neuronal excitability by facilitating the initiation and propagation of action potentials. VGSCs consist of a pore-forming, 260-kD α subunit (Nav1.1 to Nav1.9) that is associated with two of four accessory β subunits (β1 to β4) in the central nervous system (CNS). The α subunits are responsible for voltage sensing and ion conductance and the β subunits modulate channel kinetics and membrane localization.
Mutations in the SCN1A gene encoding Nav1.1 have been established as causing several subtypes of epilepsy, including Generalized Epilepsy with Febrile Seizures Plus (GEFS+, MIM 604233) and Severe Myoclonic Epilepsy of Infancy (SMEI or Dravet Syndrome, MIM 607208) (Escayg et al., 2000, Escayg et al., 2001, Claes et al., 2001, Ohmori et al., 2003, Fujiwara et al., 2003, Wallace et al., 2001, Wallace et al., 2003, Sugawara et al., 2001, Nagao et al., 2005, Mantegazza et al., 2005, Annesi et al., 2003). GEFS+ is a familial disorder characterized by febrile seizures that persist beyond the age of six, with a wide range of afebrile seizure types and severities among affected family members. SMEI is an intractable childhood epilepsy disorder that is often associated with mental retardation and ataxia. Although great strides have been made towards identifying the genetic basis for these disorders, the mechanisms that lead to seizure generation remain poorly understood.
The SCN1A mutation R1648H was first identified in a large GEFS+ family (Escayg et al., 2000). The invariant, positively charged R1648 residue is located in the voltage-sensing S4 segment of the fourth homologous domain (D4) of the α subunit. Previous electrophysiological analyses of the R1648H mutation identified different alterations in sodium channel gating kinetics, namely accelerated of recovery from inactivation and decreased use-dependent inactivation of channel activity (Spampanato et al., 2001), as well as increased persistent inward current during sustained depolarization (Lossin et al., 2002, Vanoye et al., 2006). These differences could be due to the fact that these experiments used different non-neuronal heterologous expression systems - Xenopus oocytes (Spampanato et al., 2001) and tsA201 cells (Lossin et al., 2002), a derivative of HEK293 human embryonic kidney cells. Neither system is a good model of a neuron. Mutation of R1648 to cysteine causes SMEI (Ohmori et al., 2003) and results in significant impairment of fast inactivation, including persistent, noninactivating channel activity and a depolarized shift in the voltage dependence of activation (Rhodes et al., 2004).
To study the R1648H mutant Nav1.1 channels in their native neuronal environment, we generated a mouse model of GEFS+. We used a bacterial artificial chromosome (BAC) transgenic strategy in which the R1648H mutation was introduced into the orthologous position of the mouse Scn1a gene contained in a BAC clone. The BAC was also modified to carry a FLAG epitope tag and the amino acid substitution E954Q, which confers resistance to tetrodotoxin (TTX) and saxitoxin (STX), making it possible to block the endogenous CNS sodium channels and characterize the functional properties of just the transgenic Nav1.1 channels.
Methods
Ethical approval
All experiments were performed according to guidelines established by and with the approval of the Institutional Animal Care and Use Committees of Emory University and the University of California, Irvine.
Identification of a suitable BAC clone
Examination of the assembled mouse genomic sequence from the Mouse Genome Resources database (http://www.ncbi.nlm.nih.gov/genome/guide/mouse/) and the Ensembl Mouse genome server (http://www.ensembl.org/Mus_musculus/) revealed several overlapping BAC clones that contained sequences from the mouse Scn1a gene. In silico analysis identified clone 259E13 (187 kb) from the RPCI-23 BAC library, which was predicted to contain all coding and noncoding Scn1a exons (Martin et al., 2007). Based on the alignment of the BAC end sequences from 259E13 (accession numbers AZ017834 and AZ017832) with the mouse genomic sequence, we determined that this clone extends approximately 115 kb upstream of the Scn1a translation start site and 3 kb downstream of the polyA termination sequence. There were no other genes predicted within 259E13. PCR analysis confirmed that clone 259E13 contained the 26 Scn1a coding exons and all the identified noncoding exons (Martin et al., 2007).
BAC modification and generation of transgenic mice
The BAC was modified in Escherichia coli as described by Lee et al. (2001). Briefly, DNA from clone 259E13 was isolated (Qiagen Maxiprep Kit, Qiagen) and electroporated into DY380 cells (a kind gift from Dr. Neil Copeland). DY380 cells containing clone 259E13 were grown to an optical density of 0.5 to 0.8, induced at 42°C, made electrocompetent and used immediately for transformation with each targeting DNA construct. Two modifications to clone 259E13 were made to generate the control BAC construct. First, the FLAG epitope tag (DYKDDDDK) was introduced immediately downstream of the first coding ATG (Hopp et al., 1988). Second, the E954Q substitution was introduced into exon 15 to confer TTX and STX resistance (Kontis and Goldin, 1993). The mutant BAC construct was generated by introducing the SCN1A GEFS+ mutation R1648H into the control BAC. Each modification was achieved via a PCR-generated targeting fragment that contained the desired substitution flanked by 70–80 bp of sequence with homology to the BAC.
BAC DNA was extracted using the NucleoBond Plasmid Maxi Kit (Clontech Labs), digested with NotI to release the genomic insert, and separated using pulse-field gel electrophoresis. The DNA was purified by agarase digestion and microdialysis against microinjection buffer before injection into fertilized FVB/NJ mouse oocytes.
Eleven founders were generated and screened for the level of transgene expression; two control transgenic lines, Line 1 (Scn1aTg-C1) and Line 8 (Scn1aTg-C8), and two mutant transgenic lines, Line 5 (Scn1aTg-RH5) and Line 9 (Scn1aTg-RH9), were selected for further characterization.
Genotyping of mice
DNA from tail biopsies of 12-day-old mice was used for genotyping with a FLAG-specific forward primer and a reverse primer in Scn1a exon1 (FLAG-F and FLAG-R, respectively; see Table 1 for primer sequences). To assess the integrity of the BAC transgene, mice were also screened with primers specific to the SP6 end (SP6-F and SP6-R) as well as the T7 end (T7-F and T7-R) of the BAC vector. The presence of the R1648H mutation was confirmed by PCR amplification and sequence analysis using the primer pair Scn1a-26F/Scn1a-26R that flanks the mutation site.
Table 1.
Primers for genotyping
| Primer | Sequence | Amplicon (bp) |
|---|---|---|
| FLAG-F | 5′-GGCCAAAGCTCAACATAGAGAATG-3′ | 259 |
| FLAG-R | 5′-TACAAGCACTGTTTGCTCTTTATC-3′ | |
| SP6-F | 5′-CATTAAGGTATGGAGGGGAG-3′ | 165 |
| SP6-R | 5′-AGGTGACACTATAGAAGGATCCGC-3′ | |
| T7-F | 5′-ATAGGGAGAGGATCCGCGGAATTC-3′ | 361 |
| T7-R | 5′-GGAGCATGTGTCCTTCTTAC-3′ | |
| Scn1a-26F | 5′-AGGAGGAGAGGTGCATCCTGGGAC-3′ | 575 |
| Scn1a-26R | 5′-GGGTCACAGTCAGGGGGTTTGCTG-3′ | |
| FLAGRT-F | 5′-GTGCAGGATGACAAGATGGACTAT-3′ | 492 |
| FLAGRT-R | 5′-GGGAGGGTTACTCATTGTCATAAA-3′ | |
| Scn1aF | 5′-ACCTGACAGCTTCAACTTCTTCAC-3′ | 400 |
| FLAGRT-R | 5′-GGGAGGGTTACTCATTGTCATAAA-3′ |
mRNA analysis
Total RNA was extracted from whole brains or dissected brain regions of 5-week-old mice from each line using the RNeasy Lipid Tissue Mini Kit (Qiagen). First-strand cDNA was synthesized from 5 μg of RNA using oligo(dT)12–18 and Superscript II according to the manufacturer’s instructions (Invitrogen). Expression of the BAC transgene in each line was confirmed by PCR analysis of 1 μl of first-strand cDNA using a forward primer (FLAGRT-F) located in the FLAG epitope and a reverse primer (FLAGRT-R) located in Scn1a exon 3. The expected fragment of 492 bp was found in all transgenic lines and was absent in nontransgenic littermates. PCR analysis of first-strand cDNA from dissected brain regions was performed in order to compare the expression profile of the BAC transgene with endogenous Scn1a. The primer pair FLAGRT-F/FLAGRT-R was used to specifically amplify the transgene, and endogenous Scn1a transcripts were amplified using a forward primer in Scn1a exon 1 (Scn1aF) and FLAGRT-R.
Western blotting
Total Nav1.1 protein levels were determined using an anti-Nav1.1 antibody (Chemicon). Protein levels were normalized to α tubulin levels, which were determined with an anti-α tubulin antibody (Cedarlane Labs). Briefly, membrane proteins from the brains of 5-week-old mice were extracted as previously described (Isom et al., 1995). Fifteen micrograms of protein were separated on a 7.5% SDS-polyacrylamide gel and transferred to a PVDF membrane (GE Healthcare). The membrane was incubated with either anti-Nav1.1 diluted 1:200 (Chemicon) or anti-α tubulin diluted 1:10,000 (Cedarlane Labs) and then with either HRP-conjugated anti-rabbit or anti-mouse IgG diluted 1:5000 (GE Healthcare). Signal was visualized by chemiluminescent detection with the ECL Detection System (GE Healthcare), and the intensity of each band was quantified using the Gel Logic 2200 Digital Imaging system (Kodak) and normalized to the level of α tubulin.
