Abstract
Accurate chromosome segregation depends on sister kinetochores making bioriented attachments to microtubules from opposite poles. An essential regulator of biorientation is the Ipl1/Aurora B protein kinase that destabilizes improper microtubule–kinetochore attachments. To identify additional biorientation pathways, we performed a systematic genetic analysis between the ipl1-321 allele and all nonessential budding yeast genes. One of the mutants, mcm21Δ, precociously separates pericentromeres and this is associated with a defect in the binding of the Scc2 cohesin-loading factor at the centromere. Strikingly, Mcm21 becomes essential for biorientation when Ipl1 function is reduced, and this appears to be related to its role in pericentromeric cohesion. When pericentromeres are artificially tethered, Mcm21 is no longer needed for biorientation despite decreased Ipl1 activity. Taken together, these data reveal a specific role for pericentromeric linkage in ensuring kinetochore biorientation.
INTRODUCTION
The precise regulation of chromosome segregation ensures that each daughter cell receives an entire complement of the genome. During replication, cohesion is established between sister chromatids. Microtubules (MTs) bind to these sister chromatids through kinetochores, the specialized protein complexes that assemble on centromeric DNA. To ensure that sister chromatids segregate away from each other, kinetochores must biorient and attach to MTs originating from opposite poles. If sister kinetochores make syntelic attachments to MTs from the same pole, the spindle checkpoint halts the cell cycle, allowing time for the defect to be corrected.
Although the precise mechanism by which sister kinetochores biorient is not known, tension generated by MT-pulling forces on linked sister chromatids appears to be required (for review, see Pinsky and Biggins, 2005). Micromanipulation experiments demonstrated that artificially applying tension on sister kinetochores both stabilizes and increases the number of MT–kinetochore attachments (Nicklas and Ward, 1994; King and Nicklas, 2000). These observations suggest that syntelic attachments are unstable due to a lack of tension. Consistent with this, a key regulator of biorientation and the spindle checkpoint is the conserved protein kinase Ipl1/Aurora B, which destabilizes improper MT–kinetochore attachments (for review, see Ruchaud et al., 2007).
Sister chromatid cohesion is also essential for biorientation because it keeps sisters physically associated and allows tension to be generated by MTs that pull on the linked sister kinetochores (Tanaka et al., 2000). When two kinetochores were physically connected on a single DNA molecule, they were able to biorient in an Ipl1-dependent manner (Dewar et al., 2004). These studies led to the conclusion that the critical role of cohesion was to physically link sister chromatids instead of orienting sister kinetochores toward opposite poles as previously suggested. However, these experiments did not eliminate the possibility that cohesion has an additional role in biorientation or that other biorientation pathways exist (for review, see Hauf and Watanabe, 2004).
The bulk of sister chromatid cohesion is mediated by the cohesin complex that consists of four subunits: Smc1, Smc3, Mcd1/Scc1, and Scc3 (for review, see Peters et al., 2008). Cohesin appears to form a ring-like structure that encircles and traps sister chromatids together. Although it is still not mechanistically understood how cohesion is established between sister chromatids, this multistep process is normally coupled to replication and requires cohesin-loading by the Scc2/Scc4 complex (Toth et al., 1999; Ciosk et al., 2000; Lengronne et al., 2006) and acetylation of Smc3 by the Eco1/Ctf7 acetyltransferase during S phase (Michaelis et al., 1997; Skibbens et al., 1999; Toth et al., 1999; Ben-Shahar et al., 2008; Unal et al., 2008; Zhang et al., 2008). Cohesion is then maintained until metaphase when all kinetochores come under tension and biorient. At this time, the anaphase-promoting complex degrades the anaphase inhibitor Pds1/securin, liberating the Separase protease to cleave Mcd1 and release cohesin from DNA.
Cohesin is not randomly distributed throughout the genome. In budding yeast, cohesin complexes are enriched at ∼10–15-kb intervals called cohesin-associated regions (CARs) that tend to be A+T rich or occur at sites of convergent transcription (Blat and Kleckner, 1999; Megee et al., 1999; Tanaka et al., 1999; Laloraya et al., 2000; Lengronne et al., 2004). A conserved feature of cohesin localization is a strong enrichment at pericentromeres that coincides with the large heterochromatic regions that flank the regional centromeres in most organisms (for review, see Grewal and Jia, 2007). Although budding yeast lack pericentromeric heterochromatin, cohesin is enriched in a domain that extends ∼50 kb around the 125-base pair point centromere in a kinetochore-dependent manner (Megee et al., 1999; Tanaka et al., 1999; Weber et al., 2004). The enrichment of cohesin around centromeres appears paradoxical because biorientation causes centromeres to split before anaphase (Goshima and Yanagida, 2000; He et al., 2000; Pearson et al., 2001), although cohesin may mediate intramolecular rather than intermolecular interactions in this region (Yeh et al., 2008). Regardless, the enrichment of pericentromeric cohesin appears to ensure the fidelity of chromosome segregation in most organisms. In fission yeast, the major function of pericentromeric heterochromatin in mitotic chromosome segregation is to recruit cohesin (Bernard et al., 2001; Yamagishi et al., 2008). A budding yeast chromosome that lacks pericentromeric cohesion exhibits chromosome segregation defects despite retaining cohesin along the arm (Eckert et al., 2007). Furthermore, budding yeast respond to decreased tension between sister chromatids by recruiting cohesin specifically around pericentromeres (Eckert et al., 2007; Ocampo-Hafalla et al., 2007). Taken together, these data suggest that pericentromeric cohesin may have a specific function in chromosome segregation beyond simply holding sisters together.
We reasoned that there might be pathways that facilitate biorientation whose roles have been masked by the strength of the Ipl1 error correction mechanism. We therefore isolated mutants that are required for viability when the function of the budding yeast Ipl1/Aurora B kinase is reduced. Three of the mutants identified (Mcm16, Mcm21, and Mcm22) encode components of the 12-member CTF19 kinetochore complex that was originally identified in budding yeast (Ortiz et al., 1999; Poddar et al., 1999; Cheeseman et al., 2002). COMA, a four-subunit subcomplex of the larger CTF19 complex that consists of Ctf19, Mcm21, Okp1, and Ame1, was later isolated (De Wulf et al., 2003). Here, we further analyze the function of the conserved Mcm21 protein that is a member of both complexes and find that it is required to enrich cohesin factors at pericentromeres. Strikingly, Mcm21 becomes essential for biorientation when Ipl1 function is reduced, and this appears to be related to its role in pericentromeric cohesion. When pericentromeres are artificially tethered, Mcm21 is no longer needed for biorientation despite decreased Ipl1. Taken together, these data are consistent with a specific requirement for cohesin enrichment at pericentromeres to facilitate kinetochore biorientation.
