Abstract
During yeast mating, cell fusion is followed by the congression and fusion of the two nuclei. Proteins required for nuclear fusion are found at the surface (Prm3p) and within the lumen (Kar2p, Kar5p, and Kar8p) of the nuclear envelope (NE). Electron tomography (ET) of zygotes revealed that mutations in these proteins block nuclear fusion with different morphologies, suggesting that they act in different steps of fusion. Specifically, prm3 zygotes were blocked before formation of membrane bridges, whereas kar2, kar5, and kar8 zygotes frequently contained them. Membrane bridges were significantly larger and occurred more frequently in kar2 and kar8, than in kar5 mutant zygotes. The kinetics of NE fusion in prm3, kar5, and kar8 mutants, measured by live-cell fluorescence microscopy, were well correlated with the size and frequency of bridges observed by ET. However the kar2 mutant was defective for transfer of NE lumenal GFP, but not diffusion within the lumen, suggesting that transfer was blocked at the NE fusion junction. These observations suggest that Prm3p acts before initiation of outer NE fusion, Kar5p may help dilation of the initial fusion pore, and Kar2p and Kar8p act after outer NE fusion, during inner NE fusion.
INTRODUCTION
During yeast mating, two haploid cells fuse to form a single diploid cell. Many aspects of mating have been studied in great detail, including the cell signaling pathway leading to the formation of the yeast mating projection called the “shmoo” (Marsh and Rose, 1997; Elion, 2000; Bardwell, 2005). However, less is known about the membrane fusion events occurring during mating itself, including plasma membrane fusion and nuclear membrane fusion. Recently, it was shown that nuclear membrane fusion is a three-step process, beginning with outer membrane fusion, proceeding to inner membrane fusion, and finally spindle pole body (SPB) fusion (Melloy et al., 2007). As a next step, we have set out to determine which karyogamy proteins participate in the two distinct steps of nuclear membrane fusion.
Nuclear fusion mutants are classified into two major groups (Kurihara et al., 1994). Class I mutants are defective in nuclear congression, whereas class II mutants display defects in nuclear membrane fusion. Class II mutants all arrest with closely apposed nuclei, suggesting that the genes affected all play a role in nuclear membrane fusion. The class II genes KAR2, KAR5, KAR8, and PRM3 are the focus of this study.
Kar2p has multiple functions in the endoplasmic reticulum/nuclear envelope (ER/NE). It is responsible for protein translocation into the ER (Vogel et al., 1990), protein folding (Rose et al., 1989), ER-associated degradation (Nishikawa et al., 2001; Kabani et al., 2003), and nuclear fusion (Rose et al., 1989; Latterich and Schekman, 1994; Ng and Walter, 1996; Brizzio et al., 1999). However the isolation of mutant alleles relatively specific to nuclear fusion suggest that Kar2p's role in karyogamy is separate from its role in protein translocation (Rose et al., 1989; Vogel et al., 1990).
KAR2 is known to interact genetically with KAR8/JEM1, encoding a DnaJ protein, mutation of which causes a strong defect in karyogamy (Nishikawa and Endo, 1997; Brizzio et al., 1999). Kar2p associates with several DnaJ proteins, including Sec63p, and each interaction may be important for all of its different functions (Walsh et al., 2004). Mutations in Sec63p result in partial karyogamy defects (Ng and Walter, 1996), as do mutations in the other proteins associated with the ER-chaperone complex, Sec71/Kar7p, and Sec72p (Ng and Walter, 1996; Brizzio et al., 1999). With regards to Sec63p, genetic data suggest that the ER-chaperone's role in nuclear fusion is separate from translocation (Ng and Walter, 1996). It seems likely that several NE/ER proteins are playing dual roles during mitotic growth and mating.
Although Kar2p and Kar8p are expressed throughout the yeast vegetative cycle and recruited for nuclear fusion, two proteins, Kar5p and Prm3p, are specifically expressed during mating. KAR5 is induced by mating pheromone up to ∼100-fold (Roberts et al., 2000), and kar5 mutants display a strong bilateral karyogamy defect (Kurihara et al., 1994; Beh et al., 1997; Erdman et al., 1998). On induction by pheromone, Kar5p is detected as a NE protein concentrated near the site of initiation of nuclear membrane fusion, adjacent to the SPB (Beh et al., 1997). Because of its pheromone dependent expression and localization, it has been hypothesized that Kar5p might organize a membrane fusogenic complex in preparation for membrane fusion (Beh et al., 1997; Erdman et al., 1998). PRM3 is up-regulated ∼60-fold in response to pheromone (Roberts et al., 2000) and belongs to a group of genes encoding membrane-associated proteins with pheromone-dependent expression (Heiman and Walter, 2000). Prm3 mutants are karyogamy-defective (Beilharz et al., 2003), and overexpression of KAR5 suppresses this mating defect (Shen et al., 2009). Prm3p is present on the outer NE and colocalizes with Kar5p near the SPB (Shen et al., 2009).
The proteins Prm3p, Kar2p, Kar5p, and Kar8p reside in different locations in relation to the NE; Kar5p is an integral membrane protein, Kar2p and Kar8p reside in the lumen of the NE, and Prm3p is located on the outer membrane (Rose et al., 1989; Beh et al., 1997; Nishikawa and Endo, 1997; Shen et al., 2009). Hence, these proteins are properly positioned to potentially act at different stages of nuclear fusion. Previous work showed that the karyogamy mutants block nuclear fusion with different morphologies, suggesting that they might act at different stages. Conventional electron microscopy revealed the presence of membrane bridges connecting the haploid nuclei in kar2, kar5, kar7, and kar8 mutant zygotes (Kurihara et al., 1994; Brizzio et al., 1999). The bridges in kar8 mutants were larger than the bridges in kar5 mutants. The bridges in kar5 kar8 mutant zygotes were similar to those of a kar5 mutant, suggesting that Kar8p may act downstream of Kar5p in nuclear fusion (Brizzio et al., 1999). However, the limited spatial information of conventional thin-sectioning electron microscopy and possible artifacts associated with earlier fixation conditions lead us to reinvestigate the karyogamy mutants.
We used two techniques, electron tomography (ET) and time-lapse microscopy of live cells using a fluorescently tagged NE luminal marker to analyze the karyogamy mutants. Together these techniques were necessary to demonstrate that wild-type NE fusion occurs in three steps (Melloy et al., 2007). Although ET provides resolution and structural details superior to that of conventional EM, live-cell microscopy provides information about the kinetics of inner and outer nuclear membrane fusion (Melloy et al., 2007). These observations suggest that Prm3p acts before initiation of outer NE fusion, Kar5p may help dilation of the initial fusion pore, and Kar2p and Kar8p act after outer NE fusion, during inner NE fusion.
MATERIALS AND METHODS
Strains and Yeast Methods
Strains used in this study are described in Table 1. General yeast methods including yeast transformations, and media preparation were conducted as previously described (Rose et al., 1990; Amberg et al., 2005). The mutant alleles, kar5-486 and kar8-1333, were chosen because of previous EM observations (Kurihara et al., 1994; Brizzio et al., 1999). However, the extents of their defects in nuclear fusion are similar to deletions in these genes (Beh et al., 1997; Brizzio et al., 1999). KAR2 is an essential gene, and the kar2-1 allele exhibits the strongest defect in nuclear fusion. Genomic integration of a monomeric red fluorescent protein (mRFP) fluorescent tag at the C-terminus of SPC42 was performed using a PCR-based method (Longtine et al., 1998; Melloy et al., 2007). A 3X-GFP-HDEL–containing plasmid (pMR5029) was transformed into donor strains and used for time-lapse microscopy.
