Abstract
Sensory organs are composed of neurons, which convert environmental stimuli to electrical signals, and glia-like cells, whose functions are not well-understood. To decipher glial roles in sensory organs, we ablated the sheath glial cell of the major sensory organ of Caenorhabditis elegans. We found that glia-ablated animals exhibit profound sensory deficits and that glia provide activities that affect neuronal morphology, behavior generation, and neuronal uptake of lipophilic dyes. To understand the molecular bases of these activities, we identified 298 genes whose mRNAs are glia-enriched. One gene, fig-1, encodes a labile protein with conserved thrombospondin TSP1 domains. FIG-1 protein functions extracellularly, is essential for neuronal dye uptake, and also affects behavior. Our results suggest that glia are required for multiple aspects of sensory organ function.
Glia, the largest cell population in vertebrate nervous systems, are implicated in processes governing nervous system development and function (1). However, the functions of few glial proteins are characterized. Astrocytic glia are often positioned near synapses, and can respond to and participate in synaptic activity (2, 3), influencing the response of postsynaptic cells to presynaptic stimulation (4).
Sensory neurons convert environmental stimuli into neuronal activity, and their receptive endings are often associated with glia, such as retinal pigmented epithelial cells and Müller glia or olfactory ensheathing cells. Because sensory neurons are postsynaptic to the environment, their associated glia may impact sensory activity in ways analogous to synaptic astrocytes.
Sensory organs are conserved structures, exhibiting morphological, functional, and molecular similarities among diverged species (5). To understand glial contributions to sensory neuron functions, we studied the largest sensory organ of the nematode C. elegans, the amphid. This organ mediates responses to chemical, thermal, and tactile stimuli, promoting attractive and repulsive behaviors that are easily assayed. Each of the bilateral amphids comprises twelve neurons extending ciliated dendrites to the anterior tip (5). These neurons can be grouped based on association with the single amphid sheath glial cell: the dendritic receptive endings of four neurons are entirely surrounded by this glial cell in a hand-in-glove configuration, while remaining cilia are encased in a channel formed by the same glial cell, and are exposed, through a pore, to the outside environment (5, 6) (fig. S1).
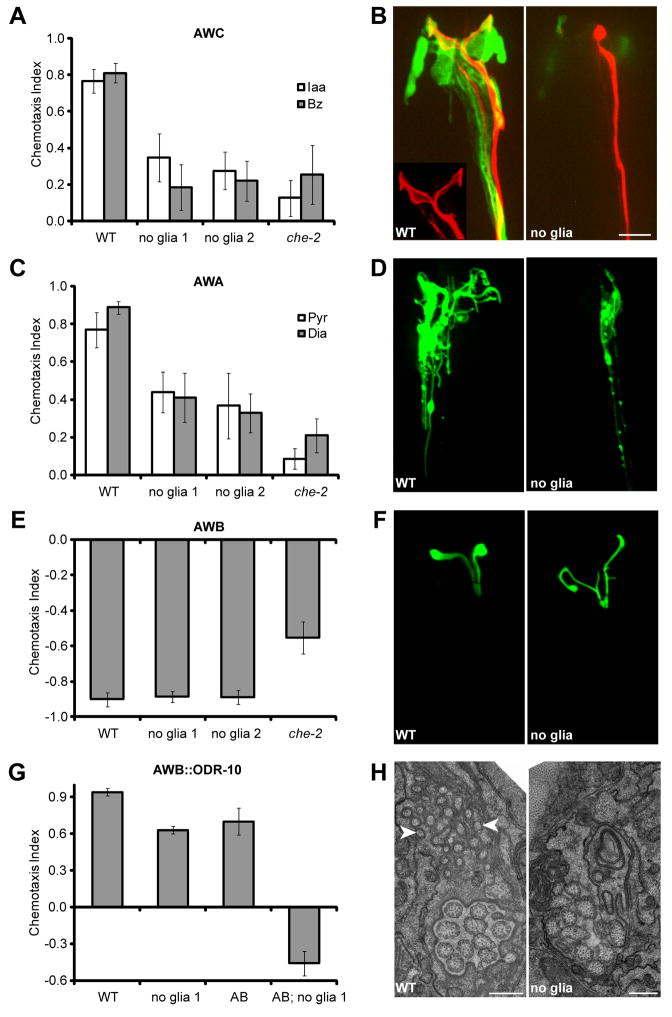
