Abstract
Accurate gene expression requires the precise control of mRNA levels, which are determined by the relative rates of nuclear (pre-)mRNA synthesis and processing, and cytoplasmic mRNA turnover. A key step in mRNA degradation is the removal of the poly(A) tail, which involves several deadenylases including components of the Ccr4–Not complex. Here, we focused on the role of the human paralogues CNOT7 (hCaf1/Caf1a) and CNOT8 (hPop2/Caf1b/Calif), which possess deadenylase activity mediated by DEDD nuclease domains. We show that efficient proliferation requires both subunits, although combined knockdown of CNOT7 and CNOT8 further reduces cell proliferation indicating partial redundancy between these proteins. Interestingly, the function of CNOT7 in cell proliferation partly depends on its catalytic activity. On the other hand, the interaction between CNOT7 and BTG2, a member of the antiproliferative BTG/Tob family involved in transcription and mRNA decay appears less important for proliferation of MCF7 cells, suggesting that CNOT7 does not function solely in conjunction with BTG2. Further analysis of gene expression profiles of CNOT7 and/or CNOT8 knockdown cells underscores the partial redundancy between these subunits and suggests that regulation of several genes, including repression of the antiproliferative genes MSMB and PMP22, by the Ccr4–Not complex contributes to cell proliferation.
INTRODUCTION
Accurate gene expression requires the precise control of mRNA levels that are determined by the relative rates of (pre-)mRNA synthesis and processing, and by mRNA turnover. Degradation of eukaryotic mRNA is initiated by the shortening and removal of the poly(A) tail by at least two different complexes containing distinct deadenylase subunits (Parker and Song, 2004; Garneau et al., 2007; Goldstrohm and Wickens, 2008). In both yeast and human cells, initial shortening of the poly(A) tail is carried out by Pan2, which forms a heterodimer with Pan3 (Brown et al., 1996; Boeck et al., 1998; Brown and Sachs, 1998; Uchida et al., 2004). Subsequent removal of poly(A) residues is achieved by Ccr4–Not, a conserved complex, which contains about 10 subunits, including several deadenylase components (Tucker et al., 2001; Temme et al., 2004; Yamashita et al., 2005). The yeast Ccr4 protein and its human orthologues CNOT6 (hCcr4/Ccr4a) and CNOT6L (hCcr4-like/Ccr4b) contain an exonuclease/endonuclease/phosphatase (EEP) domain that is responsible for the RNA nuclease activity (Dupressoir et al., 2001; Chen et al., 2002; Tucker et al., 2002; Morita et al., 2007). In addition, these proteins interact via leucine-rich motifs with yeast Caf1 (Pop2) or its human orthologues CNOT7 (hCaf1/Caf1a) and CNOT8 (hPop2/Caf1b/Calif), which contain RNA nuclease activities attributed to DEDD domains (Daugeron et al., 2001; Dupressoir et al., 2001; Clark et al., 2004; Viswanathan et al., 2004; Bianchin et al., 2005). The CNOT7 and CNOT8 proteins interact with members of the BTG/Tob family of antiproliferative proteins, which are implicated in mRNA turnover (Ezzeddine et al., 2007; Funakoshi et al., 2007; Mauxion et al., 2008) and transcription (Prevot et al., 2000; Yoshida et al., 2000; Tzachanis et al., 2001; Passeri et al., 2006; Ou et al., 2007). Consistent with their role in mRNA decay, combined depletion of CNOT7 and CNOT8 in human cells results in decreased deadenylation of bulk mRNA and stabilizes an unstable reporter mRNA (Schwede et al., 2008).
Recruitment of Ccr4–Not to cytoplasmic mRNA may involve a series of translation-coupled events (Funakoshi et al., 2007). It was proposed that competitive binding of PABPC1 results in exchange of the release factor eRF3 for Pan2-Pan3, thereby initiating deadenylation (Funakoshi et al., 2007). Subsequently, the Ccr4–Not complex may be recruited via interactions mediated by the CNOT7 and/or CNOT8 subunits and Tob, which in turn can bind to PABPC1 (Ezzeddine et al., 2007; Funakoshi et al., 2007). However, other mechanisms may also operate to recruit the Ccr4–Not complex to mRNA molecules.
Deadenylation and decay of mRNA is also associated with microRNA-mediated gene repression (Wu et al., 2006; Wakiyama et al., 2007). In Drosophila, microRNA-mediated mRNA deadenylation requires the activity of the Ccr4–Not complex (Behm-Ansmant et al., 2006). Underscoring the functional link between mRNA degradation and microRNA-mediated repression is the colocalization of microRNAs, translationally repressed mRNA, and protein factors involved in mRNA deadenylation and decapping in cytoplasmic P-bodies (Jakymiw et al., 2005; Liu et al., 2005a,b; Pillai et al., 2005; Sen and Blau, 2005).
Furthermore, and consistent with its functions in yeast, a number of protein–protein interactions point to a separate role of the Ccr4–Not complex in transcription in human cells (Collart, 2003; Collart and Timmers, 2004). The large subunit, CNOT1, is a ligand-dependent repressor of nuclear receptor-mediated transcription, including by ERα- and RXR-containing heterodimers, via interactions mediated by multiple LxxLL motifs (Winkler et al., 2006). In contrast, CNOT6, CNOT7, and RQCD1 (hCaf40/CNOT9) potentiate transcription mediated by several nuclear receptors including ERα and RARα (Prevot et al., 2001; Hiroi et al., 2002; Morel et al., 2003; Garapaty et al., 2008), which may be mediated via interactions with the nuclear receptor coactivator NIF1 (Garapaty et al., 2008). Intriguingly, mice lacking Cnot7 appear healthy, but display a male infertility phenotype similar to RARβ null mice (Nakamura et al., 2004). Together, these data suggest that the Ccr4–Not complex mediates both transcriptional activation and repression in addition to its role in mRNA decay.
Despite the increased understanding of the properties of the human Ccr4–Not complex, relatively little is known about its role in cellular biology. The deadenylase activity of CNOT6L influences cell proliferation by regulating mRNA levels of the cell cycle inhibitor p27/Kip1, although its paralogue CNOT6 does not appear to be involved in regulation of cell proliferation (Morita et al., 2007). Here, we report that CNOT7 and CNOT8 contribute to efficient proliferation of breast cancer cells. The function of CNOT7 in cell proliferation partly depends on the deadenylase catalytic activity. Further analysis of gene expression profiles of CNOT7 and/or CNOT8 knockdown cells underscores the partial redundancy between these subunits in the control of gene activity and indicates that the regulation of several genes, including PMP22 and MSMB, by the Ccr4–Not complex is important for cell proliferation.
MATERIALS AND METHODS
Plasmids, Mutagenesis, and Small Interfering RNA
The open reading frames of CNOT7 and CNOT8 were obtained by reverse transcriptase (RT)-PCR using MCF7 NKI total RNA as a template. After cloning the respective cDNAs into the SmaI site of pBluescript II KS(+) and sequence verification, the cDNAs were subcloned in pcDNA3-FLAG (BamHI-EcoRV) and pCMV5-HA (XhoI-digested cDNA ligated into SalI-digested vector). Silent mutations in the small interfering RNA (siRNA)-target sequence (four mismatches), and site-directed mutations to inactivate the active sites (D40A) and to substitute residues required for interaction with BTG2 (E247A and Y260A) were introduced using the standard QuikChange protocol, or the QuikChange Multi kit (Stratagene, La Jolla, CA). Primer sequences are available upon request.
The following siRNA duplexes were used (Dharmacon Research, Boulder, CO): CNOT7 (D-012897-01, D-012897-02, and SMARTpool L-012897-00), CNOT8 (D-018791-01, D-018791-02, and SMARTpool L-018791-00), and nontargeting control siRNA (D-001210-01, and SMARTpool D-001810-10).
Cell Culture and Transfection
MCF7 NKI and human embryonic kidney (HEK) 293T cells were routinely maintained in DMEM supplemented with 10% fetal bovine serum, l-glutamine, and penicillin/streptomycin.
Transfection of siRNA (5 nM) into MCF7 NKI cells was carried out using INTERFERin (Polyplus, Strasbourg, France) following the manufacturer's instructions.
