Abstract
Toll-like receptors (TLRs) activate the innate immune system in response to pathogens. Here we showed that TLR9 proteolytic cleavage is a prerequisite for TLR9 signaling. Inhibition of lysosomal proteolysis rendered TLR9 inactive. The C-terminal fragment of TLR9 thus generated included a portion of the TLR9 ectodomain, as well as the transmembrane and cytoplasmic domains. This cleavage fragment bound to the TLR9 ligand CpG, and, when expressed in Tlr9−/− dendritic cells, restored CpG-induced cytokine production. Although cathepsin L generated the requisite TLR9 cleavage products in a cell-free in vitro system, several proteases influenced TLR9 cleavage in intact cells. Lysosomal proteolysis thus contributes to innate immunity by facilitating specific cleavage of TLR9.
INTRODUCTION
Toll-like receptors (TLRs) are essential sensors of microbial infection in multicellular hosts, and TLR engagement promotes initiation of adaptive immune responses1,2. The TLR family consists of 10 members in humans and 13 in mice3,4. TLR1, TLR2, TLR4, TLR5 and TLR6 are localized to the plasma membrane and are involved in recognition of microbial products or lipid components5–9. TLR3, TLR7 (http://www.signaling-gateway.org/molecule/query?afcsid=A002299) and TLR9 (http://www.signaling-gateway.org/molecule/query?afcsid=A002301) are localized in intracellular compartments and recognize pathogen-derived double-stranded RNA, single-stranded RNA and unmethylated CpG DNA, respectively10–15. The intracellular localization of TLR9 is critical for the discrimination of self and non-self nucleic acids16. Upon stimulation of cells with CpG DNA, TLR9 redistributes from the endoplasmic reticulum (ER) to lysosomes, where a signaling cascade is triggered by recruitment of the MyD88 adaptor molecule14,17.
The papain-like lysosomal cysteine protease family, which includes the major thiol proteases cathepsins B and L, plays an important role in the degradation of endocytosed and intracellular proteins18. At the same time, compounds that block lysosomal acidification such as chloroquine or bafilomycin A inhibit CpG DNA-driven signaling and responses19. More recently, lysosomal proteolysis in general20 and cathepsin K specifically21 have been implicated in TLR9 signaling but the molecular mechanisms underlying their involvement remain to be elucidated.
RESULTS
Cathepsin-mediated cleavage of TLR9
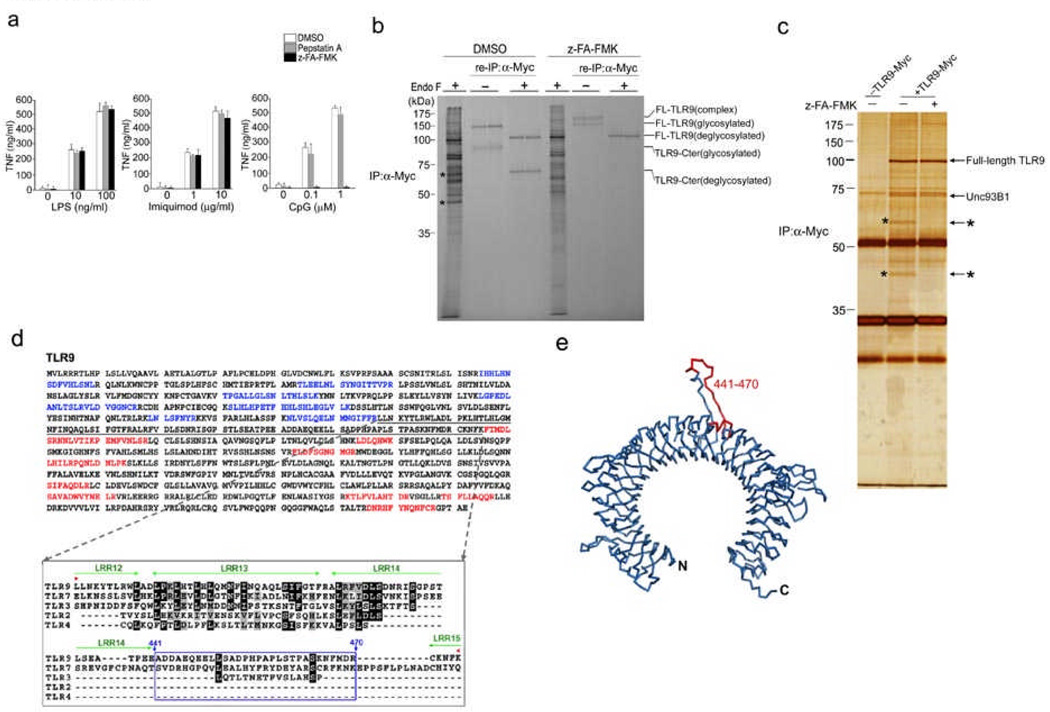
To investigate whether cathepsin activity is required for TLR responses, we exposed the RAW macrophage cell line to pepstatin A, an inhibitor of aspartic proteases, or to z-FA-FMK, a cysteine protease inhibitor that blocks cathepsin activity; we then exposed these cells to TLR4 (LPS), TLR7 (Imiquimod) and TLR9 (CpG) agonists. The z-FA-FMK inhibitor significantly inhibited tumor necrosis factor (TNF) production by RAW macrophages stimulated with CpG, but did not influence TNF release after exposure to LPS or Imiquimod (Fig. 1a). Thus, lysosomal cysteine proteases inhibited by z-FA-FMK play a role in the responsiveness of TLR9, but not TLR4 or TLR719.
Figure 1.
