Abstract
Insulin-like growth factor (IGF)-I is a critical protein for cell development and growth. Alternative splicing of the igf1 gene gives rise to multiple isoforms. In rodents, proIGF-IA and proIGF-IB have different carboxy-terminal extensions called the E-peptides (EA and EB) and upon further posttranslational processing, produce the identical mature IGF-I protein. Rodent EB has been reported to have mitogenic and motogenic effects independent of IGF-I. However, effects of EA or EB on mature IGF-I, or whether proIGF-IA and proIGF-IB have different properties, have not been addressed. To determine whether the presence of EA or EB affected the distribution and stability of mature IGF-I protein, transient transfections of cDNAs encoding murine IGF-IA, IGF-IB, and mature IGF-I were performed in C2C12 cells, a skeletal muscle cell line. IGF-I secretion was measured by enzyme-linked immunosorbent assay of the media, and did not differ between expression of proIGF-IA, proIGF-IB, or mature IGF-I expression. Next, epitope-tagged constructs were transfected to determine cellular distribution of IGF-I, EA, and EB in the cells throughout the culture. IGF-I was detected in significantly fewer nontransfected cells in cultures transfected with mature IGF-I compared with transfection of proIGF-IA or proIGF-IB. These results demonstrate that EA and EB are not required for IGF-I secretion but that they increase cell entry of IGF-I from the media. This study provides evidence that the EA and EB may modulate IGF-I in addition to having independent activity.
INTRODUCTION
Insulin-like growth factor (IGF)-I is a critical protein for development and growth in many tissues. In skeletal muscle, IGF-I not only coordinates proliferation and differentiation of myoblasts during development but also enhances regeneration, protein synthesis, and increased mass (Florini et al., 1996). Its ability to promote skeletal muscle hypertrophy has been demonstrated by several methods, including transgenic overexpression, viral gene delivery, and systemic administration of the protein (Coleman et al., 1995; Adams and McCue, 1998; Barton-Davis et al., 1998; Musaro et al., 2001; Barton, 2006b). IGF-I mediates its effects by binding to the IGF-I receptor (IGF-IR) found on the cell surface, activating the inherent tyrosine kinase activity of the receptor, and enabling internalization of the receptor–ligand complex to instigate signaling cascades, and to ultimately affect gene expression and protein synthesis (Laviola et al., 2007; Romanelli et al., 2007; Monami et al., 2008). IGF-IR activation seems to be independent of the isoform from which IGF-I was produced.
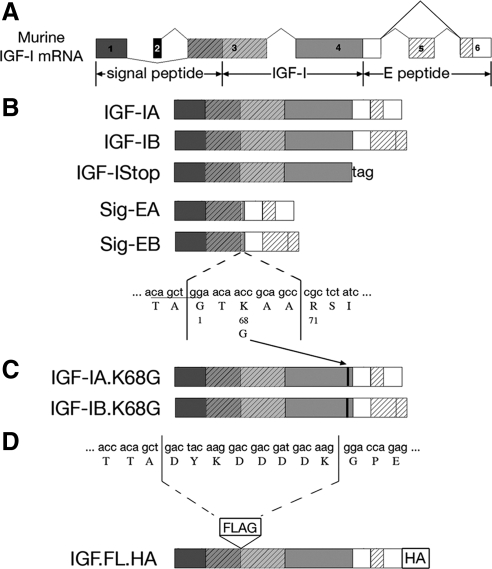
Several IGF-I isoforms are produced by alternative splicing of the igf1 gene (reviewed in Adamo et al., 1993; Lund, 1998; Barton, 2006a). The translated propeptides for all isoforms contain the identical sequence for mature IGF-I protein, but the C-terminal portions of each isoform, called E-peptides, are divergent (Figure 1A). In rodents, most IGF-I transcripts exclude exon 5 and splice exon 4 directly to exon 6, and they are defined as class A. The inclusion of exon 5, which is 52 nucleotides, causes a frame shift in the open reading frame of the subsequent exon and gives rise to a premature stop within exon 6. This splice form, class B, only occurs in up to 10% of the igf1 transcripts. In humans, exon 5 is significantly longer (515 nucleotides) (Rotwein et al., 1986), and unique splice forms occur. Human class C IGF-I is produced from an internal splice site within exon 5 that joins 49 nucleotides of exon 5 with exon 6. This insertion, like in rodent class B, causes a frame shift and premature termination in exon 6. However, human class B IGF-I, contains only exon 5, resulting in an E-peptide extension which, to date, also has been observed only in nonhuman primates (Wallis, 2009). The different E-peptides share only up to 50% sequence homology at the amino acid level.
Figure 1.
Schematic of cDNA constructs generated for this study. (A) Exons and alternative splicing in murine Igf1. Alternative splicing of exons 1 or 2 to exon 3 produces the signal peptide. The mature IGF-I protein is invariant and encoded by exons 3 and 4. The C-terminal E-peptides are encoded by exons 4, 5, and 6, and alternative splicing of exon 5 inclusion gives rise to divergent E-peptides. (B) Constructs that retain or exclude mature IGF-I or the E-peptides. IGF-IA retains mature IGF-I and the EA-Peptide. IGF-IB retains mature IGF-I and the EB-peptide. IGF-IStop retains mature IGF-I but excludes both E-peptides. SigEA and SigEB exclude mature IGF-I but retain the EA- and EB-peptides, respectively. (C) Mutagenesis of mature IGF-I/E-peptide processing site. Lysine 68 was changed to glycine to block the primary cleavage site between the mature IGF-I protein and the E-peptides. (D) Addition of epitope tags to IGF-I constructs. A FLAG tag was inserted between the signal peptide and mature IGF-I immediately after the processing site. In separate constructs, an HA tag was added to the C terminus of the EA- or EB-peptide.
Previous studies have separated the activity of the E-peptides from those of mature IGF-I by the addition of neutralizing antibodies that block IGF-IR activation and have clearly demonstrated E-peptide bioactivity that is independent of mature IGF-I. Using this approach, a unique portion of the human class B E-peptide (IBE1) was shown to cause concentration-dependent cell growth in human bronchial epithelial cells (Siegfried et al., 1992) and in neuroblastoma cells (Kuo and Chen, 2002). In addition, increased proliferation and migration of myoblasts by the human class C/rodent class B E-peptide (human EC/rodent EB) also has been observed (Yang and Goldspink, 2002; Mills et al., 2007). To date, no biological activity has been ascribed to class A E-peptide, which is the product of the predominant isoform.