Cotransfection of plasmid DNA (1.6 μg) and siRNA (20 nM) into 30,000 MCF7 NKI or HEK 293T cells in six-well plates was done using jetSI-ENDO following the manufacturer's instructions (Polyplus). Cells were lysed in 100 μl lysis buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, 0.2% Nonidet P-40, 5% (vol/vol) glycerol, 0.5 mM dithiothreitol, and protease inhibitors) followed by two freeze/thaw cycles, and 5–10 μl was analyzed by immunoblotting.
Plasmid DNA was transfected into HEK 293T cells using jetPEI (Polyplus). Cells (n = 500,000) were plated in a 60-mm culture dish and transfected with 2 μg pCMV-Flag-CNOT7 and 3 μg pCMV5-HA-BTG2. After 16 h, cells were incubated in the presence of proteasome inhibitor MG132 (10 μM) for an additional 8 h.
For the analysis of cell proliferation, MCF7 NKI cells (150,000 cells in a T25 flask) were transfected with siRNA duplexes. After 24 h, cells were trypsinized and plated in 12 wells of six-well plates (10,000 cells/well). Subsequently, cells were trypsinized and counted in a hemocytometer at 24-h intervals.
Stable transfection of vectors expressing Flag-CNOT7 (6 μg DNA) in MCF7 NKI cells (400,000 cells in a 60-mm dish) was carried out using the calcium-phosphate procedure. Selection was initiated 48 h after transfection in medium containing 800 μg/ml G418. Clonally derived cell populations were isolated and maintained in medium containing 400 μg/ml G418. Clones with comparable expression levels of wild-type CNOT7 (C5), D40A (C1), and E247A Y260A (B6) were selected by immunoblotting.
Antibodies and Immunoprecipitation
Transfected HEK 293T cells were lysed in 500 μl lysis buffer. The cell suspension was left on ice for 5–10 min, freeze-thawed twice, and spun for 10 min in a microcentrifuge (full speed, 4°C). Each immunoprecipitate contained 500 μg protein lysate adjusted to 500 μl, 20 μl of a 50% mixture of protein A/protein G-agarose, and 2 μg M2 anti-Flag antibody and was incubated overnight at 4°C. After three washes with 1.0 ml lysis buffer, bound proteins were analyzed by SDS-PAGE and immunoblotting.
Antibodies used recognized CNOT1 (Winkler et al., 2006), CNOT3 (mouse 4B8, AbNova), β-tubulin (mouse 2–28-33, Santa Cruz), FLAG (mouse M2, Sigma), and HA (rat 3F10, Santa Cruz).
Flow Cytometry
MCF-7 NKI cells (150,000 cells in a T25 flask) were transfected with siRNA as described above. After 24 h, the cells were washed once in PBS and fresh medium was added. After an additional 46 h, cells were labeled for a further 2 h in the presence of 20 μM bromodeoxyuridine (BrdU) and prepared for bivariate flow cytometry using propidium iodide and FITC-conjugated anti-BrdU antibody 3D4 (BD Biosciences, San Jose, CA). Analysis was carried out using a FACS Aria flow cytometer, FACSDiva software (BD Biosciences), and the WinMDI package (http://facs.scripps.edu).
Gene Expression Profiling
MCF7 NKI cells (1.0 × 106 cells in a 100-mm culture dish) were transfected with 5 nM siRNA pools targeting CNOT7 and/or CNOT8, and/or a nontargeting control pool (Dharmacon On-Target Plus SMARTpool; total siRNA concentration was 10 nM). DNA-free total RNA of biological triplicates was isolated (Qiagen RNeasy, Chatsworth, CA, including on-column DNAse digestion), subjected to quality control using an Agilent 2100 Bioanalyzer (Wilmington, DE), and processed using Affymetrix Human Genome U133 plus 2.0 GeneChips (Santa Clara, CA), the manufacturer's labeling protocols and equipment (Nottingham Arabidopsis Stock Centre's International Affymetrix Service). Data were normalized using the MAS5 protocol and analyzed using Excel (Microsoft, Redmond, WA), Carmaweb (Rainer et al., 2006), and GeneTrail (Backes et al., 2007).
Reverse Transcriptase-Quantitative PCR
Total RNA was isolated from (transfected) cells grown in a single well of a 6- or 12-well plate (Qiagen RNeasy kit), and cDNA was prepared using an anchored oligo-dT primer (Superscript III, Invitrogen, Carlsbad, CA). After 1:5 dilution of the cDNA reaction with TE, 1 μl diluted cDNA was analyzed in triplicates by quantitative PCR (qPCR; 20 μl reaction volume, Stratagene Brilliant (II) SYBR Green mix) using a Stratagene MX3005p cycler. GAPDH or β-actin were used as reference genes. Analysis was carried out using the Stratagene MXpro package. For analysis of mRNA stability, 5 μg/ml actinomycin D (Sigma) was added to the culture medium 72 h after siRNA transfection. For the calculation of mRNA half-lives, the data were fitted using the following equation: [mRNA] = 100 · e(−k · t), where [mRNA] = 100% at t = 0, k is the decay constant, and the half-life t1/2 is given by t1/2 = ln(2)/k. The values for k and their SE were determined using Prism 3 (GraphPad Software, San Diego, CA).
RESULTS
CNOT7 and CNOT8 Are Required for Efficient Cell Proliferation
To understand the cellular function of the CNOT7 and CNOT8 deadenylases in more detail, we transfected siRNA duplexes targeting CNOT7 or CNOT8 mRNA into human MCF7 breast cancer cells. Two duplexes targeting different regions in CNOT7 resulted in efficient knockdown of mRNA and exogenous protein levels in MCF7 cells (>90%), whereas mock treated cells, or cells treated with a nontargeting control duplex did not affect mRNA or protein levels (Figure 1A). Interestingly, we found that cells treated with siRNA duplexes targeting CNOT7 displayed a slower proliferation rate compared with cells treated with nontargeting siRNA or mock-treated cells (Figure 1B). To further understand this defect in cell proliferation, siRNA-transfected cells were pulse-labeled with the thymidine analogue BrdU to allow accurate identification of cells in S-phase. Cells were fixed, incubated with anti-BrdU antibodies conjugated to FITC and propidium iodide to stain DNA, and subjected to bivariate flow cytometry. The cell cycle profiles indicated that depletion of CNOT7 caused a significant accumulation of cells in the G1-phase and a concomitant decrease of cells present in the S-phase of the cell cycle (Figure 1C). Similar results were obtained using siRNA duplexes targeting CNOT8 (Figure 1, D–F). Again, efficient knockdown was obtained at the level of mRNA and protein (>90%, Figure 1D) and reduced proliferation was observed in the absence of CNOT8 (Figure 1E). Perhaps slightly more pronounced as was the case for CNOT7, an increase of cells in G1 with a concomitant decrease in S-phase was observed in the absence of CNOT8 (Figure 1F). Together, these results reveal a role for the paralogues CNOT7 and CNOT8 in the proliferation of MCF7 cells.
Figure 1.
The Ccr4–Not deadenylase subunits CNOT7 and CNOT8 are important for cell proliferation. (A) Knockdown of CNOT7 in MCF7 cells. Knockdown of CNOT7 mRNA levels relative to GAPDH were determined after 2 d using RT-qPCR. Indicated are mock (M), nontargeting control siRNA (Con), targeting duplex 1 (#1) and 2 (#2). Inset, knockdown of CNOT7 protein levels. A vector expressing Flag-CNOT7 was cotransfected with siRNA targeting CNOT7. After 3 d, total lysates were analyzed by immunoblotting. Antibodies recognizing β-tubulin were used to assess equal loading. (B) Inhibition of cell proliferation by knockdown of CNOT7. After siRNA transfection, cells were counted in a hemocytometer at 24-h intervals. (C) Increased G1- and reduced S-phase in MCF7 cells transfected with CNOT7 siRNA. Dot plots of bivariate cell cycle profiling using propidium iodide fluorescence to determine DNA content (horizontal) and anti-BrdU fluorescence (FITC) to identify cells in S-phase (vertical) are shown. Cells (n = 20,000) were analyzed per condition 72 h after transfection. The percentages of cells in G1-, S-, and G2/M-phase are indicated. (D) Knockdown of CNOT8 in MCF7 cells (see A for details). (E) Inhibition of cell proliferation by knockdown of CNOT8 (see B for details). (F) Increased G1- and reduced S-phase in MCF7 cells transfected with CNOT8 siRNA (see C for details).