Cortical recording-implant surgery
Adult transgenic mice from Lines 1 and 9 and nontransgenic littermates were surgically implanted with electroencephalography (EEG) and electromyography (EMG) electrodes for seizure activity recordings. All animals were anesthetized with isoflurane gas and placed on a stereotaxic frame (Cartesian Research, Oregon). Two pairs of stainless-steel recording screws were placed on the surface of the cortex. The first pair was located 2 mm anterior to Bregma and 1.2 mm right of the central suture and 1.5 mm posterior to Bregma and 1.2 mm right of the central suture, whereas the second pair was placed 0.5 mm posterior to Bregma and 2.2 mm left of the central suture and 3.5 mm posterior to Bregma and 2.2 mm left of the central suture. In addition, EMG activity was monitored using two fine wires (Cooner AS632, California) inserted bilaterally into the nuchal muscle. The EEG and EMG electrodes were attached to a connector (VMS-OHIO), and dental ceramic compound was poured over the screws and base of the skullcap. All animals were allowed to recover from surgery for at least 7 days. Each mouse was attached to a series of bioelectric amplifiers (Stellate System, Harmonie Software version 5.0b). Mouse behavior was also simultaneously monitored with digital video recording. At least 96 hours of recording was obtained for each mouse while moving freely in the monitoring cage. Epileptiform activity was defined as being greater than twice the amplitude of the baseline waveform. All recordings were manually scored.
Seizure susceptibility
R1648H mutant transgenic lines (Lines 5 and 9) and the control transgenic lines (Lines 1 and 8) on an FVB/NJ background were bred to C57BL/6J mice (Jackson Laboratories, Bar Harbor). Kainic acid (KA, 15 mg/kg, i.p., Sigma) was administered to 2- to 4-month-old F1 (FVB/NJ X C57BL/6J) transgenic mice and wild-type littermates. The mice were then placed in a clear chamber and monitored for two hours. Seizure severity was scored using a modified Racine scale (Racine, 1972) as follows: stage 0, no response; stage 1, staring; stage 2, nodding; stage 3, forelimb clonus; stage 4, rearing or falling; stage 5, generalized seizure; stage 6, death. Statistical analysis for the percentage of mice progressing to generalized seizures during seizure susceptibility testing was performed by Fisher’s exact test. The Mann-Whitney U rank sum test was used to analyze the maximal seizure stage. Differences were considered significant when P < 0.05.
Survival assay
Heterozygous control mice from Line 1 (Scn1aTg-C1) and the mutant transgenic mice from Line 9 (Scn1aTg-RH9) on a FVB/NJ background were crossed to heterozygous Scn1a knockout mice (Scn1a+/−) on a mixed CD1/FVB/NJ background to generate double heterozygous mutants. Double heterozygous mice were further crossed to Scn1a+/− mice to generate mice that carried each transgene but lacked endogenous Scn1a. The survival of the double heterozygous mutants and the mutants that carried each transgene but lacked endogenous Scn1a was compared with wild-type littermates over a six-month period.
Neuron preparation and identification
Dissociated cortical neurons were cultured as described previously (Hilgenberg et al., 1999, Li et al., 1999). Briefly, mice were decapitated after anesthesia with either ice (P5–P8) or halothane (P14–P19). Brains were quickly removed and sliced in chilled dissecting solution. The cortical region was dissected out, treated with papain for 30 min at 37°C, transferred to 2 ml of neurobasal medium supplemented with B27 at room temperature and triturated through the tip of a glass pipette. Cells were plated on coverslips coated with poly-L-lysine and cultured in glial-conditioned media. Electrophysiological recordings were performed within 24–36 h of obtaining neuronal cultures.
Excitatory pyramidal neurons and inhibitory bipolar neurons were identified by correlating shape with staining for anti-GluR1 (Glutamate Receptor 1) (1:5000, Upstate Biotechnology) and anti-GAD67 (Glutamic Acid Decarboxylase 67) (1:5000, Chemicon), respectively (Fong et al., 2005, Graf et al., 2004). Secondary antibodies Alexa 568 anti-rabbit IgG and Alexa 488 anti-mouse IgG2a (Molecular Probes) were used, respectively, for observation with epifluorescent illumination.
Electrophysiology
Freshly cultured neurons from P5–P8 and P14–P19 mice were voltage-clamped at 20°C in the whole-cell patch configuration using an Axopatch 200B patch clamp with Digidata 1322A interface and pCLAMP 8.0 software. The bath solution consisted of 150 mM NaCl, 3 mM KCl, 15 mM TEA-Cl, 4 mM BaCl2, 0.1 mM CdCl2 and 10 mM HEPES (pH 7.4), and the electrode solution consisted of 140 mM CsF, 10 mM NaCl, 5 mM EGTA and 10 mM HEPES (pH 7.3). Series resistance was compensated to 80–90%, and linear leak subtraction was used for all recordings. Corrections were made for leak and capacity currents using P/4 subtraction. Currents were filtered at 5 kHz and sampled at 20 kHz. Recordings were obtained in the presence and absence of 75–100 nM STX in the bath.
The voltage dependence of activation and inactivation, recovery from inactivation, use-dependent inactivation, kinetics of slow inactivation and percentage of persistent current were studied using protocols described by Spampanato et al. (2001). Briefly, the voltage dependence of activation was analyzed using a protocol with depolarizations from −60 mV to +15 mV in 5-mV intervals. Peak current amplitudes were converted to conductance values, normalized for comparison and plotted against voltage. The voltage dependence of inactivation was analyzed using varying depolarizations for 50 ms to allow inactivation followed by repolarization, then depolarizing to a test potential of −15 mV to elicit the remaining available current. Peak currents during the test pulse were normalized for comparison and plotted against the voltage of the conditioning pulse. Recovery from inactivation was analyzed using a protocol that first inactivated the channels by depolarization to −5 mV for 50 ms, followed by recovery at −80 mV for a variable period of time (2 ms to 60 ms) and a test pulse to −5 mV. Peak current amplitudes during the test pulse were normalized to peak current amplitudes during the conditioning pulse and plotted against recovery time. Use-dependent inactivation was examined by successive depolarizations to −5 mV from a holding potential of −80 mV at 39 Hz. Measurement of persistent current was done using prolonged 200-ms depolarizations to −10 mV. Each voltage step was followed by > 5-s recovery period at −80 mV to minimize the accumulation of use-dependent inactivation. Pulse generation, data collection and analyses were done with pCLAMP 8.0, Excel and SigmaPlot 9.0 software. Data shown are means ± SEM.
The average values of the parameters of activation, inactivation, recovery from inactivation and use-dependent inactivation that are shown in Table 3 were obtained after fitting the data with the following equations.
Table 3.
Electrophysiological parameters
| Bipolar Neurons | Pyramidal Neurons | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wild-Type | E954Q | R1648H/ E954Q |
E954Q +STX |
R1648H/ E954Q +STX |
Wild-Type | E954Q | R1648H/ E954Q |
E954Q +STX |
R1648H/ E954Q +STX |
|
| Voltage Dependence of Activation | ||||||||||
| Z (e0) | 2.7 ± 0.9 | 3.2 ± 0.8 | 4.6 ± 1.0 | 2.1 ± 0.8 | 2.6 ± 0.7 | 3.0 ± 0.8 | 4.4 ± 1.2 | 4.5 ± 0.9 | 2.6 ± 0.8 | 3.0 ± 0.9 |
| V½(mV) | −28.4 ± 4.1 | −29.7 ± 4.3 | −30.1 ± 6.2 | −24.6 ± 5.4 | −24.6 ± 5.3 | −26.7 ± 4.6 | −26.2 ± 3.9 | −28.9 ± 5.1 | −20.6 ± 5.9* | −20.0 ± 6.7* |
| n | 8 | 10 | 3 | 4 | 14 | 9 | 7 | 8 | 10 | 7 |
| Voltage Dependence of Inactivation | ||||||||||
| A (mV) | 10.0 ± 2.1 | 9.7 ± 1.9 | 10.1 ± 1.2 | 20.4 ± 2.1* | 19.9 ± 2.1* | 7.4 ± 1.2 | 8.3 ± 2.8 | 6.8 ± 2.1 | 15.0 ± 3.5* | 12.0 ± 3.9* |
| V½ (mV) | −59.7 ± 8.3 | −61.3 ± 7.3 | −57.7 ± 6.8 | −58.4 ± 7.1 | −57.1 ± 5.6 | −52.1 ± 7.3 | −51.3 ± 6.2 | −58.7 ± 8.2* | −51.9 ± 7.3 | −65.4 ± 9.3* |
| n | 7 | 21 | 12 | 9 | 18 | 6 | 4 | 6 | 4 | 6 |
| Recovery from Inactivation | ||||||||||
| A | 0.9 ± 0.21 | 0.9 ± 0.16 | 0.9 ± 0.05 | 0.9 ± 0.08 | 0.8 ± 0.08* | 0.8 ± 0.08 | 0.9 ± 0.07 | 0.9 ± 0.05 | 0.85 ± 0.08 | 0.84 ± 0.07 |
| τ(ms) | 9.4 ± 0.2 | 9.0 ± 0.4 | 12.1 ± 0.1 | 8.0 ± 0.2 | 8.0 ± 0.3 | 9.3 ± 0.1 | 11.5 ± 0.08 | 7.2 ± 0.06 | 9.2 ± 0.09 | 7.7 ± 0.3 |
| c | 0.1 ± 0.04 | 0.1 ± 0.05 | 0.1 ± 0.08 | 0.1 ± 0.02 | 0.2 ± 0.02* | 0.1 ± 0.02 | 0.1 ± 0.03 | 0.1 ± 0.01 | 0.1 ± 0.07 | 0.2 ± 0.08* |
| n | 6 | 7 | 6 | 3 | 13 | 7 | 5 | 10 | 4 | 9 |
| Use-Dependent inactivation at 39 Hz | ||||||||||
| A1 | 0.4 ± 0.05 | 0.4 ± 0.08 | 0.3 ± 0.03 | 0.4 ± 0.08 | 0.25 ± 0.09 | 0.3 ± 0.008 | 0.4 ± 0.07 | 0.4 ± 0.03 | 0.3 ± 0.02 | 0.3 ± 0.08 |
| A2 | 0.7 ± 0.06 | 1.0 ± 0.09 | 1.4 ± 0.2 | 1.3 ± 0.08 | 0.27 ± 0.04 | 0.9 ± 0.03 | 0.8 ± 0.07 | 0.2 ± 0.06 | 0.3 ± 0.02 | 0.4 ± 0.09 |
| A3 | 1.8 ± 0.2 | 2.2 ± 0.8 | 1.1 ± 0.6 | 3.5 ± 0.09 | ||||||
| τ1 (ms) | 0.9 ± 0.08 | 0.8 ± 0.07 | 1.1 ± 0.1 | 0.9 ± 0.2 | 4.5 ± 0.2 | 0.8 ± 0.07 | 0.8 ± 0.08 | 5.9 ± 0.05 | 5.0 ± 0.06 | 3.5 ± 0.57 |
| τ2 (ms) | 0.03 ± 0.004 | 0.02 ± 0.001 | 0.02 ± 0.005 | 0.02 ± 0.007 | 0.35 ± 0.07 | 0.03 ± 0.004 | 0.03 ± 0.005 | 0.3 ± 0.008 | 0.3 ± 0.002 | 0.3 ± 0.01 |
| τ3 (ms) | 0.02 ± 0.006 | 0.02 ± 0.01 | 0.03 ± 0.008 | 0.01 ± 0.007 | ||||||
| c | 0.31 ± .08 | 0.25 ± 0.01 | 0.25 ± 0.02 | 0.20 ± 0.01 | 0.25 ± 0.03 | 0.22 ± 0.04 | ||||
| n | 18 | 24 | 10 | 8 | 17 | 11 | 14 | 13 | 12 | 13 |
Parameters of voltage dependence of activation and inactivation, recovery and use-dependent inactivation of bipolar and pyramidal neurons were determined as described in the Methods.