How the E-peptides affect the actions of mature IGF-I has been a matter of debate. Increased muscle expression of the human IGF-I C/rodent IGF-IB occurs in response to eccentric exercise or damage (Yang et al., 1997; Hameed et al., 2003), and it has been postulated that this isoform is a key component of the repair process through the direct actions of the E-peptide. Comparison of the two murine IGF-I isoforms by viral gene transfer revealed that they were equivalent in promoting muscle hypertrophy in young growing mice (Barton, 2006b). However, in the same study, tissue content of total mature IGF-I after viral delivery of IGF-IB was consistently higher, suggesting that production and stability of IGF-I may be isoform specific. To address the possibility of indirect effects of the E-peptides via mature IGF-I stability, an in vitro system was designed to monitor the production and localization of IGF-I and the E-peptides. The goal of this study was to determine whether the production, distribution, or stability of mature IGF-I differed between IGF-IA and IGF-IB.
MATERIALS AND METHODS
IGF-I Constructs
The cDNA for murine Igf-1A and Igf-1B (GenBank AY878192 and AY878193, respectively) formed the basis for all constructs. IGF-IA and IGF-IB included the sequence to encode the class I signal peptide, IGF-I, and the respective E-peptide. IGF-IStop lacked E-peptide sequences, and a stop codon was inserted at the end of the mature IGF-I. SigEA and SigEB retained the signal peptide and the respective E-peptides in the absence of IGF-I. This was achieved by blunt end ligation of glycine 1 and threonine 67 of the mature IGF-I protein. These constructs possessed the recognition sites for processing between the signal peptide and mature IGF-I, as well as between mature IGF-I and the E-peptide. Site-directed mutagenesis (QuikChange II; Stratagene, La Jolla, CA) was used to mutate lysine 68 to glycine blocking the primary cleavage site between IGF-I and the E-peptides (Duguay et al., 1995) (IGF-IAK68G and IGF-IBK68G).
Fusion constructs including epitope tags and the IGF-I sequences above also were generated to enable indirect detection of the transfected gene product by immunocytochemistry. A FLAG epitope tag was inserted between the signal peptide and the IGF-I protein immediately after the processing site. This strategy has been used successfully in previous studies to monitor the processing of IGF-I (Duguay et al., 1997; Wilson et al., 2001). A hemagglutinin (HA) epitope tag was placed at the C terminus of each construct containing an E-peptide, followed by a stop codon.
All cDNA constructs were inserted into the NheI and XhoI restriction sites of pCMV.IRES.eGFP (Clontech, Mountain View, CA) for transient transfection. Schematic details each construct used in this study are shown in Figure 1.
Muscle Cell Culture
C2C12 cells were plated in growth medium (79% DMEM, 20% fetal bovine serum, 1% l-glutamine, and 0.2 ml of gentamicin) in 4.5 cm2 dishes containing a fluoropolymer film square (ACLAR; Electron Microscopy Sciences, Fort Washington, PA) within the dish. Cells were grown to 80% confluence. Transient transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). For each transfection, cells were incubated with a total of 3 μg of plasmid DNA plus 8 μl of Lipofectamine in 1 ml of Opti-MEM (Invitrogen) for a total of 4 h. Cells were switched into differentiation medium (98% DMEM, 2% horse serum, and 0.2 ml of gentamicin) for 24 h after transfection. Controls included transfection of empty vector (green fluorescent protein [GFP]), Lipofectamine only (Mock), and no transfection (control). Media was removed from the culture dish and stored at −80°C for IGF-I production measurements, and the cells adherent to ACLAR were fixed in 2% paraformaldehyde for immunocytochemistry. Each condition was performed in triplicate.
Validation of Transfection Expression
An additional set of transfections was used to confirm the expression of all constructs. Total RNA was isolated from cultures 24 h after transfection with TriZol (Invitrogen) and treated with RNase-free DNase I (Roche Diagnostics, Indianapolis, IN) (30 mg of RNA incubated with 10 U of DNase at 37°C for 20 min). Then, 1 μg of RNA was reversed transcribed, and the resultant cDNA was subjected to quantitative real-time polymerase chain reaction (qRT-PCR) to detect expression of the IGF-I construct and GFP based on previously published methods (Klein et al., 2000; Barton, 2006b). Oligonucleotides are listed in Table 1. Standard curves were generated for all expression plasmids by qRT-PCR using serial dilutions of the relevant plasmid. Normalization of expression cassettes was performed in two different ways. First, transfection efficiency of each vector was estimated by measuring GFP transcript copy number relative to the housekeeping gene 18s. The comparison of GFP expression among all transfections was normalized to expression of GFP from the empty vector. Second, expression efficiency for IGF-I was determined by measuring transcript copy numbers for GFP and for the IGF-I cDNA of interest based on the standard curves and by comparing the ratio of IGF to GFP for each transfection. Controls included cells without transfection (control), and RNA without reverse transcription.
Table 1.
Oligonucleotides and Band sizes for qRT-PCR
| Gene target | Sense primer (5′-3′) | Antisense primer (5′-3′) | Band (bp) |
|---|---|---|---|
| 18s | ctctgttccgcctagtcctg | aatgagccattcgcagtttc | 151 |
| GFP | gcagtgcttcagccgctac | aagaagatggtgcgctcctg | 96 |
| IGF-IA; IGF-IAK68G | acacctcttctacctggcgctctgc | ctagattctgtaggtcttgtttcctgc | 369 |
| IGF-IB; IGF-IBK68G | acacctcttctacctggcgctctgc | attctgtaggtcttgtttcctgc | 399 |
| IGF-IStop | acacctcttctacctggcgctctgc | ctaggctgcttttgtaggcttcagtg | 265 |
| SigEA | atggggaaaatcagcagccttcc | ctagattctgtaggtcttgtttcctg | 268 |
| SigEB | atggggaaaatcagcagccttcc | ctacttgtgttcttcaaatgtacttcc | 286 |
Detection of IGF-I Protein
Total IGF-I in the media was measured by a commercially available enzyme-linked immunosorbent assay (ELISA) kit (MG100; R&D Systems, Minneapolis, MN). This kit detects total rodent IGF-I and is not affected by IGF-I binding proteins or IGF-II. It does not detect IGF-I in horse serum but can detect endogenous IGF-I production by C2C12 cells. The assay can detect IGF-I at 30–2000 pg/ml, with an intraassay precision of 4.3% and an interassay precision of 5.9%. Data were acquired in duplicate on a microtiter-plate reader (Dynex Technologies, Chantilly, VA) at 450 nm.