Table 1.
Strains and plasmids
| Strain | Genotype | Source or reference |
|---|---|---|
| MS23 | MATα, trp1-Δ1, lys2-801, ade2-101 | Rose lab |
| MS1554 | MATa, ura3-52, leu2-3,112, ade2-101, his3-Δ200 | Rose lab |
| MS1111 | MATa, ura3-52, leu2-3,112, ade2-101, kar2-1 | Rose lab |
| MS2689 | matΔ::LEU2, ura3-52, leu2-3, 112, trp1Δ1, his3::TRP1, kar5-486, [YCpMATα] | Kurihara et al. (1994) |
| MS7804 | Same as MS2689 except SPC42:mRFP | This study |
| MS2690 | matΔ::LEU2, ura3-52, leu2-3, 112, trp1Δ1, his3::TRP1, kar5-486 | Kurihara et al. (1994) |
| MS7822 | Same as except SPC42:mRFP, ss-3X-GFP-HDEL (TRP1) | This study |
| MS2705 | matΔ::LEU2, ura3-52, leu2-3, 112, trp1Δ1, his3::TRP1, kar8-1333-1333, [YCpMATα] | Kurihara et al. (1994) |
| MS7809 | Same as MS2705 except SPC42:mRFP | This study |
| MS2706 | matα::LEU2, ura3-52, leu2-3, 112, trp1Δ1, his3::TRP1, kar8-1333-1333 | Kurihara et al. (1994) |
| MS7820 | Same as MS2706 except ss-3X-GFP-HDEL (TRP1) | This study |
| MS3533 | MATa, lys2-801, his3-Δ200, ade2-101, ura3-52, leu2-3, 112, kar5-486 | Rose lab (for tomography) |
| MS3537 | MATα, lys2-801, his3-Δ200, ade2-101, ura3-52, LEU2, kar5-486 | Rose lab (for tomography) |
| MS3898 | MATα, ura3-52, leu2-3, 112, trp1-Δ1, his3-Δ200, lys2-801, kar8-1333-1333 | Rose lab (for tomography) |
| MS3899 | MATa, ura3-52, leu2-3, 112, trp1-Δ1, his3-Δ200, lys2-801, kar8-1333-1333 | Rose lab (for tomography) |
| MS7590 | MATa, prm3Δ::HIS3, ura3-52, leu2-3,112, trp1-Δ1, his3-Δ200 | Rose lab |
| MS7591 | MATα, prm3Δ::HIS3, ura3-52, leu2-3,112, ade2-101, his3-Δ200 | Rose lab |
| MS7884 | MATa, prm3Δ::HIS3, ura3-52, leu2-3,112, ade2-101, his3-Δ200, trp1-Δ1, SPC42:mRFP-KanMX6 | This study |
| MS7885 | MATα, prm3Δ::HIS3, ura3-52, leu2-3,112, ade2-101, his3-Δ200, trp1-Δ1, SPC42:mRFP-KanMX6 | This study |
| MS7892 | MS7590 with pMR5029 | |
| MS7593 | MS7884 with pMR5029 | |
| Plasmid | Relevant markers | Source or reference |
| pMR5029 | ss-3X-GFP-HDEL in pRS414 (TRP1) | Rose lab |
| pMR5484 | pFa6a-mRFP-kanMX6 | S. Clark, R. Tsien |
Preparing Mating Mixtures for ET and Live-Cell Microscopy
Equal numbers of budding yeast of opposite mating types were mated according to published methods (Gammie and Rose, 2002). For ET, mating mixtures were placed on nitrocellulose disks for 2.5 h at 30°C and prepared for electron microscopy using high-pressure freezing (O' Toole et al., 2002; Yoder et al., 2005), as previously described (Melloy et al., 2007). Serial, 200–300-nm-thick sections were prepared, and 15-nm colloidal gold particles (Sigma-Aldrich, St. Louis, MO) were used as fiducial markers for image alignment. The details of ET tomography reconstruction, and modeling using the IMOD software package (University of Colorado, Boulder, CO) have been described in detail elsewhere (Kremer et al., 1996; Mastronarde, 1997; Melloy et al., 2007). Fifteen kar2-1× WT tomograms, 15 kar5-486× kar5-486 tomograms, 14 kar8-1333× kar8-1333 tomograms, and 13 prm3Δ× prm3Δ tomograms were reconstructed and modeled.
For live-cell microscopy, mating mixtures were transferred to a 2% agarose pad (in synthetic complete media) on a microscope slide and incubated for 1.5 h at RT. Images were captured using an Applied Precision Delta Vision Microscopy System (Issaquah, WA) based on a Nikon TE200 microscope (Melville, NY), 100× NA 1.4 objectives and a Coolsnap HQ CCD camera (Photometrics, Tucson, AZ). Image deconvolution was performed using the Applied Precision SoftWoRx application, and Adobe Photoshop and Illustrator (San Jose, CA) were used to prepare figures for publication. Time-lapse observations were initiated just after cell fusion was detected (usually 1.5–2 h) and lasted for 10–15 min. Among the wild-type zygotes, nuclear fusion occurred within 10–15 min in 48 of 61 interpretable experiments. ImageJ (NIH; http://rsb.info.nih.gov/ij/) was used to quantify the green fluorescent protein (GFP) fluorescence in donor and recipient cells for all zygotes with a strong 3X-GFP-HDEL signal. The details of determining corrected integrated fluorescence intensity was described previously (Melloy et al., 2007).
Fluorescence Recovery after Photobleaching
Measurement of fluorescence recovery after photobleaching (FRAP) on 3X-GFP-HDEL and Pap1-GFP was performed using a Zeiss LSM510 confocal system (Carl Zeiss, Jena, Germany) in the Department of Molecular Biology, Princeton University. FRAP was conducted as described previously (Melloy et al., 2007). Analysis of FRAP data was completed using MATLAB program written by Thomas Gregor (Princeton University).
On-Line Supplemental Material
Supplemental materials include tables listing all tomographic reconstructions (Supplemental Tables S1–S4).
RESULTS
Karyogamy Mutants Block Nuclear Fusion with Distinct Morphologies
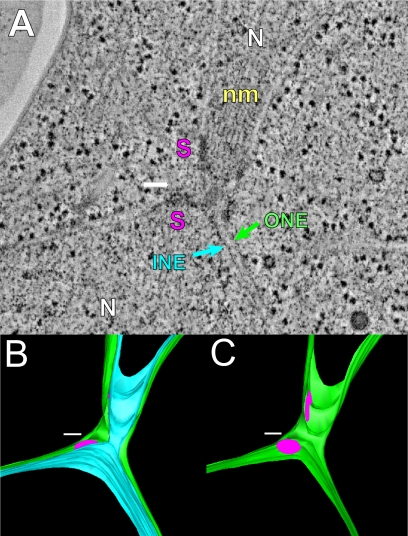
Previous work demonstrated three steps in nuclear fusion: outer membrane fusion, inner membrane fusion, and SPB fusion (Melloy et al., 2007). For comparison to the mutant zygotes in this study, Figure 1 depicts a wild-type zygote in which cell fusion is complete: the nuclei have congressed toward each other, and both outer and inner nuclear membrane fusion have occurred in the narrow region in the middle of the zygote (outer and inner membrane, green and light blue, respectively). However, SPB fusion has not occurred as indicated by the observation of two distinct spindle pole body central plaques (pink disks in model) in the zygote. The wild-type tomogram example shown in Figure 1, one of 22 imaged at different stages of nuclear fusion, has been previously published (Melloy et al., 2007). To characterize the roles of the different proteins required for nuclear fusion, we next set out to analyze selected karyogamy mutants.