Partial Redundancy of CNOT7 and CNOT8 in Cell Proliferation
To understand whether CNOT7 and CNOT8 have redundant functions, we next used combined knockdown of both genes by siRNA transfection. After verifying efficient knockdown in these cases (data not shown), we first assessed the proliferation rate of cells transfected with siRNAs targeting both CNOT7 and CNOT8 compared with cells transfected with siRNA targeting either CNOT7 or CNOT8. As shown (Figure 2A), cells transfected with siRNA targeting both CNOT7 and CNOT8 proliferated with a reproducibly slower rate when compared with cells transfected with a single targeting siRNA directed against either CNOT7 or CNOT8. This effect was observed using two different sets of targeting siRNA and indicates that CNOT7 and CNOT8 have partially redundant roles in cell proliferation.
Figure 2.
Partial redundancy of CNOT7 and CNOT8 in cell proliferation. (A) Partial redundancy of CNOT7 and CNOT8 in the proliferation of MCF7 cells. After siRNA transfection, cells were counted in a hemocytometer at 24-h intervals. Indicated are mock (M), nontargeting control siRNA (Con), duplexes 1 (#1) and 2 (#2) targeting CNOT7 and CNOT8. (B) Increased G1- and reduced S-phase in MCF7 cells transfected with both CNOT7 and CNOT8. Dot plots of bivariate cell cycle profiling using propidium iodide fluorescence to determine DNA content (horizontal) and anti-BrdU fluorescence (FITC) to identify cells in S-phase (vertical) are shown. Cells (n = 20,000) were analyzed per condition 72 h after transfection. The percentages of cells in G1-, S-, and G2/M-phase are indicated.
To characterize this proliferation defect in more detail, we determined the cell cycle profiles of cells treated with both CNOT7 and CNOT8 siRNAs using bivariate flow cytometry. Similar to what was observed when cells were transfected with siRNA targeting either CNOT7 or CNOT8 (Figure 1, C and F), the cell cycle profiles of cells transfected with both targeting siRNAs displayed an increased percentage of cells in G1 and a decreased percentage of cells in S-phase compared with mock-treated cells or cells treated with nontargeting control siRNA (Figure 2B). Thus, whereas the cell cycle defect is more pronounced when both CNOT7 and CNOT8 are knocked down compared with treatment with a single targeting siRNA, no additional cell cycle deficiency, such as the induction of apoptosis, was revealed. Together, these results indicate that CNOT7 and CNOT8 are partially redundant and are important for cell cycle progression and proliferation of MCF7 breast cancer cells.
The Deadenylase Activity of CNOT7 Contributes to Efficient Proliferation
To investigate whether the deadenylase activity of CNOT7 is involved in efficient cell proliferation, we designed an siRNA-insensitive CNOT7 cDNA by introduction of four silent mutations in the siRNA-target site (Figure 3A). In addition, a point mutation was introduced in the catalytic center resulting in amino acid substitution D40A, which is demonstrated to cause enzyme inactivity (Jonstrup et al., 2007). To verify whether the silent mutations resulted in resistance against the targeting siRNA duplexes, cells were transiently cotransfected with plasmids expressing Flag-CNOT7 and siRNA duplexes targeting CNOT7. As expected, Flag-CNOT7 protein levels were strongly reduced in cells cotransfected with wild-type cDNA and CNOT7 siRNA, but not in cells transfected with nontargeting control siRNA (Figure 3B). In contrast, cells transfected with siRNA-insensitive CNOT7 cDNA (WT* or D40A*) showed equal expression of CNOT7 in the presence of either targeting or nontargeting siRNA (Figure 3B).
Figure 3.
Involvement of the CNOT7 deadenylase activity in cell proliferation. (A) Design of an siRNA-insensitive cDNA encoding CNOT7. Four silent point mutations in the cDNA sequence targeted by siRNA duplex 2 were introduced by site-directed mutagenesis. (B) Analysis of the siRNA-insensitive cDNAs encoding CNOT7. An additional mutation was introduced resulting in the amino acid substitution D40A in the active site. Vectors expressing CNOT7, an siRNA-insensitive cDNA (WT*) and an siRNA-insensitive cDNA encoding the active site mutant (D40A*) were transiently cotransfected with siRNA duplex 2 (#2) or nontargeting control siRNA (Con) into MCF7 cells. After 3 d, total lysates were analyzed by immunoblotting. β-Tubulin was used to assess equal loading. (C) Stable expression of siRNA-insensitive CNOT7 cDNAs in MCF7 cells. Lysates from MCF7 cells stably transfected with the indicated cDNA were analyzed by immunoblotting. β-Tubulin was used to assess equal loading. (D) The catalytic activity of CNOT7 is required for efficient cell proliferation. Nontargeting control siRNA or siRNA duplexes targeting endogenous CNOT7 were transfected into MCF7 cells stably expressing siRNA-insensitive cDNAs encoding wild-type CNOT7 (WT*) or the active site mutant (D40A*). After siRNA transfection, cells were counted in a hemocytometer at 24-h intervals. (E) Increased G1- and reduced S-phase in MCF7 cells expressing catalytically inactive CNOT7. Nontargeting control siRNA or siRNA duplexes targeting endogenous CNOT7 were transfected into MCF7 cells stably expressing siRNA-insensitive cDNAs encoding wild-type CNOT7 (WT*) or the active site mutant (D40A*). Dot plots of bivariate cell cycle profiling using propidium iodide fluorescence to determine DNA content (horizontal) and anti-BrdU fluorescence (FITC) to identify cells in S-phase (vertical) are shown. Cells (n = 20,000) were analyzed per condition 72 h after transfection. The percentages of cells in G1-, S-, and G2/M-phase are indicated.
Subsequently, MCF7 cells were stably transfected with siRNA-insensitive Flag-CNOT7 cDNA expression vectors. Clonally derived cell populations were isolated and selected based on comparable expression of wild-type and D40A Flag-CNOT7 as assessed by immunoblotting using anti-Flag antibodies (Figure 3C). We noticed that the stably transfected cells divided at a reduced rate compared with nontransfected cells, which was probably because of the antibiotic selection procedure. Next, the effect on cell proliferation was determined by ablation of endogenous CNOT7 using siRNA. Thus, proliferation of MCF7 cells expressing wild-type Flag-CNOT7 was not affected when these cells were transfected with siRNA duplexes targeting endogenous CNOT7 compared with nontargeting control siRNA. In contrast, MCF7 cells expressing similar levels of Flag-CNOT7 containing the D40A amino acid substitution in the active site proliferated at a markedly slower rate when endogenous CNOT7 levels were ablated, but not when cells were treated with nontargeting control siRNA (Figure 3D). In addition, we carried out FACS analysis of these cells treated with control nontargeting siRNA, or siRNA targeting endogenous CNOT7. In support of the above data, the cell cycle profiles of MCF7 cells expressing wild-type Flag-CNOT7 were not affected by targeting endogenous CNOT7 compared with transfection with nontargeting control siRNA (Figure 3E). By contrast, although cells expressing Flag-CNOT7 (D40A) showed a similar cell cycle profile when treated with nontargeting control siRNA, transfection of these cells with siRNA targeting endogenous CNOT7 increased the number of cells present in G1 (56.3 to 63.2%), whereas the number of cells present in S-phase was significantly reduced (37.8 to 32.8%; Figure 3E). These experiments indicate that the enzymatic activity of CNOT7 contributes to efficient proliferation, although other roles of CNOT7 (e.g., structural roles in the integrity of the complex) may also be important.