The form of the secreted IGF-I also was assessed to determine whether proIGF-I or mature IGF-I peptide was secreted from the transfected cells. Media from FLAG-labeled IGF-I transfections was concentrated by centrifugation in 3000 molecular weight cut-off filters (Ultracel YM-3, Microcon; Millipore, Billerica, MA), and subjected to immunoblotting with an antibody recognizing FLAG (monoclonal antibody [mAb] FLAG M2, catalog no. F1804; Sigma-Aldrich, St. Louis, MO). Detection and analysis of band size was performed with enhanced chemiluminescence and an mm4000 detection system (Eastman Kodak, Rochester, NY).
Immunocytochemistry
An antibody recognizing GFP and conjugated to Alexa 488 (Invitrogen) was used to amplify the GFP signal after fixation. Localization of the IGF-I was achieved with antibodies against FLAG (polyclonal antibody [pAb] FLAG, catalog no. 2368; Cell Signaling Technology, Danvers, MA), and localization of the E-peptides used antibodies against HA (mAb HA-Tag [6E2], catalog no. 2367; Cell Signaling Technology; pAb HA-Tag, catalog no. H6908; Sigma-Aldrich). Secondary antibodies included anti-mouse and anti-rabbit conjugated to Alexa 488 or Alexa 555 (Invitrogen). After staining the cells were covered in aqueous mounting media containing 4,6-diamidino-2-phenylindole (DAPI) (Vectashield Laboratories, Burlingame, CA) and sealed onto a coverslip for visualization on an epifluorescence microscope (DMR; Leica Microsystems, Deerfield, IL).
Image Analysis
For each transfection and staining condition, four nonoverlapping microscopic fields were acquired at 200× using OpenLab software (Improvision, Conventry, United Kingdom). The proportion of GFP-positive cells served as an index of transfection efficiency. The proportion of FLAG positive cells with and without GFP indicated cells harboring IGF-I. The proportion of HA-positive cells with and without GFP indicated those harboring the E-peptides. Images of transfected cells (GFP positive) also were acquired at 630× and processed by nearest neighbor deconvolution by using the same software.
Statistics
One-way analysis of variance (ANOVA) was used for comparisons of all transfection conditions, followed by Tukey's multiple comparison test to determine differences between conditions. Statistical significance was accepted for p < 0.05.
RESULTS
The goal of this study was to determine whether the presence of different E-peptides affected the production, distribution, or stability of the mature IGF-I protein. In addition to expressing IGF-IA and IGF-IB, a series of expression constructs based on the IGF-IA and IGF-IB open reading frames were generated to enable the expression of mature IGF-I in the absence of either EA- or EB-peptides (IGF-IStop) or the expression of either EA- or EB-peptide in the absence of mature IGF-I (SigEA, and SigEB, respectively). In addition, cleavage mutants were generated with the intent of expressing only proIGF-IA and proIGF-IB (IGF-IA.K68G and IGF-IB.K68G, respectively), and inhibiting the ability of mature-IGF-I production.
Transfection and Expression Efficiency
The efficiency of transfection was determined for each construct by the proportion of GFP-positive cells in each dish and by qRT-PCR. The proportion of GFP-positive cells did not differ among the constructs, resulting in a combined transfection efficiency of 12.7 ± 1% (mean ± SD). Transfection efficiency also was determined by measuring the level of GFP expression with respect to a housekeeping gene (18s) and by comparing GFP expression by each plasmid to the empty vector control. As shown in Figure 2A, there was no significant difference in the relative expression of GFP between any of the transfected constructs.
Figure 2.
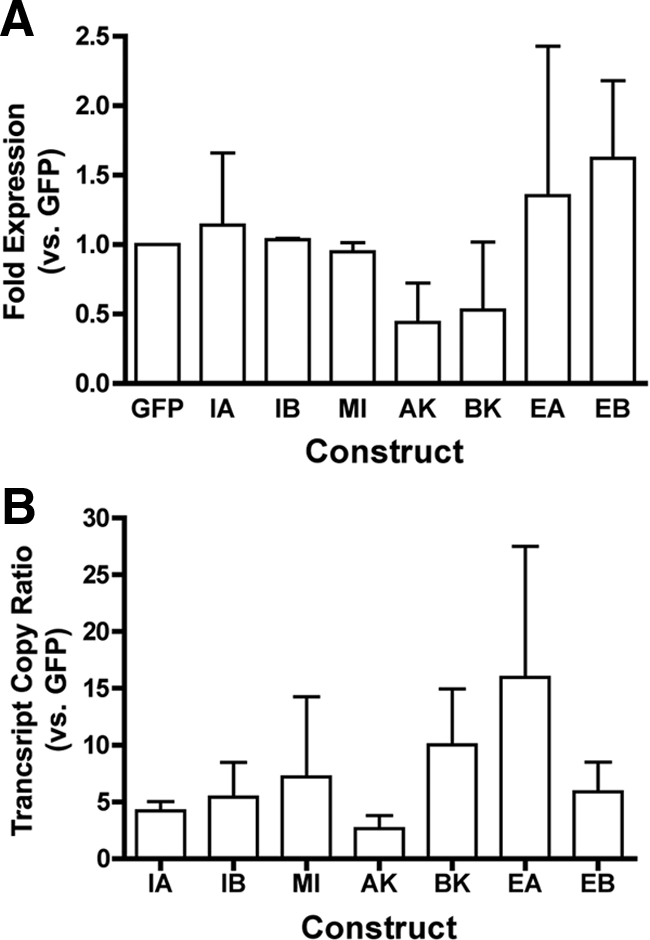
Transfection and expression efficiency for IGF-I constructs. Data are presented as mean and SEs from three independent experiments. (A) Efficiency of transfection was determined in using the relative GFP expression compared with 18s as a housekeeping gene. Data are normalized to empty vector control that expresses only GFP. No statistical difference in transfection efficiency was found among any construct used in the study. (B) Efficiency of expression of the IGF-I cDNA insert for each plasmid was determined by the transcript copy ratio of the upstream IGF-I insert to the downstream GFP insert. Transcript copies were calculated from standard curves generated for each plasmid using the primer pairs listed in Table 1. Constructs GFP (empty vector control), IA (IGF-IA), IB, (IGF-IB), MI (Mature IGF-I/IGF-Istop), AK (IGF-IAK68G), BK (IGF-IBK68G), EA (SigEA), and EB (SigEB).
Validation of expression was achieved by qRT-PCR for each construct by using primers specific to each IGF-I insert. All transfections expressed the insert of interest >3000-fold higher than in controls. The efficiency of expression in each transfection experiment was determined by comparing the calculated transcript copies for each IGF-I cDNA insert to the calculated transcript copies for GFP. As shown in Figure 2B, there was no significant difference in expression efficiency between constructs. Therefore, similar efficiencies of both transfection and expression were observed for all IGF-I constructs.