We ablated sheath glia in first-stage larvae, after the amphid had formed, by either using a laser microbeam (7), or expressing the diphtheria toxin A gene from a sheath-glia-specific promoter (8). Ablation success was monitored by disappearance of a glia-specific GFP reporter, and by electron microscopy (EM) reconstruction of amphid sensory endings. We first examined the glia-embedded sensory neurons AWC, AWA, and AWB. Animals with bilateral sheath-glia ablation display severe defects in AWC-mediated chemotaxis towards benzaldehyde or isoamyl alcohol (9) (Fig. 1A), behaving comparably to che-2(e1033) mutants, which lack functional sensory cilia (10). Similarly, ablation also reduced chemotaxis towards AWA-sensed odorants (Fig. 1C). By contrast, AWB function was not affected by sheath glia ablation (Fig. 1E).
Fig. 1.
Glia are required for behavior and cilium structure. (A) Glia-ablated animals have defective AWC-mediated odortaxis towards 1% isoamyl alcohol (Iaa) and 0.5% benzahldehyde (Bz), p < 0.001 (Student’s t-test). (B) A wild-type AWC cilium (red, odr-1::RFP) ensheathed by an amphid sheath glia (green, vap-1::GFP). Glia ablation in the ontralateral amphid results in an amorphous cilium. Anterior, up. Scale bar, 5 μm. (C) Glia-ablated animals have defective AWA-mediated odortaxis towards 1% methyl pyrazine (Pyr) and 0.1% diacetyl (Dia), p < 0.001. (D) Glia removal decreases AWA cilium branching (odr-3::ODR-3p::GFP). (E) Glia are not required for AWB function, 10% 2-nonanone avoidance. (F) AWB cilium morphology appears grossly normal, although additional branching and failure of the two cilia to spread is often observed (str-1::ODR-10::GFP). (G) Animals expressing ODR-10 in AWA and AWB (AB) are attracted to diacetyl. However, dual-sensing animals lacking glia are repelled. (H) EM showing absence of AFD microvilli-like projections (arrowheads) in glia-ablated animals. Scale bar, 0.5 μm. WT, wild type; no glia, diphtheria-toxin-ablated glia; che-2, che-2(e1033) mutants; error bars, SD of 12 or more assays.
To confirm these neuron-selective effects of glia on odortaxis, we expressed the ODR-10 diacetyl receptor, normally found only in AWA (11), also in AWB neurons. As previously described (12), animals expressing ODR-10 in both neurons are less attracted to diacetyl than wild-type animals, reflecting the opposing behavioral outputs of these neurons (Fig. 1G). However, consistent with a defect in AWA, sheath-glia ablated animals expressing ODR-10 in both neurons are repelled by diacetyl (Fig. 1G). In glia-ablated animals, the extracellular environment of AWA and AWB is identical. The normal AWB response, therefore, suggest that odorant molecules can access and interact with odorant receptors in the absence of glia, and that the presence of glia is required for integrating opposing environmental stimuli.
We also examined thermotaxis, a behavior mediated by the AFD sheath-glia embedded neuron. While wild-type animals seek their cultivation temperature on a thermal gradient (13, 14), inactivation of AFD by cell ablation or by the ttx-1(p767) mutation results in cryophilic/athermotactic behavior (14). Sheath-glia ablation does not seem to eliminate AFD function, but results in thermophilic behavior (fig. S2), suppressible by ttx-1(p767) (fig. S2D).
