Abstract
Autophagy is a major pathway of intracellular degradation mediated by formation of autophagosomes. Recently, autophagy was implicated in the degradation of intracellular bacteria, whose size often exceeds the capacity of normal autophagosomes. However, the adaptations of the autophagic machinery for sequestration of large cargos were unknown. Here we developed a yeast model system to study the effect of cargo size on the requirement of autophagy-related (Atg) proteins. We controlled the size of peroxisomes before their turnover by pexophagy, the selective autophagy of peroxisomes, and found that peroxisome size determines the requirement of Atg11 and Atg26. Small peroxisomes can be degraded without these proteins. However, Atg26 becomes essential for degradation of medium peroxisomes. Additionally, the pexophagy-specific phagophore assembly site, organized by the dual interaction of Atg30 with functionally active Atg11 and Atg17, becomes essential for degradation of large peroxisomes. In contrast, Atg28 is partially required for all autophagy-related pathways independent of cargo size, suggesting it is a component of the core autophagic machinery. As a rule, the larger the cargo, the more cargo-specific Atg proteins become essential for its sequestration.
INTRODUCTION
Macroautophagy (hereafter autophagy) is a conservative eukaryotic pathway for the degradation of intracellular material in the lysosome/vacuole via its sequestration by double-membrane vesicles called autophagosomes. Autophagy has been implicated in many physiological processes and human diseases (Huang and Klionsky, 2007; Rubinsztein et al., 2007; Mizushima et al., 2008). It is mainly a nonselective process sequestering a random portion of cytoplasm under starvation conditions. However, under certain conditions protein complexes, organelles and intracellular pathogens are selectively removed from the cytosol by specialized autophagy-related pathways. For example, the cytoplasm-to-vacuole targeting (Cvt) pathway selectively delivers certain vacuolar resident hydrolases under vegetative conditions (Stromhaug and Klionsky, 2003; Farre et al., 2007), macropexophagy (hereafter pexophagy) ensures selective turnover of peroxisomes under peroxisome repression conditions (Dunn et al., 2005; Monastyrska and Klionsky, 2006; Sakai et al., 2006), and xenophagy selectively disposes microbes that invade cells during infection (Levine and Deretic, 2007; Schmid and Munz, 2007).
Irrespective of the material being degraded by autophagy-related pathways, multiple steps are required to sequester it from the cytosol in double-membrane vesicles. These include induction of the particular pathway, cargo selection and packaging, nucleation of vesicle formation, vesicle expansion and completion, retrieval of autophagic machinery components, fusion of completed vesicles with the lysosome/vacuole, breakdown of the single-membrane vesicle and its cargo, and transport of liberated amino acids and lipids to the cytosol for reuse (Klionsky, 2005; Suzuki and Ohsumi, 2007). To date, 32 autophagy-related (Atg) proteins are known, of which 17 are components of the core autophagic machinery essential for all autophagy-related pathways and 15 are the additional components required only for certain pathways or species (Kabeya et al., 2007; Meijer et al., 2007; Xie and Klionsky, 2007; Farre et al., 2008).
The core autophagic machinery includes 1) the Atg9 cycling system (Atg1, Atg2, Atg9, Atg13, Atg18, and Atg27), 2) the phosphatidylinositol 3-kinase complex (Atg6/Vps30, Atg14, Vps15, and Vps34), and 3) the ubiquitin-like protein system (Atg3, Atg4, Atg5, Atg7, Atg8, Atg10, Atg12, and Atg16). Although the role of the core machinery in the double-membrane vesicle formation is well studied (Xie and Klionsky, 2007), much less is known about how the core machinery is adapted or modulated with additional components to accommodate the nonselective sequestration of bulk cytosol (autophagosome formation) or selective sequestration of specific cargos (Cvt vesicle, pexophagosome, or bacteria-containing autophagosome formation). For example, the Atg17 complex (Atg17, Atg29, and Atg31) constitutes the autophagosome-specific addition to the core machinery. The Cvt vesicle formation specifically requires the Atg19–Atg11 receptor–adaptor complex and both autophagosome and Cvt vesicle formation require an additional component of the Atg9 cycling system, Atg23. The pexophagosome-specific additions include the Atg30–Atg11–Atg17 receptor–adaptors complex, the coiled-coil protein Atg25, and the sterol glucosyltransferase Atg26. Both Cvt vesicle and pexophagosome formation require the coiled-coil protein Atg28 and the phosphatidylinositol 3-phosphate binding proteins Atg20, Atg21, and Atg24 (Farre et al., 2007, 2008; Kabeya et al., 2007; Xie and Klionsky, 2007). The additions for bacteria-containing autophagosomes are unknown.
Most components of the autophagic machinery colocalize at the phagophore assembly site (PAS; Noda et al., 2002), whose organization is different under growth and starvation conditions. The Cvt-specific PAS organization depends on the cargo-receptor-adaptor complex of aminopeptidase I (Ape1), Atg19, and Atg11 (Shintani and Klionsky, 2004). In contrast, the autophagy-specific PAS organization depends on two Atg17 complexes, Atg1–Atg13–Atg17 and Atg17–Atg29–Atg31 (Suzuki et al., 2007; Cao et al., 2008; Cheong et al., 2008; Kawamata et al., 2008). Although the levels of Atg1, Atg11, Atg17, and Atg19 at the PAS are the same under both growth and starvation conditions, Atg11 and Atg19 are dispensable for autophagy, whereas Atg17, Atg29, and Atg31 are dispensable for the Cvt pathway (Kamada et al., 2000; Kim et al., 2001; Scott et al., 2001; Kawamata et al., 2005; Kabeya et al., 2007; Geng et al., 2008). Interestingly, both Atg11 and Atg17 are required for pexophagy in Saccharomyces cerevisiae and interact with the peroxisome receptor Atg30 in Pichia pastoris (Kim et al., 2001; Cheong et al., 2005; Farre et al., 2008). However, how the pexophagy- or xenophagy-specific PAS is organized is unknown.
The size and number of double-membrane vesicles depends on the amount of Atg8 and Atg9 at the PAS (Yen et al., 2007; Geng et al., 2008; Xie et al., 2008). The levels of both Atg8 and Atg9 at the PAS double after the transfer of cells from vegetative to starvation conditions, which correlates with assembly of 300–900-nm diameter autophagosomes instead of 150-nm diameter Cvt vesicles (Baba et al., 1997; Geng et al., 2008). Atg8 and Atg9 are recruited to the Cvt-specific PAS in an Atg11-dependent manner. However, their recruitment to the autophagy-specific PAS does not require Atg11, but depends on Atg17 (Shintani and Klionsky, 2004; He et al., 2006; Suzuki et al., 2007; Cheong et al., 2008). Pexophagosomes are much larger than Cvt vesicles and bacteria-containing autophagosomes can be considerably larger than typical starvation-induced autophagosomes (Dunn et al., 2005; Huang and Klionsky, 2007). However, how pexophagosomes or bacteria-containing autophagosomes are formed around exceptionally large cargos is unknown.
Here we used two independent approaches to control peroxisome size and show for the first time that the requirements of Atg11 and Atg26, as well as the importance of the Atg30–Atg11 interaction, for pexophagy correlate with peroxisome size. We conclude that the larger the peroxisomes, the more pexophagy-specific Atg proteins become essential for their degradation. Therefore, the pexophagosome-specific additions to the core autophagic machinery may largely constitute the adaptations for increased cargo size and could lead us to the mechanisms generating bacteria-containing autophagosomes in humans.
MATERIALS AND METHODS
Strains, Plasmids, and Transformation
The P. pastoris strains and plasmids are listed in Table 1. Cells were transformed by electroporation (Cregg and Russell, 1998). His+-transformants were selected on SD (1.7 g/l yeast nitrogen base [YNB] without amino acids and ammonium sulfate, 2% wt/vol dextrose, 0.5% wt/vol ammonium sulfate, and 2% wt/vol agar) plates without histidine. Arginine (50 mg/l) was added, when needed. Geneticin- or zeocin-resistant transformants were selected on YPD (1% wt/vol yeast extract, 2% wt/vol bacto peptone, 2% wt/vol dextrose, and 2% wt/vol agar) plates with 0.25 mg/ml geneticin or 0.1 mg/ml zeocin, respectively. Two independent transformants of each strain with each plasmid were examined in parallel by each functional assay.
Table 1.