Secretion of IGF-I
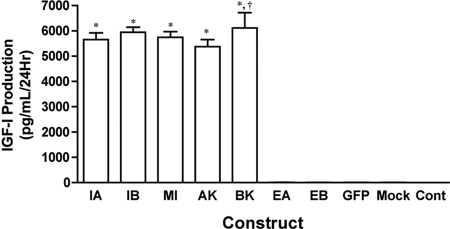
Secretion of IGF-I from cells into the media was measured by ELISA 24 h after transfection (Figure 3). IGF-I secretion was significantly higher from transfected cells than controls when the transfection construct retained the mature IGF-I protein coding sequence. Secretion was not affected by the presence or absence of EA or EB, for IGF-I secretion from cells transfected with the IGF-IA, IGF-IB, IGF-IStop constructs was equivalent. Secretion of IGF-I from cells transfected with the cleavage mutant constructs (IGF-IA.K68G and IGF-IB.K68G) did not differ from IGF-IA or IGF-IB. However, there was more IGF-I secreted from IGF-IB.K68G transfections than from cells transfected with IGF-IA.K68G. Endogenous IGF-I secretion was not altered by transfection agent (mock), the transfection vector (GFP), or the transfection of SigEA or SigEB constructs. Levels of IGF-I produced by these cultures were between 4 and 10 pg/ml and were not visible in the scale in Figure 3. Secretion of IGF-I also was determined in cell transfections of epitope-tagged constructs. The presence of FLAG or HA on the constructs did not affect the secretion of IGF-I from the cells (data not shown).
Figure 3.
IGF-I production is not affected by the presence of the E-peptides. Media content of IGF-I served as an index of IGF-I production after transfection. Production of IGF-I was significantly higher than control cells when the cDNA construct contained the sequence encoding mature IGF-I (IGF-IA, IGF-IB, IGF-IStop, IGF-IA.K68G [AK68G], and IGF-IB.K68G [BK68G]). No change in IGF-I production was afforded by the presence of the E-peptides (IGF-IA or IGF-IB compared with IGF-IStop; SigEA or SigEB compared with control). IGF-IB.K68G had higher IGF-I production than IGF-IA.K68G. The transfection conditions (mock) or the vector alone (GFP) did not affect IGF-I production. Levels of IGF-I production from Control, Mock, GFP, EA, and EB ranged from 4 to 10 pg/ml and are not apparent on the graph. Statistical comparisons from Tukey's multiple comparisons test; *p < 0.05 for comparison to control cultures; †p < 0.05 for comparisons between isoforms. Constructs: IA (IGF-IA), IB, (IGF-IB), MI (Mature IGF-I/IGF-Istop), AK (IGF-IAK68G), BK (IGF-IBK68G), EA (SigEA), EB (SigEB), GFP (empty vector control), Mock (Lipofectamine only), and Cont (no transfection).
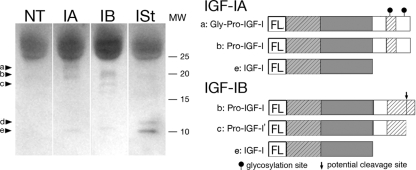
Processing of IGF-I from proIGF-I to mature IGF-I can occur both intracellularly and extracellularly in a number of cell types (Conover et al., 1989, 1993; Duguay et al., 1997; Duguay, 1999; Wilson et al., 2001). To determine the forms of IGF-I that were secreted from transfected C2C12 cells, the media from FLAG-IGF-IA, FLAG-IGF-IB, and FLAG-IGF-IStop transfected cells was subjected to immunoblotting to detect FLAG-labeled IGF-I. When the E-peptides were present in the construct, both proIGF-I and mature IGF-I could be detected in the media (Figure 4). FLAG-IGF-IA transfected cultures had an additional higher molecular weight band (Gly-ProIGF-I) consistent with glycosylation of the EA-peptide (Bach et al., 1990; Duguay et al., 1995; Wilson et al., 2001). In FLAG-IGF-IB–transfected cultures, an additional band that was smaller than proIGF-I was evident (band c, ProIGF-I'). In FLAG-IGF-Istop transfected cultures, the mature IGF-I band was apparent, as well as one higher molecular weight band that was not evident in the IGF-IA and IGF-IB lanes.
Figure 4.
Form of secreted IGF-I from C2C12 cells after transfection. Immunoblotting of concentrated media with anti-FLAG was used to distinguish between proIGF-I and mature IGF-I in the culture media (left). Both pro- (bands a and b) and fully processed (mature, band e) IGF-I were detected when IGF-IA (IA) or IGF-IB (IB) constructs were transfected. IGF-IA lanes had a higher molecular weight band (a) consistent with glycosylated proIGF-IA, shown in the right panel. IGF-IB lanes had an additional lower molecular weight band (c) that could result from protease cleavage within the EB-peptide (right). Mature IGF-I (e) could be produced by IGF-IStop (ISt), and this lane serves as a control for the size of secreted IGF-I. However, a higher molecular weight band occurred (band d), which was not evident in the IGF-IA and IGF-IB lanes. NT, media from nontransfected cultures.
Cellular Distribution of IGF-I and E-Peptides
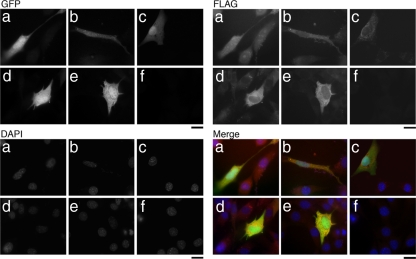
Localization of IGF-I and the E-peptides was assessed by immunocytochemistry of the FLAG and HA epitope tags 24 h after transfection (Figures 5 and 6). All GFP-positive cells were positive for FLAG or for HA when these epitope tags were in the cDNA constructs. GFP was found in the cytoplasm and nucleus of positively transfected cells. FLAG and HA staining in the GFP-positive cells was concentrated in the perinuclear region but also could be detected throughout the cell. Expression of the cleavage mutant constructs (IGF-IAK68G and IGF-IBK68G) altered cell shape (Figures 5, d and e, and 6, e and f). First, these cells had more cytoplasmic extensions than in other conditions for both FLAG- and HA-labeled constructs. Second, FLAG and HA staining in these cells seemed restricted to the perinuclear regions in contrast to the more widespread distribution found in cells transfected with the other constructs. FLAG-tagged IGF-IA, IGF-IB, and IGF-IStop had similar localization patterns in the GFP-positive cells (Figure 5, a–c, respectively). HA-tagged IGF-IA, IGF-IB, SigEA, and SigEB also had similar staining patterns in the GFP-positive cells (Figure 6, a–d). The pattern of HA staining was independent of the IGF isoform that had been transfected.