indicates statistically significant differences.
| Activation: | G=1/(1+exp[−0.03937*z*(V−V½)]), in which G is conductance, z is the apparent gating charge, V is the potential of the given pulse and V− is the potential for half-maximal activation. |
| Inactivation: | I=1/(1+exp[(V−V½)/a]), in which I is equal to the test-pulse current amplitude, V is the potential of the conditioning pulse, V½ is the voltage for half-maximal inactivation and a is the slope factor. |
| Recovery: | I =1−A·exp(−t/τ)−c, in which A is the fraction of current that recovered with the time constant τ, t is the recovery time and c is the fraction of current that did not recover. |
| Use-Dependent inactivation: | I=A1*exp(−t/τ1)+A2*exp(−t/τ2)+c; double exponential, in which A1 and A2 are the relative fractions of current that decayed with the time constants τ, and τ2, t is the decay time and c is the remaining current. I=A1*exp(−t/τ1)+A2*exp(−t/τ2)+A3*exp(−t/τ3); triple exponential, in which A1, A2 and A3 are the relative fractions of current that decayed with the time constants τ1, τ2 and τ3, and t is the decay time. |
The p values for statistical significance of each parameter were obtained by performing a pairwise t-test against corresponding values obtained from wild-type neurons.
Results
Generation and characterization of control and mutant transgenic mice
Since we wanted a model system to study the mechanism by which SCN1A missense mutations lead to GEFS+, we constructed transgenic mice using a BAC clone containing the complete mouse Scn1a gene. One advantage of this approach is that the channel protein can be modified to distinguish it from endogenous channels, while the gene remains regulated by the native promoter in the BAC transgene. Because GEFS+ is inherited in an autosomal dominant fashion, even a single mutant allele in the presence of the two wild-type alleles is likely to at least partially recapitulate the disease phenotype.
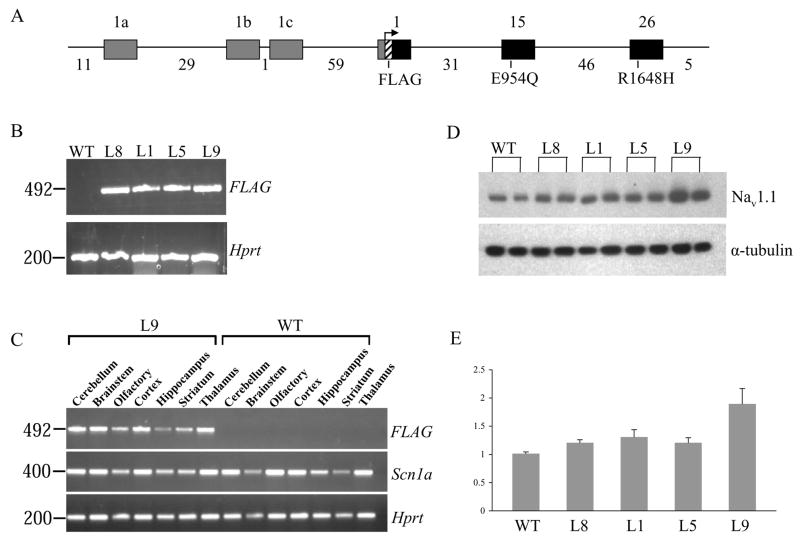
To construct the transgene, BAC clone 259E13 containing the complete genomic sequence of Scn1a was modified to contain the FLAG epitope tag as well as the E954Q substitution, which confers resistance to TTX and STX (Fig. 1A). This modified BAC construct was used to generate the control transgenic mice (Lines 1 and 8). The BAC clone was then modified to carry the GEFS+ mutation R1648H in order to generate the mutant transgenic lines (Lines 5 and 9) that served as a model of human GEFS+. To examine the expression of the transgene, RNA was prepared and purified from the brains of transgenic mice and wild-type littermates. Transgene-specific expression was confirmed in the selected lines by RT-PCR analysis using a forward primer with homology to the FLAG epitope tag (Fig. 1B). RT-PCR analysis of dissected brain regions demonstrated that the expression profile of the BAC transgene was similar to endogenous Scn1a (Fig. 1C). Comparison of total Nav1.1 protein levels between age-matched transgenic lines and wild-type littermates showed that the highest expression was observed in Line 9, which carried the mutant transgene (Fig. 1D). Nav1.1 protein levels in transgenic mice from Line 9 were approximately two-fold higher than for wild-type littermates, suggesting that the level of expression of the transgene was comparable to the two endogenous copies of Scn1a. Lines 1, 5 and 8 had comparable levels of expression that corresponded to approximately 1.2-fold higher than endogenous Nav1.1 protein levels (Fig. 1E). Despite several attempts, we were unable to obtain control transgenic lines with expression comparable to Line 9.
Figure 1. Expression of transgenes.
A. Schematic structure of the Scn1a BAC transgenes. The mouse Scn1a gene contains 26 coding exons and three noncoding exons. The three-noncoding exons (1a, 1b and 1c; grey boxes) and three of the coding exons (1, 15 and 26; black boxes) are shown. Genomic distances between exons are indicated in kilobases (kb). The FLAG epitope (striped box) is located immediately downstream of the first coding methionine in exon 1. The amino acid substitution E954Q was introduced into exon 15 to confer resistance to STX and TTX. The mutant transgene also contains the GEFS+ mutation R1648H located in exon 26. The BAC clone contains 11 kb of genomic sequence upstream of exon 1a and 10 kb of sequence downstream of the stop codon in exon 26. B. RT-PCR analysis using a forward primer specific to the FLAG epitope confirmed the expression of the transgene in the four selected lines. No PCR product was observed in the wild-type (WT) lane. PCR amplification of the mouse Hprt gene served as a control for the quality of the first-strand cDNA. First lane, 100 bp molecular weight marker. C. RT-PCR analysis of dissected brain regions demonstrated that the expression profile of the BAC transgene from Line 9 was similar to endogenous Scn1a. PCR amplification using a forward primer specific to the FLAG epitope generated PCR products from the transgenic samples but not the wild-type (WT) samples. PCR amplification using a primer pair specific to Scn1a generated PCR products from all samples. Amplification of the mouse Hprt gene served as a control for the quality of the first-strand cDNA. D. Fifteen micrograms of protein purified from whole mouse brain was separated on an SDS-polyacrylamide gel and transferred to a PVDF membrane. The membrane was probed with anti-Nav1.1 (upper panel) and anti-α tubulin (lower panel). Blots are representative of a single experiment. Lines 8, 1, 5 and 9 are shown. Two mice from each line were analyzed. WT represents an age-matched wild-type mouse. E. The bar graph represents the summarized results of three separate Western blots in which intensity of the Nav1.1 band from each mouse was normalized to the corresponding α tubulin band. The Nav1.1/α tubulin ratio from the wild-type littermate was assigned a value of 1, and the ratios from the other mice were normalized to this value.
R1648H transgenic mice show decreased seizure thresholds
To determine whether R1648H transgenic mice exhibit spontaneous seizures, cortical activity was continuously measured for 96 hours in transgenic mice from Line 1 (n= 4) and Line 9 (n= 4) and wild-type littermates from Line 1 (n= 3) and Line 9 (n= 3). We observed no electrographic or behavioral seizures.