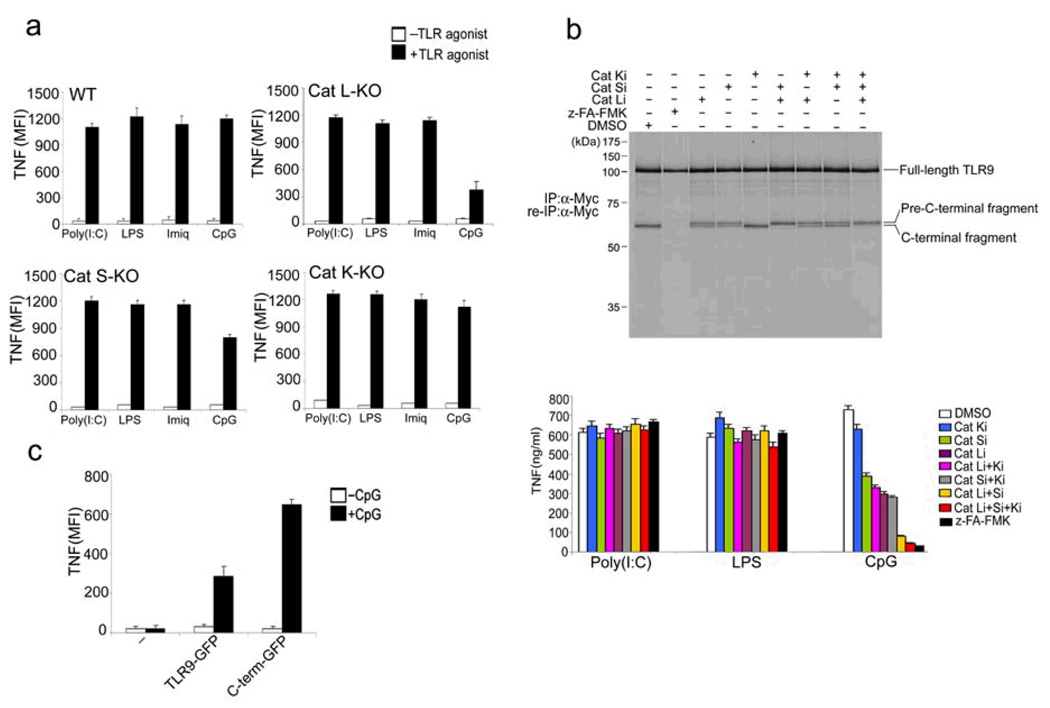
TLR9 is cleaved into two distinct polypeptides by cathepsins. (a) RAW macrophages were treated with either pepstatin A, z-FA-FMK or DMSO, followed by incubation with LPS, Imiquimod or CpG DNA. (b) RAW macrophages expressing C-terminally Myc-tagged TLR9 were pre-treated with DMSO or z-FA-FMK. Radioactively labeled proteins were subjected to immunoprecipitation with anti-Myc. One tenth of the immunoprecipitates were resolved by SDS-PAGE. After denaturation, the remainder was subjected to re-immunoprecipitation with anti-Myc and treated with EndoF where indicated. Asterisks depict 45 kDa and 65 kDa TLR9 cleavage fragments. We analyzed duplicate samples for each condition. FL: full length; Cter: C-terminal fragment (c) RAW macrophages expressing TLR9-Myc were treated with DMSO (−) or z-FA-FMK (+), anti-Myc immunoprecipitated proteins were visualized by silver staining. Polypeptides of 65 and 45 kDa (*) were analyzed by LC/MS/MS. (d) Top, peptides identified by LC/MS/MS from (c) are highlighted in blue (N-terminal) and red (C-terminal) in the murine Tlr9 sequence. No peptides were identified in the region encompassing residues 378–475 (underlined). Bottom, alignment of the region encompassing the cleavage site(s) of TLR9 (378–475) with sequences of other indicated TLRs. Residues 441 to 470 (boxed in blue) are part of a flexible loop. Leucine rich repeats (LRR) are highlighted in green. (e) Ribbon representation of a model of the TLR9 ectodomain based on the crystal structure of the TLR3 ectodomain. The predicted cathepsin cleavage site (441–470) is highlighted in red. Data are representative of four (a) or two (b–c) independent experiments (a;average, s.d.).
To establish how cathepsin activity controls TLR9 responses, we explored the ‘fate’ of newly synthesized TLR9. We generated a RAW macrophage derivative that stably expressed C-terminally Myc-tagged TLR9 and exposed it to z-FA-FMK or DMSO. Cells were then labeled with 35S methionine and cysteine for 2 h and chased for 6 h, after which we immunoprecipitated TLR9-Myc with a Myc-specific antibody. To improve electrophoretic resolution, we digested the immunoprecipitates with peptide:N-glycosidase F (EndoF), which eliminates carbohydrate heterogeneity. In the anti-Myc immunoprecipitates of DMSO-treated cells, we detected two distinct polypeptides of 65 and 45 kDa (Fig. 1b, asterisks), that were not present in immunoprecipitates of cells that were labeled in the presence of z-FA-FMK (Fig. 1b). We then denatured the initial anti-Myc immunoprecipitates and performed a re-immunoprecipitation with anti-Myc, to retrieve C-terminally Myc-tagged TLR9. In these re-immunoprecipitates, we detected the 65 kDa polypeptide in cells treated with DMSO, but not in cells exposed to z-FA-FMK (Fig. 1b). However, we observed similar rates of TLR9 synthesis during labeling. As we could retrieve the 65 kDa polypeptide with the Myc-specific antibody, we concluded that it is derived from TLR9.
To explore the nature of the 65 and 45 kDa polypeptides, we treated RAW macrophages expressing TLR9-Myc with DMSO or z-FA-FMK, immunoprecipitated TLR9 with anti-Myc, resolved glycosidase-digested proteins by SDS-PAGE and visualized proteins by silver staining. We detected full length TLR9 (~100 kDa) in DMSO- and z-FA-FMK–treated cells, and the two polypeptides of 65 kDa and 45 kDa in DMSO- but not z-FA-FMK–treated cells (Fig. 1c, asterisks). The interaction between UNC93B1 and TLR9, critical for translocation of TLR9 to lysosomes22, 23 was detected in DMSO- and z-FA-FMK–treated cells (Fig. 1c and Supplementary Fig. 1a, online). We unambiguously identified the 65 kDa and 45 kDa polypeptides as the C-terminal and N-terminal fragments of TLR9, respectively, by mass spectrometry (Fig. 1d and Supplementary Fig. 1b). The identification of the 45 kDa polypeptide as the N-terminal fragment of TLR9 explains why we did not retrieve it by re-immunoprecipitation with anti-Myc (Fig. 1b). Thus, its appearance in the initial anti-Myc immunoprecipitates suggests that the N-terminal fragment interacted with either full length TLR9, with the C-terminal cleavage fragment, or with both. Fragmentation of TLR9 was readily detectable after 3 h of chase, a timeframe consistent with delivery of TLR9 to lysosomes (Supplementary. Fig. 2, online).
Identification of cleavage site(s) in the ectodomain of TLR9
To identify the likely site(s) of cleavage in TLR9, we determined the C-terminal boundary of the N-terminal segment, and the N-terminal boundary of the C-terminal fragment based on the unambiguous presence of peptides from these fragments as identified by mass spectrometry (Fig. 1d and Supplementary Fig. 1a, online). Our analysis confined the possible cleavage site(s) to a region encompassing amino acids 378–475 of TLR9 (Fig. 1d). As the leucine-rich repeat (LRR) regions, some of which are stabilized by disulfide bonds, form a tightly packed hydrophobic core, these regions should be less susceptible to proteolysis24. Secondary structure prediction programs designated residues 441 – 470, which are situated between LRR 14 and 15, as part of a flexible loop that could render TLR9 susceptible to proteolysis (Fig. 1d). Based on published TLR structures24, 25, we assume that the LRRs of TLRs generally take on a rather similar fold, and that LRR14 and LRR15 of TLR9 can thus be used to buttress the intervening segment of TLR9. Molecular modeling showed the presence of residues 441–470, which are contained within the putative cleavage region, in an extended loop (Fig. 1e).
Only in the presence of z-FA-FMK did we detect an additional, more slowly migrating species of TLR9 (Fig. 1b and Fig. 2a, arrowhead). Glycosidase digestion showed that this form had acquired partial EndoH resistance (Fig. 2a), consistent with passage through the Golgi apparatus. Because the cathepsin inhibitor z-FA-FMK was required for its visualization, this form of TLR9 is likely cleaved in lysosomes. The fact that it is normally cleaved and thus eliminated would account for the earlier failure to detect this complex EndoH-resistant form of TLR9 22, 23.