Figure 1.
Wild-type nuclear fusion occurs in three steps. (A) A representative tomographic slice of a wild-type zygote fixed after completion of outer and inner nuclear membrane fusion. INE, inner NE; ONE, outer nuclear envelope; N, nucleus; S, SPB; nm, nuclear microtubules. (B) A model corresponding to the tomogram shown in A highlighting the outer (green) and inner membranes (light blue) of the NE. (C) Same model as in B except that only the outer membrane (green) is visible. Note that the central plaques of the spindle pole bodies (SPBs; pink) remain distinct after nuclear membrane fusion. Scale bar, 100 nm.
The four mutants that are the focus of this study all cause strong defects in nuclear fusion as determined by light microscopy (>92% karyogamy defect; Beh et al., 1997; Brizzio et al., 1999; Shen et al., 2009). With the exception of kar2-1, all the mutant zygotes were formed in matings in which both parents contained the same mutation. The kar2-1 mutation exhibits a unilateral mating defect, in which only one mating partner needs to be mutant to block karyogamy (Rose et al., 1989). By light microscopy, all the karyogamy mutants appeared to have a similar phenotype. However, by ET and live-cell microscopy, each mutant exhibited a distinct and characteristic phenotype. Tomograms were generated for 13–16 mutant zygotes of each genotype (see Supplemental Material).
PRM3
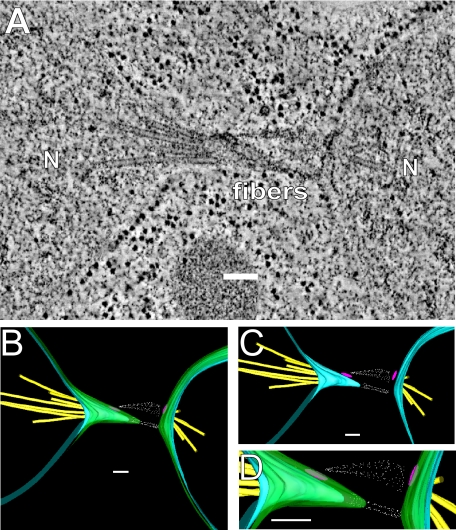
Figure 2 depicts a representative tomogram of a prm3Δ mutant zygote. In 12 of 13 prm3 mutant zygotes, the nuclei have congressed but not completed nuclear fusion. In the remaining zygote, nuclear fusion was completed. In the characteristic morphology of prm3 mutant zygotes, the NEs of the two nuclei are elongated toward one another, but the outer NEs have not fused (9 of 13). Thin, electron-dense fibers, originating from the vicinity of the SPBs (both from the face of the central plaque and the area of the half bridge), appear to connect the two nuclei (Figure 2B). Where the two NEs are pulled together, the inner and outer NEs follow roughly parallel contours (Figure 2, C and D), suggesting that the two membranes are connected in this region.
Figure 2.
prm3Δ mutant zygotes fail to form membrane bridges. (A) Tomographic slice of a prm3Δ zygote showing no membrane fusion and electron-dense material between the SPBs. The nucleus (N) and electron-dense fibers are indicated. (B–D) Models of the tomogram shown in A. Outer membranes (green), inner membranes (blue), microtubules (yellow), spindle pole bodies (pink), and fibers (white) are modeled. Scale bars, 100 nm. Zygotes are produced from matings of MS7590 × MS7591.
In three of the 13 prm3 zygotes, the outer, but not inner, NEs have fused. However, in two of the three zygotes, three or more SPBs were observed, indicating that these zygotes have entered the vegetative mitotic cycle. Therefore, it is likely that these represent membrane fusion events associated with vegetative ER and NE remodeling. After excluding the two mitotic cells, we conclude that the prm3 mutant blocks nuclear fusion before outer membrane fusion in ∼90% of zygotes.
KAR5
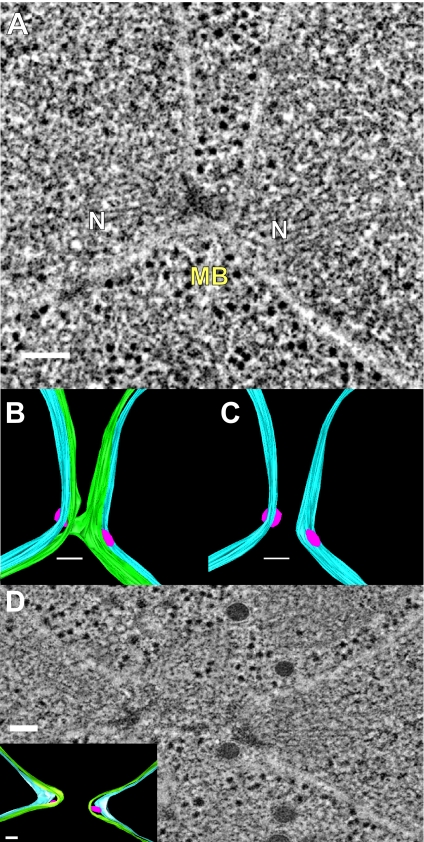
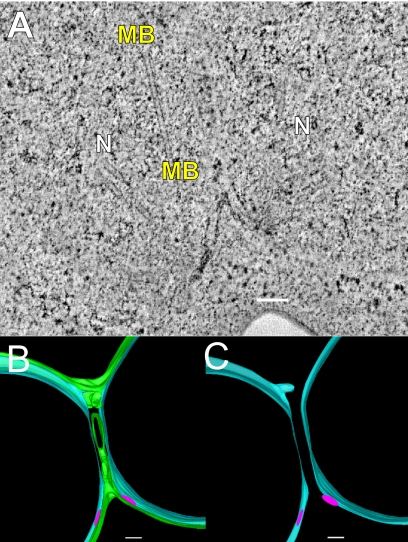
In 14 of the 15 kar5 mutant zygotes (Figure 3), the nuclei have congressed but not completed nuclear fusion (n = 15). In four zygotes ≥3 SPBs were observed, indicating that these cells had progressed into the vegetative mitotic cycle. In one additional zygote, nuclear fusion was observed. In six zygotes (including 1 with ≥3 SPBs), the outer NEs appeared to be continuous between the two nuclei, effectively connecting the two nuclei by a thin membranous bridge (Figure 3, A–C). The bridges had an average diameter of 48 ± 7 nm (mean ± SE) and were roughly circular in cross-section (average ellipticity ratio of 0.75 ± 0.05; mean ± SE; see Figure 5F).