Figure 5.
Cellular distribution of FLAG epitope-tagged IGF-I constructs FLAG-IGF-IA (a), FLAG-IGF-IB (b), FLAG-IGF-Istop (c), FLAG-IGF-IAK68G (d), FLAG-IGF-IBK68G (e), and no transfection (f). GFP serves as an indicator of positive transfection and is found in the cytoplasm and nucleus of all transfected cells. FLAG is detected in both transfected and nontransfected cells. It is found throughout the cytoplasm and concentrated in the perinuclear regions in transfected and nontransfected cells. Cleavage mutant constructs (d and e) have altered cell shape and lower intensity FLAG staining in the multiple cytoplasmic extensions. DAPI staining identifies the number of nuclei within each field. The merged images are pseudocolored, with GFP as green, FLAG as red, and DAPI as blue. Bar, 10 μm.
Figure 6.
Cellular distribution of HA epitope-tagged IGF-I constructs HA-IGF-IA (a), HA-IGF-IB (b), HA-SigEA (c), HA-SigEB (d), HA-IGF-IAK68G (e), and HA-IGF-IBK68G (f). GFP serves as an indicator of positive transfection and is found in the cytoplasm and nucleus of all transfected cells, as in Figure 5. HA is detected in both transfected and nontransfected cells. It is found throughout the cytoplasm and concentrated in the perinuclear regions in transfected and nontransfected cells. Cleavage mutant constructs (e and f) have altered cell shape and little detectable HA staining in the multiple cytoplasmic extensions. DAPI staining identifies the number of nuclei within each field. The merged images are pseudocolored, with GFP as green, HA as red, and DAPI as blue. Bar, 10 μm.
GFP-negative cells that were FLAG or HA positive served as indicators of internalization of IGF-I or E-peptide, respectively. We took advantage of the mixed population of transfected and nontransfected cells within each culture dish to evaluate cell entry of FLAG as an indicator of IGF-I entry and HA as an indicator of EA or EB entry. Within the GFP-negative cells of each transfection experiment, the percentage of FLAG- or HA-positive cells was quantified (Figure 7).
Figure 7.
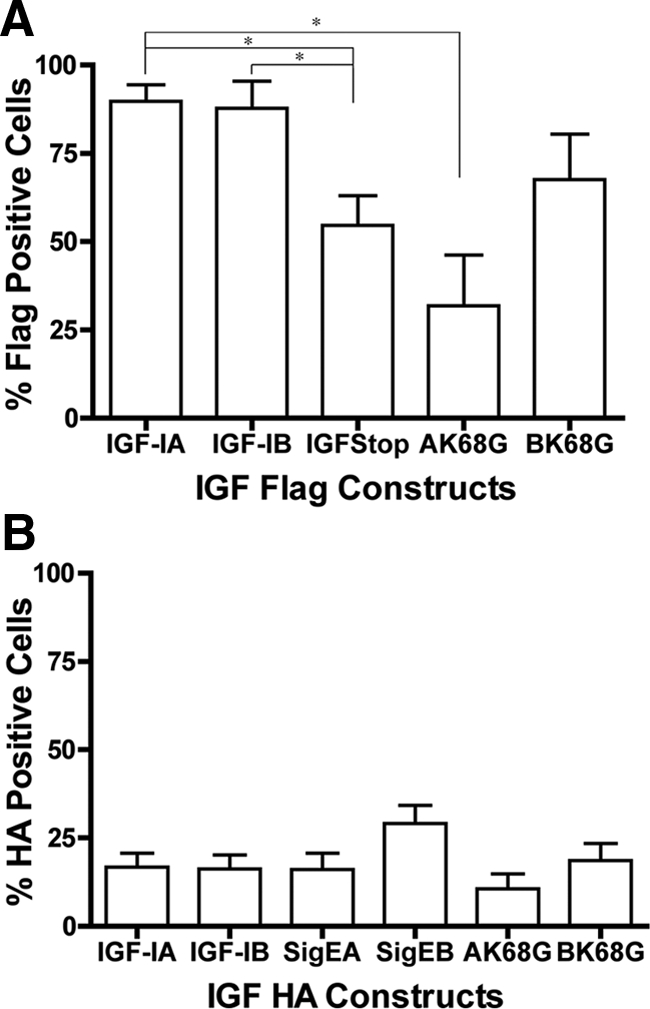
Proportion of C2C12 cells that internalize epitope tags after transfection. (A) FLAG uptake is dependent upon the transfected IGF-I construct. There was no difference in the proportion of FLAG-positive cells after transfection of FLAG-IGF-IA or FLAG-IGF-IB. Expression of mature IGF-I (FLAG-IGF-IStop) significantly reduced the proportion of FLAG-positive cells. Mutation of the primary cleavage site between mature IGF and the EA-peptide in IGF-IA (AK68G) also resulted in a significant decrease in FLAG-positive cells. (B) HA uptake is independent of the transfected IGF construct. There was no significant difference in the proportion of HA-positive cells in the presence or absence of the sequence encoding mature IGF. Mutation of the primary cleavage sites between IGF and the E-peptide (AK68G, BK68G) did not affect the proportion of HA-positive cells. Comparisons by one-way ANOVA followed by Tukey's multiple comparison test.
Most of the cells in FLAG-IGF-IA or FLAG-IGF-IB transfections were FLAG positive, and there was no statistical difference in FLAG (IGF-I) uptake between these isoforms (Figure 7A). However, FLAG-positive cells in FLAG-IGF-IStop-transfected cultures were significantly lower than the constructs retaining the E-peptides (IGF-IA and IGF-IB). Therefore, presence of the E-peptides in the cDNA construct altered the proportion of FLAG-positive cells. Transfection of cells with the cleavage mutant construct FLAG-IGF-IAK68G resulted in a significant decrease in FLAG-positive cells compared with FLAG-IGF-IA. There was no statistical difference in the proportion of FLAG-positive cells between FLAG-IGF-IB and FLAG- IGF-IBK68G. Therefore, inhibition of the native cleavage site between mature IGF-I and the EA-peptide seemed to be important for normal uptake of IGF-I into neighboring cells. HA was detected in a lower proportion of nontransfected cells than FLAG (Figure 7B). There were no statistical differences in the percentage of HA-positive cells among all of the HA epitope-tagged constructs. Therefore, E-peptide internalization seems independent of IGF-I.