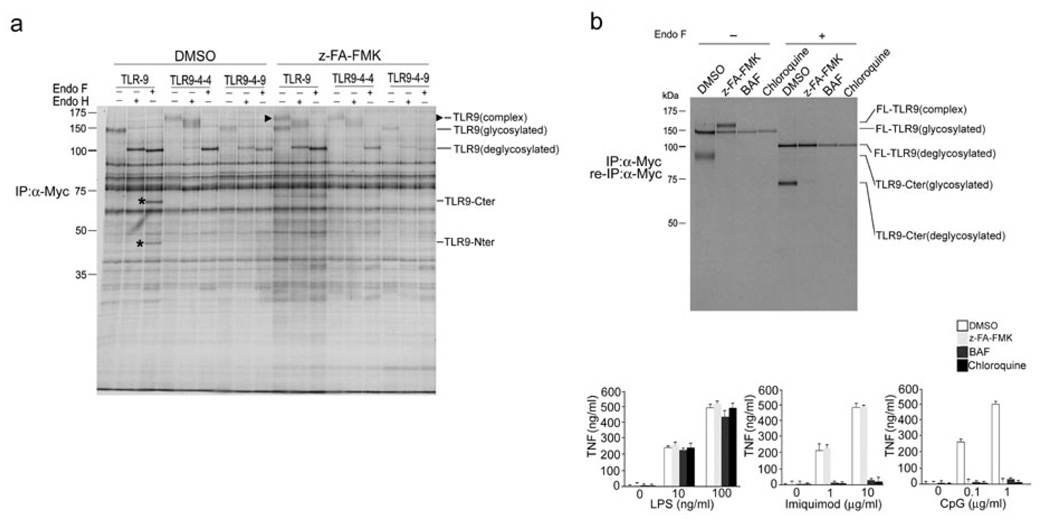
Figure 2.
Trafficking of TLR9 to the endolysosomal compartment is required for its fragmentation. (a) RAW macrophages stably expressing Myc-tagged TLR chimeras or Myc-tagged wild-type TLR9 were treated with DMSO or 10 µM z-FA-FMK for 12 h, metabolically labeled for 1.5 h and chased for 6 h. TLR proteins were recovered by immunoprecipitation with an anti-Myc, treated with glycosidases EndoH or EndoF where indicated and resolved by SDS-PAGE. The arrowhead indicates mature EndoH resistant TLR9, and the asterisks denote the C-terminal TLR9 cleavage fragments. (b) Top, RAW macrophages expressing TLR9-Myc were incubated for 10 h with DMSO (lanes 1 and 5), 10 µM z-FA-FMK (lanes 2 and 6) and for 4 h with 5 µg/ml bafilomycin (lanes 3 and 7) or 5 µM chloroquine (lanes 4 and 8) and metabolically labeled for 1 h, followed by a chase period of 6 h. Immunoprecipitation and reimmunoprecipitation were performed with an anti-Myc. Immunoprecipitates were subjected to treatment with EndoF where indicated. Bottom, RAW macrophages were incubated with DMSO or 10 µM z-FA-FMK for 12 h, 5 µg/ml bafilomycin or 5 µM chloroquine for 4 h, stimulated with increasing concentrations of LPS, Imiquimod or CpG DNA for 2 h, and TNF secretion was analyzed by ELISA. Data are representative of three independent experiments (b;average, s.d.).
Endolysosomal localization of TLR9 cleavage
To determine if the endolysosomal compartment is the location of TLR9 cleavage, we assessed the effect of the cathepsin inhibitor z-FA-FMK on the expression, maturation and stability of wild-type TLR9 and the TLR9 chimeras referred to as 9-4-4 and 9-4-9 (Fig. 2a). TLR9-4-4 contains the transmembrane and cytoplasmic region of TLR4, whereas TLR9-4-9 contains only the transmembrane region of TLR422. These TLR9 chimeras fail to interact with UNC93B1 and therefore do not reach the lysosomal compartment, the presumed location of CpG DNA recognition and TLR9 signaling22, 23. Unlike wild-type TLR9, neither TLR9-4-4 nor TLR9-4-9 yielded the two distinct fragments (Fig. 2a). These findings suggest that localization to the endolysosomal compartment is required for TLR9 cleavage.
As TLR7 is also located in the endolysosomal compartment, we sought to determine whether TLR7, like TLR9, undergoes proteolytic processing. In the absence of the cathepsin inhibitor, under conditions where the TLR9 cleavage products were readily detected, we retrieved only full length TLR7 (Supplementary Fig. 3, online). In addition, we did not detect a mature EndoH resistant TLR7 species in the presence of z-FA-FMK as we did for TLR9 (Supplementary Fig. 3, online). Next, we used the vacuolar H+ ATPase inhibitor bafilomycin A and the weak base chloroquine to block endosomal acidification and thus, indirectly, lysosomal protease activity19. Both compounds, like z-FA-FMK, blocked cleavage of TLR9, as well as CpG-driven TNF production (Fig. 2b). In contrast, bafilomycin A and chloroquine, but not z-FA-FMK, suppressed Imiquimod-driven TNF production. Thus, we concluded that fragmentation of TLR9 by lysosomal proteases is inhibited by interference with lysosomal acidification; in contrast, TLR7 shows no signs of such lysosomal cleavage.
Influence of cleavage on TLR9 function
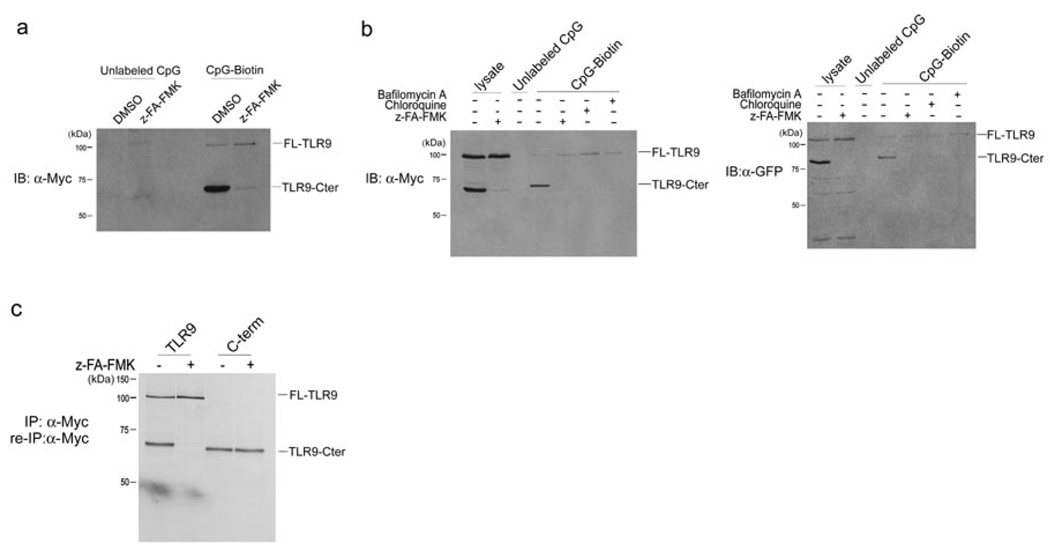
We next analyzed the interaction between CpG DNA and TLR9 in the presence and absence of z-FA-FMK (Fig. 3). TLR9-Myc–expressing RAW cells were pre-treated with z-FA-FMK, followed by incubation with biotinylated or unlabeled CpG DNA. Biotin-CpG DNA and materials bound to it were recovered with streptavidin agarose and the presence of co-precipitated TLR9-Myc was examined by immunoblotting with anti-Myc. In the absence of the cathepsin inhibitor, we observed a strong interaction of the C-terminal TLR9 fragment with CpG DNA, and weak but detectable binding between CpG and full length TLR9 (Fig. 3a). As expected in light of the absence of the C-terminal TLR9 cleavage fragment in lysates of cells treated with z-FA-FMK, we did not detect binding of a C-terminal TLR9 fragment to CpG DNA in z-FA-FMK–treated cells (Fig. 3a). Notably, z-FA-FMK had no effect on the interaction between full length TLR9 and CpG DNA, but the strength of the association between full length TLR9 and biotinylated CpG DNA appeared to be markedly weaker than with the interaction between CpG and the C-terminal TLR9 fragment (Fig. 3a). As z-FA-FMK blocked CpG DNA-induced TNF production but not binding of CpG DNA to full length TLR9, the interaction between CpG DNA and full length TLR9 is not sufficient for TLR9 activation. TLR9 molecules tagged with GFP displayed similar binding to CpG (Fig. 3b).