Analysis of the Interaction between CNOT7 and BTG2
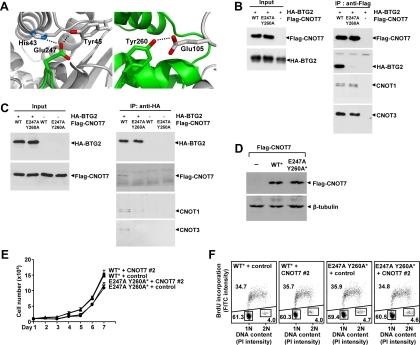
Recently, it was shown that proteins from the BTG/Tob family are involved in mRNA turnover by interacting with CNOT7 and CNOT8 (Ezzeddine et al., 2007; Funakoshi et al., 2007; Mauxion et al., 2008). To understand how CNOT7 interacts with BTG/Tob family members, we analyzed the crystal structure of CNOT7 and a fragment of Tob (PDB accession 2D5R). Based on proximity to Tob and evidence for H-bonding, two CNOT7 residues, Glu247 and Tyr260, that are located away from the active site were selected for mutagenesis (Figure 4A). To test the importance of these residues for the interaction with BTG/Tob proteins, HEK 293T cells were cotransfected with plasmids expressing Flag-CNOT7 and HA-BTG2. As expected, immunoprecipitation of FLAG-CNOT7 also precipitated HA-BTG2, as well as other Ccr4–Not subunits such as CNOT1 and CNOT3. In contrast, but as predicted by the structural analysis, Flag-CNOT7 containing two amino acid substitutions, E247A and Y260A, did not coprecipitate HA-BTG2, although it did coprecipitate CNOT1 and CNOT3, suggesting that the introduced amino acids substitutions do not interfere with Ccr4–Not complex formation (Figure 4B). When HEK 293T cells were transfected with a plasmid expressing HA-BTG2 in combination with the empty Flag-vector, no immunoprecipitation of either CNOT1, CNOT3, or HA-BTG2 was observed using anti-Flag antibodies, indicating that the analyzed immunoprecipitations were specific (Figure 4B).
Figure 4.
Analysis of the interaction of CNOT7 with BTG2/Tob family members. (A) Identification of CNOT7 residues Glu247 and Tyr260 interacting with Tob residues. The structure of the complex between CNOT7 with Tob (PDB accession number 2D5R) was analyzed and hydrogen bonding calculated using Deepview/Swiss PDB Viewer. The image was generated using PyMOL. Indicated are CNOT7 (green), Tob (white), and calculated H-bonds (dashed lines). (B) Requirement of CNOT7 residues Glu247 and Tyr260 for interaction with BTG2. Plasmids pCMV5-HA-BTG2 and a vector expressing wild-type Flag-CNOT7 (WT), CNOT7 containing amino acid substitutions (E247A Y260A), or empty vector were transiently cotransfected into HEK 293T cells. Total lysates (left) and anti-Flag immunoprecipitates (right) were analyzed using the indicated antibodies. (C) Involvement of CNOT7 residues Glu247 and Tyr260 for the interaction between BTG2, and CNOT1 and CNOT3. Plasmids pCMV5-HA-BTG2 or empty vector were transiently cotransfected into HEK293T cells with vectors expressing wild-type Flag-CNOT7 (WT) or CNOT7 containing amino acid substitutions (E247A Y260A). Total lysates (left) and anti-HA immunoprecipitates (right) were analyzed using the indicated antibodies. (D) Stable expression of siRNA-insensitive CNOT7 cDNAs in MCF7 cells. Lysates from MCF7 cells stably transfected with the indicated cDNA were analyzed by immunoblotting. β-Tubulin was used to assess equal loading. (E) Functional analysis of the interaction between CNOT7 and BTG2. Nontargeting control siRNA or siRNA duplexes targeting endogenous CNOT7 were transfected into MCF7 cells stably expressing siRNA-insensitive cDNAs encoding wild-type CNOT7 (WT*) or the BTG2-interaction mutant (E247A Y260A*). After siRNA transfection, cells were counted in a hemocytometer at 24-h intervals. (F) Functional analysis of MCF7 cells expressing CNOT7 with a perturbed BTG2-interaction surface. Nontargeting control siRNA or siRNA duplexes targeting endogenous CNOT7 were transfected into MCF7 cells stably expressing siRNA-insensitive cDNAs encoding wild-type CNOT7 (WT*) or the BTG2-interaction mutant (E247A Y260A*). Dot plots of bivariate cell cycle profiling using propidium iodide fluorescence to determine DNA content (horizontal) and anti-BrdU fluorescence (FITC) to identify cells in S-phase (vertical) are shown. Cells (n = 20,000) were analyzed per condition 72 h after transfection. The percentages of cells in G1-, S-, and G2/M-phase are indicated.
To investigate whether BTG2 can interact with the Ccr4–Not complex via interactions with other subunits, a reciprocal immunoprecipitation experiment was carried out (Figure 4C). As expected, immunoprecipitation of HA-BTG2 did coprecipitate Flag-CNOT7, CNOT1, and CNOT3. By contrast, coprecipitation of HA-BTG2 and Flag-CNOT7 (E247A Y260A) was not observed (Figure 4C). Furthermore, the coimmunoprecipitation of CNOT1 and CNOT3 with HA-BTG2 was significantly reduced under these conditions. Although the residual coprecipitation of CNOT1 and CNOT3 may be mediated via endogenous CNOT7, no immunoprecipitation of either CNOT1, CNOT3, or Flag-CNOT7 was observed in the absence of HA-BTG2, indicating that the analyzed immunoprecipitations were specific (Figure 4C). Thus, these experiments indicate that the interaction between BTG2 and CNOT7 is important for the interaction between BTG2 and other Ccr4–Not components.
To investigate the importance of the interaction between CNOT7 and BTG2 for cell proliferation, we used the siRNA-insensitive CNOT7 cDNA containing four silent mutations in the siRNA-target site in combination with mutations resulting in amino acids substitutions E247A and Y260A. Subsequently, MCF7 cells were stably transfected with siRNA-insensitive Flag-CNOT7 cDNA expression vectors. Stably transfected cells were screened for expression of Flag-CNOT7 and selected based on comparable expression of wild-type and E247A/Y260A Flag-CNOT7 as assessed by immunoblotting using anti-Flag antibodies (Figure 4D). The effect on cell proliferation was then determined by ablation of endogenous CNOT7 using siRNA or treatment with nontargeting control siRNA. As shown, proliferation of MCF7 cells expressing wild-type Flag-CNOT7 was not affected by targeting endogenous CNOT7 using siRNA compared with transfection with nontargeting control siRNA (Figure 4E). In addition, although MCF7 cells expressing Flag-CNOT7 (E247A Y260A) divided at a slightly reduced rate, proliferation of these cells was not affected after ablation of endogenous CNOT7 by siRNA compared with transfection with nontargeting control siRNA, as was observed for cells expressing wild-type Flag-CNOT7 (Figure 4E). In addition, we carried out FACS analysis of these cells treated with control nontargeting siRNA, or siRNA targeting endogenous CNOT7. In support of the above data, the cell cycle profiles of MCF7 cells expressing wild-type Flag-CNOT7 were not affected by targeting endogenous CNOT7 using siRNA compared with treatment with nontargeting control siRNA (Figure 4F). In addition, treatment of cells expressing Flag-CNOT7 (E247A Y260A) with control siRNA or siRNA targeting endogenous CNOT7 did not affect the G1- or S-phase distribution significantly (Figure 4F).
Thus, the interaction between CNOT7 and BTG2 appears less important for the efficient transition from G1- to S-phase in MCF7 cells. Although BTG2 is involved in general mRNA decay pathways (Mauxion et al., 2008), these results indicate that defects in mRNA turnover do not per se result in a proliferation deficiency. Alternatively, these results may indicate redundancy between CNOT7 and CNOT8, and/or among BTG/Tob family members.