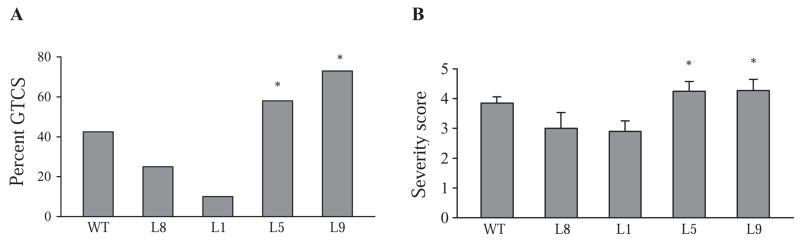
Since spontaneous seizures were not observed in the R1648H transgenic mice, we next examined their thresholds for seizures induced by the proconvulsant kainic acid (KA) (Fig. 2). We observed that more than 50% of R1648H transgenic mice (Lines 5 and 9) but fewer than 25% of control transgenic mice (Lines 1 and 8) progressed to generalized tonic-clonic seizures (GTCS) (Fig. 2A). Only 10% of Line 1 mice progressed to GTCS, which was significantly lower than for the mutant transgenic lines (Line 5: 58%; P= 0.03 and Line 9: 73%; P= 0.01) (Fig. 2A). A trend towards greater resistance to KA was also observed in the control transgenic mice from Line 8 (25% progressing to GTCS), although this difference was not statistically different from the mutant lines (Line 8 versus Line 5, P= 0.20; Line 8 versus Line 9, P= 0.07). Forty-two percent of wild-type littermates progressed to GTCS, which is higher than control transgenic mice but lower than mutant transgenic mice. However, these differences between transgenic mice and wild-type littermates were not statistically significant. The control transgenic lines also reached a lower average score on the modified Racine scale compared with the mutant lines, and the difference in severity score between Line 1 and the mutant lines was statistically significant (Line 1 versus Line 5, P= 0.02; Line 1 versus Line 9, P= 0.03) (Fig. 2B). These results suggest that the R1648H mutation leads to a more severe response to KA. In contrast, the higher levels of wild-type Nav1.1 in the control transgenic lines appear to be associated with a less severe response to KA.
Figure 2. More severe response to kainic acid (KA) in mutant transgenic mice.
A. The percentage of mice exhibiting GTCS, and B. the average severity score of control and mutant transgenic mice and age-matched wild-type littermates in response to 15 mg/kg KA are shown. Wild-type littermates, n= 33; Line 8, n= 8; Line 5, n= 12; Line 1, n= 10; Line 9, n= 11. *P < 0.05 when compared to Line 1.
Nav1.1 is expressed at three-fold higher levels in bipolar compared with pyramidal neurons
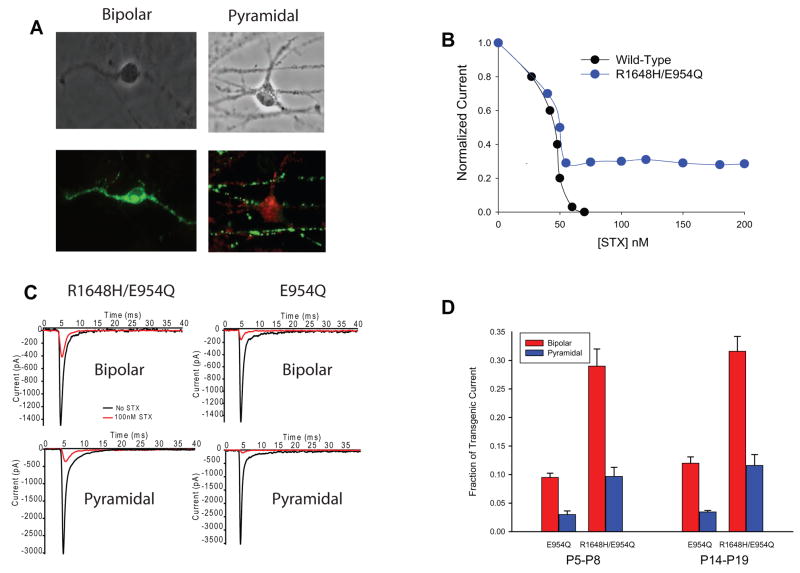
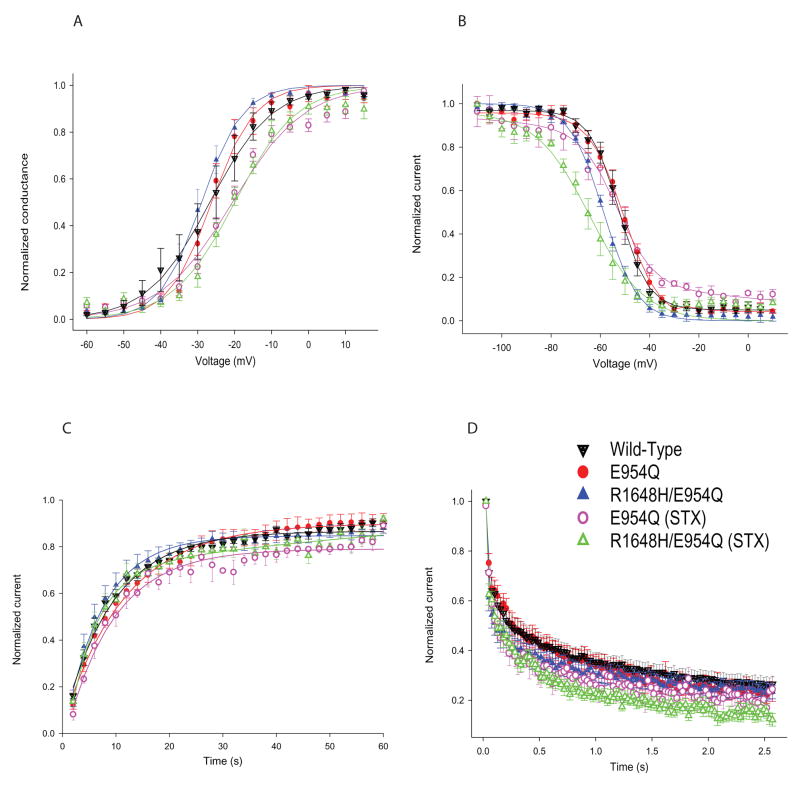
The expression of Scn1a increases during the first few postnatal weeks (Beckh et al., 1989), so we characterized the sodium currents in cortical neurons dissociated from mice at ages P5–P8 and P14–P19. Because the phenotypic effects of the R1648H mutation could be due to changes in either excitatory or inhibitory neurons in the CNS, the neurons were separated into the two major classes of cortical neurons: inhibitory bipolar-shaped and excitatory pyramidal-shaped neurons. Each type of neuron was initially identified by immunostaining, which was then correlated with morphology for patch clamping. Bipolar neurons were identified as being inhibitory by positive immunostaining for the glutamic acid decarboxylase GAD67, as shown in Figure 3A (left panel, green). Pyramidal neurons were identified as being excitatory by staining for the metabotropic glutamate receptor GluR1 (Figure 3A, right panel, red).
Figure 3. STX resistance of transgenic channels in bipolar and pyramidal neurons.
A. P5–P8 cortical bipolar and pyramidal neurons stained with anti-GAD67 (green) and anti-mGluR1 (red). B. STX dose-response relationship from wild-type and R1648H transgenic P5–P8 bipolar neurons. C. Sample traces showing sodium current recorded from P5–P8 bipolar R1648H/E954Q (top left), pyramidal R1648H/E954Q (bottom left), bipolar E954Q (top right) and pyramidal E954Q (bottom right) neurons with (red) and without (black) 100 nM STX during a depolarization to −10 mV. D. Percentage of STX-resistant current in bipolar and pyramidal neurons from R1648H/E954Q and E954Q control mice at ages P5–P8 (left) and P14–P19 (right).
We first determined whether the R1648H mutation affected the sodium current amplitudes in each cell type. In neurons from wild-type mice, there were no significant differences between the current amplitudes in bipolar-shaped neurons (1476.2 ± 123.4 pA, n=44) and those in the larger pyramidal-shaped neurons (1576.9 ± 132.1 pA, n=19). To control for cell size, the current amplitudes were normalized to cellular capacitance, which is a measure of the total cell membrane (Table 2). There was a slightly larger ratio of current density (1.05) in bipolar versus pyramidal neurons from wild-type mice, and an approximately equal ratio (0.98) in bipolar versus pyramidal neurons from mice expressing the E954Q transgene (Table 2). In contrast, the ratio of current density in bipolar versus pyramidal neurons was only 0.51 from mice expressing the R1648H/E954Q mutant transgene. The lower current density in R1648H/E954Q bipolar neurons (−69 ± 14 pA/pF) was significantly different compared to both R1648H/E954Q pyramidal neurons (−136 ± 22 pA/pF) and wild-type bipolar neurons (−116 ± 11 pA/pF) (Table 2). These results suggest that the R1648H mutation specifically decreased the total sodium current amplitude in bipolar neurons.
Table 2.
Current densities in bipolar and pyramidal neurons
| Genotype | Wild-Type | E954Q | R1648H/E954Q |
|---|---|---|---|
| Bipolar Current Density (pA/pF) | −116 ± 11 | −99 ± 14 | −69 ± 14* |
| Bipolar Number of Cells | 33 | 41 | 22 |
| Pyramidal Current Density (pA/pF) | −110 ± 19 | −101 ± 19 | −136 ± 22 |
| Pyramidal Number of Cells | 17 | 26 | 28 |
| Current Density Ratio (Bipolar/Pyramidal) | 1.05 | 0.98 | 0.51 |
Current density (total current divided by cellular capacitance) was determined as described in the Methods.
indicates statistically significant difference (p<0.05) in current density between bipolar cells from R1648H/E954Q and wild-type mice and between bipolar and pyramidal cells from R1648H/E954Qmice.