The ciliated sensory receptive endings of AWC, AWA, AFD, and to a lesser extent AWB, were defective in sheath-glia ablated animals (Fig. 1). Sheath-glia ablation resulted in complete loss of the AWC wing-like cilium structure (Fig. 1B; n >100), and expansion of this structure in dauer animals was also blocked (fig. S3; n = 2). Similarly, the highly branched processes of the AWA cilium were largely eliminated in sheath-glia ablated animals (Fig. 1D; n = 10), as were the microvilli-like extensions of the AFD sensory ending (Fig. 1H; n = 15). Ciliary localization of olfactory signaling proteins, including ODR-10 (AWB, Fig. 1F; AWA, AWC, fig. S4), the G-alpha protein ODR-3 (AWA, Fig. 1D), and the cyclic-nucleotide-gated channel subunit TAX-4 (AWC, fig. S4), was unaltered.
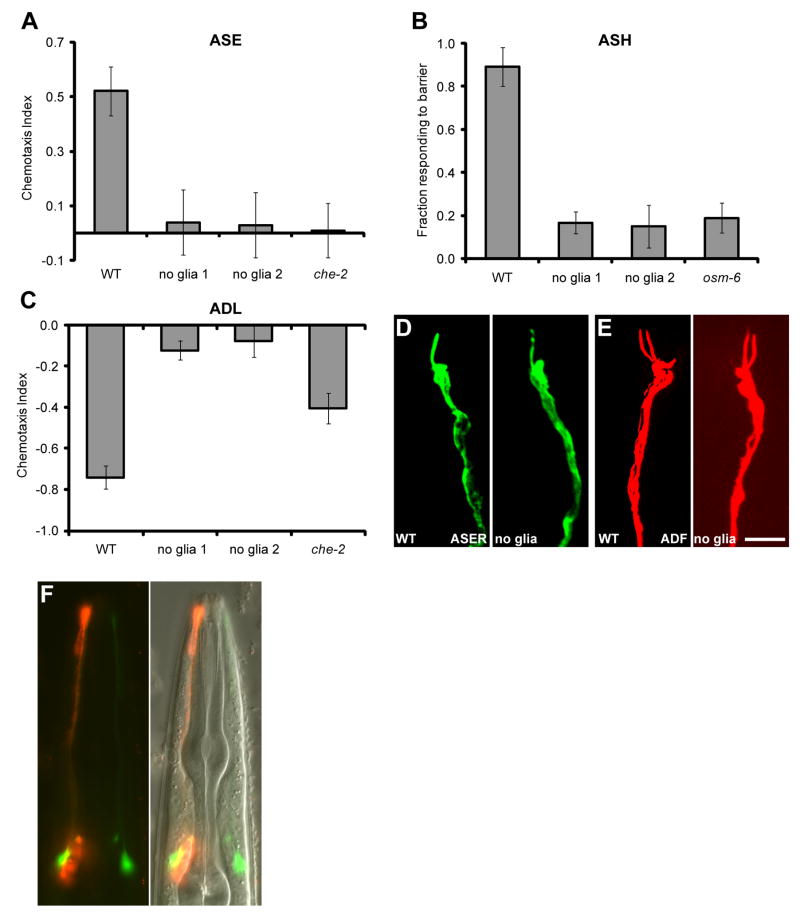
We next examined behaviors mediated by amphid channel neurons. Sheath-glia ablation completely blocked chemotaxis towards NaCl (Fig. 2A), a behavior mediated by the ASE neurons (15). Avoidance of a high osmolarity barrier, mediated by ASH (16), was also entirely abrogated (Fig. 2B), as was long-range avoidance of 1-octanol (Fig. 2C), a behavior mediated by ADL (12). Surprisingly, sensory ending morphology, length, and microtubule organization of all channel neurons appeared normal in ablated animals (Fig. 2D and E; fig. S1). Furthermore, ciliary localization of intraflagellar transport components (CHE-11, DYF-11), or of ODR-10, expressed in ASH, was not disrupted by sheath-glia ablation (fig. S4).