Gene Expression Profiling Indicates a Synergistic Effect of CNOT7 and CNOT8 Knockdown
To obtain further insight into the mechanism by which the CNOT7 and CNOT8 deadenylases influence gene expression, we carried out genome-wide expression analysis. MCF7 cells were transfected with a pool of nontargeting control siRNA or siRNA pools targeting CNOT7, CNOT8, or CNOT7 and CNOT8. Total RNA from biological triplicates was isolated, processed, and hybridized using Affymetrix Human Genome GeneChips analyzing more than 47,000 transcripts. Analysis of the expression data showed that knockdown of either CNOT7 or CNOT8 resulted in significant differential expression of only four or three genes, respectively, whereas combined knockdown of CNOT7 and CNOT8 resulted in differential regulation of 255 genes (>1.5-fold differential expression, p < 0.05; Figure 5A; Supplementary Data). Interestingly, most differentially expressed genes were up-regulated (164 genes) in CNOT7/CNOT8 double knockdown cells, consistent with their role as repressors of gene expression, although down-regulated genes (91 genes) were also identified (Figure 5A; Supplementary Data). Clustering based on genes and conditions using the subset of genes significantly affected in CNOT7/CNOT8 double knockdown cells revealed that the expression profiles of single CNOT7 and CNOT8 knockdown cells are more closely related to each other compared with the expression profile of CNOT7/CNOT8 double knockdown cells and showed that the expression profiles are highly related (Figure 5A). This notion is underscored by the calculation of Pearson's correlation coefficients of the expression profiles, which indicated that the expression profiles of CNOT7 and CNOT8 single knockdown cells and control cells correlated better compared with the profile of CNOT7/CNOT8 double knockdown cells. This is suggestive of a (partially) redundant role of these factors (Figure 5B).
Figure 5.
Gene expression profiling indicates synergistic effects of CNOT7 and CNOT8 knockdown. (A) Diagram of hierarchical clustering of gene expression profiles of MCF7 cells treated with the indicated siRNA pool. Probes are represented vertically, whereas conditions are shown horizontally. The subset of probes were selected on the basis of the CNOT7/CNOT8 double knockdown expression profile (>1.5-fold differential expression compared with control nontargeting siRNA pool, p < 0.05) and clustered alongside the profiles obtained from CNOT7 and CNOT8 siRNA treatment. The clustering was carried out using the Comprehensive R-based Microarray Analysis tool CARMAweb (https://carmaweb.genome.tugraz.at/carma/). (B) Matrix of Pearson's correlation coefficients of gene expression profiles as indicated. (C and D) Gene ontology analysis of up-regulated (C) and down-regulated (D) genes. All molecular function categories as determined by GeneTrail using standard analysis settings are shown (http://genetrail.bioinf.uni-sb.de/).
Gene ontology analysis was carried out to obtain more insight into the functions of the identified differentially expressed genes in CNOT7/CNOT8 double knockdown cells. Interestingly, a notable difference was observed when the classification of molecular function of up-regulated genes was compared with that of down-regulated genes (Figure 5, C and D, respectively). Whereas the majority of the categorized up-regulated genes are associated with DNA-binding activity and/or activities relating to transcription (Figure 5C), the majority of classified down-regulated genes were typed as nucleic acid binding and/or RNA binding and no significant category relating to a role in transcription was identified for down-regulated genes (Figure 5D). Taken together, the expression profiling data show that CNOT7 and CNOT8 appear to act synergistically. Consistent with their role as deadenylases, a significant fraction of the differentially expressed genes are repressed by CNOT7 and CNOT8 and encode transcription factors. By contrast, a notable number of genes are positively regulated by CNOT7 and CNOT8, of which a significant proportion encode nucleic acid– and/or RNA-binding characteristics.
Deregulation of Genes Modulating Cell Cycle Progression in CNOT7 and/or CNOT8 Knockdown Cells
To validate the microarray analysis and look in more detail at the mechanism by which CNOT7 and CNOT8 regulate gene expression, we used RT-qPCR to determine the expression levels of several genes in the CNOT7/CNOT8 double knockdown cells. These genes were up-regulated in CNOT7/CNOT8 double knockdown cells between 1.7-fold (CCNG2/Cyclin G2) and 3.3-fold (EGR1), as determined using genome-wide expression analysis, and were of particular interest because their overexpression is associated with a decreased proliferation rate of several cell types (Zoidl et al., 1995; Zoidl et al., 1997; Garde et al., 1999; Jetten and Suter, 2000; LaTulippe et al., 2002; Shukeir et al., 2003; Roux et al., 2005; Stossi et al., 2006). We focused on up-regulated genes, because this effect correlates well with the known roles of CNOT7 and CNOT8 as deadenylases promoting mRNA turnover or transcriptional repressors. As expected, we confirmed up-regulation of all genes tested upon knockdown of CNOT7 and CNOT8 compared with control treated cells transfected with nontargeting siRNA (Figure 6, A–E). Generally, we found stronger effects on mRNA levels in CNOT7 knockdown cells, which may be due to a generally higher knockdown efficiency using CNOT7 siRNA compared with CNOT8 siRNA (data not shown). Interestingly, however, the tested genes displayed a similar deregulation in the absence of either CNOT7 or CNOT8, with a synergistic effect after combined knockdown of CNOT7 and CNOT8 (Figure 6, A–E).
Figure 6.
Confirmation of genes regulated by CNOT7 and CNOT8. (A–F) Analysis of the indicated mRNA transcript levels in cells treated with siRNA pools targeting CNOT7 and/or CNOT8, or control. mRNA levels of the indicated genes were detected using RT-qPCR using β-actin as a reference gene. All assays were carried out in triplicate. (G) Determination of PMP22 mRNA stability. The indicated siRNA pools were transfected into MCF7 cells. Actinomycin D was added (72 h after transfection), and total mRNA was isolated at 0, 1, 3, 6, and 12 h after treatment. mRNA transcript levels were determined by RT-qPCR using β-actin as a reference gene. The PMP22 mRNA half lives (decay constants ± SE) derived from the slope of the fitted line are indicated: 6.2 h (0.112 ± 0.0218; control), 4.8 h (0.143 ± 0.0405; CNOT7), 6.5 h (0.106 ± 0.0332; CNOT8), and 41.4 h (0.0168 ± 0.00752; CNOT7+CNOT8). (H) Determination of Cyclin G2 mRNA stability. Seventy-two hours after transfection, total RNA was isolated after 0, 0.5, 1, 2, and 4 h of treatment with actinomycin D. mRNA transcript levels were determined by RT-qPCR using β-actin as a reference gene. The Cyclin G2 mRNA half-lives (decay constants ± SE) derived from the slope of the fitted line are indicated: 2.7 h (0.255 ± 0.0419; control), 2.1 h (0.329 ± 0.0312; CNOT7), 2.0 h (0.341 ± 0.0492; CNOT8), and 4.4 h (0.158 ± 0.0154; CNOT7+CNOT8). All assays were carried out in triplicate.
In addition to this set of up-regulated genes, we also analyzed a gene that was down-regulated in the absence of CNOT7 and/or CNOT8. Consistent with the results from the microarray analysis, the gene encoding the RNA-binding protein DDX6/Rck/p54 was significantly down-regulated in the absence of either CNOT7 or CNOT8. Moreover, further down-regulation was observed after combined knockdown of CNOT7 and CNOT8 (Figure 6F).
Interestingly, these data show that the CNOT7 and CNOT8 paralogues can largely compensate for each other's function in agreement with their high sequence similarity. In addition, these results identify several genes influencing cell cycle progression, such as MSMB, PMP22 and Cyclin G2, whose expression is modulated by CNOT7 and CNOT8.
Increased PMP22 and Cyclin G2 mRNA stability in CNOT7 and CNOT8 Knockdown Cells
To test whether the up-regulation of genes in CNOT7 and CNOT8 knockdown cells could be due to an increased mRNA half-live, we used the RNA polymerase II inhibitor actinomycin D in combination with RT-qPCR to determine mRNA levels. After siRNA transfection, cells were treated with actinomycin D, and total RNA was isolated at several time points. Subsequently, mRNA levels were determined by RT-qPCR. Because the mRNA of MSMB was stable after actinomycin D treatment, we first analyzed transcript stability of PMP22, which is almost fourfold up-regulated in CNOT7/CNOT8 knockdown cells as determined by RT-qPCR (Figure 6B). As shown, the decay rate of PMP22 mRNA was decreased in CNOT7/CNOT8 double knockdown cells compared with nontargeting control treated cells by more than sixfold (Figure 6G). By contrast, no significant increase in PMP22 mRNA stability was observed in CNOT7 or CNOT8 single knockdown cells (Figure 6G), which may be explained by the modest induction of PMP22 in CNOT7 or CNOT8 single knockdown cells (∼1.5-fold; Figure 6B). For Cyclin G2, which is about twofold up-regulated in CNOT7/CNOT8 double knockdown cells as determined by RT-qPCR (Figure 6E), less stabilization (∼1.5-fold) of its transcript was observed in the presence of actinomycin D compared with PMP22, as expected (Figure 6H). As was observed for PMP22, no significant increase in Cyclin G2 mRNA stability was observed in CNOT7 or CNOT8 single knockdown cells, consistent with only a minor up-regulation of Cyclin G2 mRNA in these cells (<1.5-fold; Figure 6E).