MATERIALS AND METHODS
Systematic Genetic Analysis Screen
Systematic genetic analysis (SGA) was performed at 23°C, using an array of viable yeast deletion strains in the S288c strain background (MATa xxxΔ::KAN) as previously described (Tong et al., 2001; Waples et al., 2008). The query strain YBL165b (MATα ipl1-321::NAT can1Δ) was generated by backcrossing the temperature-sensitive (ts) ipl1-321 allele nine times into the Y2454 parent strain (Tong et al., 2001), followed by integration of a NatR-MX4 cassette immediately downstream of the Ipl1 stop codon. The correct integration was verified by colony PCR and linkage analysis between the ts and NatR resistance genes before performing screens. Ipl1-321::NatR xxxΔ::KAN double mutants were systematically generated in quadruplicate, by mating the query strain in parallel to duplicate arrays containing each viable yeast gene deletion. After sporulation on plates, haploid progeny (wild type [WT], single, and double mutant) were selected through two sequential rounds of growth on SD-HIS+CAN plates based on the presence of the MFA1pr-HIS3 marker. Double mutant (ipl1-321::NatR xxxΔ::KAN) progeny were selected under the same media conditions plus antibiotics. Synthetic growth defects were identified by comparing colony sizes between haploid versus double mutant selection plates as well as versus an YCG1::NatR Δxxx::KAN control (in the presence of antibiotics). Under stringent conditions (23°C), ∼200 double mutant combinations exhibited slow or no growth phenotypes at least two to four times on the double mutant plates. Of these, 23 interactions were confirmed by standard tetrad dissection and linkage analysis to have a synthetic lethal or sick growth phenotype, and these interactions are reported in this article. Double mutant combinations with poor sporulation efficiencies or low spore viability were not considered further. Genetic interactions between ipl1-321 and mcm21, ctf8, dcc1, and bim1 were also confirmed in the w303 strain background but the others have not been analyzed in w303.
Microbial Techniques and Plasmids
Media and microbial techniques were as described (Sherman et al., 1974; Rose et al., 1990). In all synchronous cell cycle experiments reported, α-factor was used to arrest cells in G1 and cell cycle progression was monitored by scoring the budding index. For the relevant experiments, doxycycline (25 μg/ml) and nocodazole (10–15 μg/ml, 2.5–3 h) were added upon G1 release. Checkpoint arrest was confirmed by the presence of a single DNA mass. For the noncleavable-Mcd1 experiment, cells were released into media containing galactose to induce expression. Cells were shifted to 37°C after bud emergence. Yeast strains are listed in Supplemental Table S1.
The deg-ipl1 plasmid (pSB244) was constructed by PCR amplification of the IPL1 ORF using primers SB89 and SB90 with PstI and NotI sites engineered and ligated into pSB230 digested with the same enzymes. The plasmid is integrated at the ADE2 locus after digestion with AflII. The tetramerizing LacI (pSB1591) was constructed by ligating the EagI/MluI fragment from pAFS55 into pSB116. Primer sequences are available upon request.
Microscopy
Analysis of GFP-LacI was performed as described (Biggins et al., 1999). Indirect immunofluorescence was performed as described (Rose et al., 1990) with antibodies that recognize the myc tag (9E10, Covance, (Princeton, NJ) and Alexa Fluor 488–conjugated anti-green fluorescent protein (GFP; Molecular Probes, Eugene, OR) at a 1:500 dilution. Chromosome spreads were performed as described (Michaelis et al., 1997; Loidl et al., 1998) using the Alexa Fluor GFP antibody (1:1000) and anti-Cse4 (1:250; Pinsky et al., 2003). For all microscopy experiments, more than 200 cells were scored unless otherwise noted. The scale bar in all images equals 5 μm. The Bernoulli distribution was used to assess statistical significance at 95% confidence.
Protein and Immunological Techniques
Protein extracts were made and immunoblotted as described (Minshull et al., 1996). Antibodies that recognize the myc tag (9E10) and the hemagglutinin (HA) tag (12CA5) were obtained from Covance and used at a 1:10000 dilution. Anti-tubulin immunoblotting (Accurate Chemical and Scientific, Westbury, NY) was used at 1:1000 dilution as a loading control.
Chromatin Immunoprecipitation and Kinase Assays
Immunoprecipitations were performed using 3F10 anti-HA antibodies (Roche, Indianapolis, IN) or M2 anti-Flag antibodies (Sigma, St. Louis, MO). Chromatin immunoprecipitation (ChIP) was performed, and samples were quantified as described previously (Collins et al., 2005). Sequences of PCR primers (Eckert et al., 2007) are available upon request. Quantification of bound DNA was calculated as the percentage of total chromatin isolated before immunoprecipitation. ChIP experiments were performed at least three times. The Ipl1 kinase assay was performed as described (Buvelot et al., 2003; Kotwaliwale et al., 2007).
RESULTS
Ipl1-321 SGA Screen Identifies Novel Genetic Interactions
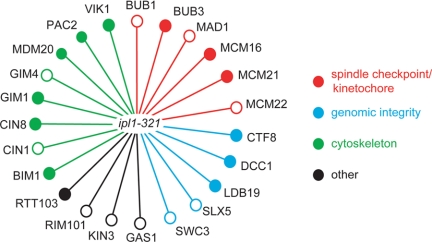
To identify additional biorientation pathways, we analyzed mutants in nonessential genes for interactions with ipl1-321, an allele that accumulates syntelic attachments due to decreased kinase activity at the nonpermissive temperature (37°C; Biggins et al., 1999). Ipl1-321 activity is also substantially reduced at lower temperatures, but these cells remain viable due to redundant pathways (Kotwaliwale et al., 2007). We therefore performed an SGA at 23°C by crossing the ipl1-321 allele to a genome-wide deletion set of all nonessential yeast genes (Tong et al., 2001; Waples et al., 2008). We confirmed 23 genetic interactions with mutants in genes broadly involved in the spindle checkpoint, chromatin structure/genome stability, and the cytoskeleton (Figure 1). Because Ipl1 has a number of functions, the genetic interactions could represent a variety of cellular defects. It will therefore be critical to directly study each interaction to understand its requirement for viability when Ipl1 function is reduced.
Figure 1.
IPL1 genetic interaction profiles. Synthetic genetic interactions between the hypomorphic allele ipl1-321::NAT (YBL165b) and indicated yeast deletion strains. Filled and open circles denote synthetic lethal and sick interactions, respectively.
Mcm21 Has a Role in Pericentromeric Sister Chromatid Cohesion
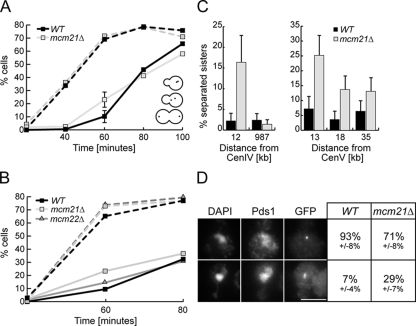
Because we were interested in pathways that regulate biorientation, we focused on the three kinetochore mutants identified in the screen that all encode components of the CTF19 kinetochore complex (mcm21Δ, mcm16Δ, and mcm22Δ). We began by monitoring the segregation of a pair of sister chromatids in an mcm21Δ strain. ChrIV was marked by the binding of a GFP-LacI fusion to Lac operator sequences inserted 12 kb from the centromere (pericentromeric), a locus that remains linked when sisters biorient (Straight et al., 1996). WT and mcm21Δ cells were released from G1 and analyzed for sister separation at 20-min intervals after bud emergence (Figure 2A). In both strains, sister separation was first detected 60 min after G1 release. Surprisingly, despite similar kinetics of bud emergence, 23% of mcm21 cells had separated sisters compared with only 10% of WT cells at this time point. The phenotype was not due to aneuploidy because two GFP foci were only observed in 2% of G1-arrested cells. We performed a similar analysis on mcm22Δ cells but found that the sister separation in mcm22Δ cells was only slightly increased over WT cells (15 vs. 10%, Figure 2B). Because mcm21Δ cells had a larger sister separation defect and a stronger genetic interaction with ipl1-321, we decided to further pursue characterization of the mcm21 mutant strain.