Figure 3.
kar5-486 mutant zygotes infrequently form small membrane bridges. (A) Tomographic slice of a kar5-486 mutant zygote that has completed cell fusion, with its nuclei unfused and closely apposed. Note the presence of a membrane bridge near the SPBs. N, nucleus; MB, membrane bridge. (B) Model corresponding to the tomogram shown in A highlighting the outer NE with the membrane bridge (green), the unfused inner NE (light blue), and the spindle pole bodies (pink). (C) Same model as in B shown without the outer NE to indicate the separation of the inner NEs (light blue) and the two distinct SPBs (pink). (D) Tomographic slice of a kar5-486 mutant zygote that has completed cell fusion, but its nuclei are unfused, closely apposed, and do not have any membrane bridges. (D, inset) Model corresponding to tomographic slice shown in D. Scale bars, 100 nm.
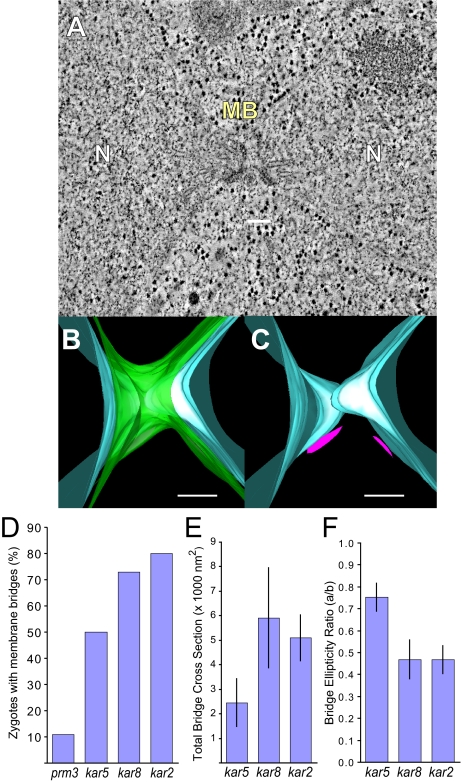
Figure 5.
kar2-1 mutant zygotes frequently form membrane bridges. (A) Tomographic slice of a kar2-1 mutant zygote in which an outer NE bridge has formed. N, nucleus; MB, membrane bridge. (B) Model corresponding to the tomogram shown in A depicting the outer NE with bridge (green) and the inner NE (light blue). (C) Same model as B except that only the two inner NEs (light blue) and two SPBs (pink) are visible. (D) Graph indicating the percent of zygotes with membrane bridges in the karyogamy mutant zygotes. (E) Graph showing the average total bridge cross-section for each type of mutant zygote. (F) Graph indicating average ellipticity ratio of the bridges in the mutant zygotes. The ellipticity is the ratio of the smallest diameter (major axis) to the largest diameter (major axis) for a cross-section across each bridge. Scale, 100 nm.
Interestingly, unlike the prm3 mutant zygotes, the inner and outer NEs were not parallel in the region of the bridges. Although the outer NEs appeared to be pulled away from the nucleus to form the bridge (green in model, Figure 3, B and C), the inner NEs retained a smoothly rounded morphology (light blue in model, Figure 3, B and C). As a consequence the distance between the inner NE and the midpoint of the outer NE bridges was ∼55 ± 6 nm (mean ± SE); significantly greater than the average distance between the inner and outer NEs elsewhere (24 ± 1 nm; mean ± SE; p = 0.0001).
A similar lack of parallel contours was observed in the kar5 mutant zygotes in which no bridge was observed (Figure 3D); the outer membranes of the apposed nuclei were drawn together, but the inner membranes remained rounded. Thus, the average distance between the inner and outer NEs near the SPBs in the kar5 zygotes was significantly expanded (38 ± 3.2 nm, n = 16), relative to the average distance elsewhere on the NE (25 ± 1 nm; p < 0.0001). For comparison, the distances in the prm3 zygotes were 26 ± 2 nm (n = 14) and 21 ± 0.8 nm close to and far from the SPB, respectively. Taken together, the morphologies of the kar5 mutants suggest that Kar5p is required for efficient initiation of outer membrane fusion and may play a role in coupling the inner and outer NEs in the vicinity of the site of membrane fusion.
KAR8
In all of the kar8 mutant zygotes, the nuclei completed nuclear congression, but not nuclear fusion (n = 14). A tomogram of a typical zygote is shown in Figure 4. In three zygotes, SPB duplication had occurred, indicating that these cells had progressed into the vegetative mitotic cycle. In the majority of zygotes (11 of 14) the outer NEs were continuous between the two nuclei forming large membranous bridges connecting the two nuclei. In four zygotes, more than one bridge was observed (e.g., Figure 4, A–C), indicating that outer NE fusion initiated at multiple sites. The bridges observed in the kar8 zygotes were markedly larger in cross-sectional area (∼4300 nm2) than those formed in the kar5 zygotes (∼2400 nm2, Figure 5E) and considerably more elongated (ellipticity ratio = 0.47 ± 0.09). The increased cross-sectional area indicates that outer NE fusion had progressed further resulting in dilation of the membrane bridge connecting the two nuclei. Taking the size and number of bridges into account, the total cross-sectional area connecting the two nuclei was markedly greater in the kar8 zygotes than the kar5 zygotes (see Figure 5E).
Figure 4.
kar8-1333 mutant zygotes frequently form large membrane bridges. (A) Tomographic slice of a kar8-1333 mutant zygote in which two membrane bridges have formed between two closely apposed nuclei. N, nucleus; MB, membrane bridge. (B) Model corresponding to the tomogram shown in A, indicating the outer and inner NEs (green and light blue, respectively) and the two outer NE bridge connections. (C) Same model as in B except only the distinct inner NEs (light blue) and distinct SPBs (pink) are visible. Scale bars, 100 nm.
Where the outer NEs were connected by more elongated bridges (e.g., the upper bridge in Figure 4, A–C), the inner NE followed a contour roughly parallel to the outer NE. The average distance between the inner NE and the midpoint of the outer NE bridges was 37 ± 3 nm; significantly less than the 55 nm observed for kar5 zygotes (p = 0.001). Thus, in the kar8 mutant zygotes, the inner and outer NEs appeared to be more closely coupled together.
KAR2
All of the kar2 zygotes completed nuclear congression but not nuclear fusion (n = 16). A typical zygote is displayed in Figure 5. In one zygote, SPB duplication had occurred, indicating that these cells had progressed into the vegetative mitotic cycle. Similar to kar8, 13 of the zygotes contained outer NEs that were continuous between the two nuclei, consistent with successful outer NE fusion. Unlike the kar8 mutant zygotes, only a single bridge was observed per zygote. Like kar8, the average cross-sectional area of the bridges in kar2 zygotes was similar to those of the kar8 zygotes, markedly larger and more elliptical than those observed in the kar5 zygotes (Figure 5, E and F). Like the prm3 and kar8 zygotes, the inner and outer NEs followed roughly parallel contours at and near the bridges. Taken together, these data suggest that the kar2 and kar8 mutants are defective for inner nuclear fusion.
To summarize the tomography data, the different mutations blocked nuclear fusion with distinct morphologies, suggesting that they act at different stages in the nuclear fusion pathway. The prm3Δ mutants were the least likely to form bridges between the outer NEs, kar5 mutants showed an intermediate frequency of outer membrane fusion and kar8 and kar2 showed outer NE fusion in most zygotes (Figure 5D). Similarly, the cross-sectional areas of the outer NE connections between the two nuclei imply an ordered pathway, with the bridges in kar5 zygotes being distinctly smaller and more circular in cross-section than those in the kar2 and kar8 zygotes (Figure 5, E and F). Interestingly, in the three examples of prm3 mutant zygotes in which outer NE fusion occurred, the average cross-sectional area and ellipticity ratio was similar to kar2 and kar8. Although two of these were in zygotes that had advanced into mitosis, these data suggest that although prm3 mutants initiate fusion only rarely, they are able to progress past the block that occurs in the kar5 mutants.