Fig. 2.
Glia affect channel neuron function but not morphology. (A) Glia-ablated animals fail to detect 0.2 M NaCl (p < 0.001), an ASE-mediated behavior. (B) Glia-ablated animals fail to avoid a 4M fructose ring (p < 0.001), an ASH-mediated behavior. osm-6, osm-6(p811) mutants. (C) Glia-ablated animals fail to avoid 1-octanol in a long-range assay (p < 0.001), an ADL-mediated behavior. (D and E) The morphology of amphid channel neurons is not affected by glia removal. ASER, gcy-5::GFP; ADF, T08G3.3::RFP). Scale bar, 5 μm. (F) Glia are required for neuronal uptake of DiI (red). Only the right amphid sheath glia is ablated. AWC (green, odr-1::YFP) indicates the location of the dendrite bundles. Error bars, SD of 12 or more assays.
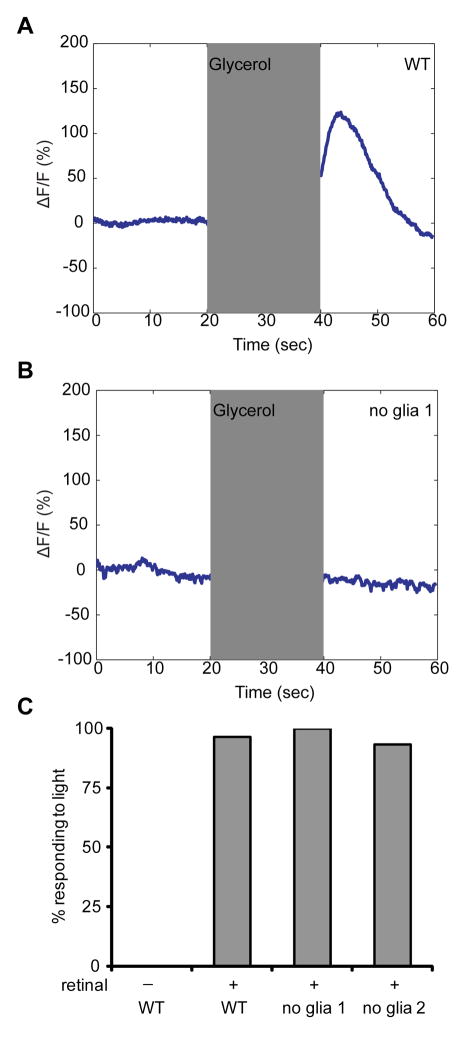
We used G-CaMP to examine Ca2+ level changes in ASH in response to high osmolarity. Whereas wild-type animals increase intracellular Ca2+ following exposure to and removal of an osmotic stimulus (Fig. 3A and fig. S5) (17), sheath-glia ablated animals lacked these responses (Fig. 3B and fig. S5). To determine whether signaling downstream of Ca2+ elevation was disrupted, we expressed the light-activated channel channelrhodopsin-2 (ChR2) (18) within ASH. In the presence of retinal, a ChR2 cofactor, glia-ablated (and wild-type) animals initiate backward locomotion (Fig. 3C), demonstrating that downstream signaling is intact and that glia are not required for ASH health/viability.
Fig. 3.
Glia are required for Ca2+ responses in ASH. (A) As determined by G-CaMP fluorescence, ASH responds to application and removal of 1M glycerol (17). Shaded region, stimulus duration. (B) ASH fails to respond to glycerol in glia-ablated animals. (C) Glia are not required for neuronal function downstream of Ca2+ entry. Activation of ASH-expressed ChR2 by light causes animals to move backwards. n = 30 for each.