Figure 2.
Mcm21 is required for pericentromeric cohesion. (A) Total sister chromatid separation (includes all categories shown in representative depictions of cells) was monitored during a synchronous cell cycle after release from G1 in WT and mcm21Δ cells (SBY818, SBY1897) at a ChrIV pericentromeric locus. Dotted lines indicate percent of budded cells. (B) Total separation of the ChrIV pericentromere was monitored in WT, mcm21Δ, and mcm22Δ cells (SBY818, SBY1897, SBY1983) after release from G1. Budding index shown as dotted lines. (C) Sister separation was monitored in nocodazole-arrested cells at various loci on ChrIV and ChrV (SBY818, SBY1897, SBY6133, SBY6134, SBY7876, SBY7877, SBY7878, SBY7879, SBY7880, and SBY7881). (D) Immunofluorescence microscopy was performed on strains in A. Cells shown are 100 min after G1 release (n > 100).
We noticed that the separated sister foci in mcm21 mutant cells were always closely spaced within the mother cell at the early time points (Supplemental Figure S1), a phenotype that is rarely seen in WT cells. Because these data suggested that the cohesion defect may be specific to pericentromeres, we monitored sister separation at various loci on ChrIV and ChrV. To eliminate potential differences in MT-pulling forces and cell cycle progression, cells were released from G1 into nocodazole to depolymerize the MTs and arrest cells in metaphase. Although there was a significant increase in separated GFP foci in mcm21 cells at the ChrIV pericentromeric locus, there was no detectable defect at the telomere (Figure 2C, left). The lower percentage of separation in the mcm21 cells observed in this experiment was presumably due to the lack of MT-pulling forces that enhance the ability to resolve separated pericentromeres. The sister separation defect was also observed with a GFP mark 13 kb from the centromere on ChrV that decreased as it was moved further away to 18 and 35 kb (Figure 2C, right). In all of these experiments, the percentage of cells with separated foci during the G1 arrest never exceeded 3%. Taken together, these data strongly suggest that there is a cohesion defect specific to all pericentromeres in mcm21Δ cells.
To test whether the pericentromeric sister separation in mcm21 cells was due to the premature initiation of anaphase, we performed immunofluorescence microscopy to localize GFP-LacI and the Pds1 protein that is degraded at anaphase onset. WT and mcm21Δ cells were quantified 100 min after release from G1 for Pds1 staining and sister separation (Figure 2D). As expected, a single GFP-LacI focus was observed in the majority of WT cells that also had high levels of Pds1 (93%). In contrast, 29% of mcm21Δ cells with strong Pds1 staining had two GFP foci in close proximity, indicating that the pericentromeric locus prematurely separated in metaphase. These data are consistent with our ability to detect separated sisters in mcm21 cells arrested in nocodazole. In addition, more than 22% of mcm21 cells arrested in metaphase by the overexpression of a nondegradable version of Pds1 or repression of the Cdc20 activator of anaphase also exhibited separated pericentromeres (data not shown).
Cohesin Loading at Pericentromeres Is Perturbed in mcm21Δ Cells
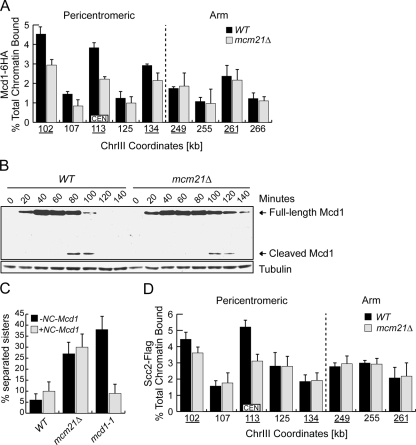
Because the Ctf19 protein is required for pericentromeric cohesin recruitment in response to nocodazole treatment that decreases kinetochore tension (Eckert et al., 2007), we examined the localization of the Mcd1 cohesin protein in mcm21 cells by ChIP. We arrested WT and mcm21Δ cells containing Mcd1-6HA in nocodazole to reduce tension and also to eliminate any cell cycle variation between the strains. The chromatin samples were analyzed by PCR at known CAR and non-CAR sites both proximal and distal to the centromere on ChrIII. Consistent with our observation that a locus near CEN3 prematurely separates (data not shown), Mcd1 binding decreased at the three CAR sites tested in the pericentromeric region of mcm21Δ cells (Figure 3A). However, there were no differences between WT and mcm21Δ cells at CAR or non-CAR sites on the chromosomal arm.
Figure 3.
Mcm21 is required for cohesin loading at pericentromeres. (A) ChIP was carried out on nocodazole-arrested WT and mcm21Δ cells containing Mcd1-6HA (SBY7002 and SBY7007). CAR (underlined) and non-CAR sites were assessed for Mcd1-6HA binding at pericentromeric and arm loci on ChrIII as shown. (B) Lysates of strains in A were immunoblotted with anti-HA antibody during a synchronous cell cycle. Tubulin is shown as a loading control. (C) Sister separation was monitored at the pericentromeric locus in WT, mcm21Δ, and mcd1-1 cells that contained (SBY6998, SBY6999, and SBY7847) or did not contain (SBY818, SBY1897, and SBY7846) NC-Mcd1 during a nocodazole arrest. (D) ChIP analysis of nocodazole-arrested WT and mcm21Δ cells containing Scc2-Flag (SBY7651 and SBY7652) was performed as in A.
To determine whether the decrease in Mcd1 at pericentromeres could be due to premature cleavage in the mcm21 mutant cells, we monitored the appearance of the Mcd1 C-terminal cleavage fragment that results from Separase cleavage as cells were released from G1 (Uhlmann et al., 1999). Although Mcd1 cleavage was first apparent in WT cells at 80 min, it was not detected until 100 min in mcm21Δ cells (Figure 3B). Consistent with this, the full-length Mcd1 protein was retained longer in mcm21Δ cells than in WT cells. The delay in cohesin cleavage and degradation is most likely due to the transient activation of the spindle checkpoint in mcm21Δ cells (see below, Figure 5E).
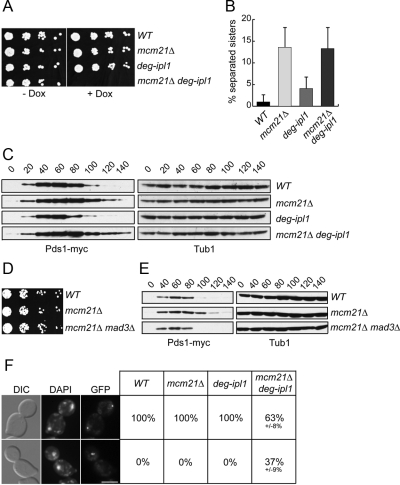
Figure 5.