Transfer of NE Lumenal Proteins in Karyogamy Mutants
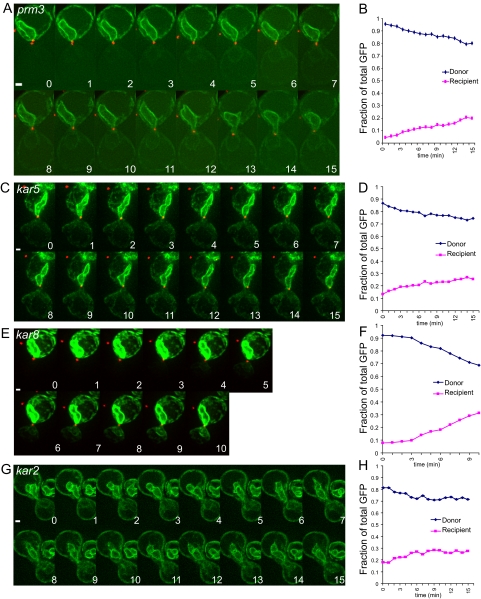
To complement the electron tomographic studies of the karyogamy mutants, we used live-cell microscopy to investigate the kinetics of NE fusion. GFP fused to a secretory signal sequence and an ER retention signal (3X-GFP-HDEL) served as a marker for the NE/ER lumen, as described previously (Gammie and Rose, 2002; Melloy et al., 2007). Using this system, previous work demonstrated the rapid transfer from a donor nucleus into a nonmarked recipient nucleus, coincident with outer NE fusion (Melloy et al., 2007). In typical experiments, matings were set up between two karyogamy mutants of opposite mating types, or, for kar2-1, the mutant was crossed to a wild-type strain. In each case, one mating partner was labeled with 3X-GFP-HDEL (donor), whereas the other was unlabeled (recipient). In most experiments, the spindle pole bodies were labeled with Spc42p-mCherryFP (or Spc42p-mRFP) as a marker for the completion of nuclear congression, although this was also apparent from the background fluorescence of the recipient nucleus. Images were captured every minute (for mutant zygotes) or more frequently (20–30 s for wild-type zygotes) over a period of 10–15 min, during which time wild-type zygotes usually completed nuclear fusion. Shown in Figure 6, A, C, E, and G, are typical time-lapse experiments for each mutant strain. For all images, the rates of 3X-GFP-HDEL transfer were calculated from the slope of the fraction of total fluorescent in the recipient nucleus (Figure 6, B, D, F, and H). For cases where the rate of transfer exhibited a significant inflection (e.g., Figure 6E) the maximal rate was calculated from the steepest part of the slope. For wild-type zygotes the rate of transfer before outer envelope fusion slope was also measured. The aggregate data for all wild-type and mutant zygotes are displayed in Figure 7.
Figure 6.
Transfer of a NE/ER luminal marker as a measure of outer NE fusion. (A) A time-lapse experiment (1-min time points) of two mating prm3Δ cells after cell fusion is complete. The SPBs have merged into a single dot (Spc42-RFP in red) by time point 2 indicating nuclear congression is complete. For A, C, E, and G, the NE/ER lumen is tagged with an ss-3X-GFP-HDEL NE/ER marker (green), with the donor nucleus shown at the top of the image and the potential recipient nucleus at the bottom. Note that in A, there is a slight increase in fluorescence in the recipient nucleus over time. (B) Graph of the fraction of total GFP fluorescence found in the recipient nucleus over time. The graph indicates donor GFP fluorescence in green and recipient GFP fluorescence in red. prm3 mutant zygotes were produced from MS7892 × MS7591 and MS7893 × MS7591 crosses. (C) Time-lapse analysis of kar5-486 mutant zygotes (1-min time points) after the NE/ER luminal marker. (D) Graph showing the fraction of total GFP in the donor and recipient cell during the time course in C. (E) Time-lapse analysis of a kar8-1333 mutant zygote (1-min time points) tracking the NE/ER luminal marker. (F) A graph showing fraction of total GFP in the donor and recipient during the time course showing a higher rate of transfer of GFP fluorescence into the recipient over time in these zygotes. (G) Time-lapse analysis of the transfer of a GFP-tagged ER luminal marker after cell fusion in a kar2-1 mutant zygote (1-min time points). (H) The graph to the right shows the fraction of total GFP in the donor and recipient during the time course.
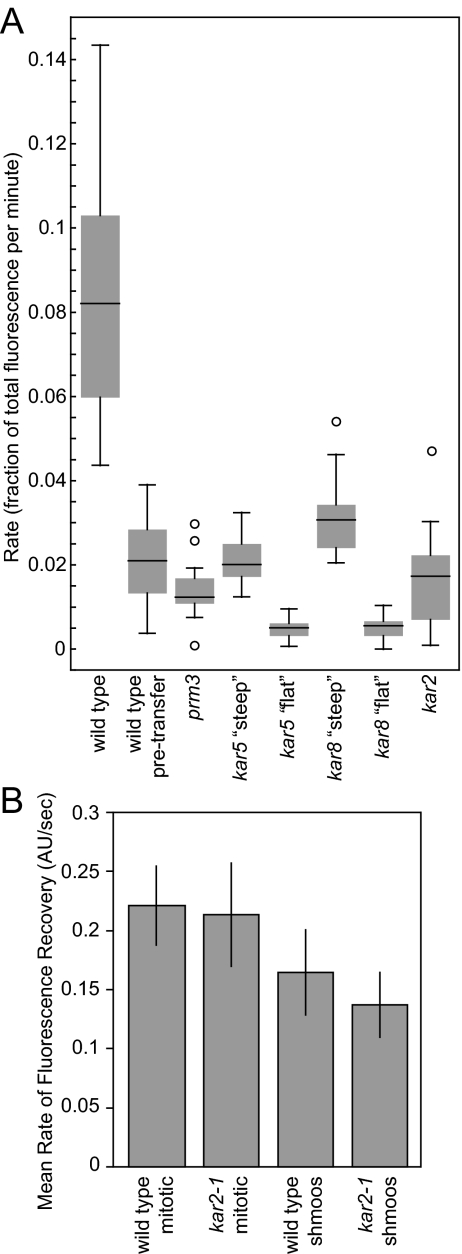
Figure 7.
Analysis of NE/ER luminal marker transfer rates and FRAP analysis in wild-type and mutant NEs. (A) Graph of the fractional rate of the lumenal NE marker 3X-GFP-HDEL. The rate of transfer in wild-type cells is shown both after and before (pretransfer) NE fusion for comparison to the mutant zygotes. The kar5 and kar8 mutants contained two different populations of zygotes with distinctly different rates of transfer labeled “steep” and “flat.” For each mutant a box and whisker plot is shown. The box contains the inner quartiles, separated by the median. The whiskers depict the upper and lower quartiles. Outliers (more than 1.5 times the inner quartile range from the upper or lower quartiles) are depicted as circles. (B) FRAP analysis of wild-type and kar2-1 cells indicating rate of mobility of the NE/ER lumenal marker for wild-type mitotic, wild-type shmooing, kar2-1 mitotic, and kar2-1 shmooing cells as evidenced by the rate of fluorescence recovery. Shown are the average rates in arbitrary units/s. Error bars, SD. The number of cells of each type ranged from 11 to 23.