DISCUSSION
This study used transient transfection of C2C12 cells to determine whether the presence of the carboxy-terminal extensions in proIGF-I could affect the actions of mature IGF-I. We found that IGF-I expression, production, and secretion are independent of the EA and EB. However, using an indirect method of detection, we found diminished uptake of IGF-I into neighboring cells occurs when only mature IGF-I is expressed. Uptake of EA and EB occurs in a small proportion of neighboring cells, and in contrast to IGF-I uptake, this process does not seem to be dependent upon the presence of mature IGF-I. After secretion from the transfected cells, both proIGF-I and mature IGF-I occur in the media, confirming that a proportion of IGF-I is secreted with an E-peptide attached. However, visualization of epitope-tagged IGF-I, EA, and EB show that these elements are found in proportions of cells, suggesting that they act independently upon cell entry.
Bioactivity of the E-Peptides
Significant research effort from many different groups has established that the E-peptides, specifically rodent EB and human EB, have bioactivity that is independent of IGF-I receptor activation, which include E-peptide effects on proliferation and cell migration (Siegfried et al., 1992; Kuo and Chen, 2002; Yang and Goldspink, 2002; Mills et al., 2007). This has provided an important conceptual shift in identifying the functions of igf1 gene products that includes not only mature IGF-I but also multiple E-peptides that are cleaved from proIGF-I. By tracking the fate of IGF-I, EA, and EB in culture, we have found an additional property of the E-peptides, where they enhance the uptake of IGF-I into cells.
Both EA- and EB-peptides were equivalent in potentiating the uptake of IGF-I into neighboring cells. This suggests that proIGF-I, regardless of isoform, may help to stabilize IGF-I in the media, prevent binding protein interaction, or enhance ligand-receptor binding. At this point, the mechanism is unknown, but it seems to occur extracellularly. Neither EA nor EB affects IGF secretion, so the earliest possible step for modulation occurs in the media. EA and EB could act as chaperones for cell entry of an IGF-I subpopulation either as the proIGF-I species or associate with mature IGF-I after cleavage. However, because the E-peptides enter neighboring cells in the same proportion regardless of the presence of IGF-I (Figure 7B), it is unlikely that E-peptide serves as a cofactor for IGF-I. An alternative explanation is that the E-peptides aid in the release of IGF-I from IGF-I binding proteins, either directly or indirectly, enabling the mature IGF-I ligand to bind to its receptor. It will require further testing to determine how the E-peptides modulate IGF-I uptake.
IGF-I Can Be Secreted as Mature IGF-I and proIGF-I
Secretion of IGF-I from the transfected cells seemed to be independent of the isoform expressed. Previous studies have investigated the secretion products after IGF-IA expression (Conover et al., 1989, 1993; Duguay et al., 1997; Duguay, 1999; Wilson et al., 2001). Processing of IGF-IB has not been studied to the same extent as the dominant isoform. Results from the media show that both pro- and mature IGF-I are present similar to IGF-IA (Figure 4). However, there is an additional lower molecular weight band found in the media of IGF-IB–treated cells (proIGF-I'), suggesting that there is a second cleavage site within the EB-peptide. Indeed, a consensus sequence for protease cleavage exists in the unique portion of rodent IGF-IB (Arg-Arg-Arg-Lys) (Barr, 1991), which could result in the smaller proIGF-I band. Mutation of the primary cleavage site in IGF-IB (K86G) did not affect IGF-I production or uptake, unlike the same mutation in IGF-IA. Previous identification of post-translational processing of IGF-IA demonstrated several tandem cleavage sites between IGF-I and the E-peptide (Duguay et al., 1997), and when the primary site was eliminated by mutagenesis, cleavage occurred downstream at subsequent intact sites. These sites are conserved in all IGF-I isoforms, and the redundancy in protease targets suggests that separation of IGF-I from the E-peptide is critical for function of mature IGF-I. However, why IGF-IB, but not IGF-IA, could withstand mutation of the first cleavage site in our cell uptake assay is not clear. One possible explanation is that the posttranslational processing of IGF-IA and IGF-IB differ due to the presence of glycosylation sites on the EA-peptide (Bach et al., 1990; Duguay et al., 1995; Wilson et al., 2001) and IGF-IA may be more sensitive to mutations, but this remains to be tested.
Our quantification method for IGF-I secretion is based on detection by ELISA, which is specific for rodent IGF-I, and the measurement is independent of IGF binding protein sites. At this point, it has not been directly determined if the ELISA is equally sensitive for mature IGF-I and proIGF-I, which means we may have underestimated the level of IGF-I secretion from many of the transfected constructs. In related preliminary studies, mutations of all sites directly involved with cleavage between mature IGF-I and the EB peptide have been generated, and the secreted product is still detectable by the ELISA assay (unpublished observations). This suggests that proIGF-I can be measured with this method, but further testing is needed to confirm the sensitivity of our assay.
Identification of Cellular Localization of Mature IGF-I, EA, and EB
Our approach for tracking the IGF-I and E-peptides produced by transient transfection is based on previously published methods (Duguay et al., 1995; Wilson et al., 2001). In the former studies, it was demonstrated that the FLAG epitope is retained with mature IGF-I after cleavage from the signal peptide and serves as highly sensitive marker to monitor protein processing and localization of IGF-I. Furthermore, there is no loss in specificity of mature IGF-I for its receptor when FLAG is attached (Zhang et al., 1994). The FLAG epitope can identify both pro- and mature IGF-I by immunoblotting (Figure 4); however, it cannot distinguish between these forms by immunocytochemical staining. We know from the IGF-IStop construct transfections that mature IGF-I can be found inside nontransfected cells, but in the proIGF-IA and proIGF-IB constructs, it is possible that both pro- and mature IGF-I could enter cells. For example, the significant difference in cell uptake between IGF-IStop and IGF-IA or IGF-IB may be caused by the absence of proIGF-I in the IGF-IStop transfections. Given that the IGF-IA cleavage mutant (K68G) also had diminished cell uptake, it suggests that the preferred form for cell entry is mature IGF-I and that separation of the E-peptide from IGF-I occurs in the media before cell entry. Although the form of IGF-I that enters nontransfected cells cannot be determined from our results, these tools can be used in future studies to clarify which IGF-I species are taken up by muscle cells.
We extended the general methodology to include an epitope tag for the E-peptides so that both EA- and EB-peptides also could be monitored. The secretion of the EA-peptide has been demonstrated in past studies of IGF-I processing and supports that the EA peptide is present in the extracellular space in both glycosylated and nonglycosylated forms (Bach et al., 1990; Duguay et al., 1997; Wilson et al., 2001). Our observation of HA-positive cells also support that the EA- and EB-peptides enter a subset of nontransfected cells with and without the presence of IGF-I (Figure 7B). The size of the EB-peptide that was endocytosed could not be determined with this approach. As implied by the FLAG labeling results, it is possible that the entire EB-peptide or the eight-residue C-terminal peptide produced by protease cleavage was the HA-detectable fragment within nontransfected cells.