Figure 3.
The C-terminal TLR9 fragment directly interacts with CpG DNA. (a) RAW macrophages expressing TLR9-Myc were pretreated with DMSO or 10 µM z-FA-FMK for 12 h and then incubated with 3 µM unlabeled or biotinylated CpG DNA for 3 h at 37°C. CpG DNA and materials bound to it were recovered on streptavidin agarose, resolved by SDS-PAGE and probed with anti-Myc. (b) RAW macrophages expressing TLR9-Myc or TLR9-GFP were treated with DMSO or 10 µM z-FA-FMK for 12 h, followed by incubation with 5 µg/ml bafilomycin or 5 µM chloroquine for 4 h and incubated with 3 µM biotinylated or unlabeled CpG DNA for 3 h at 37°C. CpG DNA and materials bound to it were retrieved on streptavidin agarose, subjected to digestion with EndoF, resolved by SDS-PAGE next to total input lysate and probed with anti-Myc or anti-GFP. (c) RAW macrophages expressing either wild-type TLR9-Myc or Myc-tagged C-terminal fragment of TLR9 (471–1032) were treated with z-FA-FMK (+) or DMSO (−) and metabolically labeled. Lysates were subjected to immunoprecipitation with anti-Myc, digested with EndoF and resolved by SDS-PAGE. Data are representative of three independent experiments.
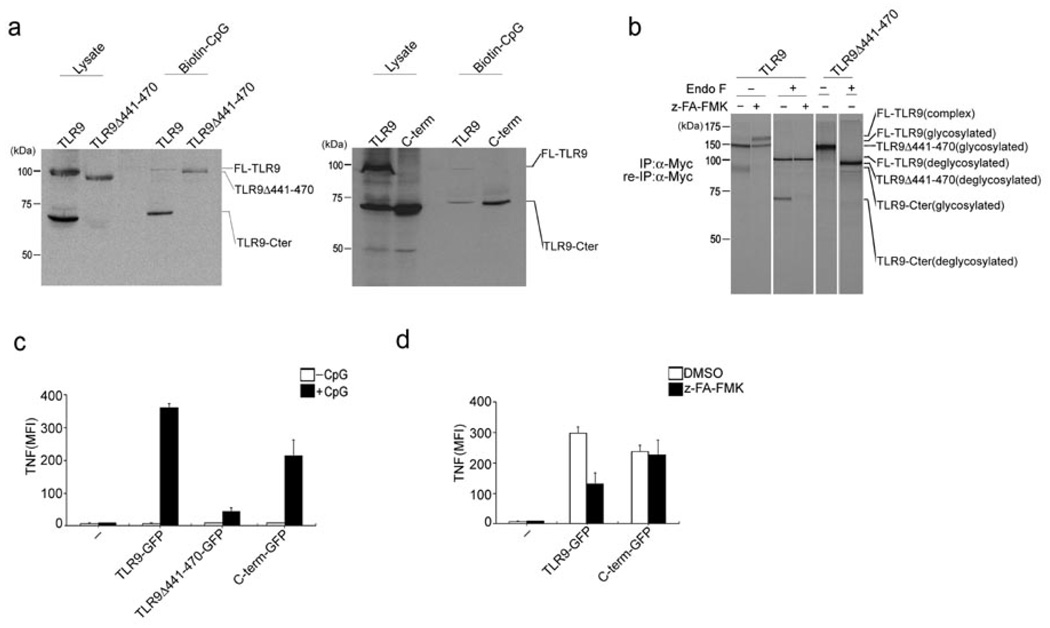
A recombinant Myc-tagged TLR9 C-terminal fragment encompassing residues 471–1032, when expressed in RAW macrophages, was detected, even in the presence of z-FA-FMK (Fig. 3c). This recombinant C-terminal fragment, when expressed in isolation, also bound to biotinylated CpG, to a degree more strongly than full length Myc-tagged TLR9 (Fig. 4a).
Figure 4.
The C-terminal TLR9 fragment is the active form responsible for binding CpG DNA and subsequent TLR9 signal transduction. (a) RAW macrophages stably expressing TLR9Δ441–470 or the C-terminal TLR9 fragment (471–1032) were incubated with 3 µM biotinylated CpG DNA for 3 h at 37°C. CpG DNA and materials bound to it were precipitated with streptavidin agarose, subjected to digestion with EndoF and precipitates and input lysates were immunoblotted with anti-Myc. (b) Myc-tagged wild-type TLR9 and Myc-tagged TLR9Δ441–470 were immunoprecipitated and reimmunoprecipitated with anti-Myc from DMSO- or z-FA-FMK-treated and metabolically labeled RAW macrophages, digested with EndoF where indicated, and visualized by SDS-PAGE. (c,d) BMDCs from Tlr9−/− mice were retrovirally transduced with vectors encoding GFP-tagged wild-type TLR9, TLR9Δ441–470 or the C-terminal TLR9 fragment (471–1032). (c) Cells were stimulated with CpG DNA (1 µM) for 4 h in the presence of brefeldin A at day 5 of BMDC culture. Cells were fixed and stained with anti-TNF, and TNF-expressing GFP+ cells were quantified by flow cytometry. Data were generated from three independent experiments and are expressed as mean fluorescence intensity (MFI). (d) Cells were stimulated with CpG (1 µM) for 4 h in the presence of DMSO or 1 µM z-FA-FMK for 8 h at day 5 of BMDC culture. TNF expression was measured as in (c). Data are representative of three independent experiments (c,d; average, s.d.).
Next, we sought to determine if fragmentation of TLR9 by cathepsin L is critical for subsequent TLR9 signal transduction. We investigated the fragmentation of TLR9 and its interaction of CpG DNA in RAW cells that stably express either Myc-tagged wild-type TLR9 or TLR9Δ441–470, a TLR9 deletion mutant lacking the proposed cleavage region (Fig. 4a, b). TLR9Δ441–470 was not cleaved, but still showed capacity to bind biotin-CpG DNA (Fig. 4a, b). These findings suggest no gross structural alteration in the structure of the TLR9Δ441–470 mutant protein. To analyze the functional capacity of TLR9Δ441–470, we measured its ability to restore CpG-induced TNF production in bone marrow-derived dendritic cells (BMDCs) from Tlr9−/− mice. Wild-type TLR9, the C-terminal cleavage fragment of TLR9, or TLR9Δ441–470, each tagged with GFP, were expressed by retroviral transduction in Tlr9−/− BMDCs; these cells were then exposed to CpG DNA, and TNF production in GFP+ cells was measured by flow cytometry (Fig. 4c). BMDCs transduced with the TLR9Δ441–470 mutant showed reduced TNF production in response to CpG DNA stimulation, compared to cells expressing wild-type TLR9 (Fig. 4c). Since the transmembrane segment and cytoplasmic tail of TLR9Δ441–470 are identical to those of full length TLR9, and UNC93B1 bound wild-type TLR9 and TLR9Δ441–470 to a similar degree (data not shown), we believe that trafficking properties were unaffected by the deletion of residues 441–470.