Wild-type zygotes exhibited a slow initial phase of transfer with an average fractional rate of transfer of 0.021 ± 0.003/min (mean ± SE; n = 18). The rate of transfer increased dramatically to 0.088 ± 0.007/min (n = 25), presumably as regions of NE fusion expanded to allow rapid diffusion into the recipient NE lumen. When nuclear congression was prevented by including the kar1-1 mutation in one parent, the rate of transfer was significantly slower than the wild-type initial pretransfer phase (0.0056 ± 0.0012/min; n = 6).
In prm3Δ mutant zygotes, a constant slow rate of transfer was observed (Figures 6, A and B, and 7A); shift into a rapid phase rate of transfer was never observed. The rate for prm3Δ (0.016 ± 0.003/min; n = 17) was not significantly different from the pretransfer rate for wild-type zygotes (p = 0.2). Similarly, when congression was blocked in crosses of prm3Δ to prm3Δ kar1-1, the rate of transfer (0.0066 ± 0.0013/min; n = 10) was not significantly different from the wild-type crossed to kar1-1.
Both the kar5 and the kar8 mutant zygotes exhibited two different rates of transfer (Figure 7A). For each mutant, a substantial number of zygotes exhibited very low rates, significantly slower than the wild-type initial phase (kar5: 0.0048 ± 0.0007/min, n = 13, p < 0.0001; kar8: 0.0052 ± 0.0014/min, n = 6, p < 0.0001). The remaining kar5 mutant zygotes exhibited a rate of fusion indistinguishable from the wild-type pretransfer rate (0.021 ± 0.002/min; n = 11, Figures 6, C and D, and 7A). The remaining kar8 zygotes exhibited a significantly faster rate of transfer (0.032 ± 0.004/min; n = 9, p = 0.02); in most cases showing a shift from the very slow initial phase to the faster rate (Figure 6, E and F). The faster rate of transfer observed for some of the kar8 mutant zygotes is correlated with the formation of larger membrane bridges seen by ET, which should allow more rapid transfer of lumenal contents. Support for the hypothesis that faster rate of transfer in the kar8 mutants is due to membrane bridges comes from observation of zygotes formed from matings to a kar1 kar8 double mutant. In all 12 zygotes examined none exhibited rapid transfer, suggesting that the rapid phase of transfer in kar8 zygotes requires nuclear congression. The rate of transfer in the kar1 kar8 zygotes (0.0032 ± 0004/min; n = 12) was substantially slower than for wild-type kar1 and kar1 prm3 matings.
In general, the kar2 mutant zygotes showed a slow rate of transfer (0.017/min ± 0.002, n = 25) similar to that seen for the wild-type initial phase and prm3 zygotes (Figures 6, G and H, and 7A). Two additional zygotes showed wild-type rates of transfer (0.083/min) for a short period of time, consistent with a 5–10% frequency of complete nuclear fusion (Beh et al., 1997), although in neither case did transfer go to completion.
The slow transfer in kar2 zygotes was surprising, given the extended regions of outer NE fusion observed by tomographic analysis. Given that Kar2p is the Hsp70 chaperone resident in the NE/ER lumen (Rose et al., 1989), the presence of misfolded proteins might reduce the diffusion of the marker protein. Alternatively, Kar2p may play a specific role in protein assembly/disassembly at the site of nuclear fusion. To distinguish between these two alternatives, we conducted FRAP experiments to measure the mobility of the 3X-GFP-HDEL marker protein in mutant and wild-type mitotic and shmooing cells (Figure 7B). The rate of recovery was significantly faster in mitotic cells (0.22 ± 0.01/s, n = 19 for wild type and 0.21 ± 0.01/s, n = 23 for kar2) than in shmoos (0.16 ± 0.01/s, n = 20 and 0.14 ± 0.01/s, n = 11 for wild type and kar2, respectively). However, no significant difference was seen between wild-type and kar2 cells, in either condition. Therefore the slow rate of transfer through the membrane bridge in kar2 mutants is not due to overall reduce mobility and is most likely is due to a specific defect at the site of nuclear membrane fusion.
DISCUSSION
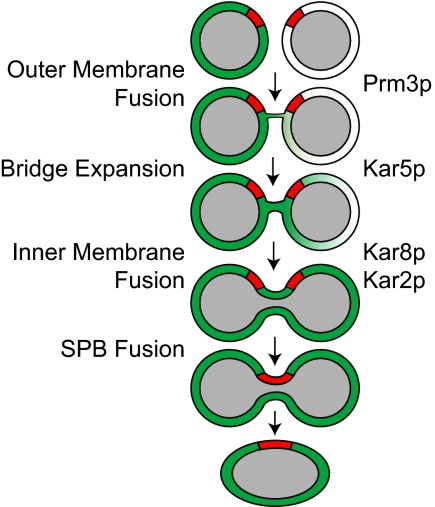
In this work, we describe the phenotypes of several type II karyogamy mutants revealed by ET and time-lapse fluorescence microscopy. The mutants differ in the morphology of unfused nuclei, the frequency and size of membrane bridges connecting the two NEs, and the rate of transport of NE lumenal material between the two nuclei. Given previous work showing that NE fusion occurs in at least two sequential steps, the characteristic mutant phenotypes, together with the localization of the proteins, provide a natural placement for their functions in the pathway of nuclear fusion (Figure 8).
Figure 8.
A pathway for nuclear fusion: The diagram depicts the pathway for nuclear fusion indicating possible steps for each protein. The nucleoplasm is shown in gray, the SPB is shown in red and contents of the nuclear lumen is shown is shown in green.
The prm3 mutant zygotes failed to form membranous connections between the outer NEs and the rate of transfer of lumenal proteins between the nuclei is similar to that of wild-type cells before nuclear fusion. Prm3p is localized to the cytoplasmic face of the NE (Shen et al., 2009), consistent with a function in the initiation of nuclear fusion. The observation of membrane bridges in two zygotes that had progressed into mitosis suggests that prm3 nuclei are not defective for all types of ER/NE fusion. The possibility that prm3 mutants are specifically defective for the mating-specific NE fusion event is supported by the enrichment of Prm3p near the SPB dependent on Kar5p (Shen et al., 2009). The specific function of Prm3p in the initiation of membrane fusion remains unclear. Lacking the characteristic motifs of a SNARE protein, it seems unlikely that Prm3p directly catalyzes fusion. Possibly, Prm3p facilitates NE fusion by recruiting SNARE proteins in concert with the localization of Kar5p.