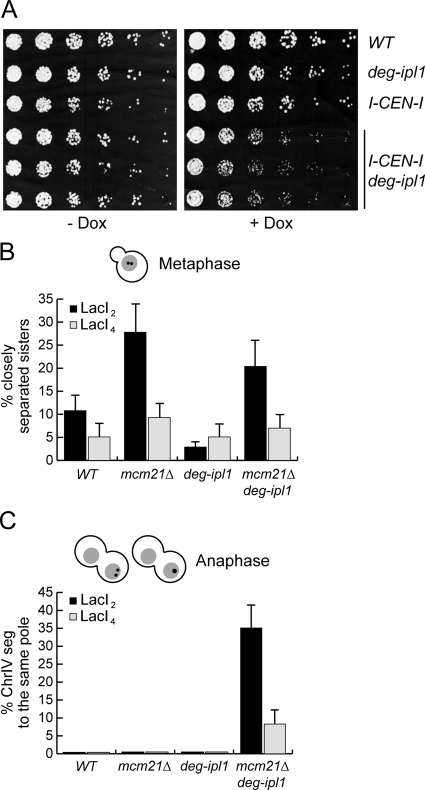
Mcm21 is required for kinetochore biorientation when Ipl1 function is impaired. (A) Serial dilutions (fivefold) of WT, mcm21Δ, deg-ipl1, and mcm21Δ deg-ipl1 (SBY818, SBY1897, SBY6940, and SBY5551) cells were plated in the presence or absence of doxycycline. (B) Sister separation of the ChrIV pericentromeric locus was monitored in strains in A released from G1 into a nocodazole arrest in the presence of doxycycline. (C) Lysates from strains in A were immunoblotted against Pds1-myc and tubulin during a synchronous cell cycle. (D) Fivefold dilutions of WT, mcm21Δ, and mcm21Δ mad3Δ cells (SBY818, SBY1897, and SBY7656) were plated for viability. (E) Lysates of cells in D were immunoblotted for Pds1-myc and tubulin during a synchronous cell cycle. (F) ChrIV segregation was assessed in strains in A that reached anaphase during a synchronous cell cycle in the presence of doxycycline (n > 100).
Because a population of cleaved cohesin might have escaped detection, we also tested whether the expression of an ectopic copy of noncleavable Mcd1 (NC-Mcd1) could suppress the cohesion defect (Uhlmann et al., 1999). WT and mcm21Δ cells with or without NC-Mcd1 were arrested in G1 and released into nocodazole under conditions that induce NC-Mcd1. Sister separation was quantified at the pericentromeric locus on ChrIV (Figure 3C). Although NC-Mcd1 was sufficient to link sisters in mcd1-1 cells, it did not suppress the separation defect in mcm21Δ cells, consistent with the data indicating that cohesin is not prematurely cleaved (Figure 3B).
We therefore asked whether cohesion establishment was defective in mcm21 cells by analyzing the binding of the Scc2 cohesin-loading factor by ChIP. Cells containing Scc2-Flag were arrested in nocodazole and the chromatin bound to Scc2 was analyzed by PCR at the same CAR and non-CAR loci analyzed for Mcd1 binding. Similar to our findings for Mcd1-HA binding, Scc2 enrichment at the centromeric CAR site decreased in the absence of Mcm21 but was unperturbed at arm sites (Figure 3D). However, in contrast to Mcd1, we did not observe changes in Scc2 binding at the pericentromeric CAR sites we assayed. We also analyzed the Eco1 cohesion establishment factor by ChIP but did not detect any differences in enrichment around centromeres in mcm21 cells under similar growth conditions (data not shown). Therefore, the absence of Mcm21 may prevent proper pericentromeric cohesion due to the inability to fully recruit the Scc2 cohesin-loading factor to the centromere.
Mcm21 Has a Function in Kinetochore Biorientation
We next addressed the nature of the defect that leads to the synthetic lethality between mcm21 and ipl1-321. We considered the possibility that the loss of Mcm21 alters Ipl1 localization, resulting in synthetic lethality between mcm21 and ipl1 mutants. To test this, we carried out chromosome spreads to visualize kinetochores on nocodazole-arrested WT and mcm21Δ cells containing Ipl1-GFP. Ipl1 colocalized with the Cse4 kinetochore protein in both WT and mcm21Δ cells (Figure 4A). In addition, we did not detect any obvious changes in GFP intensity in the absence of Mcm21, suggesting that Ipl1 localizes to kinetochores normally. Similar results were observed for chromosome spreads performed on cells from asynchronous cultures (data not shown), as well as when we visualized Ipl1-GFP by live microscopy (Supplemental Figure S2). To further analyze Ipl1 localization to centromeres, we attempted ChIP. However, Ipl1-Flag was not enriched at the centromere in WT cells compared with an untagged control strain (data not shown), consistent with previously reported ChIP data (He et al., 2001). Taken together, our data suggest that Ipl1 localizes to kinetochores normally in the absence of Mcm21.
Figure 4.
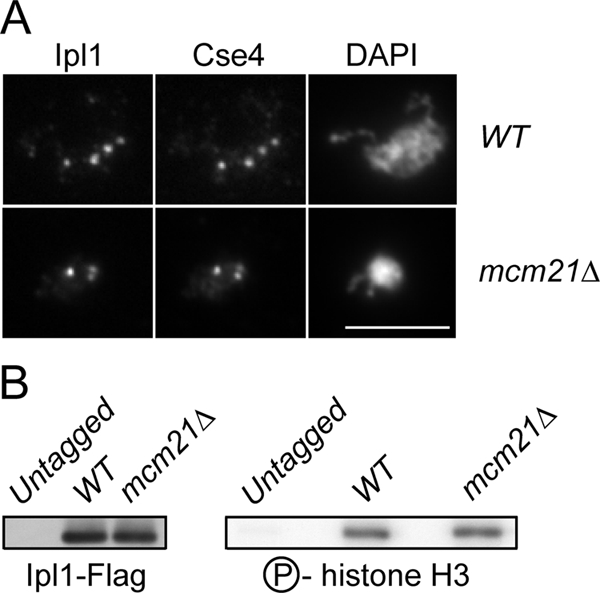
Ipl1 localization and function are not altered in the absence of Mcm21. (A) Chromosome spreads were performed on nocodazole-arrested WT and mcm21Δ cells (SBY8352, SBY8353) to localize Cse4 and Ipl1-GFP. (B) Ipl1-FLAG was immunoprecipitated from untagged, WT, and mcm21Δ cultures (SBY3, SBY7018, and SBY7019). The immunoprecipitates were immunoblotted with anti-Flag (left) or subjected to in vitro kinase assays (right).
We next tested whether Ipl1 kinase activity was altered in the absence of Mcm21. Ipl1-Flag was immunoprecipitated from asynchronously growing cultures of WT and mcm21Δ cells and incubated with histone H3 and 32P-ATP in a kinase assay in vitro. We did not detect any change in histone H3 phosphorylation in mcm21Δ cells compared with WT cells (Figure 4B), indicating that overall Ipl1 kinase activity is not altered in mcm21Δ cells.