Together, these results indicate that up-regulation in CNOT7/CNOT8 double knockdown cells of several genes involved in cell cycle regulation, including PMP22 and Cyclin G2, may be due to increased mRNA half-lives, consistent with the role of CNOT7 and CNOT8 in deadenylation and the involvement of the deadenylase activity of CNOT7 in efficient cell cycle progression (Figure 3, D and E).
DISCUSSION
CNOT7 and CNOT8 Are Important for Efficient Cell Proliferation of MCF7 Cells
Several published observations led us to investigate the role of the CNOT7 and CNOT8 deadenylase subunits of the Ccr4–Not complex in cell proliferation. First, we recently identified an interaction between CNOT1, the largest subunit of the Ccr4–Not complex, and estrogen receptor α, a well-characterized mitogenic factor in breast cancer cells (Winkler et al., 2006). Furthermore, the CNOT7 and CNOT8 paralogues physically interact with members of the BTG/Tob family of antiproliferative proteins that inhibit cell cycle progression from G1- to S-phase (Bogdan et al., 1998; Ikematsu et al., 1999; Prevot et al., 2001; Yoshida et al., 2001). Also, it was reported that overexpression of CNOT7 reduces colony formation in NIH3T3 and U2OS cells (Bogdan et al., 1998).
Here, we show that CNOT7 and CNOT8 are important for efficient cell proliferation and the catalytic activity is involved in cell cycle progression. Interestingly, CNOT7 appears to be mostly nuclear in serum-starved diploid human fibroblasts and partially relocates to the cytoplasm during S-phase entry, which may suggest the involvement of the deadenylase activity during this phase of the cell cycle (Morel et al., 2003). In contrast to the catalytic activity, however, the interaction with BTG/Tob family members may not be essential for their role in cell division.
The CNOT7/CNOT8 deadenylases may affect the expression of a number of genes involved in cell cycle regulation, both positively and negatively, the summation of which resulting in decreased proliferation. At least two mechanisms may be involved. First, CNOT7 and CNOT8 can repress the expression of genes associated with inhibition of cell proliferation, such as MSMB and PMP22. The MSMB gene encodes the tumor suppressor PSP94 (prostatic secretory protein of 94 amino acids; β-microseminoprotein), which inhibits prostatic cancer growth and whose loss of expression is characteristic of metastatic tumors compared with primary prostate tumors (Garde et al., 1999; LaTulippe et al., 2002; Shukeir et al., 2003). MSMB is also expressed in other tissues including female breast and reproductive tissues, suggesting a role in cell proliferation in these organs as well (Baijal-Gupta et al., 2000). Increased expression of PMP22 (also known as growth arrest–specific/GAS 3), which shares significant homology with EMP1, is involved in several neuropathies and can influence epithelial cell morphology and proliferation by influencing the G1-to-S-phase progression (Zoidl et al., 1995; Zoidl et al., 1997; Jetten and Suter, 2000; Roux et al., 2005).
A second mechanism by which CNOT7 and CNOT8 contribute to cell proliferation may be by positively regulating the expression of genes required for cell proliferation, such as the DEAD-box RNA helicase DDX6/Rck/p54 (Akao et al., 2006). Interestingly, DDX6 is a homologue of yeast Dhh1, which interacts with the Ccr4–Not complex (Coller et al., 2001; Maillet and Collart, 2002). Moreover, like CNOT7 and CNOT8, DDX6 is a component of cytoplasmic P-bodies (Cougot et al., 2004), which are enriched in translationally repressed mRNA molecules and proteins participating in microRNA-mediated translational repression and mRNA decay (Garneau et al., 2007; Parker and Sheth, 2007). Whether the interaction between DDX6 and Ccr4–Not is conserved and/or whether the reduced expression of DDX6 mRNA in the absence of CNOT7 and/or CNOT8 is mechanistically linked remains to be investigated. Interestingly, comparison of our gene expression data with published studies investigating yeast caf1Δ cells (Cui et al., 2008; Azzouz et al., 2009) indicated that human DDX6 (yeast DHH1) may be an evolutionary conserved target gene whose correct expression level is dependent on the presence of human CNOT7/CNOT8 (yeast CAF1).
The CNOT7 and CNOT8 Paralogues Display Overlapping Functions
Because of the high amino acid sequence similarity of CNOT7 and CNOT8 (76% identity and 89% similarity), it was not unexpected to find that these proteins can partially compensate each other's function. This was evident from both the phenotypic analysis and the microarray analysis of gene expression profiles, although both CNOT7 and CNOT8 also appear to have unique functions as demonstrated in the latter experiment. This is in marked contrast, however, to the CNOT6 and CNOT6L deadenylase subunits of the Ccr4–Not complex (78% identity and 88% similarity at the amino acid level). In this case, CNOT6L is required for cell proliferation of NIH 3T3 cells, in contrast to CNOT6, which does not appear to be involved (Morita et al., 2007).
The partially overlapping function of CNOT7 and CNOT8 may explain the relatively mild phenotype observed in Cnot7 null mice. These mice have a normal appearance, although males are sterile because of a defect in spermatogenesis (Berthet et al., 2004; Nakamura et al., 2004). In addition, Cnot7 mice display an increased bone mass caused by elevated osteoblast activity because of enhanced BMP signaling (Washio-Oikawa et al., 2007). Interestingly, a similar observation was made by observing mutant mice lacking Tob, which can interact with CNOT7 and CNOT8 (Yoshida et al., 2000). However, Tob mice are prone to spontaneous tumorigenesis, which is not observed in Cnot7 mice (Yoshida et al., 2003).
Mechanism of Gene Regulation by the CNOT7 and CNOT8 Subunits of the Ccr4–Not Complex
On knockdown of CNOT7 and CNOT8, most differentially expressed genes were up-regulated in agreement with a role for these proteins in mRNA turnover. Consistent with this notion, we found that transcript levels of PMP22 and—to a lesser extent—Cyclin G2 were stabilized in the absence of CNOT7 and CNOT8. To address whether particular motifs were enriched in the 3′ untranslated regions of the genes up-regulated in the absence of CNOT7 and CNOT8, we searched for the presence of AU-rich elements (ARE) in the 3′ untranslated regions of all differentially expressed genes using the ARED database (http://rc.kfshrc.edu.sa/ARED). However, although a higher proportion of predicted AREs were identified in genes up-regulated in CNOT7/CNOT8 double knockdown cells compared with down-regulated genes, no clear preference was identified.
In addition to a role in mRNA turnover, the Ccr4–Not complex is also implicated in the regulation of transcription by both positively and negatively modulating gene expression. In this light, the expression of certain genes may also be increased upon siRNA transfection, because knockdown of CNOT7 and/or CNOT8 may interfere with the function of the Ccr4–Not complex as a transcriptional repressor. In addition, down-regulation of genes, such as DDX6, may be due to a defect in the Ccr4–Not function that positively affects transcription. Indeed, a positive role in regulating transcription mediated by the nuclear receptors ERα and RARβ is also reported for both CNOT7 and CNOT8 (Prevot et al., 2001; Morel et al., 2003; Nakamura et al., 2004).