Although prm3 mutants do not initiate NE fusion, the two nuclei became connected by fibers associated with the SPBs (spindle plaque and half-bridge). Similar fibers were seen connecting the SPBs in kar5 and kar8 zygotes (e.g., Figure 3D), showing that they are not unique to prm3. The identity of the fibers is not readily apparent, but their association with the SPB suggests that they may be derived from the amorphous “pericentriolar” material (PCM) found near microtubule-organizing centers. The identities of proteins in the yeast PCM are not well established, but include components of the microtubule-nucleating γ-tubulin complex (Jaspersen and Winey, 2004). Interestingly, the γ-tubulin complexes become localized to the half-bridge during mating (Pereira et al., 1999), where they nucleate the cytoplasmic microtubules required for nuclear congression. Thus the fibers may be derived from microtubule-associated proteins, although their length (up to 200 nm) would seemingly preclude them being derived directly from the γ-tubulin complex. Similarly, the fibers may contain protein components also found in the half-bridge. The half-bridge includes Sfi1p, Cdc31p, Kar1p, and Mps3p (Jaspersen and Winey, 2004). Although Cdc31p and Sfi1p are known to form fibrous complexes, these are still significantly smaller (60 nm) than the observed fibers (Li et al., 2006). Yet another possible source of the fibers may be high molecular weight “tethering” proteins associated with the initial docking steps in membrane fusion (Whyte and Munro, 2002). In either case, the fibers may help bring the membranes close together, facilitating SNARE-dependent outer membrane fusion. Furthermore, the linking of the SPBs may facilitate their eventual spatial registration and fusion.
The phenotype of the kar5 mutants was more pleiotropic than other mutants. In half of the zygotes, no membranous bridges between the nuclei were observed, suggesting that Kar5p is required to initiate membrane fusion. In zygotes with membrane bridges, their narrow cross-section suggests that Kar5p plays a role in the expansion of the initial outer membrane fusion event. Possibly, these two functions are not distinct, and in the absence of bridge expansion, the initial fusion event is reversible.
The kar5 zygotes exhibited an increased spacing between the inner and outer nuclear membranes in regions where the two nuclei were pulled together. This observation suggests that Kar5p may play a role in the linkage of the two membranes in the vicinity of the SPBs during mating. As the only integral membrane protein that is strongly required for nuclear fusion, Kar5p is an excellent candidate for this role. Kar5p contains two carboxy-terminal transmembrane domains and a lumenal domain including regions predicted to form a coiled coil (Beh et al., 1997). Thus by interacting with itself or with another unidentified protein, Kar5p could link the inner and outer nuclear membranes. Coupling of the two membranes might then facilitate the subsequent fusion of the two inner NEs. A more direct role for Kar5p in inner NE fusion cannot be determined from these experiments because of the earlier prior in bridge formation and expansion.
Another candidate protein for playing a role in linking the inner and outer NEs is Mps3p, a member of the SUN family of proteins (Tzur et al., 2006). In metazoan cells SUN proteins are thought to form diverse bridging connections between the outer and inner NEs and between the NE and nuclear-associated cytoskeletal elements (Tzur et al., 2006). In budding yeast, Mps3p also connects the NE and the SPB (Jaspersen et al., 2006). Mps3p (Nep98p) has been shown to interact with Kar8p by yeast two-hybrid analysis and play a role in nuclear fusion (Nishikawa et al., 2003).
The kar2 and kar8 zygotes exhibited high rates of outer NE fusion, but failed to perform inner NE fusion, suggesting that these two proteins act specifically at this second step. Similarities in their phenotype were expected because Kar2p is the DnaK/Hsp70 chaperone resident in the lumen of ER/NE and Kar8p is a DnaJ homolog with Kar2p interacts (Nishikawa and Endo, 1997; Brizzio et al., 1999). A significant difference between the two mutants concerned their rates of lumenal GFP transfer. The very slow rate of transfer in the kar2 zygotes suggests that diffusion through the bridge connecting the nuclei was impeded. FRAP experiments showed that protein movement within the NE lumen proper was not affected. Therefore it is likely that movement is impeded specifically near or within the membrane bridge. As a chaperone, Kar2p may play a role in the disassembly of proteins present at the site of outer NE fusion, some of which may have facilitated fusion and whose disassembly would be needed for progression to inner NE fusion. In contrast, most of the kar8 zygotes did allow significant lumenal protein transfer, suggesting that reduced diffusion across the bridge is not the basis of the mutant defect. Instead, it suggests that Kar8p, presumably in association with Kar2p, is required for the subsequent initiation of inner NE fusion.
The different rates of lumenal protein transfer of the in kar2 and kar8 mutants explain a puzzling difference in their genetic behavior. Although they are all recessive, kar2 mutations are unilateral (only one parent must be mutant to manifest the nuclear fusion defect), whereas kar8 mutations are bilateral (both parents must be mutant). The behavior of the kar8 mutant in matings to wild type can be explained by Kar8p in the lumen of the wild-type NE being able to diffuse into the mutant nucleus and thereby help facilitate inner NE fusion. In contrast, diffusion from the wild-type nuclear lumen would be blocked in kar2 zygotes, preventing complementation.
The initial slow phase of transfer observed in wild-type zygotes may be due to an initial membrane fusion event before dilation of the bridge or transfer via recycling between the ER and Golgi compartments or some combination of both processes. In support of the recycling hypothesis, we found that the rate of transfer into peripheral areas of the ER occurred at the same rate as the initial phase of transfer into the NE (data not shown). This suggests that, before outer NE fusion, transfer into the lumens of the peripheral ER and NE occurs by the same mechanism. The absence of membrane bridges in prm3Δ zygotes, taken together with the similarity between the prm3Δ transfer rate and the wild-type initial phase, supports the hypothesis that the slow initial phase is due to recycling.
If the initial slow phase of transfer is due to the initial NE membrane fusion events, then preventing the nuclei from coming into close proximity should completely block transfer. In contrast, if the initial slow phase is due to recycling, then blocking congression may reduce, but not eliminate, transfer by decreasing the probability that recycling vesicles containing 3X-GFP-HDEL fuse with a more distant recipient nucleus. Blocking congression in otherwise wild-type zygotes with the kar1-1 mutation eliminated transition to the rapid phase of transfer and reduced the overall rate of transfer to 25–30% of the slow initial phase. Similarly, introduction of kar1-1 into prm3Δ zygotes, in which no membrane bridges were observed, reduced the rate of transfer by the same degree as in wild-type zygotes. Thus the effect of kar1-1 on the initial slow phase of transfer is most consistent with an effect on recycling because of decreased nuclear proximity.
Assuming that the slow initial phase is due to recycling, then the rate of transfer in zygotes without NE bridges should be the same in all mutants and be similar to the wild-type initial transfer rate. However, a subset of both kar5 and kar8 zygotes exhibited rates substantially slower than the wild-type initial transfer rate. On the basis of the tomography data, we interpret these as being the zygotes without membrane bridges. We speculate that the kar5 and kar8 mutants may also affect recycling, independent of the nuclear fusion defect. Support for this hypothesis comes from the observation that the rate of transfer in kar1 kar8 mutant zygotes, in which congression is blocked and therefore transfer should only proceed by recycling, was significantly slower than the kar1 prm3 zygotes (p = 0.016).
Previous work using an in vitro ER membrane fusion assay suggested that Kar2p, Kar5p, and Kar8p are required for outer nuclear membrane fusion during karyogamy (Kurihara et al., 1994; Latterich and Schekman, 1994). The ET results cast some doubt on this conclusion. Because the in vitro assays depended on transfer of lumenal components between ER-derived membranes, one explanation for the in vitro defect may be that the assay was measuring aspects of the recycling pathway.