P. pastoris strains and plasmids used in this study
| Name | Description | Genotype and plasmid | Source |
|---|---|---|---|
| GS115 | WT | his4 | Cregg et al. (1985) |
| GS200 | WT | arg4 his4 | Waterham et al. (1996) |
| PDG2d | Δatg28 | GS200 Δatg28::ScARG4 | Stasyk et al. (2006) |
| PDG3d | Δatg26 | GS200 Δatg26::ScARG4 | Stasyk et al. (2003) |
| PPY12h | WT | arg4 his4 | Gould et al. (1992) |
| PPY12m | WT | arg4 his4 | Subramani laboratory |
| R8 | Δatg11 | GS115 atg11-2::ZeocinR | Kim et al. (2001) |
| R12 | Δatg1 | GS115 atg1-1::ZeocinR | Stromhaug et al. (2001) |
| SJCF44 | Δatg30 | PPY12m Δatg30::ZeocinR (pJCF56) | Farre et al. (2008) |
| SJCF51 | WT | PPY12m his4::pTW51 (PAOX1-GFP-SKL, HIS4) | This study |
| SJCF332 | Δatg30 | SJCF44 PAOX1::pJCF143 (PAOX1-BFP-SKL, BlasticidinR) | Farre et al. (2008) |
| SJCF385 | WT | SJCF332 his4::pJCF213 (PATG30-ATG30-GFP, HIS4) | Farre et al. (2008) |
| SJCF498 | Δatg11 Δatg8 | R8 Δatg8::GeneticinR (pJCF182) | Farre et al. (2007) |
| SJCF550 | Δatg30 | SJCF44 his4::pTW51 (PAOX1-GFP-SKL, HIS4) | This study |
| SJCF652 | Δatg30 | SJCF44 his4::pTW74 (PGAPDH-GFP-SKL, HIS4) | This study |
| SJCF757 | ATG30(S112A) | SJCF332 his4::pJCF369 (PATG30-ATG30(S112A)-GFP, HIS4) | Farre et al. (2008) |
| SJCF925 | Δatg8 | PPY12h Δatg8::GeneticinR (pJCF182) | This study |
| SJCF929 | Δatg17 | PPY12h Δatg17::GeneticinR (pKB7) | This study |
| SJCF936 | Δatg30 | PPY12h Δatg30::ZeocinR (pJCF56) | This study |
| SJCF948 | Δatg11 Δatg17 | R8 Δatg17::GeneticinR (pKB7) | This study |
| SMY261 | WT | PPY12m his4::pTW74 (PGAPDH-GFP-SKL, HIS4) | This study |
| STN18 | WT | GS115 his4::pTW74 (PGAPDH-GFP-SKL, HIS4) | Nazarko et al. (2007b) |
| STN20 | Δatg26 | PDG3d his4::pTW74 (PGAPDH-GFP-SKL, HIS4) | Nazarko et al. (2007b) |
| STN22 | Δatg1 | R12 his4::pTW74 (PGAPDH-GFP-SKL, HIS4) | Nazarko et al. (2007b) |
| STN26 | WT | GS115 his4::pTW51 (PAOX1-GFP-SKL, HIS4) | Nazarko et al. (2007b) |
| STN28 | Δatg26 | PDG3d his4::pTW51 (PAOX1-GFP-SKL, HIS4) | Nazarko et al. (2007b) |
| STN29 | Δatg1 | R12 his4::pTW51 (PAOX1-GFP-SKL, HIS4) | Nazarko et al. (2007b) |
| STN37 | Δatg11 | R8 his4::pTW74 (PGAPDH-GFP-SKL, HIS4) | This study |
| STN38 | Δatg28 | PDG2d his4::pTW74 (PGAPDH-GFP-SKL, HIS4) | This study |
| STN44 | Δatg11 | R8 his4::pTW51 (PAOX1-GFP-SKL, HIS4) | This study |
| STN46 | Δatg28 | PDG2d his4::pTW51 (PAOX1-GFP-SKL, HIS4) | This study |
| STN66 | Δatg1 | R12 his4::pJCF208 (PATG8-GFP-ATG8, HIS4) | This study |
| STN68 | Δatg26 | PDG3d his4::pJCF208 (PATG8-GFP-ATG8, HIS4) | This study |
| STN70 | WT | GS115 his4::pJCF208 (PATG8-GFP-ATG8, HIS4) | This study |
| STN75 | Δatg11 | R8 his4::pJCF208 (PATG8-GFP-ATG8, HIS4) | This study |
| STN77 | Δatg28 | PDG2d his4::pJCF208 (PATG8-GFP-ATG8, HIS4) | This study |
| STN80 | Δpex11 | STN18 Δpex11::ZeocinR (pMYΔ11′) #4 | This study |
| STN81 | Δpex11 | STN18 Δpex11::ZeocinR (pMYΔ11′) #5 | This study |
| STN82 | Δatg26 Δpex11 | STN20 Δpex11::ZeocinR (pMYΔ11′) #1 | This study |
| STN83 | Δatg26 Δpex11 | STN20 Δpex11::ZeocinR (pMYΔ11′) #3 | This study |
| STN89 | Δatg30 | SJCF936 his4::pJCF208 (PATG8-GFP-ATG8, HIS4) | This study |
| STN92 | Δatg17 | SJCF929 his4::pTW51 (PAOX1-GFP-SKL, HIS4) | This study |
| STN94 | Δatg11 Δatg17 | SJCF948 his4::pTW51 (PAOX1-GFP-SKL, HIS4) | This study |
| STN189 | WT | PPY12h his4::pJCF208 (PATG8-GFP-ATG8, HIS4) | This study |
| STN223 | WT | PPY12h arg4::pJCF340 (PATG11-FLAG-ATG11, ARG4) | This study |
Peroxisome and Pexophagy Induction Conditions
Cells were pregrown to the late exponential-stationary phase in the first YPD culture, diluted 25–50-fold with fresh YPD medium and pregrown to the early-midexponential phase in the second YPD culture, and then washed twice with YNB solution (1.7 g/l YNB without amino acids and ammonium sulfate) and inoculated into peroxisome-induction medium at an OD600 of 0.3–0.6 (for growth in most media) or 1.5–2.0 (for 15-h induction in (-C)/methylamine medium or 0.5- and 1.5-h inductions in methanol/ammonia medium). Then, cells were washed twice with YNB solution and inoculated into pexophagy-induction medium. The sources of carbon/nitrogen in peroxisome- and pexophagy-induction media as well as incubation times are shown in the figures. All peroxisome- and pexophagy-induction media were prepared in YNB solution. Histidine (50 mg/l) and/or arginine (50 mg/l) were added, when needed. All peroxisome-, but not pexophagy-induction, media contained 0.05% wt/vol yeast extract. The concentration of carbon sources were as follows: 0.5% vol/vol methanol, 0.5% vol/vol oleate, 0.5% vol/vol ethanol, or 2% wt/vol glucose. The oleate stock emulsion contained 20% vol/vol oleate and 0.5% vol/vol Tween-80. The concentration of nitrogen sources were 0.25 or 0.5% wt/vol ammonium sulfate or 0.1% wt/vol methylamine hydrochloride.
Electron Microscopy of Peroxisomes
Peroxisomes were induced as described above, and cells were processed as described previously (Harper et al., 2002). Images were captured on a transmission electron microscope (1200 EX II, JEOL, Peabody, MA) coupled to a digital camera (Orius 600, Gatan, Pleasanton, CA) and processed using the Gatan Digital Micrograph and Adobe Photoshop 7.0 software (San Jose, CA). Areas of individual peroxisomes were measured using ImageJ software (http://rsb.info.nih.gov/ij/).
Biochemical Studies of Pexophagy
Peroxisomes were induced as described above, cells were washed twice with YNB solution from peroxisome-induction medium and transferred to fresh glucose/(-N) or ethanol/(-N) medium at an OD600 of 1 or 2, respectively, to induce pexophagy. Cells from 1-ml culture samples were collected by centrifugation after 0, 6, 12, and 24 h (for methanol- or oleate-induced cells) or 0, 3, 6, and 12 h (for methylamine-induced cells) of adaptation to the favorable carbon source. Crude extracts were prepared in the presence of TCA (Baerends et al., 2000). SDS-PAGE and immunoblotting were performed as described previously (Laemmli, 1970; Kyhse-Andersen, 1984). Antigen–antibody complexes were detected by enhanced chemiluminescence.
Fluorescence Microscopy of Pexophagy
Peroxisomes were induced as described above in the presence of 2.5 or 5 μg/ml FM 4-64, diluted from 1 mg/ml stock solution in DMSO. Then, cells were washed twice with YNB solution, aliquots were placed on ice until observation, and the rest of the cells were transferred to fresh glucose/(-N) or ethanol/(-N) medium at an OD600 of 1 or 2, respectively, to induce pexophagy. After 2 h (for methylamine-induced cells), 9 h (for oleate-induced cells), or 12 h (for methanol-induced cells) of adaptation to the favorable carbon source, cell cultures were placed on ice until observation. The progress of pexophagy was determined immediately after each time point. Images were captured on a motorized fluorescence microscope (Axioskop 2 MOT, Carl Zeiss MicroImaging, Thornwood, NY) coupled to a monochrome digital camera (AxioCam MRm, Carl Zeiss MicroImaging) and processed using the AxioVision 4.5 and Adobe Photoshop 7.0 software.
Ape1 Maturation Assay
Cells were pregrown twice in YPD as described for pexophagy studies, washed twice with YNB solution, and transferred to glucose/ammonia and glucose/(-N) media at an OD600 of 0.03 and 0.5, respectively, to study maturation of Ape1 under growth and starvation conditions. The 1.5 OD600 of cells from each culture sample were collected by centrifugation after 14 and 15 h of adaptation to glucose/ammonia and glucose/(-N), respectively.