When C. elegans are soaked in lipophilic dyes (e.g. DiI), some channel neurons, and AWB, take up and concentrate the dye. DiI uptake was eliminated in all amphid neurons in glia-ablated animals (Fig. 2F). Thus, dye filling (defective in channel neurons and AWB), ciliary morphology (defective mainly in AWA, AWC, and AFD), and behavior generation (not defective in AWB) are independent properties of amphid sensory neurons, each requiring the presence of sheath glia.
To uncover glial factors controlling these neuronal properties, we compiled a list of amphid sheath-glia-enriched transcripts. mRNA from cultured GFP-expressing amphid-sheath glia was compared to mRNA from other cultured embryonic cells by hybridizing each population to an oligonucleotide gene array. We identified 298 unique transcripts with greater than four-fold enrichment (table S1), including the known glial genes daf-6 and vap-1 (19). 159/298 transcripts are predicted to encode transmembrane or secreted proteins that could potentially interact with amphid sensory neurons. These secreted proteins include Ca2+ binding proteins and a KCl cotransporter, which may explain glial contributions to Ca2+ elevation in ASH.
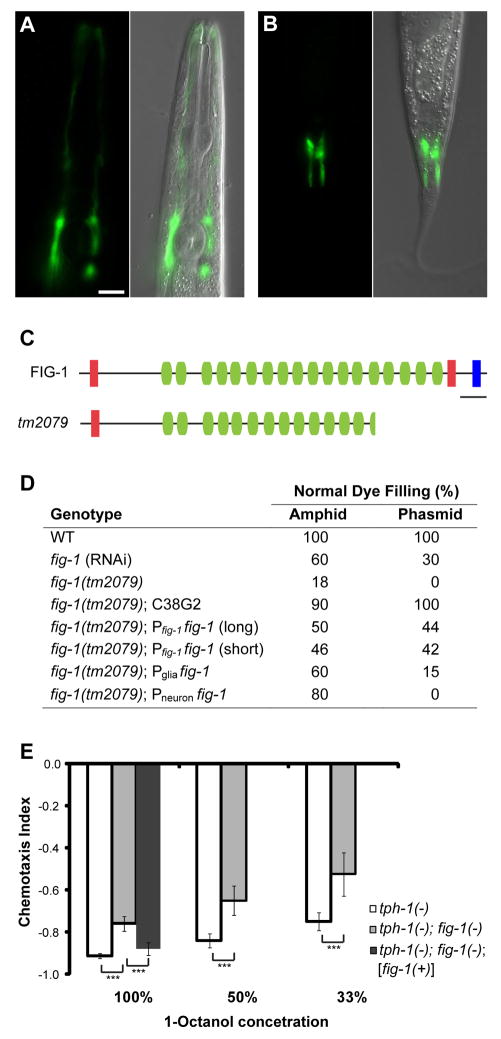
To validate our results, we generated GFP reporter fusion constructs to promoters of seven genes. Five were expressed exclusively in amphid sheath glia and phasmid sheath glia (an amphid-like tail sensory organ) (Fig. 4A and B; fig. S6).
Fig. 4.
Glial fig-1 is required for neuronal dye filling and function. (A and B) fig-1 is expressed in amphid (A) and phasmid (B) sheath glia. Anterior, up. Scale bar, 5 μm. (C) FIG-1 domain structure. Red, thrombospondin type 1 domain; green, C6 repeats; blue, EGF-like type II domain; bar, 200 amino acids. The predicted protein in the fig-1(tm2079) deletion is shown. (D) fig-1 is required for DiI accumulation. One representative line shown for each condition: C38G2, cosmid containing fig-1; glial promoter, T02B11.3; neuronal promoter, sra-6; n > 40 for each. (E) fig-1 is required for 1-octanol avoidance. Assays were performed in the tph-1(mg280) background, which suppresses movement defects of fig-1(tm2079) animals. fig-1(tm2079) mutants perform worse at all three concentrations, and these defects can be rescued by fig-1(+). Asterisks, p < 0.001. Error bars, 95% confidence intervals. At least 24 assays for each condition.