On the basis of these results and analysis, we propose a model for the nuclear fusion pathway in which Prm3p is involved in initiation of outer membrane fusion, Kar5p facilitates the progression of outer membrane fusion, and both Kar2p and Kar8p facilitate inner membrane fusion (Figure 8). Future studies will address how the Kar proteins work together to control NE fusion, and how the distinct events of outer and inner NE fusion are coordinated.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Winey, M. Morphew, and E. O'Toole for help with the tomography studies. We especially thank Richard McIntosh for his continued support and encouragement. We also thank to N. Erdeniz (Princeton University), S. Clark (Princeton University), R. Tsien (University of California, San Diego), and M. Longtine (Oklahoma State University) for constructs and D. Welsh for computer support. M.D.R. was supported by National Institutes of Health (NIH) Grant R01 GM37739 during this project, and P.M. was supported by NIH grants T32 CA009528 and F32 GM069288. The electron tomography was supported by a grant from the National Center for Research Resources to the Laboratory for 3D Electron Microscopy of Cells (RR 00592).
Abbreviations used:
- NE
nuclear envelope
- ET
electron tomography.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-02-0163) on July 1, 2009.
REFERENCES
- Amberg D. C., Burke D. J., Strathern J. N. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- Bardwell L. A walk-through of the yeast mating pheromone response pathway. Peptides. 2005;26:339–350. doi: 10.1016/j.peptides.2004.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beh C. T., Brizzio V., Rose M. D. KAR5 encodes a novel pheromone-inducible protein required for homotypic nuclear fusion. J. Cell Biol. 1997;139:1063–1076. doi: 10.1083/jcb.139.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz T., Egan B., Silver P. A., Hofmann K., Lithgow T. Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:8219–8223. doi: 10.1074/jbc.M212725200. [DOI] [PubMed] [Google Scholar]
- Brizzio V., Khalfan W., Huddler D., Beh C. T., Andersen S.S.L., Latterich M., Rose M. D. Genetic interactions between KAR7/SEC71, KAR8/JEM1, KAR5, KAR2 during nuclear fusion. Mol. Biol. Cell. 1999;10:609–626. doi: 10.1091/mbc.10.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E. A. Pheromone response, mating and cell biology. Curr. Opin. Microbiol. 2000;3:573–581. doi: 10.1016/s1369-5274(00)00143-0. [DOI] [PubMed] [Google Scholar]
- Erdman S., Lin L., Malczynski M., Snyder M. Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol. 1998;140:461–483. doi: 10.1083/jcb.140.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie A. E., Rose M. D. Assays of cell and nuclear fusion. Methods Enzymol. 2002;351:477–498. doi: 10.1016/s0076-6879(02)51866-8. [DOI] [PubMed] [Google Scholar]
- Heiman M. G., Walter P. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J. Cell Biol. 2000;151:719–730. doi: 10.1083/jcb.151.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S. L., Martin A. E., Glazko G., Giddings T. H., Jr., Morgan G., Mushegian A., Winey M. The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J. Cell Biol. 2006;174:665–675. doi: 10.1083/jcb.200601062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S. L., Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Kabani M., Kelley S. S., Morrow M. W., Montgomery D. L., Sivendran R., Rose M. D., Gierasch L. M., Brodsky J. L. Dependence of endoplasmic reticulum-associated degradation on the peptide binding domain and concentration of BiP. Mol. Biol. Cell. 2003;14:3437–3448. doi: 10.1091/mbc.E02-12-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J. R., Mastronarde D. N., McIntosh J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kurihara L. J., Beh C. T., Latterich M., Schekman R., Rose M. D. Nuclear congression and membrane fusion: two distinct events in the yeast karyogamy pathway. J. Cell Biol. 1994;126:911–923. doi: 10.1083/jcb.126.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latterich M., Schekman R. The karyogamy gene KAR2 and novel proteins are required for ER-membrane fusion. Cell. 1994;78:87–98. doi: 10.1016/0092-8674(94)90575-4. [DOI] [PubMed] [Google Scholar]
- Li S., Sandercock A. M., Conduit P., Robinson C. V., Williams R. L., Kilmartin J. V. Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication. J. Cell Biol. 2006;173:867–877. doi: 10.1083/jcb.200603153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Marsh L., Rose M. D. The pathway of cell and nuclear fusion during mating in S. cerevisiae. In: Pringle J. R., Jones E. W., editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology. Vol. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 827–888. [Google Scholar]
- Mastronarde D. N. Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- Melloy P., Shen S., White E., McIntosh J. R., Rose M. D. Nuclear fusion during yeast mating occurs by a three-step pathway. J. Cell Biol. 2007;179:659–670. doi: 10.1083/jcb.200706151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D.T.W., Walter P. ER Membrane protein complex required for nuclear fusion. J. Cell Biol. 1996;132:499–509. doi: 10.1083/jcb.132.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S., Endo T. The yeast JEM1p is a DnaJ-like protein of the endoplasmic reticulum membrane required for nuclear fusion. J. Biol. Chem. 1997;272:12889–12892. doi: 10.1074/jbc.272.20.12889. [DOI] [PubMed] [Google Scholar]
- Nishikawa S., Terazawa Y., Nakayama T., Hirata A., Makio T., Endo T. Nep98p is a component of the yeast spindle pole body and essential for nuclear division and fusion. J. Biol. Chem. 2003;278:9938–9943. doi: 10.1074/jbc.M210934200. [DOI] [PubMed] [Google Scholar]
- Nishikawa S. I., Fewell S. W., Kato Y., Brodsky J. L., Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 2001;153:1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O' Toole E. T., Winey M., McIntosh J. R., Mastronarde D. N. Electron tomography of yeast cells. Methods Enzymol. 2002;351:81–95. doi: 10.1016/s0076-6879(02)51842-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Grueneberg U., Knop M., Schiebel E. Interaction of the yeast gamma-tubulin complex-binding protein Spc72p with Kar1p is essential for microtubule function during karyogamy. EMBO J. 1999;18:4180–4195. doi: 10.1093/emboj/18.15.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts C. J., et al. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Misra L. M., Vogel J. P. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Shen S., Tobery C. E., Rose M. D. Prm3p is a pheromone-induced peripheral nuclear envelope protein required for yeast nuclear fusion. Mol. Biol. Cell. 2009;20:2438–2450. doi: 10.1091/mbc.E08-10-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur Y. B., Wilson K. L., Gruenbaum Y. SUN-domain proteins: ‘Velcro’ that links the nucleoskeleton to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2006;7:782–788. doi: 10.1038/nrm2003. [DOI] [PubMed] [Google Scholar]
- Vogel J. P., Misra L. M., Rose M. D. Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J. Cell Biol. 1990;110:1885–1895. doi: 10.1083/jcb.110.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P., Bursac D., Law Y. C., Cyr D., Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte J. R., Munro S. Vesicle tethering complexes in membrane traffic. J. Cell Sci. 2002;115:2627–2637. doi: 10.1242/jcs.115.13.2627. [DOI] [PubMed] [Google Scholar]
- Yoder T. J., McElwain M. A., Francis S. E., Bagley J., Muller E.G.D., Pak B., O'Toole E. T., Winey M., Davis T. N. Analysis of a spindle pole body mutant reveals a defect in biorientation and illuminates spindle forces. Mol. Biol. Cell. 2005;16:141–152. doi: 10.1091/mbc.E04-08-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.