GFP-Atg8 Processing Assay
Cells were pregrown twice in YPD as described for pexophagy studies, washed twice with YNB solution, and transferred to glucose/ammonia medium at an OD600 of 0.03 for 15 h. Then, cells were washed twice with YNB solution and transferred to glucose/(-N) medium. Cells from 1-ml culture samples were collected by centrifugation after 0, 1, 2, 4, and 6 h of nitrogen starvation.
RESULTS
Peroxisome Size in P. pastoris Is Easily Manipulated by Peroxisome Inducers and Induction Time
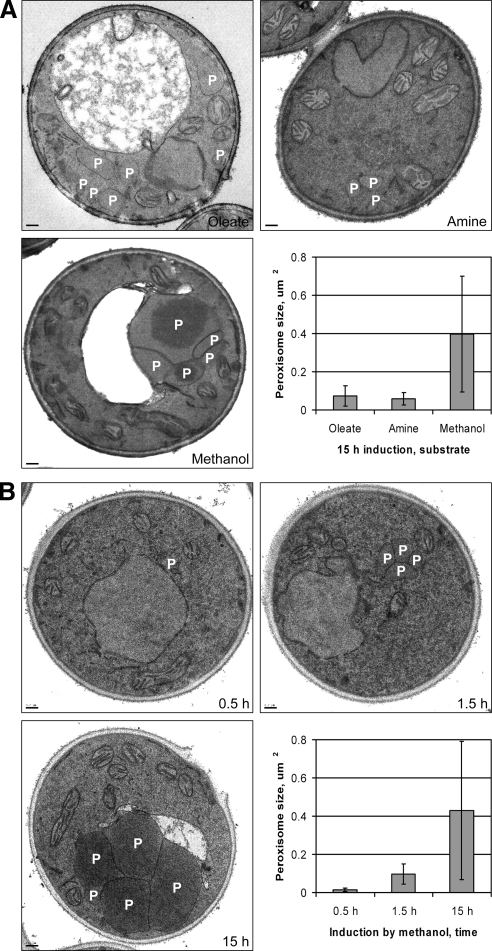
Recently, we found that Atg26 is essential for degradation of methanol-induced peroxisomes, but only partially required for degradation of oleate- or amine-induced peroxisomes (Nazarko et al., 2007a,b). We hypothesized that the larger size of methanol-, relative to oleate- or amine-induced, peroxisomes resulted in the enhanced requirement of Atg26 for pexophagy. To clarify the role of Atg26, we studied the sizes of methanol-, oleate-, or amine-induced peroxisomes by electron microscopy. The WT cells induced for 15 h in oleate (O15), amine (A15), or methanol (M15) medium had on average 5.9, 1.7, and 3.6 peroxisomes/cell, respectively. The average area of O15 (0.074 ± 0.053 μm2) and A15 (0.059 ± 0.032 μm2) peroxisomes was 5.4- and 6.7-fold smaller than that of M15 (0.397 ± 0.303 μm2) peroxisomes, respectively (Figure 1A).
Figure 1.
Manipulation of peroxisome size in P. pastoris by peroxisome inducers and induction time. The WT (GS115) cells were induced: (A) for 15 h in oleate (oleate/ammonia), amine ((-C)/methylamine) or methanol (methanol/ammonia) medium and (B) for 0.5, 1.5, or 15 h in methanol/ammonia medium. P, peroxisome. Bar, 0.2 μm.
To discriminate between the effects of peroxisome inducer and size on pexophagy, we manipulated the size of methanol-induced peroxisomes by varying the induction time. The WT cells induced in methanol medium for 0.5 (M0.5), 1.5 (M1.5), or 15 (M15) h contained on average 1.6, 3.0, and 3.4 peroxisomes/cell, respectively. The average area of M0.5 (0.014 ± 0.008 μm2) and M1.5 (0.097 ± 0.053 μm2) peroxisomes was 30.7- and 4.4-fold smaller than that of M15 peroxisomes (0.430 ± 0.362 μm2), respectively (Figure 1B). Interestingly, the sizes of O15 and A15 peroxisomes fell in the size range of M0.5–M1.5 peroxisomes, allowing us to study the role of Atg26 in degradation of the differently sized peroxisomes with a single inducer, methanol.
Atg26 Is Not Essential for Pexophagy of Small Methanol-induced Peroxisomes
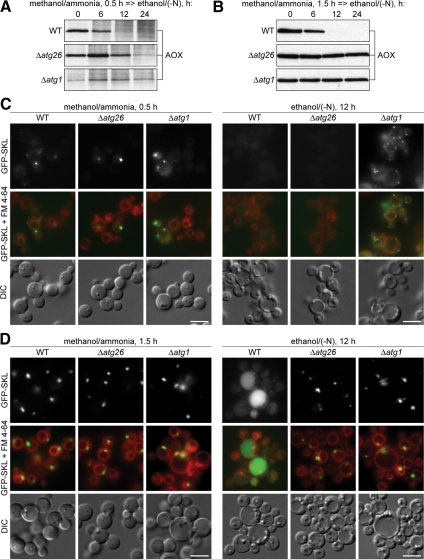
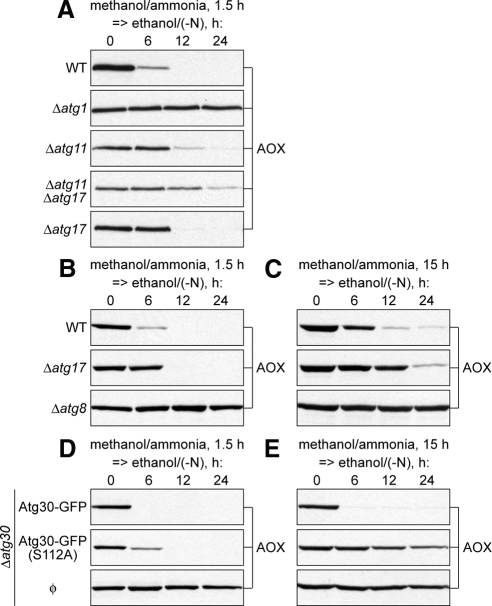
The pexophagy of M15 peroxisomes is completely blocked in the P. pastoris Δatg26 cells (Nazarko et al., 2007b). To check whether Atg26 is indeed a pexophagy enhancer essential for degradation of only large (M15) peroxisomes, the WT, Δatg26, and Δatg1 cells were induced in methanol medium for 0.5 and 1.5 h and transferred to ethanol medium to induce pexophagy (Figure 2, A and B). In the WT cells with both small and medium peroxisomes, peroxisomal alcohol oxidase (AOX) was completely degraded after 12 h of ethanol adaptation. However, it was stable for at least 24 h in the Δatg1 mutant, independent of the size of methanol-induced peroxisomes, in agreement with the essential role of Atg1 in all autophagy-related pathways in P. pastoris (Mukaiyama et al., 2002; Farre et al., 2007; Nazarko et al., 2007b). Remarkably, Atg26 was only partially required for pexophagy of small peroxisomes (Figure 2A), but became essential for pexophagy of medium peroxisomes (Figure 2B).
Figure 2.
Atg26 is not essential for pexophagy of small methanol-induced peroxisomes. (A and B) The WT (GS115), Δatg26 (PDG3d), and Δatg1 (R12) cells were induced in methanol medium for (A) 0.5 h or (B) 1.5 h and transferred to ethanol medium. At the indicated time points culture samples were collected and immunoblotted with antibodies against peroxisomal AOX. (C and D) The WT (STN26), Δatg26 (STN28), and Δatg1 (STN29) cells were induced in methanol medium for (C) 0.5 h or (D) 1.5 h and transferred to ethanol medium for 12 h. Pexophagy, as judged by a reduction in peroxisome number per cell and/or by the vacuolar delivery of GFP-SKL, was monitored by fluorescence microscopy. Peroxisomes were labeled with GFP-SKL and vacuolar membranes with FM 4-64. Bar, 5 μm.
To confirm that small (but not medium) peroxisomes are degraded in the Δatg26 cells, we labeled peroxisomes with a fusion of the green fluorescent protein (GFP) to the peroxisomal targeting signal 1, Ser-Lys-Leu (SKL), expressed under control of promoter of the P. pastoris AOX1 gene (Figure 2, C and D). M0.5 cells of all strains already contained a single GFP-SKL labeled peroxisome. However, these peroxisomes were no longer present in the majority of the WT and Δatg26 cells after 12 h of ethanol adaptation. At the same time, the Δatg1 mutant retained its peroxisomes (Figure 2C). M1.5 cells of all strains developed clusters of medium peroxisomes. However, only peroxisomes of the WT strain were degraded in the vacuole (Figure 2D). Altogether, our biochemical and microscopy data show that small methanol-induced peroxisomes can be degraded without Atg26 and its role might be peroxisome size-specific.