To understand why mcm21 ipl1 mutants are inviable, we constructed a conditional Ipl1 allele by fusing a degron tag to its N-terminus (Deg-Ipl1) to destabilize the protein and target it for degradation via the proteasome (Cormack and Struhl, 1992). The expression of the allele is also repressed by doxycycline, so we analyzed the growth of WT, mcm21Δ, deg-ipl1, and mcm21Δ deg-ipl1 cells in the presence and absence of doxycycline. All of the strains grew similarly on plates without doxycycline, indicating that the cells retain enough Ipl1 function for viability (Figure 5A). As expected, WT and mcm21Δ cells also grew normally in the presence of doxycycline. However, doxycycline did not inhibit the growth of deg-ipl1 cells, so these cells retain enough Ipl1 function to support viability even when its expression is inhibited. In contrast, the mcm21Δ deg-ipl1 strain was inviable on doxycycline. These data indicate that deg-ipl1 depends on MCM21 function for viability, fortuitously creating a conditional strain. We confirmed that the mcm21Δ deg-ipl1 cells lose viability upon release from G1 (Supplemental Figure S3).
Although the precise reason why the deg-ipl1 allele is hypomorphic is not known, it gave us a way to analyze the requirement for Mcm21 in these cells. First, we assayed cohesion at the pericentromeric locus 12 kb from CEN4 in mcm21Δ deg-ipl1 cells. We performed these experiments in WT, mcm21Δ, deg-ipl1, and mcm21Δ deg-ipl1 cells treated with nocodazole to eliminate potential differences in MT–kinetochore interactions and cell cycle progression (Figure 5B). Consistent with previous data suggesting that Ipl1 is not required for cohesion in budding yeast (Biggins et al., 1999), we did not detect any exacerbation of the mcm21Δ cohesion defect in the double mutant.
We then assessed whether mcm21 mutant cells activate the spindle checkpoint in an Ipl1-dependent manner. We analyzed Pds1 levels in WT, mcm21Δ, deg-ipl1, and mcm21Δ deg-ipl1 cells that were released from G1 into doxycycline (Figure 5C). There was a transient delay in Pds1 destruction in mcm21 cells that was not abolished when deg-ipl1 was repressed. These data suggest that the inviability of these cells is not due to a defect in spindle checkpoint activation, so we directly examined the requirement of the checkpoint for the viability of mcm21 mutant cells by constructing an mcm21 mad3 double mutant. This strain grew normally (Figure 5D), but completely abolished the delay in Pds1 destruction in mcm21 cells (Figure 5E). Therefore, mcm21 cells transiently activate the spindle checkpoint but do not require checkpoint activity for viability. These data are consistent with the observation that cohesin mutants also transiently activate the spindle checkpoint (Skibbens et al., 1999; Biggins et al., 2001). Although it was previously concluded that the mcm21 mad3 cells are viable because Mad3 is not required to detect defects in tension at kinetochores, the status of the spindle checkpoint was never analyzed in the double mutant cells (Lee and Spencer, 2004). Instead, our data show that Mad3 is required for the spindle checkpoint delay induced by a lack of the Mcm21 protein.
Because the essential function of Ipl1 is to biorient kinetochores, we tested whether there is an increased dependency on Mcm21 for biorientation when Ipl1 function is reduced. We were unable to monitor biorientation by directly analyzing sister centromere splitting at metaphase because we could not distinguish it from the pericentromeric cohesion defect in mcm21Δ cells. As a result, we analyzed sister chromatid segregation at anaphase by releasing WT, mcm21Δ, deg-ipl1, and mcm21Δ deg-ipl1 cells from G1 into media containing doxycycline. Cells were analyzed for ChrIV segregation when >50% of the cells had proceeded into anaphase and segregated DNA masses to opposite poles (Figure 5F). Consistent with the viability data, sister chromatids segregated to opposite poles in all of the WT, mcm21Δ, and deg-ipl1 cells. However, both copies of ChrIV segregated to the same pole in ∼37% of the mcm21Δ deg-ipl1 cells, suggesting that the kinetochores were mono-oriented. In support of this, the sisters were always in close proximity to the spindle poles, and we sometimes observed cells where both sisters were pulled into the bud (see Figure 5F). Assuming that all 16 chromosomes in budding yeast have an equal probability of mono-orienting in mcm21Δ deg-ipl1 double mutant cells, cells would rarely segregate all of their chromosomes properly. Therefore, although we were not able to directly visualize the biorientation process, these data are most consistent with the possibility that Mcm21 is required for proper biorientation when Ipl1 function is down-regulated.
Pericentromeric Sister Chromatid Cohesion Facilitates Biorientation
Although a variety of data suggest that pericentromeric cohesin is important for chromosome segregation, its precise role has not yet been elucidated (Allshire et al., 1995; Kellum and Alberts, 1995; Peters et al., 2001; Eckert et al., 2007). While our work was in preparation, data were published indicating that it facilitates biorientation in fission yeast (Sakuno et al., 2009). We had also been working on the possibility that pericentromeric cohesin aids biorientation during mitosis. In this case, a decrease in pericentromeric cohesin should reduce the efficiency of biorientation and therefore increase the dependency on Ipl1 to correct mal-oriented attachments. To test this, we utilized the I-CEN-I strain that prevents cohesin recruitment around CEN3 (Eckert et al., 2007). WT, I-CEN-I, deg-ipl1, and I-CEN-I deg-ipl1 cells were plated for viability in the presence and absence of doxycycline (Figure 6A). Although all of the strains grew similarly in the absence of doxycycline (Eckert et al., 2007), I-CEN-I deg-ipl1 cells grew more slowly when Ipl1 function was reduced by addition of doxycycline. Therefore, the disruption of pericentromeric cohesin on a single chromosome sensitizes cells to a slight reduction in Ipl1 activity, supporting the idea that pericentromeric cohesion aids biorientation.
Figure 6.
Pericentromeric linkage promotes biorientation. (A) Serial dilutions (threefold) of WT, deg-ipl1, I-CEN-I and three independent spores of I-CEN-I deg-ipl1 cells (SBY3, SBY6993, SBY8102, SBY8103, SBY8104, and SBY8105) were plated in the presence and absence of doxycycline. (B) WT, mcm21Δ, deg-ipl1, and mcm21Δ deg-ipl1 cells containing LacI2 (SBY818, SBY1897, SBY6940, and SBY5551) or LacI4 (SBY7871, SBY7872, SBY7873, and SBY7874) were monitored for closely separated sisters 80 min after G1 release in cells containing a single nucleus. (C) ChrIV segregation was monitored in the same experiment when the majority of cells had entered anaphase and segregated DNA masses to opposite poles (n > 100). The corresponding budding index is reported in Supplemental Figure S7.
As an alternative test of the relationship between pericentromeric cohesion and biorientation, we asked whether the biorientation defect in mcm21Δ deg-ipl1 cells could be suppressed when pericentromeric linkage is restored. To this end, we utilized a tetramerizing version of LacI (LacI4) that is sufficient to hold sister chromatids together in the absence of MT-pulling forces and was used to argue that kinetochore geometry has an important role during meiosis I (Straight et al., 1996; Lacefield and Murray, 2007). Nontetramerizing GFP-LacI used in the previous experiments (LacI2) or tetramerizing GFP-LacI (LacI4) were expressed in WT, mcm21Δ, deg-ipl1, and mcm21Δ deg-ipl1 cells that contained LacO sequences 12 kb from CEN4. Sister separation was quantified 80 min after release from G1, a time when mcm21 cells exhibit premature separation at the pericentromere (Figure 6B). Strikingly, the cohesion defect decreased in both mcm21Δ and mcm21Δ deg-ipl1 cells, indicating that the tetramerizing LacI fusion can restore pericentromeric linkage.