We screened enriched genes by RNA interference (RNAi) for defects in amphid neuron dye filling (Dyf phenotype), and identified the gene F53B7.5, which we renamed fig-1 (Dyf, expressed in glia). RNAi against fig-1 resulted in dye filling defects in amphids and phasmids (Fig. 4D). An 1117 bp deletion in fig-1, tm2079, also perturbed dye filling (Fig. 4C), and this defect was rescued by a cosmid spanning the fig-1 locus. Interestingly, fig-1(tm2079) mutants exhibited normal neuronal and amphid sheath glia structure (fig. S1I), demonstrating that access to dye is not sufficient for dye filling, and that glia-dependent neuronal properties are required for dye filling.
fig-1(RNAi) defects could be induced at all developmental stages and were observed within 24 hours of dsRNA exposure (table S2), suggesting that while fig-1 mRNA is highly expressed (table S1), FIG-1 protein must be labile, consistent with a non-structural role.
Expression of a fig-1 promoter::GFP reporter was detected exclusively within amphid and phasmid sheath glia (Fig. 4A and B), and was first evident in late embryos, continuing throughout adulthood. Thus, FIG-1 expression may be required continuously for neuronal dye filling.
fig-1 is predicted to generate two alternatively-spliced mRNAs encoding proteins of 3095 (long) and 2892 (short) amino acids, the short isoform being sufficient for rescue (Fig. 4D). Both proteins contain an N-terminal signal sequence, a TSP1 thrombospondin domain, 18 C6 domains, and a second TSP1 domain (Fig. 4C). The larger protein is also predicted to contain an EGF-like type II motif at its C-terminus (see Supporting Online Material). TSP1 and EGF-like motifs are characteristic domains found in astrocyte-secreted thrombospondin proteins implicated in synapse development (20).
To determine whether FIG-1 protein can act cell non-autonomously, we expressed a fig-1(short) cDNA transgene under either sheath glia (T02B11.3) or sensory neuron (sra-6; ASH, and weakly in ASI, PHA, and PHB) promoters. Both transgenes rescued fig-1(tm2079) mutants (Fig. 4D), as expected if FIG-1 acted extracellularly.
Finally, although fig-1(tm2079) mutants exhibited normal behavior towards most stimuli tested (fig. S7), we identified a modest but significant defect in ADL-dependent 1-octanol avoidance (Fig. 4E), suggesting that fig-1 also contributes to behavior generation.
We have demonstrated that C. elegans amphid sheath glia provide associated neurons with at least three separate activities, and have identified a molecular mediator contributing to two of these functions. Recent studies suggest that C. elegans glia share developmental similarities with vertebrate glia (21). At least some of the glial functions we describe might, therefore, be conserved in other sensory systems.
Astrocyte-secreted thrombospondins play important postsynaptic roles in synapse assembly and function (20). Our studies of FIG-1, which contains domains also present in thrombospondins, demonstrate that this glial factor plays a key role in modulating sensory neuron properties. The rapid turnover of FIG-1 protein is intriguing, suggesting possible dynamic roles. Could FIG-1 and thrombospondins have related molecular functions? Sensory receptive endings share some similarities with postsynaptic neuronal endings. Both respond to diffusible cues by activating G protein-coupled receptors (11) or ligand-gated ion channels (22); postsynaptic dendritic spines are highly malleable in shape and size (23, 24), as are sensory neuron receptive endings (25); and many vertebrate excitatory synapses are ensheathed by glia, as are sensory neuron receptive endings. These observations, together with the domain structure of FIG-1, suggest the highly speculative notion that analogies between the ‘sensory synapse’ and true synapses might, in part, reflect molecular homologies. Our results provide strong evidence for essential glial contributions to sensory organ function.