Atg26 Becomes Essential for Pexophagy of Enlarged Oleate-induced Peroxisomes
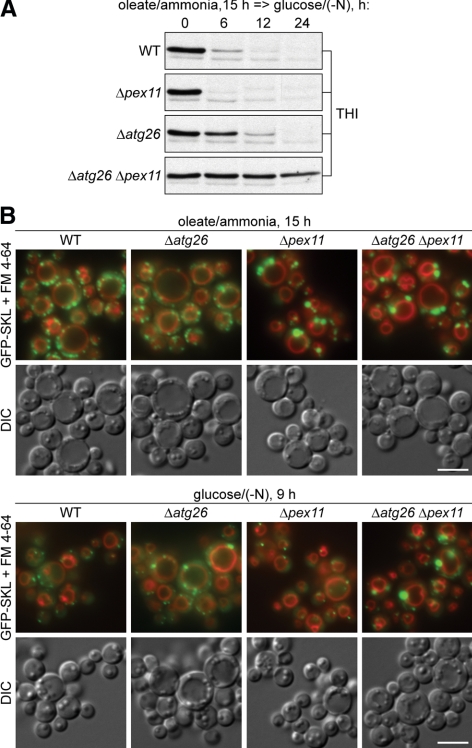
Recently, we showed that pexophagy of small-to-medium O15 peroxisomes occurs in the P. pastoris Δatg26 cells (Nazarko et al., 2007b). We sought to verify our hypothesis that Atg26 is essential for degradation of only large peroxisomes using an alternative strategy, by increasing the size of oleate-induced peroxisomes. We constructed the Δpex11 and Δatg26 Δpex11 mutants by deleting the PEX11 gene in the WT and Δatg26 strains that already expressed GFP-SKL under the control of the constitutive promoter of the P. pastoris GAPDH gene. Yeast and mammalian cells lacking Pex11 have fewer and larger peroxisomes than normal (Yan et al., 2005). Among the P. pastoris proteins that control the number and size of oleate-induced peroxisomes (Pex11, Pex30, and Pex31), the absence of Pex11 has the most severe phenotype, with enlarged O15 peroxisomes reaching the size of large M15 peroxisomes (Yan et al., 2008).
The WT, Δpex11, Δatg26, and Δatg26 Δpex11 cells were induced in oleate medium for 15 h and transferred to glucose medium to induce pexophagy (Figure 3A). In the Δpex11, WT, and Δatg26 cells, there was almost complete degradation of peroxisomal thiolase (THI) after 6, 12, and 24 h of glucose adaptation, respectively. Although the delayed degradation of small-to-medium O15 peroxisomes in the Δatg26 was expected (Nazarko et al., 2007b), the reproducibly more efficient degradation of enlarged O15 peroxisomes in the Δpex11 was surprising. More interestingly, there was an almost complete block of degradation of enlarged O15 peroxisomes in the Δatg26 Δpex11 double mutant (Figure 3A), indicating that the requirement of Atg26 increases as the size of oleate-induced peroxisomes increases.
Figure 3.
Atg26 becomes essential for pexophagy of enlarged oleate-induced peroxisomes. (A) The WT (STN18), Δpex11 (STN80), Δatg26 (STN20), and Δatg26 Δpex11 (STN83) cells were induced in oleate medium for 15 h and transferred to glucose medium. (B) The WT (STN18), Δatg26 (STN20), Δpex11 (STN80), and Δatg26 Δpex11 (STN82) cells were induced in oleate medium for 15 h and transferred to glucose medium for 9 h. Bar, 5 μm.
These biochemical results were confirmed by fluorescence microscopy (Figure 3B). O15 cells of the Δpex11 and Δatg26 Δpex11 had much fewer and larger peroxisomes than WT and Δatg26. However, after 9 h of glucose adaptation, the vast majority of enlarged O15 peroxisomes disappeared in the Δpex11 strain, but not in the Δatg26 Δpex11 double mutant (Figure 3B). The Δatg26 strain exhibited delayed degradation of small-to-medium O15 peroxisomes relative to the WT strain, as described (Nazarko et al., 2007b). Taken together, our biochemical and microscopy data demonstrate that Atg26 becomes essential for pexophagy of enlarged O15 peroxisomes. In summary, experimenting with the sizes of methanol- and oleate-induced peroxisomes led us to conclude that Atg26 is a pexophagy enhancer essential for degradation of medium-to-large peroxisomes independent of the nature of peroxisome inducers.
Similar-sized Oleate- and Amine-induced Peroxisomes Have Similar Requirements of Pexophagy-specific Atg Proteins
Atg26 together with Atg11, Atg28, and Atg30 is required for the early stages of pexophagy, but not for autophagy in yeasts. All four proteins are at least partially localized at the PAS and required for pexophagosome formation in P. pastoris (Kim et al., 2001; Oku et al., 2003; Stasyk et al., 2003; Stasyk et al., 2006; Yamashita et al., 2006; Farre et al., 2008; unpublished data). Therefore, Atg11, Atg26, Atg28, and Atg30 were suggested to provide peroxisome recognition and guidance of the sequestering membranes around peroxisomes (Dunn et al., 2005; Farre et al., 2008). But like Atg26, these pexophagy-specific Atg proteins might just facilitate pexophagosome formation and be essential for pexophagy of only large peroxisomes.
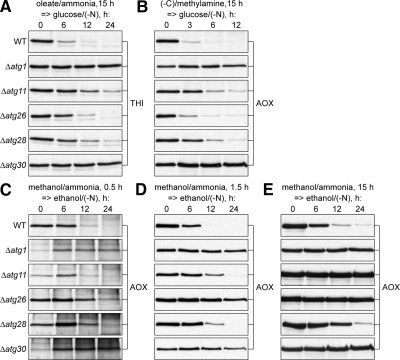
To address this possibility we studied the roles of Atg11, Atg28, and Atg30 in degradation of small-to-medium O15 and A15 peroxisomes, which are about the same size, but induced by different substrates. The WT, Δatg1, Δatg11, Δatg26, Δatg28, and Δatg30 cells were induced in oleate (Figure 4A) or amine (Figure 4B) medium for 15 h and transferred to glucose medium to induce pexophagy. As expected, in the WT cells peroxisomal THI (Figure 4A) or AOX (Figure 4B) was degraded, whereas in the Δatg1 cells it was stable. Surprisingly, Atg11 and Atg28 were only partially required for degradation of both O15 and A15 peroxisomes, like Atg26. However, Atg30 was essential. Fluorescence microscopy confirmed these observations (Supplemental Figures S1–3). In summary, similar-sized (small-to-medium), but differently induced (O15 and A15) peroxisomes have similar requirements of pexophagy-specific Atg proteins.
Figure 4.
Atg11 and Atg28 regulate the magnitude of pexophagic response in peroxisome size-dependent and -independent manner, respectively. The WT (GS115), Δatg1 (R12), Δatg11 (R8), Δatg26 (PDG3d), Δatg28 (PDG2d), and Δatg30 (SJCF936 in A and B or SJCF44 in C–E) cells were induced in (A) oleate or (B) amine medium for 15 h and transferred to glucose medium or induced in methanol medium for (C) 0.5 h, (D) 1.5 h, or (E) 15 h and transferred to ethanol medium.
Differently sized Methanol-induced Peroxisomes Have Different Requirements of Atg11
The link between peroxisome size and the requirement of Atg proteins was explored further by studying the roles of Atg11, Atg28, and Atg30 in the degradation of M0.5, M1.5, and M15 peroxisomes, which are of different sizes, but are induced by the same substrate, methanol. The WT, Δatg1, Δatg11, Δatg26, Δatg28, and Δatg30 cells were induced in methanol medium for 0.5, 1.5, and 15 h and transferred to ethanol medium to induce pexophagy (Figure 4, C–E). Importantly, the kinetics of AOX degradation in WT cells was comparable, irrespective of peroxisome size. The negative control Δatg1 was completely blocked in pexophagy. Interestingly, Atg11 was only partially required for degradation of small and medium peroxisomes (Figure 4, C and D), but was essential for degradation of large peroxisomes (Figure 4E). The pattern of Atg11 requirement resembled that of Atg26 with the only difference that Atg26 was already essential for degradation of medium peroxisomes (Figure 4D). In contrast, Atg28 was partially required and Atg30 was essential for degradation of methanol-induced peroxisomes of any size. Collectively (Figure 4), these results suggest that Atg28 regulates the magnitude of the pexophagic response and Atg30 tags peroxisomes for degradation regardless of peroxisome size and the nature of peroxisome inducers. In summary, 1) similarly induced, but differently sized peroxisomes (M0.5, M1.5, and M15) have different requirements of at least two selective Atg proteins, Atg11 and Atg26; 2) the larger the peroxisomes, the more selective Atg proteins become essential for their degradation (neither Atg26, nor Atg11, is essential for pexophagy of small peroxisomes; Atg26, but not Atg11, is essential for pexophagy of medium peroxisomes, and both Atg26 and Atg11 are essential for pexophagy of large peroxisomes).