The difference in total sodium current density might reflect selective expression of the transgenic channels in the two classes of neurons. To determine if the transgenic channels were expressed at different levels in bipolar and pyramidal neurons, we took advantage of the E954Q substitution that makes the transgenic channels resistant to TTX and STX, because endogenous CNS sodium channels are blocked by nanomolar concentrations of either toxin. Sodium channels with the E954Q substitution have an apparent Kd of 5.4 μM for TTX and 54.1 μM for STX, compared with 18.6 nM for TTX and 2.8 nM for STX for wild-type Nav1.2 (Kontis and Goldin, 1993). The E954Q substitution did not affect any of the functional properties of the sodium channel (Kontis and Goldin, 1993). The more than 1,000-fold decrease in sensitivity to STX in channels with the E954Q mutation makes it easy to block all of the natively expressed wild-type sodium channels with STX without affecting the transgenic channels, allowing study of transgenic channels in isolation from the endogenous channels.
Sodium currents were recorded from bipolar R1648H/E954Q neurons in the presence of increasing concentrations of STX to determine the concentration that blocked endogenous sodium channels without affecting the transgenic E954Q channels (Figure 3B). All of the sodium current in neurons from wild-type mice was blocked by 70 nM STX, whereas approximately 30% of the current remained in neurons from R1648H/E954Q mice, even at the highest STX concentration of 200 nM. Based on these results, we used 100 nM STX to record currents through the transgenic channels while completely blocking the endogenous wild-type channels. Figure 3C shows sample traces recorded from bipolar (top) and pyramidal (bottom) neurons from R1648H/E954Q (Line 9, left) and E954Q (Line 1, right) control mice at P5–P8 in the absence (black) and the presence of 100 nM STX (red). For these studies, we only used neurons expressing large sodium current amplitudes, which increased the reliability of measuring and analyzing the current remaining in the presence of STX. We divided the current remaining in the presence of 100 nM STX by the total sodium current amplitude to determine the percentage of STX-resistant transgenic channels expressed in bipolar versus pyramidal neurons. The results are shown in Figure 3D for E954Q control (Line 1) and R1648H/E954Q (Line 9) transgenic mice at ages P5–P8 (left). In both transgenic lines, the fraction of transgene-derived sodium current was approximately three times higher in bipolar neurons (red) compared to pyramidal neurons (blue). To determine whether expression of the transgenic channel increased with development, we performed a similar analysis using bipolar and pyramidal neurons from P14–P19 mice. The percentage of STX-resistant current increased slightly for both the R16485/E954Q and E954Q channels in bipolar and pyramidal neurons (Figure 3D, right). However, the ratio of STX-resistant current in bipolar neurons (red) to STX-resistant current in pyramidal neurons (blue) remained the same, indicating that the Scn1a gene is still preferentially expressed in bipolar neurons in P14–P19 mice. Since the transgenic sodium channel is regulated by endogenous transcription factors acting on the native promoter in the BAC, the relative expression in bipolar and pyramidal cells should reflect the native expression of Nav1.1 in cortical bipolar and pyramidal cells.
R1648H sodium channels have slower recovery from inactivation and increased use-dependent inactivation in bipolar neurons
Although the effects of the R1648H mutation on sodium channel properties have been examined in a variety of heterologous expression systems (Spampanato et al., 2001, Lossin et al., 2002, Alekov et al., 2000), those results may not accurately reflect the effects of the mutation in native neurons. To determine how the mutation alters sodium channel properties in P5–P8 cortical bipolar neurons, we compared the properties of R1648H/E954Q transgenic channels (Line 9) with those of control transgenic channels containing only the E954Q substitution (Line 1). In both cases, currents were recorded in the presence of 100 nM STX to determine the effects of the mutation on the channel and in the absence of STX to determine whether the 30% of sodium current carried by the mutant transgenic channels altered the properties of the total sodium current in the neuron.
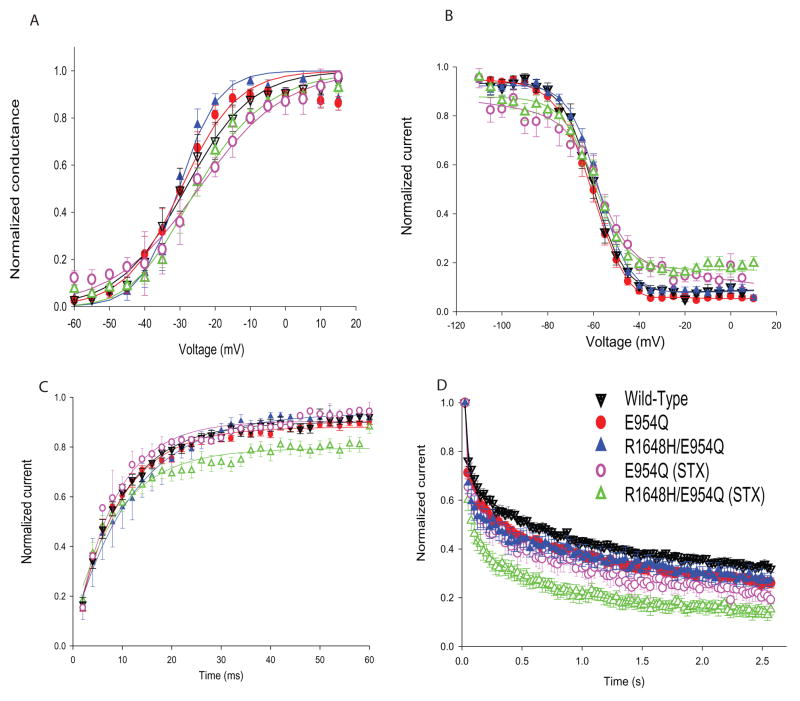
We characterized the voltage dependence of activation and inactivation, use-dependent inactivation, recovery from inactivation and the percentage of persistent sodium current. Alterations in each of these properties have been seen using heterologous expression systems to study SCN1A mutations that cause epilepsy (Lossin et al., 2002, Spampanato et al., 2001, Spampanato et al., 2003, Barela et al., 2006). There were no significant differences in the voltage dependence of either activation or inactivation in either the presence or absence of STX (Figures 4A and B, Table 3). There were small differences in the slope factor for activation (Table 3), particularly for currents recorded in the presence of STX, but these did not correlate with the presence of the R1648H mutation. These differences most likely represent variability in the recordings due to the very low current amplitudes in those cells (only 30% of the total current).
Figure 4. Properties of the sodium channels in bipolar interneurons.
A. Voltage dependence of activation. Sodium currents were recorded from a holding potential of −80 mV during 20 ms depolarizations to a range of potentials from −60 to +15 mV in 5 mV increments. Conductance values were calculated by dividing the peak current amplitude by the driving force at each potential and normalizing to the maximum conductance. B. Voltage dependence of inactivation. The voltage dependence of inactivation was determined using a two-step protocol in which a conditioning pulse was applied from a holding potential of −80 mV, consisting of 100 msec depolarizations to a range of potentials from −110 to +10 mV in 5 mV increments, followed by a test pulse to −15 mV. The peak current amplitude during each test pulse was normalized to the current amplitude during the first test pulse and plotted as a function of the conditioning pulse potential. The data were fit with a two-state Boltzmann equation. C. Recovery from inactivation. The recovery protocol was performed from a holding potential of −80 mV and consisted of a conditioning depolarization to −5 mV for 50 msec (which inactivated > 95% of the channels), a variable recovery time interval at −80 mV and a test depolarization to −5 mV for 20 ms. The recovery period was decreased by 2 ms each time, with the maximum recovery time being 60 ms. Fractional recovery was calculated by dividing the maximum current amplitude of the test pulse by the maximum current amplitude of the corresponding conditioning pulse. Fractional recovery is plotted as a function of time. D. Use-dependent inactivation. Currents were elicited from a holding potential of −80 mV by 17.5-msec depolarizations to −5 mV at 39 Hz. The protocol was performed for 2.56 sec until the current reached equilibrium. Peak current amplitudes were normalized to the initial peak current amplitude and plotted against pulse number. Solid symbols represent recordings in the absence of STX, and open symbols represent recordings in the presence of 100 nM STX. Values represent means, and error bars indicate SEM. Sample sizes (n) and the parameters of the fits are shown in Table 3.
Once sodium channels become inactivated, they undergo a latency period during which they cannot open until they recover from the inactivated state to the closed state. The isolated R1648H channels in the presence of STX showed slower and more incomplete recovery from inactivation than the control transgenic or endogenous wild-type channels (Figure 4C). The R1648H channels recovered to only approximately 80%, compared with 90% for the wild-type channels (Table 3). We observed no differences in the absence of STX, indicating that the alteration in recovery was too small to distinguish when only 30% of the sodium current was carried by the mutant channels.