These results bring into question the identity of the bioactive peptide produced by IGF-IB. Mechano growth factor (MGF) is based on the unique portion of the IGF-IB E-peptide encoded by exons 5 and 6 and has been used in many studies to increase cell proliferation, migration, and survival (Yang and Goldspink, 2002; Mills et al., 2007). In several cases, cells have been exposed to synthesized MGF peptide for extended periods to evoke the observed effects. This raises the possibility for protease cleavage to produce different peptides, either of which could be bioactive. A careful and systematic investigation is warranted to clarify what sequence within MGF possesses activity.
A previous study examined the intracellular localization of the human IGF-I isoforms and found that expression of GFP fusion constructs containing human EB had distinct nucleolar localization, whereas those containing human EA or EC (similar to murine EB) were found throughout the cytoplasm and nucleus (Tan et al., 2002). In our study, neither E-peptide was found concentrated in the nucleus, although positive staining in the nucleoli of cells in IGF-IA–transfected culture was evident (Figure 6A), which was different from the results obtained by Tan et al. (2002). Further investigation of nuclear localization is warranted to determine whether this observation is isoform specific.
Do EA and EB alter IGF-I activity? If the presence of EA and EB increase the proportion of FLAG-IGF-I-positive cells, this implies that one property of the E-peptides is to potentiate IGF-I activity. The level of IGF-I (and E-peptide) production in the current study precludes addressing this question. Transient transfection of the IGF-I constructs produced ∼5000 pg/ml IGF-I in the media, which is quantifiable by ELISA but not by a bioassay. If we assume that all detected proteins are stable, this amounts to concentrations in the subnanomolar range, which is not anticipated to the sufficient to measurably activate IGF-I receptors, and we were unable to detect a change in phosphorylation of pathways associated with receptor activation. The calculated binding affinity of IGF-I for the IGF-IR is 1.5 nM (Kristensen et al., 1999). In adipocytes, no measurable phosphorylation of the receptor by recombinant IGF-I was apparent at <10 nM (Entingh-Pearsall and Kahn, 2004). Therefore, alternate complementary methods are being developed to determine if IGF-I actions are modulated by EA or EB. In conclusion, we have demonstrated that the E-peptides can promote increased IGF-I uptake into cells, which may enhance the activity of IGF-I. These studies can form the basis for examining independent and synergistic effects of IGF-I and the E-peptides on cellular functions.
ACKNOWLEDGMENTS
We are grateful for technical expertise contributed by Rong-Ine Ma and Jessie Feng. A portion of this work was presented at the 2007 American Dental Association Annual Session. This research was supported by National Institutes of Health grant R21 AR-056480 (to E.R.B.). L.A.P. was supported by a summer research fellowship from the University of Pennsylvania School of Dental Medicine.
Glossary
Abbreviations used:
- IGF
insulin-like growth factor.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-12-1202) on July 15, 2009.
REFERENCES
- Adamo M. L., Neuenschwander S., LeRoith D., Roberts C. T. Structure, expression, and regulation of the IGF-1 gene. Adv. Exp. Med. Biol. 1993;343:1–11. doi: 10.1007/978-1-4615-2988-0_1. [DOI] [PubMed] [Google Scholar]
- Adams G. R., McCue S. A. Localized infusion of IGF-I results in skeletal muscle hypertrophy in rats. J. Appl. Physiol. 1998;84:1716–1722. doi: 10.1152/jappl.1998.84.5.1716. [DOI] [PubMed] [Google Scholar]
- Bach M. A., Roberts C. T., Jr., Smith E. P., LeRoith D. Alternative splicing produces messenger RNAs encoding insulin-like growth factor-I prohormones that are differentially glycosylated in vitro. Mol. Endocrinol. 1990;4:899–904. doi: 10.1210/mend-4-6-899. [DOI] [PubMed] [Google Scholar]
- Barr P. J. Mammalian subtilisins: the long-sought dibasic processing endoproteases. Cell. 1991;66:1–3. doi: 10.1016/0092-8674(91)90129-m. [DOI] [PubMed] [Google Scholar]
- Barton E. R. The ABCs of IGF-I isoforms: impact on muscle hypertrophy and implications for repair. Appl. Physiol. Nutr. Metab. 2006a;31:791–797. doi: 10.1139/h06-054. [DOI] [PubMed] [Google Scholar]
- Barton E. R. Viral expression of insulin-like growth factor-I isoforms promotes different responses in skeletal muscle. J. Appl. Physiol. 2006b;100:1778–1784. doi: 10.1152/japplphysiol.01405.2005. [DOI] [PubMed] [Google Scholar]
- Barton-Davis E. R., Shoturma D. I., Musaro A., Rosenthal N., Sweeney H. L. Viral mediated expression of insulin-like growth factor I blocks the aging-related loss of skeletal muscle function. Proc. Natl. Acad. Sci. USA. 1998;95:15603–15607. doi: 10.1073/pnas.95.26.15603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman M. E., DeMayo F., Yin K. C., Lee H. M., Geske R., Montgomery C., Schwartz R. J. Myogenic vector expression of insulin-like growth factor I stimulates muscle cell differentiation and myofiber hypertrophy in transgenic mice. J. Biol. Chem. 1995;270:12109–12116. doi: 10.1074/jbc.270.20.12109. [DOI] [PubMed] [Google Scholar]
- Conover C. A., Baker B. K., Bale L. K., Clarkson J. T., Liu F., Hintz R. L. Human hepatoma cells synthesize and secrete insulin-like growth factor Ia prohormone under growth hormone control. Regul. Pept. 1993;48:1–8. doi: 10.1016/0167-0115(93)90330-b. [DOI] [PubMed] [Google Scholar]
- Conover C. A., Baker B. K., Hintz R. L. Cultured human fibroblasts secrete insulin-like growth factor IA prohormone. J. Clin. Endocrinol. Metab. 1989;69:25–30. doi: 10.1210/jcem-69-1-25. [DOI] [PubMed] [Google Scholar]