Dual Interaction of Atg30 with Atg11 and Atg17 Is Required to Organize the Pexophagy-specific PAS and Efficient Pexophagy
The partial requirement of Atg11 for the degradation of small-to-medium peroxisomes could be explained by redundancy with either Atg8 or Atg17. In S. cerevisiae the Ape1 receptor, Atg19, independently interacts with Atg11 and Atg8 for cytoplasm-to-vacuole targeting of Ape1 (Shintani et al., 2002). In growing cells Atg11 plays a major role in bridging the Ape1–Atg19 complex with the PAS. However, upon nitrogen starvation, the increased levels of Atg8 cause a partial bypass of the Atg11-dependent step (Chang and Huang, 2007). In P. pastoris Atg30 also interacts with Atg11 (Farre et al., 2008). Because, in our experiments, pexophagy was induced in medium lacking nitrogen, we tested whether the partial requirement of Atg11 for degradation of medium peroxisomes was due to the presence of Atg8. The WT, Δatg1, Δatg11, Δatg8, and Δatg11 Δatg8 cells were induced in methanol medium for 1.5 h and transferred to ethanol medium to induce pexophagy (Supplemental Figure S4A). Both Δatg8 and Δatg11 Δatg8 cells were completely blocked in the degradation of AOX showing that Atg8 is essential for pexophagy of medium peroxisomes and cannot be redundant with Atg11.
We also studied the maturation of Ape1 in the same strains in glucose medium (Supplemental Figure S4B). Under growth conditions the precursor of Ape1 (prApe1) is delivered to the vacuole by the Cvt pathway and under nitrogen starvation conditions prApe1 is delivered to the vacuole by autophagy. In the vacuole prApe1 is processed to the mature forms (mApe1) by vacuolar hydrolases (Farre et al., 2007). After 14 h of growth in glucose medium both the prApe1, and two processed forms of mApe1 were present in the WT strain. In contrast, 15 h in glucose medium without nitrogen led to complete maturation of Ape1 and degradation of the higher molecular weight mApe1. As expected (Farre et al., 2007), Δatg1 was affected in both Cvt- and autophagy-dependent maturation of Ape1, and Δatg11 was only impaired in the Cvt-dependent maturation of Ape1. However, in contrast to S. cerevisiae, where Atg8 is essential for the Cvt pathway and only partially required for autophagy (Kirisako et al., 1999; Abeliovich et al., 2000; Chang and Huang, 2007), PpAtg8 was essential for both processes (Supplemental Figure S4B). These results (Supplemental Figure S4, A and B) show that PpAtg8 is essential for all autophagy-related pathways and is not redundant with Atg11 in bridging the receptor–cargo complexes with the PAS.
The S. cerevisiae prApe1–Atg19–Atg11 complex is critical for Cvt-specific PAS organization under growth conditions (Shintani and Klionsky, 2004). However, the Atg1–Atg13–Atg17 and Atg17–Atg29–Atg31 complexes are essential for autophagy-specific PAS organization under nitrogen starvation conditions (Cao et al., 2008; Cheong et al., 2008; Kawamata et al., 2008). It was suggested that ScAtg11 and ScAtg17 serve as alternative scaffold proteins that organize the PAS in the Cvt and autophagy pathways, respectively. Consistently, the PAS localization of Atg proteins and autophagosome formation are severely affected in the S. cerevisiae Δatg11 Δatg17 double mutant (Suzuki et al., 2007; Cheong et al., 2008). Despite the fact that ScAtg11 is dispensable for starvation-induced autophagy and ScAtg17 is dispensable for the Cvt pathway, both proteins are required for glucose-induced pexophagy under nitrogen starvation conditions (Kamada et al., 2000; Kim et al., 2001; Cheong et al., 2005). Moreover, both Atg11 and Atg17 interact with Atg30 in P. pastoris (Farre et al., 2008). Therefore, we tested whether the partial requirement of Atg11 for degradation of medium peroxisomes was due to the presence of Atg17. The WT, Δatg1, Δatg11, Δatg17, and Δatg11 Δatg17 cells were induced in methanol medium for 1.5 h and transferred to ethanol medium to induce pexophagy (Figure 5A). Interestingly, Atg17 was also partially required for normal degradation of AOX, but less so than Atg11. However, the Δatg11 Δatg17 double mutant was almost completely blocked in pexophagy (Figure 5A). Fluorescence microscopy confirmed these observations (Supplemental Figure S5). Therefore, Atg11 and Atg17 are redundant for pexophagy.
Figure 5.
Dual interaction of Atg30 with Atg11 and Atg17 is required to organize the pexophagy-specific PAS and efficient pexophagy. (A) The WT (GS115), Δatg1 (R12), Δatg11 (R8), Δatg11 Δatg17 (SJCF948), and Δatg17 (SJCF929) cells were induced in methanol medium for 1.5 h and transferred to ethanol medium. (B and C) The WT (PPY12h), Δatg17 (SJCF929), and Δatg8 (SJCF925) cells were induced in methanol medium for (B) 1.5 h or (C) 15 h and transferred to ethanol medium. (D and E) The Δatg30 (SJCF332) and Δatg30 complemented with Atg30-GFP (SJCF385) and Atg30(S112A)-GFP (SJCF757) cells were induced in methanol medium for (D) 1.5 h or (E) 15 h and transferred to ethanol medium.
Because Atg17, like Atg11, was only partially required for degradation of medium peroxisomes, there was a possibility that the requirement of Atg17 increases with an increase of peroxisome size. To test this the WT, Δatg17 and Δatg8 cells were induced in methanol medium for 1.5 and 15 h and transferred to ethanol medium to induce pexophagy (Figure 5, B and C). Peroxisomes of both medium and large sizes were efficiently degraded in the WT strain, but completely escaped degradation in the Δatg8 mutant, as expected. However, the Δatg17 strain was partially affected in pexophagy irrespective of peroxisome size (Figure 5, B and C). These results place Atg17 together with Atg28 in the group of proteins that regulate the magnitude of the pexophagic response independent of peroxisome size.
Our findings indicate that 1) Atg30 is unconditionally essential for pexophagy, 2) its interacting partners Atg11 and Atg17 are redundant and 3) stimulate pexophagy in a peroxisome size-dependent and -independent manner, respectively. However, do the Atg30 adaptor proteins cooperate with Atg30 to mediate the peroxisome-PAS interaction during pexophagy? If the dual interaction of Atg30 with Atg11 and Atg17 was indeed required to organize the pexophagy-specific PAS, then the inability of Atg30 to interact with Atg11 would phenotypically mimic the lack of Atg11 and pexophagy would solely depend on the interaction of Atg30 and Atg17. To address this question we studied the degradation of medium and large peroxisomes in the Δatg30 mutant complemented with normal and mutated (S112A) forms of Atg30-GFP expressed from its endogenous promoter. Previously, we showed that phosphorylation of Atg30 at S112 is essential for its interaction with Atg11, but not with Atg17 (Farre et al., 2008). The Δatg30 mutant and complemented cells were induced in methanol medium for 1.5 and 15 h and transferred to ethanol medium to induce pexophagy (Figure 5, D and E). The Atg30-GFP fusion protein did restore pexophagy of both medium and large peroxisomes in the Δatg30 mutant. However, Atg30(S112A)-GFP rescued the pexophagy of medium (Figure 5D), but not large (Figure 5E) peroxisomes. Moreover, the phenotype of Δatg30 complemented with Atg30(S112A)-GFP resembled that of the Δatg11 mutant (Figure 4, D and E). These data demonstrate that Atg17 and/or its interaction with Atg30 are sufficient for degradation of medium peroxisomes, but both Atg11, and its interaction with Atg30, are essential for degradation of large peroxisomes. In summary, the dual interaction of Atg30 with Atg11 and Atg17 is required to organize the pexophagy-specific PAS and efficient pexophagy.
Atg28 Regulates the Magnitude of Autophagic Response in All Autophagy-related Pathways
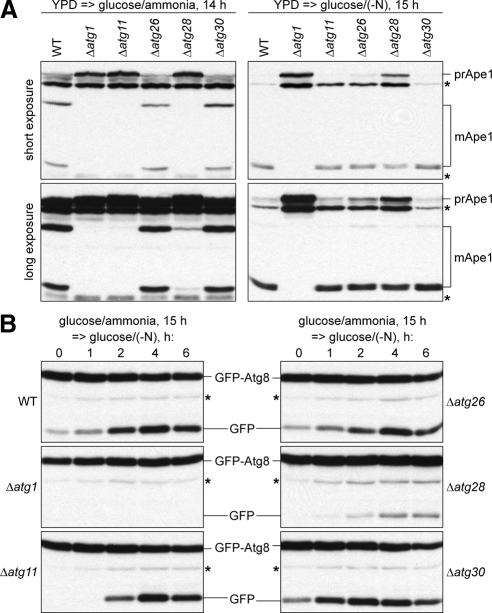
In contrast to Atg11 and Atg26, Atg28 regulates the magnitude of the pexophagic response independent of peroxisome size (Figure 4). This result is more consistent with its role in pexophagosome nucleation, rather than in elongation. If so, Atg28 might also be partially required for the nucleation of double-membrane vesicles in other autophagy-related pathways. Recently, it was reported that Atg28 is required for the Cvt pathway, but not for autophagy in P. pastoris (Stasyk et al., 2006; Farre et al., 2007). Because new biochemical autophagy assays became available for P. pastoris, namely maturation of Ape1 and the GFP-Atg8 processing assay (Farre et al., 2007, 2008), we reexamined the role of Atg28 in these pathways.