The C-terminal cleavage product of TLR9 restores function to Tlr9−/− BMDCs
We next investigated whether the C-terminal fragment, which retains a portion of the ectodomain encompassing leucine rich repeats 15–26 and contains the full transmembrane and cytoplasmic domains, is on its own sufficient for binding to CpG DNA and initiating TLR9 signal transduction. In RAW macrophages, recombinant C-terminal TLR9 fragments tagged C-terminally with either Myc or GFP, interacted strongly with biotinylated CpG DNA, as demonstrated by the recovery of the C-terminal TLR9 fragment on streptavidin agarose (Fig. 4b). Neither the expression nor the molecular weight of the recombinant C-terminal fragment were affected by z-FA-FMK (Fig. 3c). Tlr9−/− BMDCs transduced with a retrovirus encoding the recombinant GFP-tagged C-terminal fragment of TLR9 regained responsiveness to CpG DNA stimulation, as indicated by TNF̣production (Fig. 4c). TNF synthesis elicited by CpG DNA stimulation of Tlr9−/− BMDCs that express the GFP-tagged C-terminal fragment of TLR9 was not affected by treatment with z-FA-FMK (Fig. 4d). In contrast, TNF̣production facilitated by expression of GFP-tagged wild-type TLR9 in Tlr9−/− BMDCs was inhibited by z-FA-FMK (Fig. 4d). The greater sensitivity of BMDCs to cytotoxic effects of z-FA-FMK did not allow us to use the inhibitor concentrations readily tolerated by RAW cells and their derivatives to achieve full inhibition of TNF̣productioñ Nevertheless, we conclude that the C-terminal TLR9 fragment on its own is sufficient to bind CpG DNA and initiate signaling.
Multistep TLR9 celavage
If indeed residues 441–470 are correctly modeled as a flexible loop (Fig. 1e), then any of a number of proteases could presumably cleave it, as has been observed for other proteins activated by proteolysis, such as flu hemagglutinin, cholera toxin and various pro-enzymes, including lysosomal hydrolases. Similarly, although the involvement of lysosomal proteolysis in major histocompatibility class II-restricted antigen presentation is immediately evident from the inhibitory effects of a broad protease blockade, with very few exceptions, it has been difficult to implicate any particular protease in the processing of a specific antigen26,27. For the same reasons, it may be difficult to determine which members of the cathepsin family of lysosomal proteases cleave TLR9. In the case of genetic ablation or pharmacological inhibition of a single cathepsin, any of a number of cysteine proteases could ‘pick up the slack’, thereby explaining the greater inhibitory potencyof the broadly specific inhibitors over more narrowly targeted compounds28, 29. Also, mice lacking multiple cathepsins show more severe phenotypes than mice lacking a single cathepsin 30. Nonetheless, we examined the TLR responsiveness of BMDCs from mice lacking individual cathepsins. Cathepsin L-deficient BMDCs and, to a lesser extent, Cathepsin S-deficient BMDCs, showed reduced TNF production in response to CpG (Fig. 5a). We observed normal TNF production in response to CpG by BMDCs from Cathepsin K-deficient mice, seemingly at variance with a published report21 in which Cathepsin K-deficient BMDCs failed to respond to CpG stimulation, as measured by reduced IL-12 production.
Figure 5.
Multiple lysosomal proteases are required for TLR9 cleavage. (a) BMDCs from wild-type mice or mice lacking cathepsin L (Cat L-KO), cathepsin S (Cat S-KO) or cathepsin K (Cat K-KO) were stimulated with poly I:C (100 µg/ml), LPS (1 µg/ml), Imiquimod (10 µg/ml) or CpG DNA (1 µM) for 4 h in the presence of brefeldin A at day 6 of BMDC culture. Cells were fixed and stained with anti-TNF, and intracellular TNF was measured by flow cytometry. (b) Top, RAW macrophages expressing TLR9-Myc were pretreated for 12 h with DMSO, z-FA-FMK (10 µM) or the selective cathepsin L inhibitors Clik195 and Clik148 (Cat Li, 10 µM), the cathepsin S inhibitor LHVS (Cat Si, 10 nM), the cathepsin K inhibitor II (Cat Ki, 1 µM), or combinations thereof. Cells were metabolically labeled for 1.5 h followed by a 5 h chase period. TLR9 was immunoprecipitated and re-immunoprecipitated from lysates with anti-Myc, and was digested with EndoF. Bottom, RAW macrophages were treated for 12 h with the inhibitors listed and stimulated for 2 h with the indicated TLR agonists. Secreted TNF was analysed by ELISA. (c) BMDCs from Cat L-KO mice were retrovirally transduced with vectors encoding GFP-tagged wild-type TLR9 or GFP-tagged C-terminal TLR9 fragment (471–1032) or were left untransduced (−) at day 1 of BMDC culture. At day 6, cells were stimulated with CpG (1µM) for 4 h in the presence of brefeldin A, fixed and stained with anti-TNF. TNF was measured by flow cytometry in GFP+ (transduced) cells. Data are representative of two (a,c) or three (b) independent experiments (a-c; average, s.d.).
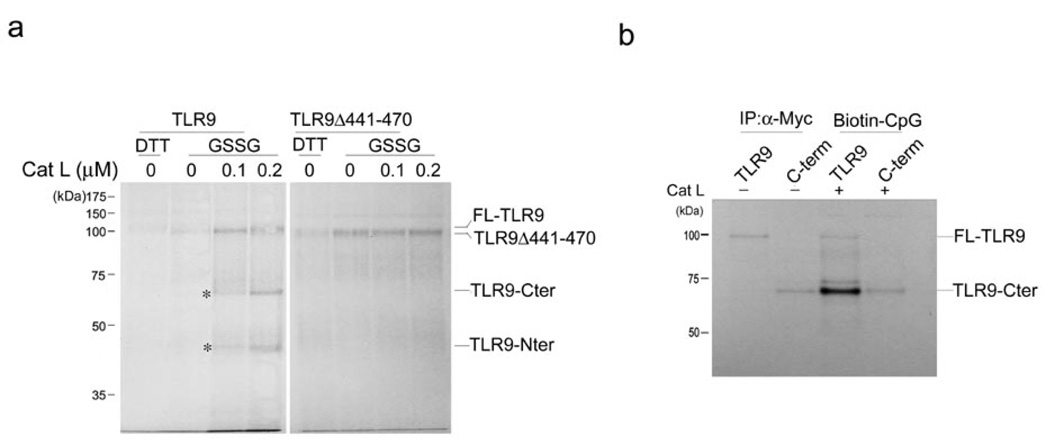
We next examined the effects of a series of selective cathepsin inhibitors on the pattern of TLR9 cleavage in and TNF production by RAW cells. z-FA-FMK, but not any of the more narrowly specific cathepsin inhibitors, completely suppressed formation of the 65 kDa C-terminal TLR9 fragment itself (Fig. 5b). Cleavage in the presence of the cathepsin K inhibitor was indistinguishable from cleavage in the presence of DMSO (Fig. 5b). Selective inhibition of cathepsin L or cathepsin S resulted in the appearance of a doublet comprised of a slightly larger band in addition to the 65 kDa band (Fig. 5b). We designated the upper band the pre-C terminal fragment. The cathepsin K inhibitor, when combined with other inhibitors, failed to show an additive effect (Fig. 5b). However, combining the cathepsin S- and cathepsin L-selective inhibitors resulted in enrichment of the pre-C terminal fragment compared to the C-terminal fragment (Fig. 5b). The pre-C-terminal fragment appeared to be inactive, as we noted a strict inverse correlation between its presence and TNF production (Fig. 5b). Partial conversion of full length TLR9 to the pre-C-terminal and C-terminal fragments yielded intermediate amounts of TNF. The slight difference in apparent molecular weight suggests that at most a few dozen residues distinguish the active C-terminal fragment from the inactive TLR9 pre-C-terminal fragment (Fig. 5b). In addition, introduction of the C-terminal TLR9 fragment into cathepsin L-deficient BMDCs restored CpG DNA-driven TNF production (Fig. 5c). Wild-type TLR9 restored TNF production to a limited extent, consistent with the limited inhibition of TNF production seen in untransduced cathepsin L−/− BMDCs (Fig. 5a, c). We conclude that more than a single lysosomal protease is involved in cleavage of TLR9, with an important role for cathepsin L. To further support the notion that cathepsin L can cleave TLR9 at the proposed cleavage site, we incubated radiochemically pure TLR9, produced by in vitro translation in the presence of properly redox-buffered microsomes31, with purified cathepsin L (Fig. 6a). Cathepsin L produced the C-terminal fragment of TLR9 in vitro (Fig. 6a), and this C-terminal fragment bound to biotinylated CpG (Fig. 6b). However, the TLR9 deletion mutant TLR9Δ441–470 was not cleaved by cathepsin L in vitro (Fig. 6a). Combined, our results establish that TLR9 undergoes cleavage in an endolysosomal compartment, a step important for TLR9 to execute its proper function.