We next analyzed biorientation in the same experiment by monitoring sister chromatid segregation at anaphase. We quantified GFP foci at the time point in each strain where the majority of cells had pulled their DNA to opposite poles (Figure 6C). As expected, WT, mcm21Δ, and deg-ipl1 cells did not exhibit a biorientation defect and always segregated ChrIV to opposite poles. Remarkably, although ∼35% of the mcm21Δ deg-ipl1 cells segregated sisters to a single pole in the presence of LacI2, only ∼8% of the cells exhibited this phenotype when LacI4 was expressed. Therefore, restoring pericentromeric linkage in mcm21Δ deg-ipl1 cells was sufficient to partially suppress the biorientation defect. As a control, we analyzed biorientation in an mcm21Δ deg-ipl1 mutant where the LacO sequences were moved to the telomere and compared that with the pericentromeric strain used in the previous experiment (Supplemental Figure S4). Although there was slight variation in the biorientation defects within the pericentromeric-marked strain between experiments, the differences are not significant. More importantly, artificially linking the telomeric locus did not suppress the biorientation defect, strongly suggesting that tethering the pericentromeres, specifically, restores proper biorientation in the absence of normal levels of pericentromeric cohesion.
DISCUSSION
We performed an SGA analysis with the ipl1-321 allele and found that at least one component of the CTF19 kinetochore subcomplex becomes important for biorientation when Ipl1 function is impaired. Our characterization of the mcm21 mutant revealed that it is involved in establishing pericentromeric cohesion. The role of Mcm21 in kinetochore biorientation can be partially bypassed if linkage is restored to pericentromeres, strongly suggesting that the enrichment of sister chromatid cohesin at pericentromeres serves to physically link them together to facilitate kinetochore biorientation.
The CTF19 Complex Ensures Proper Pericentromeric Cohesion
It was previously shown that the Ctf19 protein contributes to cohesin enrichment around centromeres when MTs are depolymerized (Eckert et al., 2007). Our work and others (A. Marston, personal communication) extend these observations by showing that components of the CTF19 complex (Mcm21, Iml3, and Chl4) are not only required for this enrichment, but also contribute to physically linking pericentromeres together, consistent with the strong genetic interactions between mutants in CTF19 components and an allele of the MCD1 gene (Supplemental Figure S5). Furthermore, Smc3-GFP fluorescence at pericentromeres was diminished in the absence of Mcm21 during a normal cell cycle (K. Bloom, personal communication), indicating that the loss of pericentromeric cohesin enrichment we observed by ChIP is not an artifact of the nocodazole arrest.
We analyzed the possible causes of the cohesion defect and found that mcm21 cells do not prematurely cleave cohesin, but that there is a decrease in the Scc2 loading factor at the centromere, suggesting that cohesion establishment may be defective. To date, we have not detected a physical interaction between Mcm21 and Scc2 (unpublished data), so it is not clear how Mcm21 contributes to cohesin establishment. The CTF19 complex may specify a chromatin modification around centromeres that enriches cohesin, similar to the γ-H2AX phosphorylation that surrounds a DNA double-strand break and serves to signal the de novo loading of cohesin there (Strom et al., 2004; Unal et al., 2004). Another possibility is that Mcm21 ensures a higher order pericentromeric chromosome structure that involves cohesin, such as the proposed cruciform structure (Yeh et al., 2008). The localization of Mcm21 is restricted to within 2 kb of the centromere (Supplemental Figure S6), so it is unlikely to directly mediate pericentromeric cohesion. Because budding yeast lack pericentromeric heterochromatin, the CTF19 kinetochore complex may fulfill this role in establishing pericentromeric cohesion. Mcm21 is conserved (McAinsh et al., 2006; McClelland et al., 2007), so it will be interesting to determine if it and other conserved CTF19 components are involved in the establishment of cohesin domains around neocentromeres that lack heterochromatin in multicellular eukaryotes (for review, see Cheeseman and Desai, 2008).
Pericentromeric Cohesion Is Important for Biorientation
We analyzed the requirement for Mcm21 function when Ipl1 function was decreased and found a defect in kinetochore biorientation. The missegregation in the mcm21Δ deg-ipl1 strain allowed us to specifically assess the role of pericentromeric sister chromatid cohesion in biorientation. Remarkably, artificially linking the pericentromeres was sufficient to suppress both the cohesion and the biorientation defects in mcm21Δ deg-ipl1 cells. Consistent with this, we found a genetic interaction between a small reduction in Ipl1 levels and a defect in pericentromeric cohesion on just a single chromosome. Although we cannot eliminate the possibility that Mcm21 directly regulates Ipl1 function in some manner, the simplest interpretation of this data is that pericentromeric cohesion directly aids biorientation, consistent with recent work in fission yeast (Sakuno et al., 2009). Although we did not detect a biorientation defect in mcm21Δ cells, the mutant was originally identified due to an increase in the nondisjunction of a nonessential minichromosome (Poddar et al., 1999). We assume that the strength of the Ipl1 correction system ensures that chromosomes biorient in mcm21 mutant cells. Furthermore, a number of the genes we identified in the SGA screen have been previously implicated in sister chromatid cohesion (Mayer et al., 2001; Kenna and Skibbens, 2003; Mayer et al., 2004), underscoring the critical connection between Ipl1 and cohesion.
We were unable to test whether suppression of the cohesion defect was sufficient to restore viability to mcm21 deg-ipl1 cells because we could not establish a way to restore linkage to all pericentromeres. Therefore, additional defects in mcm21 mutant cells may contribute to creating lethality with ipl1-321. However, many other kinetochore mutants do not exhibit synthetic interactions with ipl1-321 (Pinsky et al., 2006), suggesting specific connections to the CTF19 complex. In conclusion, our data strongly suggest that the biorientation defect in the double mutant cells is due to the lack of proper pericentromeric cohesion.