After 14 h of growth in glucose medium the WT, Δatg26, and Δatg30 strains were characterized by normal maturation of Ape1, but it was completely abolished in the Δatg1 and Δatg11 mutants (Figure 6A, left panel), as expected (Farre et al., 2007, 2008). In contrast, the Δatg28 cells were only partially affected in the Cvt pathway, because a small amount of prApe1 was processed to the mApe1 forms (Figure 6A, left panel, long exposure). After 15 h of nitrogen starvation in glucose medium, prApe1 almost completely matured to mApe1 in the WT, Δatg11, Δatg26, and Δatg30 strains, and Ape1 maturation was completely blocked only in the Δatg1 mutant (Figure 6A, right panel). These results confirmed that Atg11 and Atg30 are dispensable for autophagy (Farre et al., 2007, 2008) and provided evidence that Atg26 is not required for autophagy. However, in the Δatg28 strain, prApe1 was only partially processed to mApe1 (Figure 6A, right panel, short exposure), indicating that Atg28 is partially required for both Cvt and autophagy pathways.
Figure 6.
Atg28 regulates the magnitude of autophagic response in all autophagy-related pathways. (A) The WT (GS115), Δatg1 (R12), Δatg11 (R8), Δatg26 (PDG3d), Δatg28 (PDG2d), and Δatg30 (SJCF936) cells were grown in YPD medium and transferred to glucose medium with or without nitrogen for 14 or 15 h, respectively. (B) The WT (STN70), Δatg1 (STN66), Δatg11 (STN75), Δatg26 (STN68), Δatg28 (STN77), and Δatg30 (STN89) cells were grown in glucose medium for 15 h and transferred to glucose medium without nitrogen. *Nonspecific band.
The requirement of Atg28 in autophagy was verified by the GFP-Atg8 processing assay. During nitrogen starvation a part of GFP-Atg8 is trapped in the completed autophagosomes and degraded in the vacuole. Because of the higher resistance of GFP, versus Atg8, to the vacuolar hydrolases, the GFP moiety accumulates in the vacuole reflecting the Atg8 delivery or autophagy rates (Shintani and Klionsky, 2004; Farre et al., 2008). To study the autophagy rates, the WT, Δatg1, Δatg11, Δatg26, Δatg28, and Δatg30 cells with GFP-Atg8 expressed under its endogenous promoter were grown in glucose medium for 15 h and transferred to glucose medium without nitrogen (Figure 6B). During the time course of nitrogen starvation, GFP accumulated in the WT, Δatg11, Δatg26, and Δatg30 strains with a peak at 4 h. In contrast, GFP-Atg8 was not processed in the Δatg1 mutant. These results are in agreement with maturation of Ape1 under nitrogen starvation conditions. Both prApe1 and GFP-Atg8 processing assays prove that Atg26 is not required for autophagy, as was previously suggested based on Δatg26 survival rates under nitrogen starvation conditions (Oku et al., 2003; unpublished data). Despite some GFP accumulation in Δatg28, this strain was severely affected in the GFP-Atg8 delivery to the vacuole (Figure 6B). In summary, the results of two independent biochemical assays demonstrate that Atg28 is indeed partially required for both Cvt and autophagy pathways. Therefore, Atg28 is not specific for selective autophagy as suggested previously (Stasyk et al., 2006; Farre et al., 2007). Rather, it regulates the magnitude of autophagic response in all autophagy-related pathways.
DISCUSSION
In this article we studied the role of several Atg proteins as a function of cargo size during pexophagy. We established two reliable approaches to control peroxisome size by manipulating peroxisome inducers and induction time that provided us with information on the autophagic machinery that otherwise would be difficult to access studying selective autophagy with a cargo of fixed size (like the Cvt pathway) or nonselective autophagy that enwraps a random portion of cytoplasm. We show for the first time that Atg26 and Atg11 stimulate pexophagy in a manner that depends on cargo size. Namely, Atg26 and Atg11 are essential for scaling up pexophagy to degrade medium and large peroxisomes, respectively. On the other hand, Atg17 and Atg28 stimulate autophagy independent of cargo size, providing a basic autophagic response sufficient to sequester small-to-medium peroxisomes. Finally, both Atg11 and Atg17, and their interaction with the peroxisome receptor Atg30, are functionally important for efficient pexophagy.
Peroxisome Size Determines the Requirement of Pexophagy-specific Atg Proteins
Autophagy is a central player in the immunological control of bacterial, parasitic, and viral infections (Levine and Deretic, 2007). For example, some bacteria are specifically recognized by autophagy and targeted for degradation as cargos. However, the size of autophagosomes that sequester intracellular bacteria is often considerably larger than those formed under starvation conditions (Huang and Klionsky, 2007). For example, the area of autophagosomes enclosing a cluster of Streptococcus pyogenes (also known as group A Streptococcus) is 25–100-fold larger than the area of typical starvation-induced autophagosomes (Nakagawa et al., 2004). This raises the question regarding how such huge bacteria-containing autophagosomes arise and what factors are essential for this process. Our yeast model system that studies the effect of cargo size on the requirement of Atg proteins, the autophagy of differently sized peroxisomes, may be relevant to the question of how autophagy of large structures is achieved. Indeed, degradation of both peroxisomes in yeast and bacteria in mammalian cells are selective processes in contrast to the bulk degradation of cytosol during starvation (Nakagawa et al., 2004; Nazarko et al., 2007b). Additionally, peroxisome size in P. pastoris can vary significantly depending on the peroxisome inducer and induction time. For example, the area of M15 peroxisomes is 5.4- and 6.7-fold larger than that of O15 and A15 peroxisomes, respectively. The difference in peroxisome area is even bigger during the time course of methanol induction, because M15 peroxisomes are 4.4- and 30.7-fold larger than M1.5 and M0.5 peroxisomes, respectively. Both selectivity and peroxisome size range make pexophagy an excellent model system to study the effects of cargo size on autophagosome biogenesis.
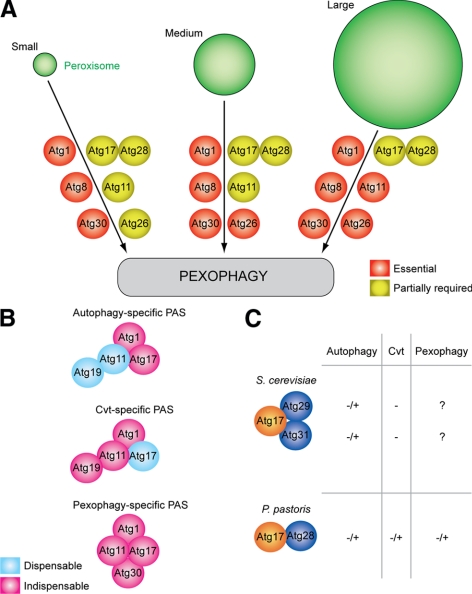
Here, we examined the requirement of Atg1, Atg8, Atg11, Atg17, Atg26, Atg28, and Atg30 proteins for pexophagy of small (M0.5), medium (M1.5), small-to-medium (O15 or A15), and large (M15) peroxisomes. The Atg proteins were divided into three groups: 1) those essential for peroxisome degradation independent of peroxisome size (Atg1, Atg8, and Atg30), 2) those that are partially required for pexophagy independent of peroxisome size (Atg17 and Atg28), and 3) others whose requirement during pexophagy depends on peroxisome size (Atg11 and Atg26; Figure 7A). We suggest that the proteins from the first two groups are essential or partially required for cargo selection and/or nucleation of the double-membrane vesicle formation according to the scheme proposed for autophagy-related pathways (Klionsky, 2005; Suzuki and Ohsumi, 2007). In contrast, the proteins from the last group are more important at the vesicle expansion step. Moreover, the larger the peroxisomes, the more Atg proteins become essential for pexophagosome formation. For example, Atg26 is essential to scale up pexophagy for degradation of medium peroxisomes and Atg11 is essential for further scale up of pexophagy for degradation of large peroxisomes (Figure 7A). Besides, Atg11 and Atg26 might regulate pexophagy in a dose-dependent manner, because both proteins are specifically induced by methanol, like the peroxin Pex12 (Supplemental Figure S6).
Figure 7.
Peroxisome size provides insights into the function of autophagy-related proteins. (A) Peroxisome size determines the requirement of pexophagy-specific Atg proteins. Atg1, Atg8, and Atg30 are essential, and Atg17 and Atg28 are partially required for pexophagy independent of peroxisome size. In contrast, the requirement of Atg11 and Atg26 increases with peroxisome size. (B) Comparison of autophagy-, Cvt-, and pexophagy-specific PASs. Atg11 and Atg17 are the structural components of any PAS. Atg11 is indispensable for Cvt vesicle formation, and Atg17 is indispensable for autophagosome formation. However, both of them are functionally important for pexophagosome formation at the pexophagy-specific PAS, organized by the peroxisome receptor Atg30. The PASs may also contain other proteins not studied here. (C) P. pastoris Atg28 is a structural and functional homolog of two S. cerevisiae proteins, Atg29 and Atg31. Atg28 interacts with Atg17, and is partially required for all autophagy-related pathways. −/+, partially required; −, not required; ?, the requirement is not known.