Supplementary Material
References and Notes
- 1.Haydon PG. Nat Rev Neurosci. 2001;2:185. doi: 10.1038/35058528. [DOI] [PubMed] [Google Scholar]
- 2.Perea G, Araque A. Science. 2007;317:1083. doi: 10.1126/science.1144640. [DOI] [PubMed] [Google Scholar]
- 3.Perea G, Araque A. J Neurosci. 2005;25:2192. doi: 10.1523/JNEUROSCI.3965-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robitaille R. Neuron. 1998;21:847. doi: 10.1016/s0896-6273(00)80600-5. [DOI] [PubMed] [Google Scholar]
- 5.Ward S, Thomson N, White JG, Brenner S. J Comp Neurol. 1975;160:313. doi: 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- 6.Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Dev Biol. 1986;117:456. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- 7.Bargmann CI, Avery L. Methods Cell Biol. 1995;48:225. doi: 10.1016/s0091-679x(08)61390-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Materials and methods are available as supporting material on Science Online.
- 9.Bargmann CI, Hartwieg E, Horvitz HR. Cell. 1993;74:515. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- 10.Fujiwara M, Ishihara T, Katsura I. Development. 1999;126:4839. doi: 10.1242/dev.126.21.4839. [DOI] [PubMed] [Google Scholar]
- 11.Sengupta P, Chou JH, Bargmann CI. Cell. 1996;84:899. doi: 10.1016/s0092-8674(00)81068-5. [DOI] [PubMed] [Google Scholar]
- 12.Troemel ER, Kimmel BE, Bargmann CI. Cell. 1997;91:161. doi: 10.1016/s0092-8674(00)80399-2. [DOI] [PubMed] [Google Scholar]
- 13.Hedgecock EM, Russell RL. Proc Natl Acad Sci USA. 1975;72:4061. doi: 10.1073/pnas.72.10.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori I, Ohshima Y. Nature. 1995;376:344. doi: 10.1038/376344a0. [DOI] [PubMed] [Google Scholar]
- 15.Bargmann CI, Horvitz HR. Neuron. 1991;7:729. doi: 10.1016/0896-6273(91)90276-6. [DOI] [PubMed] [Google Scholar]
- 16.Bargmann CI, Thomas JH, Horvitz HR. Cold Spring Harb Symp Quant Biol. 1990;55:529. doi: 10.1101/sqb.1990.055.01.051. [DOI] [PubMed] [Google Scholar]
- 17.Hilliard MA, et al. EMBO J. 2005;24:63. doi: 10.1038/sj.emboj.7600493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagel G, et al. Proc Natl Acad Sci USA. 2003;100:13940. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perens EA, Shaham S. Dev Cell. 2005;8:893. doi: 10.1016/j.devcel.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 20.Christopherson KS, et al. Cell. 2005;120:421. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 21.Yoshimura S, Murray JI, Lu Y, Waterston RH, Shaham S. Development. 2008;135:2263. doi: 10.1242/dev.019547. [DOI] [PubMed] [Google Scholar]
- 22.Sato K, Pellegrino M, Nakagawa T, Vosshall LB, Touhara K. Nature. 2008;452:1002. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- 23.Trachtenberg JT, et al. Nature. 2002;420:788. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- 24.Murai KK, Nguyen LN, Irie F, Yamaguchi Y, Pasquale EB. Nat Neurosci. 2003;6:153. doi: 10.1038/nn994. [DOI] [PubMed] [Google Scholar]
- 25.Mukhopadhyay S, Lu Y, Shaham S, Sengupta P. Dev Cell. 2008;14:762. doi: 10.1016/j.devcel.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.We thank C. Bargmann and S. Chalasani for their generous help with G-CAMP imaging, N. Pokala for the ASH-ChR2 strain and advice on assays, H. Fares for a DT-A plasmid, S. Mazel for help with FACS, S. Mitani for the tm2079 allele, A. North for help with microscopy, E. Nudleman for the thermotaxis apparatus, Shaham lab members for comments on the project and manuscript, and C. Bargmann, P. Sengupta, O. Hobert, and E. Jorgensen for strains. S.S. is a Klingenstein Fellow in the Neurosciences.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.