Models for the Role of Pericentromeric Cohesion in Biorientation
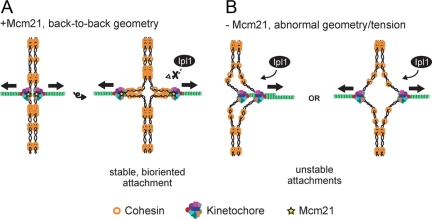
Although any linkage between sister chromatids may be sufficient to generate the tension required for biorientation (Dewar et al., 2004), our data reveal that pericentromeric cohesion significantly enhances the fidelity of biorientation. We propose two possible models that are not mutually exclusive (Figure 7). First, pericentromeric cohesion might impose a kinetochore orientation that facilitates proper chromosome segregation, consistent with recent evidence suggesting a special kinetochore geometry (Indjeian and Murray, 2007; Loncarek et al., 2007; Yeh et al., 2008; Sakuno et al., 2009). This geometry would aid in biorientation and result in stable, bioriented attachments that are not substrates for Ipl1 (Figure 7A). In the absence of this geometry, there might be an enhanced requirement for Ipl1 function to achieve biorientation, or Ipl1 could have an unidentified role in kinetochore geometry (Figure 7B). Second, a certain level of pericentromeric cohesion may be required to satisfy the tension-sensing mechanism (Figure 7B). This could be through a variety of mechanisms, such as attaining the proper spatial regulation of Ipl1 from its substrates (Tanaka et al., 2002; Liu et al., 2009; Shimogawa et al., 2009) or achieving the appropriate interkinetochore or intrakinetochore stretch (Maresca and Salmon, 2009; Uchida et al., 2009). Because defects in pericentromeric cohesion could alter these properties, MT attachments may not be properly stabilized in mcm21 mutant cells. In this case, MTs may detach from kinetochores even when they biorient, consistent with our observation that mcm21 mutant cells transiently activate the spindle checkpoint and other reports that the CTF19 complex contributes to MT–kinetochore interactions (Hyland et al., 1999; De Wulf et al., 2003; Pot et al., 2005; Tanaka et al., 2005; McAinsh et al., 2006). Therefore, mcm21 mutant cells may become very sensitive to slight decreases in Ipl1 function due to difficulty in detecting appropriate levels of tension.
Figure 7.
Model for the role of pericentromeric cohesion in biorientation. (A) Cohesion establishment at pericentromeres mediated in part by Mcm21 helps to impose steric constraints on kinetochore orientation to ensure biorientation. Ipl1 senses tension and the bioriented attachments are stabilized. The pericentromeric region is depicted as the looped out regions of the chromosome. (B) In the absence of Mcm21, the decrease in pericentromeric cohesion may alter kinetochore geometry, making it more difficult for kinetochores to biorient and increasing the requirement for Ipl1 (left). The defect in pericentromeric cohesion may also reduce the cell's ability to sense tension properly and affect the regulation of Ipl1 and/or the stability of the MT–kinetochore attachments (left and right).
In sum, although tension may be sufficient for biorientation, pericentromeric cohesion facilitates biorientation to ensure proper chromosome segregation, and thereby reduces the need for Ipl1 function. Future studies of the other genes that are required for viability when Ipl1 function is decreased may lead to the identification of other biorientation pathways or functions for the Ipl1/Aurora kinase. In the long-term, the requirement for these genes to maintain viability when Ipl1/Aurora function is reduced could be exploited to make cancer cells more sensitive to Aurora kinase inhibitors.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Brenda Andrews and Charlie Boone (University of Toronto) for use of their pinning robots and analysis software during SGA. We thank Igor Kogut and Paul Megee for information about Scc2-Flag ChIP and helpful discussions. We thank Andrew Murray (Harvard University), Peter Sorger (Harvard Medical School), and Frank Uhlmann (Cancer Research UK) for strains. We thank Adele Marston, Josefin Fernius, Kerry Bloom, and Yoshi Watanabe for communicating results and helpful comments, and the Biggins lab, Chitra Kotwaliwale, and Sue Jaspersen for critical reading of the manuscript and discussions. T.M.N. was supported in part by Public Health Service, National Research Service Award, T32 GM07270, from the National Institute of General Medical Sciences. This work was supported by National Institutes of Health grant GM064386 to S.B. and funded by Canadian Institutes of Health Research operating (FRN 57913) and salary (MSH 63646) grants to B.D.L. S.B. is a Scholar of the Leukemia and Lymphoma Society.
Glossary
Abbreviations used:
- MT
microtubule
- NC-Mcd1
noncleavable Mcd1
- SGA
systematic genetic analysis.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-04-0330) on July 15, 2009.
REFERENCES
- Allshire R. C., Nimmo E. R., Ekwall K., Javerzat J. P., Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- Ben-Shahar T. R., Heeger S., Lehane C., East P., Flynn H., Skehel M., Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- Bernard P., Maure J. F., Partridge J. F., Genier S., Javerzat J. P., Allshire R. C. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Biggins S., Bhalla N., Chang A., Smith D. L., Murray A. W. Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics. 2001;159:453–470. doi: 10.1093/genetics/159.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Severin F. F., Bhalla N., Sassoon I., Hyman A. A., Murray A. W. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat Y., Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Buvelot S., Tatsutani S. Y., Vermaak D., Biggins S. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 2003;160:329–339. doi: 10.1083/jcb.200209018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Anderson S., Jwa M., Green E. M., Kang J., Yates J. R., Chan C. S., Drubin D. G., Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Ciosk R., Shirayama M., Shevchenko A., Tanaka T., Toth A., Nasmyth K. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- Collins K. A., Castillo A. R., Tatsutani S. Y., Biggins S. De novo kinetochore assembly requires the centromeric histone H3 variant. Mol. Biol. Cell. 2005;16:5649–5660. doi: 10.1091/mbc.E05-08-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack B. P., Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- De Wulf P., McAinsh A. D., Sorger P. K. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar H., Tanaka K., Nasmyth K., Tanaka T. U. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature. 2004;428:93–97. doi: 10.1038/nature02328. [DOI] [PubMed] [Google Scholar]
- Eckert C. A., Gravdahl D. J., Megee P. C. The enhancement of pericentromeric cohesin association by conserved kinetochore components promotes high-fidelity chromosome segregation and is sensitive to microtubule-based tension. Genes Dev. 2007;21:278–291. doi: 10.1101/gad.1498707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- Grewal S. I., Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Hauf S., Watanabe Y. Kinetochore orientation in mitosis and meiosis. Cell. 2004;119:317–327. doi: 10.1016/j.cell.2004.10.014. [DOI] [PubMed] [Google Scholar]
- He X., Asthana S., Sorger P. K. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- He X., Rines D. R., Espelin C. W., Sorger P. K. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- Hyland K. M., Kingsbury J., Koshland D., Hieter P. Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J. Cell Biol. 1999;145:15–28. doi: 10.1083/jcb.145.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indjeian V. B., Murray A. W. Budding yeast mitotic chromosomes have an intrinsic bias to biorient on the spindle. Curr. Biol. 2007;17:1837–1846. doi: 10.1016/j.cub.2007.09.056. [DOI] [PubMed] [Google Scholar]
- Kellum R., Alberts B. M. Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J. Cell Sci. 1995;108(Pt 4):1419–1431. doi: 10.1242/jcs.108.4.1419. [DOI] [PubMed] [Google Scholar]
- Kenna M. A., Skibbens R. V. Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different replication factor C complexes. Mol. Cell. Biol. 2003;23:2999–3007. doi: 10.1128/MCB.23.8.2999-3007.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. M., Nicklas R. B. Tension on chromosomes increases the number of kinetochore microtubules but only within limits. J. Cell Sci. 2000;113(Pt 21):3815–3823. doi: 10.1242/jcs.113.21.3815. [DOI] [PubMed] [Google Scholar]