Interestingly, a subset of genes differentially expressed upon CNOT7/CNOT8 knockdown are known to be regulated in response to estrogen signaling. This could be expected, because we recently identified a ligand-dependent interaction between CNOT1, the large subunit of the Ccr4–Not complex, and ERα (Winkler et al., 2006). Currently, we are investigating the mechanisms by which Ccr4–Not components contribute to estrogen-dependent regulation of gene expression in more detail.
Note added in proof. The microarray data have been deposited in the ArrayExpress database (European Bioinformatics Institute, accession number E-MEXP-2218).
Supplementary Material
ACKNOWLEDGMENTS
We thank the Nottingham Arabidopsis Stock Centre for microarray hybridization and processing, David Heery and members of the Gene Regulation Group for invaluable support, and David Heery, Nicole Clarke, and Klaas Mulder for critical reading of the manuscript. This work was supported by grants from the Association for International Cancer Research (AICR 07-0494) and the Biotechnology and Biological Sciences Research Council (BB/E02338X/1).
Glossary
Abbreviations used:
- RT-qPCR
reverse transcriptase-quantitative PCR
- BrdU
5-bromo-2-deoxyuridine.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-02-0146) on July 15, 2009.
REFERENCES
- Akao Y., Matsumoto K., Ohguchi K., Nakagawa Y., Yoshida H. Human DEAD-box/RNA unwindase rck/p54 contributes to maintenance of cell growth by affecting cell cycle in cultured cells. Int. J. Oncol. 2006;29:41–48. [PubMed] [Google Scholar]
- Azzouz N., Panasenko O. O., Deluen C., Hsieh J., Theiler G., Collart M. A. Specific roles for the Ccr4-Not complex subunits in expression of the genome. RNA. 2009;15:377–383. doi: 10.1261/rna.1348209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes C., Keller A., Kuentzer J., Kneissl B., Comtesse N., Elnakady Y. A., Muller R., Meese E., Lenhof H. P. GeneTrail—advanced gene set enrichment analysis. Nucleic Acids Res. 2007;35:W186–W192. doi: 10.1093/nar/gkm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baijal-Gupta M., Clarke M. W., Finkelman M. A., McLachlin C. M., Han V. K. Prostatic secretory protein (PSP94) expression in human female reproductive tissues, breast and in endometrial cancer cell lines. J. Endocrinol. 2000;165:425–433. doi: 10.1677/joe.0.1650425. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4, NOT deadenylase and DCP1, DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet C., Morera A. M., Asensio M. J., Chauvin M. A., Morel A. P., Dijoud F., Magaud J. P., Durand P., Rouault J. P. CCR4-associated factor CAF1 is an essential factor for spermatogenesis. Mol. Cell. Biol. 2004;24:5808–5820. doi: 10.1128/MCB.24.13.5808-5820.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchin C., Mauxion F., Sentis S., Seraphin B., Corbo L. Conservation of the deadenylase activity of proteins of the Caf1 family in human. RNA. 2005;11:487–494. doi: 10.1261/rna.7135305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeck R., Lapeyre B., Brown C. E., Sachs A. B. Capped mRNA degradation intermediates accumulate in the yeast spb8–2 mutant. Mol. Cell. Biol. 1998;18:5062–5072. doi: 10.1128/mcb.18.9.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan J. A., Adams-Burton C., Pedicord D. L., Sukovich D. A., Benfield P. A., Corjay M. H., Stoltenborg J. K., Dicker I. B. Human carbon catabolite repressor protein (CCR4)-associative factor 1, cloning, expression and characterization of its interaction with the B-cell translocation protein BTG1. Biochem. J. 1998;336(Pt 2):471–481. doi: 10.1042/bj3360471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. E., Sachs A. B. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol. 1998;18:6548–6559. doi: 10.1128/mcb.18.11.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. E., Tarun S. Z., Jr, Boeck R., Sachs A. B. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:5744–5753. doi: 10.1128/mcb.16.10.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Chiang Y. C., Denis C. L. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark L. B., Viswanathan P., Quigley G., Chiang Y. C., McMahon J. S., Yao G., Chen J., Nelsbach A., Denis C. L. Systematic mutagenesis of the leucine-rich repeat (LRR) domain of CCR4 reveals specific sites for binding to CAF1 and a separate critical role for the LRR in CCR4 deadenylase activity. J. Biol. Chem. 2004;279:13616–13623. doi: 10.1074/jbc.M313202200. [DOI] [PubMed] [Google Scholar]
- Collart M. A. Global control of gene expression in yeast by the Ccr4-Not complex. Gene. 2003;313:1–16. doi: 10.1016/s0378-1119(03)00672-3. [DOI] [PubMed] [Google Scholar]
- Collart M. A., Timmers H. T. The eukaryotic Ccr4-not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog. Nucleic Acid Res. Mol. Biol. 2004;77:289–322. doi: 10.1016/S0079-6603(04)77008-7. [DOI] [PubMed] [Google Scholar]
- Coller J. M., Tucker M., Sheth U., Valencia-Sanchez M. A., Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., Babajko S., Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Ramnarain D. B., Chiang Y. C., Ding L. H., McMahon J. S., Denis C. L. Genome wide expression analysis of the CCR4-NOT complex indicates that it consists of three modules with the NOT module controlling SAGA-responsive genes. Mol. Genet. Genomics. 2008;279:323–337. doi: 10.1007/s00438-007-0314-1. [DOI] [PubMed] [Google Scholar]
- Daugeron M. C., Mauxion F., Seraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–2455. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A., Morel A. P., Barbot W., Loireau M. P., Corbo L., Heidmann T. Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics. 2001;2:9. doi: 10.1186/1471-2164-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzeddine N., Chang T. C., Zhu W., Yamashita A., Chen C. Y., Zhong Z., Yamashita Y., Zheng D., Shyu A. B. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol. Cell Biol. 2007;27:7791–7801. doi: 10.1128/MCB.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi Y., Doi Y., Hosoda N., Uchida N., Osawa M., Shimada I., Tsujimoto M., Suzuki T., Katada T., Hoshino S. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 2007;21:3135–3148. doi: 10.1101/gad.1597707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garapaty S. R., Mahajan M. A., Samuels H. H. Components of the CCR4-not complex function as nuclear hormone receptor coactivators via association with the NRC interacting factor, NIF-1. J. Biol. Chem. 2008;283:6806–6816. doi: 10.1074/jbc.M706986200. [DOI] [PubMed] [Google Scholar]
- Garde S. V., et al. Prostate secretory protein (PSP94) suppresses the growth of androgen-independent prostate cancer cell line (PC3) and xenografts by inducing apoptosis. Prostate. 1999;38:118–125. doi: 10.1002/(sici)1097-0045(19990201)38:2<118::aid-pros5>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Garneau N. L., Wilusz J., Wilusz C. J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Goldstrohm A. C., Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- Hiroi N., Ito T., Yamamoto H., Ochiya T., Jinno S., Okayama H. Mammalian Rcd1 is a novel transcriptional cofactor that mediates retinoic acid-induced cell differentiation. EMBO J. 2002;21:5235–5244. doi: 10.1093/emboj/cdf521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikematsu N., Yoshida Y., Kawamura-Tsuzuku J., Ohsugi M., Onda M., Hirai M., Fujimoto J., Yamamoto T. Tob2, a novel anti-proliferative Tob/BTG1 family member, associates with a component of the CCR4 transcriptional regulatory complex capable of binding cyclin-dependent kinases. Oncogene. 1999;18:7432–7441. doi: 10.1038/sj.onc.1203193. [DOI] [PubMed] [Google Scholar]
- Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J. C., Fritzler M. J., Chan E. K. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Jetten A. M., Suter U. The peripheral myelin protein 22 and epithelial membrane protein family. Prog. Nucleic Acid Res. Mol. Biol. 2000;64:97–129. doi: 10.1016/s0079-6603(00)64003-5. [DOI] [PubMed] [Google Scholar]