Another way to evaluate recovery from inactivation is to examine use-dependent inactivation, defined as a reduction in sodium current amplitudes during a series of depolarizations. If there is insufficient time for complete recovery between successive depolarizations, then the magnitude of the current decreases with each depolarization. Consistent with the slower recovery, the isolated R1648H channels in the presence of STX demonstrated increased use-dependent inactivation at 39 Hz (Figure 4D). Current through the mutant channels decreased to approximately 15% of the initial peak current by the end of the series of depolarizations, whereas current through the control transgenic and wild-type channels decreased to approximately 25% of the initial value. Again, the effect was not apparent when recording from both endogenous and mutant channels in the absence of STX. Finally, the percentage of persistent current was very small (0.7 to 0.8 %) and comparable for wild-type, control transgenic and R1648H mutant channels (Table 4). However, the absolute magnitudes of the persistent current were so small (12 to 39 pA) that it would be difficult to reliably detect any small differences. The properties of the mutant channels in bipolar neurons from P14–P19 mice were similar to those from P5–P8 mice (data not shown). Sodium currents from the interneurons of the control transgenic mice were indistinguishable from wild-type mice in the absence and presence of STX. This result demonstrates that the introduction of the FLAG epitope tag and the E954Q substitution did not alter channel function.
Table 4.
Persistent current
| Cell Type | % Persistent Current | n |
|---|---|---|
| Wild-type bipolar | 0.7 ± 0.1 | 5 |
| E954Q/FLAG bipolar | 0.8 ± 0.2 | 15 |
| R1648H/E954Q/FLAG bipolar | 0.7 ± 0.3 | 5 |
| Wild-type pyramidal | 0.9 ± 0.1 | 7 |
| E954Q/FLAG pyramidal | 0.9 ± 0.2 | 10 |
| R1648H/E954Q/FLAG pyramidal | 0.9 ± 0.1 | 12 |
Measurement of persistent current was performed using prolonged 200-ms depolarizations at −10 mV in the absence of STX. The mean amount of persistent current between 125 ms and 175 ms was determined and normalized to the peak current. Data shown are average normalized persistent current ± SEM. Transgenic neurons expressing wild-type Nav1.1 carrying the E954Q STX-resistant substitution (E954Q) were compared with wild-type neurons in the absence of STX and were used as controls for transgenic R1648H mice also carrying the substitution for STX resistance (R1648H/E954Q).
R1648H sodium channels have a more negative voltage dependence of inactivation in pyramidal neurons
Although only 10% of the sodium current in pyramidal neurons is conducted through the transgenic channels, it is possible that alterations in channel function in these cells might still have physiological effects. We therefore determined the effects of the R1648H mutation on sodium channel properties in pyramidal neurons. As was the case with bipolar neurons, the mutation had no effect on the voltage dependence of activation (Figure 5A and Table 3). There was a slight positive shift in the presence of STX, but this shift was observed for both the R1648H and control transgenic channels, and again probably reflects the difficulty of recording small currents (less than 10% of the total in pyramidal cells). In contrast, the R1648H mutation caused a negative shift in the voltage dependence of inactivation (Figure 5B). The shift was approximately −13 mV when recording from isolated mutant channels in the presence of STX, and it was approximately −7 mV in the absence of STX (Table 3). In contrast to the results in bipolar neurons, there was no significant difference in recovery from inactivation in pyramidal neurons (Figures 5C, Table 3). The mutant channels demonstrated slightly more of a use-dependent decrease in current compared to wild-type channels when recorded in the presence of STX (Figure 5D, Table 3), but this effect was not statistically significant. The percentage of persistent current was very low (0.9%) and comparable for wild-type, control transgenic and R1648H mutant channels (Table 4). Once again the persistent current levels were too small to detect small differences. As was the case with the bipolar neurons, the properties of the mutant channels in P14–P19 mice were similar to those in P5–P8 mice (data not shown).
Figure 5. Properties of the sodium channels in pyramidal neurons.
A. Voltage dependence of activation. B. Voltage dependence of inactivation. C. Recovery from inactivation. D. Use-dependent inactivation. All the protocols are the same as those used for bipolar neurons and are described in the legend to Figure 4. Solid symbols represent recordings in the absence of STX, and open symbols represent recordings in the presence of 100 nM STX. Values represent means, and error bars indicate SEM. Sample sizes (n) and the parameters of the fits are shown in Table 3.
R1648H transgenic channels can partially rescue Scn1a knockout mice
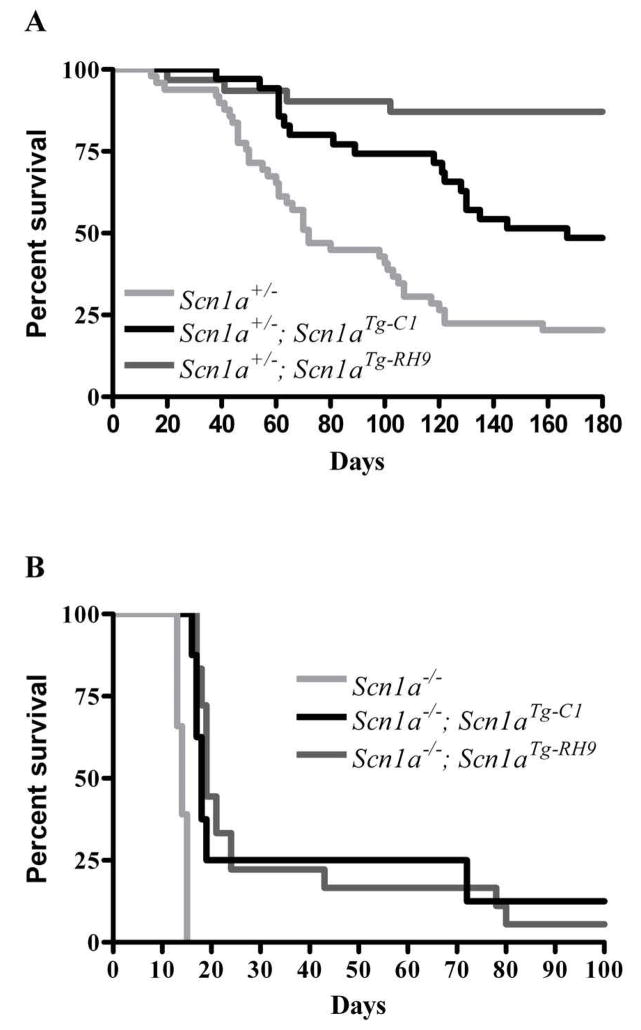
R1648H and other missense SCN1A GEFS+ mutations are associated with milder clinical phenotypes than those that arise from the loss of one SCN1A allele, as occurs in most cases of SMEI. Consistent with this difference, complete loss of Nav1.1 function in an Scn1a knockout mouse model of SMEI causes death by P15 in homozygous mice (Scn1a−/−), and heterozygous Scn1a+/− mice exhibit spontaneous seizures and reduced life spans (Yu et al., 2006). To determine whether the R1648H transgene could at least partially compensate for the loss of one Scn1a allele, we tested the ability of the control (Line 1) and mutant (Line 9) transgenes to rescue the survival of heterozygous (Scn1a+/−) and homozygous (Scn1a−/−) Scn1a knockout mice. We saw a substantial improvement in life span for Scn1a+/− mice that carried either the mutant or control transgenes (Figure 6a). At six months of age, survival rates were 25% for Scn1a+/− heterozygous mice, and 50% and 87% for Scn1a+/−; Scn1aTg-C1 and Scn1a+/−; Scn1aTg-RH9 double heterozygous mutants, respectively. The control and the R1648H transgenes also prolonged the life spans of Scn1a−/− mice, which do not survive beyond P15 (Figure 6b). Of the eight Scn1a−/− mice carrying the control transgene, six died at P16–P19, one died at P78 and one lived to more than six months. Similarly, of the 19 Scn1a−/− mice carrying the R1648H mutant transgene, 12 died at P17–P21, two died at P24, one died at P43, two died at P80 and two lived more than six months. These results demonstrate that both transgenes are functional.
Figure 6. Improved survival of Scn1a+/− and Scn1a−/− mice with the control and mutant transgenes.
A. The survival of Scn1a+/− mice (n= 50) compared with Scn1a+/−; Scn1aTg-C1 (n= 35) and Scn1a+/−; Scn1aTg-RH9 (n= 31) double heterozygous mutants. B. The survival of Scn1a−/− mice (n= 41) compared with Scn1a−/−; Scn1aTg-C1 (n= 8) and Scn1a−/−; Scn1aTg-RH9 (n= 19) mutants.
Discussion
We have created a model for studying the pathophysiology of sodium channel dysfunction in GEFS+ by expressing the Nav1.1 R1648H mutation in transgenic mice. We observed no significant differences in the overall properties of sodium channels in transgenic and wild-type bipolar neurons in the absence of STX, however small differences in sodium channel gating were detected by isolating the mutant transgenic channels in the presence of STX. The larger population of endogenous wild-type Nav1.1, Nav1.2 and Nav1.6 sodium channels probably masked these biophysical defects in the absence of STX.
In the R1648H transgenic mice, slower and incomplete recovery from inactivation and increased use-dependent inactivation would be expected to reduce channel availability in inhibitory neurons, most likely leading to partial loss of function. In addition, the smaller sodium current density in bipolar neurons from R1648H transgenic mice compared to wild-type or E954Q mice might indicate that the mutation specifically decreases sodium currents in those neurons. These results suggest that decreased sodium channel function in inhibitory neurons may lead to hyperexcitability in the CNS. This finding is consistent with observations from Scn1a knockout mice, in which reduced sodium currents in interneurons are observed (Kalume et al., 2007, Yu et al., 2006, Ogiwara et al., 2007). However, we also observed a hyperpolarized shift in the voltage dependence of inactivation in pyramidal neurons in R1648H mice. This would cause the mutant sodium channels to inactivate at more negative potentials, thereby reducing the activity of excitatory neurons, which should decrease seizure susceptibility. Since the expression of the Nav1.1 transgene is higher in bipolar neurons, we speculate that the defect in these neurons is more biologically important than the defect in pyramidal neurons.