Figure 6.
Cathepsin L cleaves TLR9 in vitro but fails to cleave the TLR9 deletion mutant lacking the region encompassing the putative cathepsin cleavage site(s). (a) Wild-type TLR9 or the TLR9 deletion mutant TLR9Δ441–470 were transcribed and translated in vitro in the presence of microsomes and 35S-methionine and either 2 mM DTT or 2 mM oxidized glutathione (GSSG). Microsomes were pelleted, lysed and recombinant cathepsin L was added for 2 h at 37°C. 10% of the total volume of the in vitro transcription and translation reaction was loaded per lane and separated by SDS-PAGE. Asterisks indicate the C- and N-terminal fragment of TLR9. (b) Myc-tagged wild-type TLR9 or C-terminal TLR9 fragment (471–1032) were transcribed and translated in vitro in the presence of microsomes and 35S-methionine. Microsomes were pelleted, lysed and 10% was subjected to immunoprecipitation with anti-Myc antibody, digested with EndoF and resolved by SDS-PAGE. 90% was incubated with 0.2 µM recombinant cathepsin L for 2 h at 37°C followed by a 2 h incubation with 5 µM biotinylated CpG DNA. Samples were then incubated with streptavidin agarose, digested with EndoF and resolved by SDS-PAGE. Data are representative of two independent experiments.
DISCUSSION
The involvement of the endolysosomal compartment in proper functioning of the nucleic acid sensing TLRs (TLR3, 7, 9) is well-documented 11–16, but the exact manner in which this environment contributes to the receipt and transmission of signals via these TLRs has not been established. We now demonstrate that TLR9 undergoes a proteolytic cleavage, executed by cysteine proteases, in the endolysosomal compartment. On a molar basis the C-terminal cleavage fragment of TLR9 binds CpG more strongly than full length TLR9, although full length TLR9 does bind CpG. The broadly specific inhibitor z-FA-FMK is most effective at blocking this cleavage, and accordingly abrogates production of TNF in cells exposed to the TLR9 agonist CpG. Our data are consistent with the involvement of more than one protease in this cleavage reaction, because we observe a blunted, rather than a completely blocked TNF response to CpG in Cathepsin L-deficient or Cathepsin S-deficient BMDCs. Furthermore, the combination of Cathepsin L - and Cathepsin S -selective inhibitors not only blocks TNF production in response to CpG, but also yields a cleavage intermediate, the pre-C-terminal fragment, the abundance of which is inversely correlated with TNF production. The expression of only the C-terminal fragment in Tlr9−/− BMDCs restores their ability to produce TNF, as seen also for Cathepsin L-deficient BMDCs. We hypothesize that absence of cleavage in presence of chloroquine and bafilomycin A is best explained by elevated pH or reduced proteolytic activity.
The delivery of TLR9 to endolysosomal compartments requires its interaction with UNC93B1, mediated via the transmembrane segment of TLR922. Eliminating this interaction, either by substituting the transmembrane segment of TLR9 or by deleting UNC93B1, causes TLR9 to traffic incorrectly to the cell surface16,23. The capacity of a chimera consisting of the extracellular domain of TLR9 and the transmembrane and cytoplasmic domains of TLR4–which localizes to the cell surface–to induce signal transduction upon CpG binding might be attributed to the recruitment of adaptors other than Myd88 to the cytoplasmic tail of TLR416,32.
We did not observe any inhibition of TNF production in Cathepsin K-deficient BMDCs or by imposition of a Cathepsin K blockade, in contrast to an earlier study that showed reduced IL12 production by Cathepsin K-deficient BMDCs in response to CpG21. We further determined that responses requiring engagement of TLR7 were not affected by z-FA-FMK (Fig. 1a), unlike the blockade in upregulation of CD86 and B cell proliferation reported earlier20. IL12 production may require the involvement of Cathepsin K at steps downstream of TLR9 engagement, which could explain the observed discrepancy. Likewise, the surface expression of CD86 and steps that control B cell proliferation may require lysosomal protease involvement for reasons other than cleavage of the TLRs involved.
We have no evidence that TLR7 is cleaved in a z-FA-FMK sensitive manner, and we have been unable to detect cleavage fragments of TLR7. This raises the question of why signaling via TLR7 or via TLR3, for that matter, is nonetheless sensitive to the inclusion of lysosomotropic agents. Several of the TLRs are known to act in cooperation with partner proteins such as CD14 and MD2 for TLR433, 34, or CD36 for TLRs 1, 2 and 635, or Dectin-1 for TLR236. We hypothesize that the activity of TLR3 and TLR7 may require cofactors whose involvement necessitates a proteolytic cleavage, or at least includes a pH sensitive step, affected by the inclusion of lysosomotropic agents.
The evolutionary significance of TLR9 cleavage is as yet unclear. Why should TLR9 be unique among lysosomal TLRs in requiring this mode of activation? The C-terminal cleavage fragment of TLR9 is capable of binding CpG DNA and signal transduction even when synthesized in the absence of the N-terminal portion of TLR9. A search for interaction partners of the N-terminal cleavage fragment might illuminate yet other aspects of TLR9 biology. Regardless of the identity of the TLR9 convertase(s) and the exact role of the cleavage products generated, full innate immunity requires lysosomal proteolysis no less than adaptive immunity.