Our findings that the level and requirement of Atg26 increase with an increase of the size of peroxisomes (Figure 7A and Supplemental Figure S6) are in agreement with the recently proposed role of Atg26 in elongation of the micropexophagic apparatus (MIPA), the cup-shaped isolation membrane formed along the peroxisome surface to bridge the opposing vacuolar sequestering membranes during micropexophagy (Yamashita et al., 2006). Mechanistically, biogenesis of the MIPA and pexophagosome are very similar, except for the fact that MIPA is fused with the vacuolar sequestering membranes before it is completed into pexophagosome. Therefore, the increased demand for Atg26 during pexophagy of medium-to-large peroxisomes most likely reflects the increased demand for elongation of the pexophagosome membrane.
Pexophagy-specific PAS and Requirements for Efficient Pexophagosome Formation
Interestingly, both Atg30 and Atg1 are essential for pexophagy irrespective of peroxisome size and the nature of the peroxisome inducer, despite the fact that Atg30, unlike Atg1, is completely dispensable for the Cvt and autophagy pathways. These results support the view that Atg30 is indeed a peroxisome receptor and Atg1 is a component of the core autophagic machinery (Xie and Klionsky, 2007; Farre et al., 2008). Atg11 and Atg17, the interacting partners of Atg30 in P. pastoris and components of Atg1 complex in S. cerevisiae, stimulate pexophagy in peroxisome size-dependent and -independent manners, respectively (Figure 7A). Both Atg11 and Atg17 are also required for pexophagy in S. cerevisiae, despite the fact that Atg11 is dispensable for nonselective autophagy and Atg17 is dispensable for the selective Cvt pathway (Kamada et al., 2000; Kim et al., 2001; Cheong et al., 2005). Moreover, we found that Atg11 and Atg17 are redundant in the degradation of the medium peroxisomes and the interaction of Atg11 with Atg30 is essential to scale up pexophagy for the degradation of large peroxisomes. The above data favor the existence of a pexophagy-specific PAS, organized by the dual interaction of Atg30 (and/or the serine-threonine protein kinase, Atg1) with functionally active Atg11 and Atg17. Therefore, the pexophagy-specific PAS combines the features of the Cvt- and autophagy-specific PASs, because Atg11 and Atg17 are not just structurally present, as in other PASs (Geng et al., 2008), but are both functionally important for pexophagosome formation (Figure 7B).
The stronger requirement of Atg11 versus Atg17, combined with their redundancy and the interactions of both proteins with Atg30, suggests that Atg11 supplies the majority and Atg17 the minority of the same functions required for pexophagosome formation. In S. cerevisiae, the delivery capacity of the Cvt and autophagy pathways depends on the amount and proper localization of Atg8 and Atg9. Artificially increased amounts of Atg11 at the PAS enhance the recruitment of Atg8 and Atg9 to this site and lead to the formation of more Cvt vesicles under growth conditions. Similarly, the naturally enhanced recruitment of Atg8 and Atg9 to the PAS under starvation conditions causes the formation of numerous and large autophagosomes (Yen et al., 2007; Geng et al., 2008; Xie et al., 2008). However, in contrast to the growth conditions, Atg8 and Atg9 are recruited to the starvation-induced PAS independent of Atg11 by two Atg17 complexes (Atg1–Atg13–Atg17 and Atg17–Atg29–Atg31; Shintani and Klionsky, 2004; He et al., 2006; Suzuki et al., 2007; Cao et al., 2008; Cheong et al., 2008; Kawamata et al., 2008). Although the same amounts of Atg11 and Atg17 are present at the PAS at all times, Atg11 and Atg17 are crucial only under the growth and starvation conditions, respectively (Kamada et al., 2000; Geng et al., 2008). Our experiments show that enormously large cargos, like peroxisomes, might require both Atg11- and Atg17-dependent pathways of Atg8 and Atg9 recruitment to the PAS to supply enough membrane for efficient autophagosome formation.
The dual interaction of Atg30 with functionally active Atg11 and Atg17 might also require the dual induction mechanism. In all our experiments, pexophagy was induced by the switch of the carbon source (e.g., methanol-to-ethanol or oleate-to-glucose) under the nitrogen starvation conditions, as described (Nazarko et al., 2007b). We speculate that Atg11- and Atg17-dependent pathways of Atg8 and Atg9 recruitment to the PAS were induced by the favorable carbon source and starvation, respectively. Although the ability of starvation to induce the Atg17-dependent pathway of PAS organization was extensively proven recently (Suzuki et al., 2007; Cao et al., 2008; Cheong et al., 2008; Kawamata et al., 2008), the effect of the change of the carbon source on the amount and localization of Atg11 requires further study. It is known that entry of S. pyogenes into the cytoplasm of human nonphagocytic cells induces selective autophagy by increasing the amount of lipidated Atg8 (Nakagawa et al., 2004). Our results predict that the induction of bulk autophagy during streptococcal infection would costimulate the selective degradation of bacteria in humans. This prediction is also supported by the fact that amino acid starvation significantly increased the number of Shigella flexneri cells associated with Atg8 in canine kidney epithelial cells (Ogawa et al., 2005).
Atg28, the Missing Piece of the PAS Puzzle
Among five Atg proteins (Atg1, Atg13, Atg17, Atg29, and Atg31) that coorganize the autophagy-specific PAS in S. cerevisiae, Atg17, Atg29 and Atg31 are required for autophagy, but not for the Cvt pathway (Kamada et al., 2000; Kawamata et al., 2005; Kabeya et al., 2007). These three proteins interact with each other and colocalize at the PAS in an Atg17-dependent manner (Kabeya et al., 2007; Kawamata et al., 2008). However, Atg29 and Atg31 have no homologues in P. pastoris and were not tested for their role in pexophagy in S. cerevisiae. We found that in P. pastoris, Atg17 is partially required for degradation of peroxisomes independent of their size, as is Atg28. Our data indicate that Atg28 interacts with Atg17 (Supplemental Figure S7A), colocalizes with Atg17 at the PAS (Stasyk et al., 2006), and is partially required for other autophagy-related pathways (Figure 6). However, Atg28 has no homologues in S. cerevisiae. Interestingly, just like P. pastoris and S. cerevisiae, other yeasts that belong to the order Saccharomycetales have also either Atg28 or Atg29 (Meijer et al., 2007). We extended this analysis to Atg31 and found that 1) Candida albicans, Debaryomyces hansenii, P. pastoris, and Hansenula polymorpha have Atg28, but not Atg29 or Atg31 and 2) S. cerevisiae, Candida glabrata, Ashbya gossypii, and Kluyveromyces lactis have Atg29 and Atg31, but not Atg28 (Supplemental Figure S7B). Consistently, the alignment of these three proteins suggested that the N- and C-terminal parts of PpAtg28 have a weak homology to ScAtg29 and ScAtg31, respectively (Supplemental Figure S7C). Therefore, we hypothesize that P. pastoris Atg28 is a structural and functional equivalent of S. cerevisiae Atg29 and Atg31 combined (Figure 7C). Further studies are required to support this finding. Summarizing, Atg28 has no specific role in the cargo recognition during selective autophagy, as previously suggested (Dunn et al., 2005; Stasyk et al., 2006; Farre et al., 2007), but together with Atg17 participates in the nucleation and expansion of the double-membrane vesicles in all autophagy-related pathways.
Using valuable new methods for the manipulation of peroxisome size we have been able to investigate the function of pexophagy-specific Atg proteins. We found that peroxisome size determines the requirement of Atg11 and Atg26. We defined the pexophagy-specific PAS and found the requirements for efficient pexophagy. Finally, we described the function of Atg28 and positioned this protein in the yeast autophagic machinery. These results demonstrate that the comparative pexophagy of the differently sized peroxisomes will provide further insights into the function of Atg proteins.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Mingda Yan (University of California, San Diego, La Jolla, CA) for the strain SMY261 and plasmid pMYΔ11′, Dr. Oleh Stasyk (Institute of Cell Biology, Lviv, Ukraine) for the antibodies against Atg26, and Dr. Marilyn Farquhar for the use of the electron microscope facility. This work was supported by a National Institutes of Health Grant GM069373 to S.S.
Abbreviations used:
- A15
amine-induced for 15 h
- AOX
alcohol oxidase
- Ape1
aminopeptidase I
- Atg
autophagy-related
- Cvt
cytoplasm-to-vacuole targeting
- O15
oleate-induced for 15 h
- M0.5
methanol-induced for 0.5 h
- M1.5
methanol-induced for 1.5 h
- M15
methanol-induced for 15 h
- mApe1
mature Ape1
- MIPA
micropexophagic apparatus
- PAS
phagophore assembly site
- prApe1
precursor of Ape1
- THI
thiolase
- YNB
yeast nitrogen base.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-03-0221) on July 15, 2009.