It is possible that the effects of the R1648H mutation may be age-specific, so that the electrophysiological differences that we observe at ages P5–P8 and P14–P19 may not directly correlate with the phenotypic effects that we observe in adult mice. For technical reasons, we were unable to obtain electrophysiological data using neurons from adult mice. However, we did not observe any differences in the properties of sodium channels from P14–P19 mice compared to P5–P8 mice, suggesting that sodium channel expression was no longer changing with development by P19. Therefore, we believe that the electrophysiological results from P14–P19 mice indicate the effects of the R1648H mutation in adult mice.
There are a number of possible explanations for the observed differences in channel gating in interneurons versus pyramidal cells. First, each cell type could express a different population of β subunits. Sodium channel α subunits in the CNS are associated with either β1 or β3 and β2 or β4. The specific repertoire of β subunits could alter the effects of the mutation. Second, each cell type may have different splicing or post-translational processing, and sodium channel properties are known to be altered by post-translational processing, such as glycosylation and phosphorylation (Astman et al., 1998, Astman et al., 2006, Chen et al., 2005, Cantrell et al., 1997). Third, there could be differential compensatory upregulation of other sodium channels in each cell type. For example, Nav1.3 was found to be upregulated in the interneurons of Scn1a knockout mice (Yu et al., 2006). Finally, the discrepancy may arise from differences in intracellular localization of the channel in bipolar versus pyramidal neurons. The expression pattern of Nav1.1 is broad, with the channel being present in the soma and possibly other regions of inhibitory neurons (Westenbroek et al., 1989, Yu et al., 2006, Whitaker et al., 2000, Whitaker et al., 2001b, Whitaker et al., 2001a). There is also evidence that Nav1.1 might be expressed in the axon and axon initial segment of interneurons, thus directly regulating action potential generation in those cells (Ogiwara et al., 2007). In pyramidal neurons, Nav1.1 is expressed in the soma and is thought to regulate subthreshold excitability (Whitaker et al., 2001b, Westenbroek et al., 1989, Gong et al., 1999). Careful analysis of the spatial and temporal distribution of Nav1.1, its splice isoforms and its regulation will be necessary to better understand how SCN1A mutations lead to GEFS+.
The effects that we observed for the R1648H mutation in either bipolar or pyramidal neurons were dramatically different from those observed previously with expression in non-neuronal heterologous cells. The R1648H mutation increased the percent of persistent current in tsA201 cells (Lossin et al., 2002, Vanoye et al., 2006) and accelerated recovery from inactivation with a concomitant decrease in use-dependent inactivation in Xenopus oocytes (Spampanato et al., 2001). In contrast, we observed delayed recovery from inactivation with increased use-dependent inactivation in bipolar neurons with no apparent increase in persistent current. These differences could be due to any of the reasons listed above for the differences between pyramidal and bipolar neurons. More importantly, the differences suggest that the characterization of disease-causing mutations using heterologous expression systems may not be physiologically relevant, underscoring the need to determine the effects of the mutations in native neurons.
Interestingly, the control transgenic mice had a less severe response to KA than wild-type mice. Although this difference did not achieve statistical significance, it suggests that overexpression of wild-type Scn1a may be associated with higher seizure thresholds. This may be attributed to greater inhibitory tone due to increased excitability of interneurons overexpressing Scn1a. In contrast, R1648H mutant transgenic mice from Line 9 exhibited the most severe response to KA. It is clear that this was not simply a result of high expression of the mutant transgene, because a second mutant transgenic line with low expression (Line 5) had a comparable phenotype. The severity of the responses to KA for Line 9 and Line 5 were statistically different compared to Line 1, but not to Line 8 or wild-type mice.
In Line 9, the level of Nav1.1 protein was approximately two times higher than in wild-type mice, which means that homozygous Scn1a−/− mice with the mutant transgene from Line 9 would be predicted to have a comparable level of Nav1.1 protein to wild-type mice. However, only 25% (5/19) of homozygous Scn1a−/− mice with this mutant transgene lived more than 25 days. This result demonstrates that the mutant transgene cannot fully recapitulate the normal function of the endogenous Nav1.1 protein and suggests that rescue was the result of increased Nav1.1 current density in Line 9 mice. In contrast, the control transgene from Line 1, with only about 20–30% of endogenous Nav1.1 protein expression, was also able to extend the survival of 25% (2/8) of Scn1a−/− mice beyond 25 days. This is consistent with the expectation that the control transgene can recapitulate more normal Scn1a function.
In summary, we have generated a BAC transgenic mouse model that made it possible to determine the effects of a human GEFS+ mutation in native neurons. These mice revealed that altered interneuron excitability likely contributes to seizure generation. We have tried to reduce the limitations typically associated with transgenic models by using a BAC transgenic approach in which the expression of the transgenic Scn1a gene would be expected to be regulated by endogenous transcription factors acting on the native promoter in the BAC. However, we cannot exclude the possibility that the transgene may still not fully recapitulate the temporal and spatial expression of the endogenous Scn1a gene. Another potential confound in the interpretation of our results was the dissimilarity between the expression levels of the transgenes in control and mutant transgenic lines. We are currently evaluating the phenotype of a knock-in GEFS+ mouse model, in which the R1648H mutation was introduced into the endogenous mouse Scn1a gene by homologous recombination. A knock-in approach will likely yield a more accurate disease model. However, the presence of the E954Q substitution in the transgene makes the BAC transgenic mouse uniquely suited for studying the biophysical effects of SCN1A GEFS+ mutations in native neurons. In addition, the speed and cost-effectiveness of the transgenic approach means we can compare the effects of multiple SCN1A mutations on the biophysical properties of the channel. Such analyses will be critical to establish whether reduced interneuron Nav1.1 sodium channel activity is a consistent effect of all SCN1A GEFS+ mutations.
Acknowledgments
This study was supported by NIH Research Grants NS046484, NS051834 (A.E.) and NS48336 (A.L.G.), and grants from the March of Dimes Birth Defects Foundation (#5-FY02-250) (A.E.) and the McKnight Endowment Fund for Neuroscience (A.L.G.), the European Integrated Project EPICURE (M.M.) and the Italian Telethon grant GGP07277 (M.M.). K.D. was supported by a fellowship from the Epilepsy Foundation. L.P. was supported fellowships from AFIP and FAPESP (07-50534-5). ST was supported by FAPESP (CEPID#98/14303-3). We are grateful to Cheryl Strauss for critically reading this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alekov AK, Rahman MM, Mitrovic N, Lehmann-Horn F, Lerche H. A sodium channel mutation causing epilepsy in man exhibits defects in fast inactivation and activation in vitro. J Physiol (Lond) 2000;529:533–539. doi: 10.1111/j.1469-7793.2000.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annesi G, Gambardella A, Carrideo S, Incorpora G, Labate A, Pasqua AA, Civitelli D, Polizzi A, Annesi F, Spadafora P, Tarantino P, Cirò Candiano IC, Romeo N, De Marco EV, Ventura P, LePiane E, Zappia M, Aguglia U, Pavone L, Quattrone A. Two novel SCN1A missense mutations in generalized epilepsy with febrile seizures plus. Epilepsia. 2003;44:1257–1258. doi: 10.1046/j.1528-1157.2003.22503.x. [DOI] [PubMed] [Google Scholar]
- Astman N, Gutnick MJ, Fleidervish IA. Activation of protein kinase C increases neuronal excitability by regulating persistent Na+ current in mouse neocortical slices. J Neurophysiol. 1998;80:1547–1551. doi: 10.1152/jn.1998.80.3.1547. [DOI] [PubMed] [Google Scholar]