METHODS
Reagents
Imiquimod (R837) and poly (I:C) were purchased from Invivogen, 1826-CpG DNA and 1826-Biotin-CpG DNA (5’- Bio-TsCsCsAsTsgsAsCsgsTsTsCsCsTsgsAsCsgsTsT) from TIB Molbiol, and LPS (E. coli 026:B6) and brefeldin A from Sigma. Endoglycosidase H (Endo H) and PNGase F were purchased from New England Biolabs. The monoclonal Myc-(9B11), GFP-(LGB-1) and rabbit TLR7-specific antibodies were obtained from Cell signaling, Abcam and Imgenex, respectively. Streptavidin agarose beads were from Pierce. Bafilomycin, chloroquine, z-FA-FMK and glutathione disulfide (GSSG) were purchased from Sigma, and CA-074Me from Calbiochem. The selective cathepsin inhibitors Clik195 and Clik148 (Cathepsin L), LHVS (Cathepsin S), and Cathepsin K inhibitor II (Cathepsin K, Calbiochem) were generously given to us by Guo-Ping Shi (Harvard Medical School, MA)37.
Mice and cell lines
C57BL/6 mice were purchased from Charles River Laboratories. Tlr9−/− mice were obtained from A. Marshak-Rothstein (Boston University, Boston, MA). All animals were maintained under specific pathogen-free conditions according to guidelines by the committee for animal care at the Whitehead Institute. Murine RAW 264.7 macrophages (ATCC TIB-71) and human embryonic kidney (HEK) cells 293-T (ATCC CRL-11268) were cultured in DME supplemented with 10% heat inactivated fetal calf serum (IFS) and penicillin/streptomycin. Cells were grown at 37°C in humidified air with 5% CO2. Cathepsin L-deficient, cathepsin S-deficient and cathepsin K-deficient mice were kindly provided by Guo-Ping Shi (Harvard Medical School, MA).
DNA cloning
All murine TLR9 constructs were C-terminally fused to Myc or GFP. Wild-type TLR9 and TLR9 chimeras were already described22. The TLR9 deletion mutant lacking residues 441–470 was generated by sequential PCR with primers 5’-CTGTCAGAAGCCACCCCTGAAGAGTGTAAGAACTTCAAGTTCACCATGGACCTG (forward) and 5’-CAGGTCCATGGTGAACTTGAAGTTCTTACACTCTTCAGGGGTGGCTTCTGAGAC (reverse). The recombinant C-terminal TLR9 fragment encompassing residues 471–1032 was generated by PCR with primers 5’-GGGGTACCTGTAAGAACTTCAAGTTCACCATGGACCTG (forward) and 5’-AGCTTTGTTTAAACTTTACAAGTCCTCTTCAGAAATGAGCTTTTGCTCTTCTGCTGTAGGTCCCCGGCAGA (reverse). The C-terminal TLR9 fragment TLR9 471–1032 was N-terminally fused with the Kb signal sequence (MVPCTLLLLLAAALAPTQTRA). All constructs were cloned into pcDNA3.1(+) (Invitrogen) or the retroviral pMSCV vector (Clontech) and verified by sequencing.
Preparation of BMDCs
BMDCs were prepared as previously described38.
Pulse-chase analysis, immunoprecipitation and Endo H/F assay
RAW macrophages (1 × 107) were starved for 50 min in medium lacking methionine and cysteine (starvation medium), labeled with 0.1 mCi/ml [35S]methionine and cysteine (Perkin Elmer) in starvation medium supplemented with dialyzed IFS and chased in complete medium with or without 10 µM of indicated inhibitor. Cells were lysed with 1% NP-40 in phosphate-buffered saline (PBS) supplemented with protease inhibitors (Roche) for 1 h at 4°C. After preclearing lysates with protein G-Sepharose (Sigma-Aldrich), primary antibodies and protein G-Sepharose were added to supernatants and incubated at 4°C. The protein G-Sepharose beads were washed five times with 0.1% NP-40 in PBS. Proteins were eluted from the beads by boiling in SDS sample buffer, separated by SDS-PAGE and polypeptides were visualized by fluorography. Digestion with Endoglycosidase H and PNGase F was performed, where indicated, at 37 °C for 3 h.
Co-immunoprecipitation and immunoblotting
Cells were lysed in 1% digitonin (Calbiochem) in buffer containing 25 mM HEPES, 100 mM NaCl, 10 mM CaCl2 and 5 mM MgCl2 (pH 7.6) supplemented with 0.5 mM phenyl-methyl-sulfonyl-fluoride (PMSF) and leupeptin. Lysates were precleared with protein G-Sepharose (Sigma-Aldrich) for 1 h at 4°C. For immunoprecipitation, samples were incubated with the appropriate antibodies for 12 h at 4°C before protein G-beads were added for 3 h. Beads were washed four times with 0.1% digitonin, and bound proteins were eluted by boiling in SDS sample buffer or 1% SDS. Proteins were separated by 10% SDS-PAGE, transferred onto a nitrocellulose membrane, blocked with 5% skim milk in PBS with 0.1% Tween-20 for 2 h and probed with the appropriate antibodies for 4 h. Membranes were washed three times with PBS with 0.1% Tween-20 and incubated with horseradish peroxidase-conjugated streptavidin for 1 h. The immunoblots were visualized with ECL detection reagent.
Large-scale affinity purification and mass spectrometry (MS)
After co-immunoprecipitation, the eluted samples were separated by SDS-PAGE and polypeptides were revealed by silver staining. The bands of interest were excised, subjected to trypsinolysis and analyzed by tandem mass spectrometry (MS/MS).
Retroviral transduction
HEK 293-T cells were transfected with plasmids encoding VSV-G and Gag-Pol, as well as either pMSCV-TLR9-Myc, pMSCV-TLR9-4-4-Myc, pMSCV-TLR9-4-9-Myc, pMSCV-TLR9Δ441–470, or pMSCV-TLR9 471–1032. 24 h and 48 h post transfection, medium containing viral particles was collected, filtered through a 0.45 µm membrane and added to RAW macrophages or BMDCs at day 1 of BMDC culture. The following day, cells were fed with fresh media.
In vitro transcription and translation
1 µg of Myc-tagged wild-type TLR9, TLR9Δ441–470 or the C-terminal TLR9 fragment TLR9 471–1032 cloned into the pcDNA3.1(+) vector was transcribed and translated in vitro with the TNT T7 Quick coupled TT system (Promega) in the presence of microsomes and 10 µCi 35S methionine (Perkin Elmer) for 1 h at 30°C in a total volume of 50 µl. Microsomes were pelleted by centrifugation at 13,000 r.p.m. for 4 min and were lysed in 20 µl of cathepsin activation buffer (1% NP40, 50 mM Na-actetate pH 5.5, 3 mM cysteine, 1 mM EDTA). Recombinant cathepsin L (Sigma) was immediately added at 0.1–0.2 µM to the microsomes and incubated for 2 h at 37°C, followed by an incubation with biotinylated CpG DNA (5 µM) for 2 h at 37°C. Reactions were diluted to a final volume of 1 ml with 1% NP40 lysis buffer with protease inhibitors and TLR9 proteins were immunoprecipitated with anti-Myc or incubated with streptavidin agarose and separated by SDS-PAGE.
Intracellular TNF assay
Tlr9−/− BMDCs expressing either wild-type or mutant TLR9 were cultured for 5 days and were stimulated with 1 µM CpG DNA for 4 h in the presence of 10 µg/ml brefeldin A. The cells were fixed with 4% formaldehyde for 10 min at room temperature and permeabilized with 0.5% saponin in FACS buffer (PBS with 2% BSA and 0.05% sodium azide) for 10 min. Cells were stained with Alexa Fluor 647-conjugated anti-TNF (clone MP6-XT22) for 30 min. Fluorescence intensity was measured with LSR I flow cytometer (BD Biosciences). Data were collected with CellQuest (BD Biosciences) and analyzed with FlowJo (Tree Star).