- Kotwaliwale C. V., Frei S. B., Stern B. M., Biggins S. A pathway containing the Ipl1/aurora protein kinase and the spindle midzone protein Ase1 regulates yeast spindle assembly. Dev. Cell. 2007;13:433–445. doi: 10.1016/j.devcel.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacefield S., Murray A. W. The spindle checkpoint rescues the meiotic segregation of chromosomes whose crossovers are far from the centromere. Nat. Genet. 2007;39:1273–1277. doi: 10.1038/ng2120. [DOI] [PubMed] [Google Scholar]
- Laloraya S., Guacci V., Koshland D. Chromosomal addresses of the cohesin component Mcd1p. J. Cell Biol. 2000;151:1047–1056. doi: 10.1083/jcb.151.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S., Spencer F. A. Bipolar orientation of chromosomes in Saccharomyces cerevisiae is monitored by Mad1 and Mad2, but not by Mad3. Proc. Natl. Acad. Sci. USA. 2004;101:10655–10660. doi: 10.1073/pnas.0404102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A., Katou Y., Mori S., Yokobayashi S., Kelly G. P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A., McIntyre J., Katou Y., Kanoh Y., Hopfner K. P., Shirahige K., Uhlmann F. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol. Cell. 2006;23:787–799. doi: 10.1016/j.molcel.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M. J., Lampson M. A., Lens S. M. Sensing chromosome bi-orientation by spatial separation of Aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J., Klein F., Engebrecht J. Genetic and morphological approaches for the analysis of meiotic chromosomes in yeast. Methods Cell Biol. 1998;53:257–285. doi: 10.1016/s0091-679x(08)60882-1. [DOI] [PubMed] [Google Scholar]
- Loncarek J., Kisurina-Evgenieva O., Vinogradova T., Hergert P., La Terra S., Kapoor T. M., Khodjakov A. The centromere geometry essential for keeping mitosis error free is controlled by spindle forces. Nature. 2007;450:745–749. doi: 10.1038/nature06344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca T. J., Salmon E. D. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J. Cell Biol. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M. L., Gygi S. P., Aebersold R., Hieter P. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol. Cell. 2001;7:959–970. doi: 10.1016/s1097-2765(01)00254-4. [DOI] [PubMed] [Google Scholar]
- Mayer M. L., et al. Identification of protein complexes required for efficient sister chromatid cohesion. Mol. Biol. Cell. 2004;15:1736–1745. doi: 10.1091/mbc.E03-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh A. D., Meraldi P., Draviam V. M., Toso A., Sorger P. K. The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation. EMBO J. 2006;25:4033–4049. doi: 10.1038/sj.emboj.7601293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland S. E., Borusu S., Amaro A. C., Winter J. R., Belwal M., McAinsh A. D., Meraldi P. The CENP-A NAC/CAD kinetochore complex controls chromosome congression and spindle bipolarity. EMBO J. 2007;26:5033–5047. doi: 10.1038/sj.emboj.7601927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee P. C., Mistrot C., Guacci V., Koshland D. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell. 1999;4:445–450. doi: 10.1016/s1097-2765(00)80347-0. [DOI] [PubMed] [Google Scholar]
- Michaelis C., Ciosk R., Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Minshull J., Straight A., Rudner A., Dernburg A., Belmont A., Murray A. W. Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr. Biol. 1996;6:1609–1620. doi: 10.1016/s0960-9822(02)70784-7. [DOI] [PubMed] [Google Scholar]
- Nicklas R. B., Ward S. C. Elements of error correction in mitosis: microtubule capture, release, and tension. J. Cell Biol. 1994;126:1241–1253. doi: 10.1083/jcb.126.5.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo-Hafalla M. T., Katou Y., Shirahige K., Uhlmann F. Displacement and re-accumulation of centromeric cohesin during transient pre-anaphase centromere splitting. Chromosoma. 2007;116:531–544. doi: 10.1007/s00412-007-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz J., Stemmann O., Rank S., Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G., Maddox P. S., Salmon E. D., Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A. H., et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Peters J. M., Tedeschi A., Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- Pinsky B. A., Biggins S. The spindle checkpoint: tension vs. attachment. Trends Cell Biol. 2005;15(9):486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Pinsky B. A., Kung C., Shokat K. M., Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat. Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- Pinsky B. A., Tatsutani S. Y., Collins K. A., Biggins S. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev. Cell. 2003;5:735–745. doi: 10.1016/s1534-5807(03)00322-8. [DOI] [PubMed] [Google Scholar]
- Poddar A., Roy N., Sinha P. MCM21 and MCM22, two novel genes of the yeast Saccharomyces cerevisiae are required for chromosome transmission. Mol. Microbiol. 1999;31:349–360. doi: 10.1046/j.1365-2958.1999.01179.x. [DOI] [PubMed] [Google Scholar]
- Pot I., Knockleby J., Aneliunas V., Nguyen T., Ah-Kye S., Liszt G., Snyder M., Hieter P., Vogel J. Spindle checkpoint maintenance requires Ame1 and Okp1. Cell Cycle. 2005;4:1448–1456. doi: 10.4161/cc.4.10.2106. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Heiter P. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W. C. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Sakuno T., Tada K., Watanabe Y. Kinetochore geometry defined by cohesion within the centromere. Nature. 2009;458:852–858. doi: 10.1038/nature07876. [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink G., Lawrence C. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1974. [Google Scholar]
- Shimogawa M. M., Widlund P. O., Riffle M., Ess M., Davis T. N. Bir1 is required for the tension checkpoint. Mol. Biol. Cell. 2009;20:915–923. doi: 10.1091/mbc.E08-07-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R. V., Corson L. B., Koshland D., Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A. F., Belmont A. S., Robinett C. C., Murray A. W. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Strom L., Lindroos H. B., Shirahige K., Sjogren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Mukae N., Dewar H., van Breugel M., James E. K., Prescott A. R., Antony C., Tanaka T. U. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Cosma M. P., Wirth K., Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Fuchs J., Loidl J., Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- Tanaka T. U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M. J., Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Tong A. H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Toth A., Ciosk R., Uhlmann F., Galova M., Schleiffer A., Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida K. S., Takagaki K., Kumada K., Hirayama Y., Noda T., Hirota T. Kinetochore stretching inactivates the spindle assembly checkpoint. J. Cell Biol. 2009;184:383–390. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F., Lottspeich F., Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Unal E., Arbel-Eden A., Sattler U., Shroff R., Lichten M., Haber J. E., Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Unal E., Heidinger-Pauli J. M., Kim W., Guacci V., Onn I., Gygi S. P., Koshland D. E. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- Waples W. G., Chahwan C., Ciechonska M., Lavoie B. D. Putting the brake on FEAR: Tof2 promotes the biphasic release of Cdc14 phosphatase during mitotic exit. Mol. Biol. Cell. 2008;20:245–255. doi: 10.1091/mbc.E08-08-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S. A., Gerton J. L., Polancic J. E., DeRisi J. L., Koshland D., Megee P. C. The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2004;2:E260. doi: 10.1371/journal.pbio.0020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y., Sakuno T., Shimura M., Watanabe Y. Heterochromatin links to centromeric protection by recruiting shugoshin. Nature. 2008;455:251–255. doi: 10.1038/nature07217. [DOI] [PubMed] [Google Scholar]
- Yeh E., Haase J., Paliulis L. V., Joglekar A., Bond L., Bouck D., Salmon E. D., Bloom K. S. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell. 2008;31:143–151. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.