REFERENCES
- Abeliovich H., Dunn W. A., Jr, Kim J., Klionsky D. J. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J. Cell Biol. 2000;151:1025–1034. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M., Osumi M., Scott S. V., Klionsky D. J., Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerends R. J., Faber K. N., Kram A. M., Kiel J. A., van der Klei I. J., Veenhuis M. A stretch of positively charged amino acids at the N terminus of Hansenula polymorpha Pex3p is involved in incorporation of the protein into the peroxisomal membrane. J. Biol. Chem. 2000;275:9986–9995. doi: 10.1074/jbc.275.14.9986. [DOI] [PubMed] [Google Scholar]
- Cao Y., Cheong H., Song H., Klionsky D. J. In vivo reconstitution of autophagy in Saccharomyces cerevisiae. J. Cell Biol. 2008;182:703–713. doi: 10.1083/jcb.200801035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. Y., Huang W. P. Atg19 mediates a dual interaction cargo sorting mechanism in selective autophagy. Mol. Biol. Cell. 2007;18:919–929. doi: 10.1091/mbc.E06-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Nair U., Geng J., Klionsky D. J. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 2008;19:668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Yorimitsu T., Reggiori F., Legakis J. E., Wang C. W., Klionsky D. J. Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg J. M., Barringer K. J., Hessler A. Y., Madden K. R. Pichia pastoris as a host system for transformations. Mol. Cell. Biol. 1985;5:3376–3385. doi: 10.1128/mcb.5.12.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg J.M., Russell K.A. Transformation. Methods Mol. Biol. 1998;103:27–39. doi: 10.1385/0-89603-421-6:27. [DOI] [PubMed] [Google Scholar]
- Dunn W. A., Jr, Cregg J. M., Kiel J. A., van der Klei I. J., Oku M., Sakai Y., Sibirny A. A., Stasyk O. V., Veenhuis M. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- Farre J. C., Manjithaya R., Mathewson R. D., Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre J. C., Vidal J., Subramani S. A cytoplasm to vacuole targeting pathway in. P. pastoris. Autophagy. 2007;3:230–234. doi: 10.4161/auto.3905. [DOI] [PubMed] [Google Scholar]
- Geng J., Baba M., Nair U., Klionsky D. J. Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. J. Cell Biol. 2008;182:129–140. doi: 10.1083/jcb.200711112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., McCollum D., Spong A. P., Heyman J. A., Subramani S. Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast. 1992;8:613–628. doi: 10.1002/yea.320080805. [DOI] [PubMed] [Google Scholar]
- Harper C. C., South S. T., McCaffery J. M., Gould S. J. Peroxisomal membrane protein import does not require Pex17p. J. Biol. Chem. 2002;277:16498–16504. doi: 10.1074/jbc.M111728200. [DOI] [PubMed] [Google Scholar]
- He C., Song H., Yorimitsu T., Monastyrska I., Yen W. L., Legakis J. E., Klionsky D. J. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J. Cell Biol. 2006;175:925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Klionsky D. J. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Kawamata T., Suzuki K., Ohsumi Y. Cis1/Atg31 is required for autophagosome formation in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2007;356:405–410. doi: 10.1016/j.bbrc.2007.02.150. [DOI] [PubMed] [Google Scholar]
- Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T., Kamada Y., Kabeya Y., Sekito T., Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol. Biol. Cell. 2008;19:2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T., Kamada Y., Suzuki K., Kuboshima N., Akimatsu H., Ota S., Ohsumi M., Ohsumi Y. Characterization of a novel autophagy-specific gene, ATG29. Biochem. Biophys. Res. Commun. 2005;338:1884–1889. doi: 10.1016/j.bbrc.2005.10.163. [DOI] [PubMed] [Google Scholar]
- Kim J., Kamada Y., Stromhaug P. E., Guan J., Hefner-Gravink A., Baba M., Scott S. V., Ohsumi Y., Dunn W. A., Jr, Klionsky D. J. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levine B., Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer W. H., van der Klei I. J., Veenhuis M., Kiel J. A. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–116. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastyrska I., Klionsky D. J. Autophagy in organelle homeostasis: peroxisome turnover. Mol. Aspects Med. 2006;27:483–494. doi: 10.1016/j.mam.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaiyama H., Oku M., Baba M., Samizo T., Hammond A. T., Glick B. S., Kato N., Sakai Y. Paz2 and 13 other PAZ gene products regulate vacuolar engulfment of peroxisomes during micropexophagy. Genes Cells. 2002;7:75–90. doi: 10.1046/j.1356-9597.2001.00499.x. [DOI] [PubMed] [Google Scholar]
- Nakagawa I., et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Nazarko T. Y., Farre J. C., Polupanov A. S., Sibirny A. A., Subramani S. Autophagy-related pathways and specific role of sterol glucoside in yeasts. Autophagy. 2007a;3:263–265. doi: 10.4161/auto.3907. [DOI] [PubMed] [Google Scholar]
- Nazarko T. Y., Polupanov A. S., Manjithaya R. R., Subramani S., Sibirny A. A. The requirement of sterol glucoside for pexophagy in yeast is dependent on the species and nature of peroxisome inducers. Mol. Biol. Cell. 2007b;18:106–118. doi: 10.1091/mbc.E06-06-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T., Suzuki K., Ohsumi Y. Yeast autophagosomes: de novo formation of a membrane structure. Trends Cell Biol. 2002;12:231–235. doi: 10.1016/s0962-8924(02)02278-x. [DOI] [PubMed] [Google Scholar]
- Ogawa M., Yoshimori T., Suzuki T., Sagara H., Mizushima N., Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- Oku M., Warnecke D., Noda T., Muller F., Heinz E., Mukaiyama H., Kato N., Sakai Y. Peroxisome degradation requires catalytically active sterol glucosyltransferase with a GRAM domain. EMBO J. 2003;22:3231–3241. doi: 10.1093/emboj/cdg331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein D. C., Gestwicki J. E., Murphy L. O., Klionsky D. J. Potential therapeutic applications of autophagy. Nat. Rev. Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- Sakai Y., Oku M., van der Klei I. J., Kiel J. A. Pexophagy: autophagic degradation of peroxisomes. Biochim. Biophys. Acta. 2006;1763:1767–1775. doi: 10.1016/j.bbamcr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- Schmid D., Munz C. Innate and adaptive immunity through autophagy. Immunity. 2007;27:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S. V., Guan J., Hutchins M. U., Kim J., Klionsky D. J. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol. Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Huang W. P., Stromhaug P. E., Klionsky D. J. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Klionsky D. J. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasyk O. V., Nazarko T. Y., Stasyk O. G., Krasovska O. S., Warnecke D., Nicaud J. M., Cregg J. M., Sibirny A. A. Sterol glucosyltransferases have different functional roles in Pichia pastoris and Yarrowia lipolytica. Cell Biol. Int. 2003;27:947–952. doi: 10.1016/j.cellbi.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Stasyk O. V., Stasyk O. G., Mathewson R. D., Farre J. C., Nazarko V. Y., Krasovska O. S., Subramani S., Cregg J. M., Sibirny A. A. Atg28, a novel coiled-coil protein involved in autophagic degradation of peroxisomes in the methylotrophic yeast Pichia pastoris. Autophagy. 2006;2:30–38. doi: 10.4161/auto.2226. [DOI] [PubMed] [Google Scholar]
- Stromhaug P. E., Bevan A., Dunn W. A., Jr GSA11 encodes a unique 208-kDa protein required for pexophagy and autophagy in Pichia pastoris. J. Biol. Chem. 2001;276:42422–42435. doi: 10.1074/jbc.M104087200. [DOI] [PubMed] [Google Scholar]
- Stromhaug P. E., Klionsky D. J. Cytoplasm to vacuole targeting. In: Klionsky D. J., editor. Autophagy. Georgetown: Landes Bioscience; 2003. pp. 84–106. [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- Waterham H. R., de Vries Y., Russel K. A., Xie W., Veenhuis M., Cregg J. M. The Pichia pastoris PER6 gene product is a peroxisomal integral membrane protein essential for peroxisome biogenesis and has sequence similarity to the Zellweger syndrome protein PAF-1. Mol. Cell. Biol. 1996;16:2527–2536. doi: 10.1128/mcb.16.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Klionsky D. J. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Xie Z., Nair U., Klionsky D. J. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S., Oku M., Wasada Y., Ano Y., Sakai Y. PI4P-signaling pathway for the synthesis of a nascent membrane structure in selective autophagy. J. Cell Biol. 2006;173:709–717. doi: 10.1083/jcb.200512142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Rachubinski D. A., Joshi S., Rachubinski R. A., Subramani S. Dysferlin domain-containing proteins, Pex30p and Pex31p, localized to two compartments, control the number and size of oleate-induced peroxisomes in Pichia pastoris. Mol. Biol. Cell. 2008;19:885–898. doi: 10.1091/mbc.E07-10-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Rayapuram N., Subramani S. The control of peroxisome number and size during division and proliferation. Curr. Opin. Cell Biol. 2005;17:376–383. doi: 10.1016/j.ceb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Yen W. L., Legakis J. E., Nair U., Klionsky D. J. Atg27 is required for autophagy-dependent cycling of Atg9. Mol. Biol. Cell. 2007;18:581–593. doi: 10.1091/mbc.E06-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.