TNF ELISA
RAW macrophages were stimulated for 2 h with increasing concentrations of TLR agonists LPS (TLR4), imiquimod (TLR7) or CpG DNA (TLR9). The conditioned medium was collected and analysed by ELISA using hamster anti-mouse/rat TNF (BD Biosciences) as a capture antibody and biotin-labeled rabbit anti-mouse as a secondary antibody (BD Biosciences).
Homology modeling
A multiple sequence alignment containing the target sequence (Mus musculus TLR9, NP_112455) and the template sequence (Mus musculus TLR3, NP_569054) was generated by ClustalW. The resulting alignment and the structure of the Mus musculus TLR3 ectodomain (PDB accession code 3ciy, chain A) were used to generate a structural model of the mus musculus TLR9 ectodomain with Swiss-Model (http://swissmodel.expasy.org/SWISS-MODEL.html)39.
Supplementary Material
Acknowledgements
We are grateful to GP. Shi (Brigham and Women’s Hospital and Harvard, Medical School, Boston, MA) for providing us with the selective cathepsin S, L and K inhibitors and cathepsin K-deficient, cathepsin S-deficient and cathepsin L-deficient mice. We thank S. Akira (Osaka University, Japan), A. Marshak-Rothstein and K. Kiefer (Boston University, Boston, MA) for providing Tlr9−/− mice, S.K. Dougan and C. Schlieker for critical reading of the manuscript, and T. DiCesare for graphical design.
This study was supported by grants from NIH as well as Novartis (to H.L.P.), the Charles A. King Trust, Bank of America, Co-Trustee (to M.M.B.) and a Landon Clay fellowship (to B.P.)
Footnotes
Author Information
The authors declare no competing financial interests.
References
- 1.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 2.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 3.Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13:816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 4.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 5.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2028. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 6.Takeuchi O, et al. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 7.Takeuchi O, et al. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int Immunol. 2001;13:933–940. doi: 10.1093/intimm/13.7.933. [DOI] [PubMed] [Google Scholar]
- 8.Ozinsky A, et al. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc Natl Acad Sci U S A. 2000;97:13766–13771. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayashi F, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 10.Hemmi H, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 11.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 12.Heil F, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 13.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 14.Latz E, et al. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 15.Nishiya T, Kajita E, Miwa S, Defranco AL. TLR3 and TLR7 are targeted to the same intracellular compartments by distinct regulatory elements. J Biol Chem. 2005;280:37107–37117. doi: 10.1074/jbc.M504951200. [DOI] [PubMed] [Google Scholar]
- 16.Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat Immunol. 2006;7:49–56. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- 17.Leifer CA, et al. TLR9 is localized in the endoplasmic reticulum prior to stimulation. J Immunol. 2004;173:1179–1183. doi: 10.4049/jimmunol.173.2.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kominami E, Ueno T, Muno D, Katunuma N. The selective role of cathepsins B and D in the lysosomal degradation of endogenous and exogenous proteins. FEBS Lett. 1991;287:189–192. doi: 10.1016/0014-5793(91)80048-8. [DOI] [PubMed] [Google Scholar]
- 19.Honda K, et al. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 2005;434:1035–1040. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto F, et al. Cathepsins are required for Toll-like receptor 9 responses. Biochem Biophys Res Commun. 2008;367:693–699. doi: 10.1016/j.bbrc.2007.12.130. [DOI] [PubMed] [Google Scholar]
- 21.Asagiri M, et al. Cathepsin K-dependent toll-like receptor 9 signaling revealed in experimental arthritis. Science. 2008;319:624–627. doi: 10.1126/science.1150110. [DOI] [PubMed] [Google Scholar]
- 22.Brinkmann MM, et al. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol. 2007;177:265–275. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 24.Choe J, Kelker MS, Wilson IA. Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science. 2005;309:581–585. doi: 10.1126/science.1115253. [DOI] [PubMed] [Google Scholar]
- 25.Jin MS, Lee JO. Structures of the Toll-like Receptor Family and Its Ligand Complexes. Immunity. 2008;29:182–191. doi: 10.1016/j.immuni.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Villadangos JA, Ploegh HL. Proteolysis in MHC class II antigen presentation: who's in charge? Immunity. 2000;12:233–239. doi: 10.1016/s1074-7613(00)80176-4. [DOI] [PubMed] [Google Scholar]
- 27.Lennon-Dumenil AM, Bakker AH, Wolf-Bryant P, Ploegh HL, Lagaudriere-Gesbert C. A closer look at proteolysis and MHC-class-II-restricted antigen presentation. Curr Opin Immunol. 2002;14:15–21. doi: 10.1016/s0952-7915(01)00293-x. [DOI] [PubMed] [Google Scholar]
- 28.Greenbaum DC, et al. Small molecule affinity fingerprinting. A tool for enzyme family subclassification, target identification, and inhibitor design. Chem Biol. 2002;9:1085–1094. doi: 10.1016/s1074-5521(02)00238-7. [DOI] [PubMed] [Google Scholar]
- 29.Bogyo M, Verhelst S, Bellingard-Dubouchaud V, Toba S, Greenbaum D. Selective targeting of lysosomal cysteine proteases with radiolabeled electrophilic substrate analogs. Chem Biol. 2000;7:27–38. doi: 10.1016/s1074-5521(00)00061-2. [DOI] [PubMed] [Google Scholar]
- 30.Felbor U, et al. Neuronal loss and brain atrophy in mice lacking cathepsins B and L. Proc Natl Acad Sci U S A. 2002;99:7883–7888. doi: 10.1073/pnas.112632299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huppa JB, Ploegh HL. In vitro translation and assembly of a complete T cell receptor-CD3 complex. J Exp Med. 1997;186:393–403. doi: 10.1084/jem.186.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawai T, et al. Lipopolysaccharide stimulates the MyD88-independent pathway and results in activation of IFN-regulatory factor 3 and the expression of a subset of lipopolysaccharide-inducible genes. J Immunol. 2001;167:5887–5894. doi: 10.4049/jimmunol.167.10.5887. [DOI] [PubMed] [Google Scholar]
- 33.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 34.Shimazu R, et al. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J Exp Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Triantafilou M, et al. Membrane sorting of toll-like receptor (TLR)-2/6 and TLR2/1 heterodimers at the cell surface determines heterotypic associations with CD36 and intracellular targeting. J Biol Chem. 2006;281:31002–31011. doi: 10.1074/jbc.M602794200. [DOI] [PubMed] [Google Scholar]
- 36.Rogers NC, et al. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity. 2005;22:507–517. doi: 10.1016/j.immuni.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 37.Yang M, et al. Cathepsin L activity controls adipogenesis and glucose tolerance. Nat Cell Biol. 2007;9:970–977. doi: 10.1038/ncb1623. [DOI] [PubMed] [Google Scholar]
- 38.Maehr R, et al. Asparagine endopeptidase is not essential for class II MHC antigen presentation but is required for processing of cathepsin L in mice. J Immunol. 2005;174:7066–7074. doi: 10.4049/jimmunol.174.11.7066. [DOI] [PubMed] [Google Scholar]
- 39.Liu L, et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.