- Duguay S. J. Post-translational processing of insulin-like growth factors. Horm. Metab. Res. 1999;31:43–49. doi: 10.1055/s-2007-978697. [DOI] [PubMed] [Google Scholar]
- Duguay S. J., Lai-Zhang J., Steiner D. F. Mutational analysis of the insulin-like growth factor I prohormone processing site. J. Biol. Chem. 1995;270:17566–17574. doi: 10.1074/jbc.270.29.17566. [DOI] [PubMed] [Google Scholar]
- Duguay S. J., Milewski W. M., Young B. D., Nakayama K., Steiner D. F. Processing of wild-type and mutant proinsulin-like growth factor-IA by subtilisin-related proprotein convertases. J. Biol. Chem. 1997;272:6663–6670. doi: 10.1074/jbc.272.10.6663. [DOI] [PubMed] [Google Scholar]
- Entingh-Pearsall A., Kahn C. R. Differential roles of the insulin and insulin-like growth factor-I (IGF-I) receptors in response to insulin and IGF-I. J. Biol. Chem. 2004;279:38016–38024. doi: 10.1074/jbc.M313201200. [DOI] [PubMed] [Google Scholar]
- Florini J. R., Ewton D. Z., Coolican S. A. Growth hormone and the insulin-like growth factor system in myogenesis. Endocr. Rev. 1996;17:481–517. doi: 10.1210/edrv-17-5-481. [DOI] [PubMed] [Google Scholar]
- Hameed M., Orrell R. W., Cobbold M., Goldspink G., Harridge S. D. Expression of IGF-I splice variants in young and old human skeletal muscle after high resistance exercise. J. Physiol. 2003;547:247–254. doi: 10.1113/jphysiol.2002.032136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D., Bugl B., G̈unzburg W. H., Salmons B. Accurate estimation of transduction efficiency necessitates a multiplex real-time PCR. Gene Ther. 2000;7:458–463. doi: 10.1038/sj.gt.3301112. [DOI] [PubMed] [Google Scholar]
- Kristensen C., Wiberg F. C., Andersen A. S. Specificity of insulin and insulin-like growth factor I receptors investigated using chimeric mini-receptors. Role of C-terminal of receptor alpha subunit. J. Biol. Chem. 1999;274:37351–37356. doi: 10.1074/jbc.274.52.37351. [DOI] [PubMed] [Google Scholar]
- Kuo Y. H., Chen T. T. Novel activities of proIGF-I E peptides: regulation of morphological differentiation and anchorage-independent growth in human neuroblastoma cells. Exp. Cell Res. 2002;280:75–89. doi: 10.1006/excr.2002.5628. [DOI] [PubMed] [Google Scholar]
- Laviola L., Natalicchio A., Giorgino F. The IGF-I signaling pathway. Curr. Pharm. Des. 2007;13:663–669. doi: 10.2174/138161207780249146. [DOI] [PubMed] [Google Scholar]
- Lund P .K. Handbook of Physiology. The Endocrine System: Cellular Endocrinology. Vol. 7. Bethesda, MD: American Physiology Society; 1998. Insulin-like growth factors: gene structure and regulation; pp. 537–571. [Google Scholar]
- Mills P., Lafreniere J. F., Benabdallah B. F., El Fahime M., Tremblay J. P. A new pro-migratory activity on human myogenic precursor cells for a synthetic peptide within the E domain of the mechano growth factor. Exp. Cell Res. 2007;313:527–537. doi: 10.1016/j.yexcr.2006.10.032. [DOI] [PubMed] [Google Scholar]
- Monami G., Emiliozzi V., Morrione A. Grb10/Nedd4-mediated multiubiquitination of the insulin-like growth factor receptor regulates receptor internalization. J. Cell. Physiol. 2008;216:426–437. doi: 10.1002/jcp.21405. [DOI] [PubMed] [Google Scholar]
- Musaro A., McCullagh K., Paul A., Houghton L., Dobrowolny G., Molinaro M., Barton E. R., Sweeney H. L., Rosenthal N. Localized Igf-1 transgene expression sustains hypertrophy and regeneration in senescent skeletal muscle. Nat. Genet. 2001;27:195–200. doi: 10.1038/84839. [DOI] [PubMed] [Google Scholar]
- Romanelli R. J., LeBeau A. P., Fumer C. G., Lazzarino D. A., Hochberg A., Wood T. L. Insulin-like growth factor type-I receptor internalization and recycling mediate the sustained phosphorylation of Akt. J. Biol. Chem. 2007;282:22513–22524. doi: 10.1074/jbc.M704309200. [DOI] [PubMed] [Google Scholar]
- Rotwein P., Pollock K. M., Didier D. K., Krivi G. G. Organization and sequence of the human insulin-like growth factor I gene. Alternative RNA processing produces two insulin-like growth factor I precursor peptides. J. Biol. Chem. 1986;261:4828–4832. [PubMed] [Google Scholar]
- Siegfried J. M., Kasprzyk P. G., Treston A. M., Mulshine J. L., Quinn K. A., Cuttitta F. A mitogenic peptide amide encoded within the E peptide domain of the insulin-like growth factor IB prohormone. Proc. Natl. Acad. Sci. USA. 1992;89:8107–8111. doi: 10.1073/pnas.89.17.8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D. S., Cook A., Chew S. L. Nucleolar localization of an isoform of the IGF-I precursor. BMC Cell Biol. 2002;3:17. doi: 10.1186/1471-2121-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis M. New insulin-like growth factor (IGF)-precursor sequences from mammalian genomes: the molecular evolution of IGFs and associated peptides in primates. Growth Horm. IGF Res. 2009;19:12–23. doi: 10.1016/j.ghir.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Wilson H. E., Westwood M., White A., Clayton P. E. Monoclonal antibodies to the carboxy-terminal Ea sequence of pro-insulin-like growth factor-IA (proIGF-IA) recognize proIGF-IA secreted by IM9 B-lymphocytes. Growth Horm. IGF Res. 2001;11:10–17. doi: 10.1054/ghir.2000.0182. [DOI] [PubMed] [Google Scholar]
- Yang H., Alnaqeeb M., Simpson H., Goldspink G. Changes in muscle fibre type, muscle mass and IGF-I gene expression in rabbit skeletal muscle subjected to stretch. J. Anat. 1997;190:613–622. doi: 10.1046/j.1469-7580.1997.19040613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. Y., Goldspink G. Different roles of the IGF-I Ec peptide (MGF) and mature IGF-I in myoblast proliferation and differentiation. FEBS Lett. 2002;522:156–160. doi: 10.1016/s0014-5793(02)02918-6. [DOI] [PubMed] [Google Scholar]
- Zhang W., Gustafson T. A., Rutter W. J., Johnson J. D. Positively charged side chains in the insulin-like growth factor-1 C- and D-regions determine receptor binding specificity. J. Biol. Chem. 1994;269:10609–10613. [PubMed] [Google Scholar]