- Astman N, Gutnick MJ, Fleidervish IA. Persistent sodium current in layer 5 neocortical neurons is primarily generated in the proximal axon. J Neurosci. 2006;26:3465–3473. doi: 10.1523/JNEUROSCI.4907-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barela AJ, Waddy SP, Lickfett JG, Hunter J, Anido A, Helmers SL, Goldin AL, Escayg A. An epilepsy mutation in the sodium channel SCN1A that decreases channel excitability. J Neurosci. 2006;26:2714–2723. doi: 10.1523/JNEUROSCI.2977-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckh S, Noda M, Lübbert H, Numa S. Differential regulation of three sodium channel messenger RNAs in the rat central nervous system during development. EMBO J. 1989;8:3611–3636. doi: 10.1002/j.1460-2075.1989.tb08534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell AR, Smith RD, Goldin AL, Scheuer T, Catterall WA. Dopaminergic modulation of sodium current in hippocampal neurons via cAMP-dependent phosphorylation of specific site(s) in the sodium channel α subunit. J Neurosci. 1997;17:7330–7338. doi: 10.1523/JNEUROSCI.17-19-07330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Cantrell AR, Messing RO, Scheuer T, Catterall WA. Specific modulation of Na+ channels in hippocampal neurons by protein kinase Cε. J Neurosci. 2005;25:507–513. doi: 10.1523/JNEUROSCI.4089-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claes L, Del-Favero J, Cuelemans B, Lagae L, Van Broeckhoven C, De Jonghe P. De novo mutations in the sodium-channel gene SCN1A cause Severe Myoclonic Epilepsy of Infancy. Am J Hum Genet. 2001;68:1327–1332. doi: 10.1086/320609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escayg A, Heils A, MacDonald BT, Haug K, Sander T, Meisler MH. A novel SCN1A mutation associated with generalized epilepsy with febrile seizures plus and prevalence of variants in patients with epilepsy. Am J Hum Genet. 2001;68:866–873. doi: 10.1086/319524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escayg A, MacDonald BT, Meisler MH, Baulac S, Huberfeld G, An-Gourfinkel I, Brice A, LeGuern E, Moulard B, Chaigne D, Buresi C, Malafosse A. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat Genet. 2000;24:343–345. doi: 10.1038/74159. [DOI] [PubMed] [Google Scholar]
- Fong AY, Stornetta RL, Foley CM, Potts JT. Immunohistochemical localization of GAD67-expressing neurons and processes in the rat brainstem: subregional distributino in the nucleus tractus solitarius. J Comp Neurol. 2005;493:274–290. doi: 10.1002/cne.20758. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Sugawara T, Mazaki-Miyazaki E, Takahashi Y, Fukushima K, Watanabe M, Hara K, Morikawa T, Yagi K, Yamakawa K, Inoue Y. Mutations of sodium channel α subunit type 1 (SCN1A) in intractable childhood epilepsies with frequent generalized tonic-clonic seizures. Brain. 2003;126:531–546. doi: 10.1093/brain/awg053. [DOI] [PubMed] [Google Scholar]
- Gong B, Rhodes KJ, Bekele-Arcuri Z, Trimmer JS. Type I and type II Na+ channel α-subunit polypeptides exhibit distinct spatial and temporal patterning, and association with auxiliary subunits in rat brain. J Comp Neurol. 1999;412:342–352. [PubMed] [Google Scholar]
- Graf ER, Zhang XZ, Jin SX, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenberg LGW, Hoover CL, Smith MA. Evidence of an agrin receptor in cortical neurons. J Neurosci. 1999;19:7384–7393. doi: 10.1523/JNEUROSCI.19-17-07384.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopp TP, Prickett KS, Price VL, Libby RT, March CJ, Cerretti DP, Urdal DL, Conlon PJ. A short polypeptide marker sequence useful for recombinant protein identification and purification. Bio/Technology. 1988;6:1204–1210. [Google Scholar]
- Isom LL, Scheuer T, Brownstein AB, Ragsdale DS, Murphy BJ, Catterall WA. Functional co-expression of the β1 and type IIA α subunits of sodium channels in a mammalian cell line. J Biol Chem. 1995;270:3306–3312. doi: 10.1074/jbc.270.7.3306. [DOI] [PubMed] [Google Scholar]
- Kalume F, Yu FH, Westenbroek RE, Scheuer T, Catterall WA. Reduced sodium current in Purkinje neurons from Nav1,1 mutant mice: implications for ataxia in severe myoclonic epilepsy in infancy. J Neurosci. 2007;27:11065–11074. doi: 10.1523/JNEUROSCI.2162-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontis KJ, Goldin AL. Site-directed mutagenesis of the putative pore region of the rat IIA sodium channel. Mol Pharmacol. 1993;43:635–644. [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Li Z, Hilgenberg LGW, O’Dowd DK, Smith MA. Formation of functional synaptic connections between cultured cortical neurons from agrin-deficient mice. J Neurobiol. 1999;39:547–557. doi: 10.1002/(sici)1097-4695(19990615)39:4<547::aid-neu8>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Lossin C, Wang DW, Rhodes TH, Vanoye CG, George AL., Jr Molecular basis of an inherited epilepsy. Neuron. 2002;34:877–884. doi: 10.1016/s0896-6273(02)00714-6. [DOI] [PubMed] [Google Scholar]
- Mantegazza M, Gambardella A, Rusconi R, Schiavon E, Annesi F, Cassulini RR, Labate A, Carrideo S, Chifari R, Canevini MP, Canger R, Franceschetti S, Annesi G, Wanke E, Quattrone A. Identification of an Nav1.1 sodium channel (SCN1A) loss-of-function mutations associated with familial simple febrile seizures. Proc Natl Acad Sci USA. 2005;102:18177–18182. doi: 10.1073/pnas.0506818102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MS, Tang B, Ta N, Escayg A. Characterization of 5′ untranslated regions of the voltage-gated sodium channels SCN1A, SCN2A, and SCN3A and identificatinon of cis-conserved noncoding sequences. Genomics. 2007;90:225–235. doi: 10.1016/j.ygeno.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao Y, Mazaki-Miyazaki E, Okamura N, Takagi M, Igarashi T, Yamakawa K. A family of generalized epilepsy with febrile seizures plus type 2 - a new missense mutation of SCN1A found in the pedigree of several patients with complex febrile seizures. Epilepsy Res. 2005;63:151–156. doi: 10.1016/j.eplepsyres.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Ogiwara I, Miyamoto H, Morita N, Atapour N, Mazaki E, Inoue I, Takeuchi T, Itohara S, Yanagawa Y, Obata K, Furuichi T, Hensch TK, Yamakawa K. Nav1.1 localizes to axons of parvalbumin-positive inhibitory interneurons: a circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. J Neurosci. 2007;27:5903–5914. doi: 10.1523/JNEUROSCI.5270-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori I, Ohtsuka Y, Ouchida M, Ogino T, Maniwa S, Shimizu K, Oka E. Is phenotype difference in severe myoclonic epilepsy in infancy related to SCN1A mutations? Brain Dev. 2003;27:488–493. doi: 10.1016/s0387-7604(03)00038-x. [DOI] [PubMed] [Google Scholar]
- Rhodes TH, Lossin C, Vanoye CG, Wang DW, George AL., Jr Noninactivating voltage-gated sodium channels in severe myoclonic epilepsy of infancy. Proc Natl Acad Sci USA. 2004;101:11147–11152. doi: 10.1073/pnas.0402482101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spampanato J, Escayg A, Meisler MH, Goldin AL. Functional effects of two voltage-gated sodium channel mutations that cause generalized epilepsy with febrile seizures plus type 2. J Neurosci. 2001;21:7481–7490. doi: 10.1523/JNEUROSCI.21-19-07481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spampanato J, Escayg A, Meisler MH, Goldin AL. The generalized epilepsy with febrile seizures plus type 2 mutation W1204R alters voltage-dependent gating of Nav1.1 sodium channels. Neuroscience. 2003;116:37–48. doi: 10.1016/s0306-4522(02)00698-x. [DOI] [PubMed] [Google Scholar]
- Sugawara T, Tsurubuchi Y, Agarwala KL, Ito M, Fukuma G, Mazaki-Miyazaki E, Nagafuji H, Noda M, Imoto K, Wada K, Mitsudome A, Kaneko S, Montal M, Nagata K, Hirose S, Yamakawa K. A missense mutation of the Na+ channel αII subunit gene Nav1.2 in a patient with febrile and afebrile seizures causes channel dysfunction. Proc Natl Acad Sci USA. 2001;98:6384–6389. doi: 10.1073/pnas.111065098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoye CG, Lossin C, Rhodes TH, George AL., Jr Single-channel properties of human Nav1.1 and mechanism of channel dysfunction in SCN1A-associated epilepsy. J Gen Physiol. 2006;127:1–14. doi: 10.1085/jgp.200509373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RH, Hodgson BL, Grinton BE, Gardiner RM, Robinson R, Rodriguez-Casero V, Sadleir L, Morgan J, Harkin LA, Dibbens LM, Yamamoto T, Andermann E, Mulley JC, Berkovic SF, Scheffer IE. Sodium channel α1-subunit mutations in severe myoclonic epilepsy of infancy and infantile spasms. Neurology. 2003;61:765–769. doi: 10.1212/01.wnl.0000086379.71183.78. [DOI] [PubMed] [Google Scholar]
- Wallace RH, Scheffer IE, Barnett S, Richards M, Dibbens L, Desai RR, Lerman-Sadie T, Lev D, Mazarib A, Brand N, Ben-Zeev B, Goikhman I, Singh R, Kremmidiotis G, Gardner A, Sutherland GR, George AL, Jr, Mulley JC, Berkovic SF. Neuronal sodium-channel α1-subunit mutations in generalized epilepsy with febrile seizures plus. Am J Hum Genet. 2001;68:859–865. doi: 10.1086/319516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westenbroek RE, Merrick DK, Catterall WA. Differential subcellular localization of the RI and RII Na+ channel subtypes in central neurons. Neuron. 1989;3:695–704. doi: 10.1016/0896-6273(89)90238-9. [DOI] [PubMed] [Google Scholar]
- Whitaker WRJ, Clare JJ, Powell AJ, Chen YH, Faull RLM, Emson PC. Distribution of voltage-gated sodium channel α-subunit and β-subunit mRNAs in human hippocampal formation, cortex, and cerebellum. J Comp Neurol. 2000;422:123–139. doi: 10.1002/(sici)1096-9861(20000619)422:1<123::aid-cne8>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Whitaker WRJ, Faull RLM, Dragunow M, Mee EW, Emson PC, Clare JJ. Changes in the mRNAs encoding voltage-gated sodium channel types II and III in human epileptic hippocampus. Neuroscience. 2001a;106:275–285. doi: 10.1016/s0306-4522(01)00212-3. [DOI] [PubMed] [Google Scholar]
- Whitaker WRJ, Faull RLM, Waldvogel HJ, Plumpton CJ, Emson PC, Clare JJ. Comparative distribution of voltage-gated sodium channel proteins in human brain. Mol Brain Res. 2001b;88:37–53. doi: 10.1016/s0169-328x(00)00289-8. [DOI] [PubMed] [Google Scholar]
- Yu FH, Mantegazza M, Westenbroek RE, Robbins CA, Kalume F, Burton KA, Spain WJ, McKnight GS, Scheuer T, Catterall WA. Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat Neurosci. 2006;9:1142–1149. doi: 10.1038/nn1754. [DOI] [PubMed] [Google Scholar]