- Jonstrup A. T., Andersen K. R., Van L. B., Brodersen D. E. The 1.4-A crystal structure of the S. pombe Pop2p deadenylase subunit unveils the configuration of an active enzyme. Nucleic Acids Res. 2007;35:3153–3164. doi: 10.1093/nar/gkm178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaTulippe E., Satagopan J., Smith A., Scher H., Scardino P., Reuter V., Gerald W. L. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- Liu J., Rivas F. V., Wohlschlegel J., Yates J. R., 3rd, Parker R., Hannon G. J. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005a;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Valencia-Sanchez M. A., Hannon G. J., Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005b;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet L., Collart M. A. Interaction between Not1p, a component of the Ccr4-not complex, a global regulator of transcription, and Dhh1p, a putative RNA helicase. J. Biol. Chem. 2002;277:2835–2842. doi: 10.1074/jbc.M107979200. [DOI] [PubMed] [Google Scholar]
- Mauxion F., Faux C., Seraphin B. The BTG2 protein is a general activator of mRNA deadenylation. EMBO J. 2008;27:1039–1048. doi: 10.1038/emboj.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel A. P., Sentis S., Bianchin C., Le Romancer M., Jonard L., Rostan M. C., Rimokh R., Corbo L. BTG2 antiproliferative protein interacts with the human CCR4 complex existing in vivo in three cell-cycle-regulated forms. J. Cell Sci. 2003;116:2929–2936. doi: 10.1242/jcs.00480. [DOI] [PubMed] [Google Scholar]
- Morita M., Suzuki T., Nakamura T., Yokoyama K., Miyasaka T., Yamamoto T. Depletion of mammalian CCR4b deadenylase triggers elevation of the p27Kip1 mRNA level and impairs cell growth. Mol. Cell Biol. 2007;27:4980–4990. doi: 10.1128/MCB.02304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., et al. Oligo-astheno-teratozoospermia in mice lacking Cnot7, a regulator of retinoid X receptor beta. Nat. Genet. 2004;36:528–533. doi: 10.1038/ng1344. [DOI] [PubMed] [Google Scholar]
- Ou Y. H., Chung P. H., Hsu F. F., Sun T. P., Chang W. Y., Shieh S. Y. The candidate tumor suppressor BTG3 is a transcriptional target of p53 that inhibits E2F1. EMBO J. 2007;26:3968–3980. doi: 10.1038/sj.emboj.7601825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Parker R., Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Passeri D., Marcucci A., Rizzo G., Billi M., Panigada M., Leonardi L., Tirone F., Grignani F. Btg2 enhances retinoic acid-induced differentiation by modulating histone H4 methylation and acetylation. Mol. Cell. Biol. 2006;26:5023–5032. doi: 10.1128/MCB.01360-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R. S., Bhattacharyya S. N., Artus C. G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Prevot D., Morel A. P., Voeltzel T., Rostan M. C., Rimokh R., Magaud J. P., Corbo L. Relationships of the antiproliferative proteins BTG1 and BTG2 with CAF1, the human homolog of a component of the yeast CCR4 transcriptional complex: involvement in estrogen receptor alpha signaling pathway. J. Biol. Chem. 2001;276:9640–9648. doi: 10.1074/jbc.M008201200. [DOI] [PubMed] [Google Scholar]
- Prevot D., Voeltzel T., Birot A. M., Morel A. P., Rostan M. C., Magaud J. P., Corbo L. The leukemia-associated protein Btg1 and the p53-regulated protein Btg2 interact with the homeoprotein Hoxb9 and enhance its transcriptional activation. J. Biol. Chem. 2000;275:147–153. doi: 10.1074/jbc.275.1.147. [DOI] [PubMed] [Google Scholar]
- Rainer J., Sanchez-Cabo F., Stocker G., Sturn A., Trajanoski Z. CARMAweb: comprehensive R- and bioconductor-based web service for microarray data analysis. Nucleic Acids Res. 2006;34:W498–W503. doi: 10.1093/nar/gkl038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K. J., Amici S. A., Fletcher B. S., Notterpek L. Modulation of epithelial morphology, monolayer permeability, and cell migration by growth arrest specific 3/peripheral myelin protein 22. Mol. Biol. Cell. 2005;16:1142–1151. doi: 10.1091/mbc.E04-07-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede A., Ellis L., Luther J., Carrington M., Stoecklin G., Clayton C. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res. 2008;36:3374–3388. doi: 10.1093/nar/gkn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen G. L., Blau H. M. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- Shukeir N., Arakelian A., Kadhim S., Garde S., Rabbani S. A. Prostate secretory protein PSP-94 decreases tumor growth and hypercalcemia of malignancy in a syngenic in vivo model of prostate cancer. Cancer Res. 2003;63:2072–2078. [PubMed] [Google Scholar]
- Stossi F., Likhite V. S., Katzenellenbogen J. A., Katzenellenbogen B. S. Estrogen-occupied estrogen receptor represses cyclin G2 gene expression and recruits a repressor complex at the cyclin G2 promoter. J. Biol. Chem. 2006;281:16272–16278. doi: 10.1074/jbc.M513405200. [DOI] [PubMed] [Google Scholar]
- Temme C., Zaessinger S., Meyer S., Simonelig M., Wahle E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 2004;23:2862–2871. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M., Staples R. R., Valencia-Sanchez M. A., Muhlrad D., Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M., Valencia-Sanchez M. A., Staples R. R., Chen J., Denis C. L., Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- Tzachanis D., Freeman G. J., Hirano N., van Puijenbroek A. A., Delfs M. W., Berezovskaya A., Nadler L. M., Boussiotis V. A. Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nat. Immunol. 2001;2:1174–1182. doi: 10.1038/ni730. [DOI] [PubMed] [Google Scholar]
- Uchida N., Hoshino S., Katada T. Identification of a human cytoplasmic poly(A) nuclease complex stimulated by poly(A)-binding protein. J. Biol. Chem. 2004;279:1383–1391. doi: 10.1074/jbc.M309125200. [DOI] [PubMed] [Google Scholar]
- Viswanathan P., Ohn T., Chiang Y. C., Chen J., Denis C. L. Mouse CAF1 can function as a processive deadenylase/3′-5′-exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J. Biol. Chem. 2004;279:23988–23995. doi: 10.1074/jbc.M402803200. [DOI] [PubMed] [Google Scholar]
- Wakiyama M., Takimoto K., Ohara O., Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washio-Oikawa K., et al. Cnot7-null mice exhibit high bone mass phenotype and modulation of BMP actions. J. Bone Miner. Res. 2007;22:1217–1223. doi: 10.1359/jbmr.070411. [DOI] [PubMed] [Google Scholar]
- Winkler G. S., Mulder K. W., Bardwell V. J., Kalkhoven E., Timmers H. T. Human Ccr4-Not complex is a ligand-dependent repressor of nuclear receptor-mediated transcription. EMBO J. 2006;25:3089–3099. doi: 10.1038/sj.emboj.7601194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Fan J., Belasco J. G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Chang T. C., Yamashita Y., Zhu W., Zhong Z., Chen C. Y., Shyu A. B. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- Yoshida Y., Hosoda E., Nakamura T., Yamamoto T. Association of ANA, a member of the antiproliferative Tob family proteins, with a Caf1 component of the CCR4 transcriptional regulatory complex. Jpn. J. Cancer Res. 2001;92:592–596. doi: 10.1111/j.1349-7006.2001.tb01135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y., et al. Mice lacking a transcriptional corepressor Tob are predisposed to cancer. Genes Dev. 2003;17:1201–1206. doi: 10.1101/gad.1088003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y., et al. Negative regulation of BMP/Smad signaling by Tob in osteoblasts. Cell. 2000;103:1085–1097. doi: 10.1016/s0092-8674(00)00211-7. [DOI] [PubMed] [Google Scholar]
- Zoidl G., Blass-Kampmann S., D'Urso D., Schmalenbach C., Muller H. W. Retroviral-mediated gene transfer of the peripheral myelin protein PMP22 in Schwann cells: modulation of cell growth. EMBO J. 1995;14:1122–1128. doi: 10.1002/j.1460-2075.1995.tb07095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoidl G., D'Urso D., Blass-Kampmann S., Schmalenbach C., Kuhn R., Muller H. W. Influence of elevated expression of rat wild-type PMP22 and its mutant PMP22Trembler on cell growth of NIH3T3 fibroblasts. Cell Tissue Res. 1997;287:459–470. doi: 10.1007/s004410050770. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.