Abstract
Ssd1 is an RNA-binding protein that affects literally hundreds of different processes and is polymorphic in both wild and lab yeast strains. We have used transcript microarrays to compare mRNA levels in an isogenic pair of mutant (ssd1-d) and wild-type (SSD1-V) cells across the cell cycle. We find that 15% of transcripts are differentially expressed, but there is no correlation with those mRNAs bound by Ssd1. About 20% of cell cycle regulated transcripts are affected, and most show sharper amplitudes of oscillation in SSD1-V cells. Many transcripts whose gene products influence longevity are also affected, the largest class of which is involved in translation. Ribosomal protein mRNAs are globally down-regulated by SSD1-V. SSD1-V has been shown to increase replicative life span¤ and we show that SSD1-V also dramatically increases chronological life span (CLS). Using a new assay of CLS in pure populations of quiescent prototrophs, we find that the CLS for SSD1-V cells is twice that of ssd1-d cells.
INTRODUCTION
Ssd1 is an RNA-binding protein (Uesono et al., 1997; Hogan et al., 2008) that can be distinguished from the hundreds of other RNA-binding proteins by its unusual properties. First of all, ssd1 is highly pleiotropic. This locus has at least nine different names because of its having been identified in genetic screens for its effect on minichromosome stability, stress tolerance, membrane trafficking, and cell wall integrity, among other things (Uesono et al., 1994; Kosodo et al., 2001; Vannier et al., 2001; Reinke et al., 2004). Systematic global screens have identified ∼200 genes that show genetic or physical interactions with Ssd1 (Reguly et al., 2006). These genes show a striking enrichment (Hong et al., 2008) for posttranslational modifiers (p = 10−14), including 19 kinases and nine histone deacetylases, and genes involved in the cell cycle and cell morphogenesis (p = 10−8). ssd1 mutants display sensitivity to high osmolarity, caffeine, fungicides¤ and numerous other compounds, which suggests a role for this protein in the maintenance of cell wall integrity (Ibeas et al., 2001; Parsons et al., 2004), but its mechanism of action remains obscure.
Despite, or perhaps because of, its impact on so many different cellular functions, SSD1 is a common site of variation in both laboratory strains and natural populations of budding yeast (Wheeler et al., 2003). One possible explanation is that SSD1-V, which encodes the full-length “wild-type” protein, reduces the virulence of Saccharomyces cerevisiae in one mouse model (Wheeler et al., 2003), but SSD1-V is critical in Candida (Gank et al., 2008) and several fungal pathogens of plants for evading their hosts' defense systems (Tanaka et al., 2007). In most cases, genetic interactions with SSD1-V are positive, whereas the premature termination alleles (ssd1-d) cause lethality or a more extreme phenotype. One important exception is the RAM signaling pathway mutants, tao3 (Du and Novick, 2002), and cbk1 (Tong et al., 2001; Jorgensen et al., 2002), whose loss of function is lethal if the SSD1-V allele is present (Jorgensen et al., 2002). Ssd1-v physically interacts with Cbk1 (Racki et al., 2000)¤ and there are consensus phosphorylation sites for Cbk1 kinase in the Ssd1 protein (Mazanka et al., 2008). These observations suggest that Cbk1 may negatively regulate Ssd1 and that unregulated Ssd1 function is deleterious. Cbk1 is a highly conserved kinase that is localized in the daughter cell, where it influences asymmetric cell fate decisions (Mazanka et al., 2008).
In most cases studied, the ssd1-d alleles exhibit properties similar to ssd1 deletions, which indicates that the ssd1 truncation results in a loss of function and perhaps a null phenotype (Kaeberlein and Guarente, 2002). However, partial function has been observed with the ssd1-d2 allele from W303 with respect to cell size, caffeine sensitivity¤ and interaction with cyclin-dependent kinase mutant cdc28-13 (Sutton et al., 1991). A recent study also showed that SSD1-V is required for Hsp104-mediated protein disaggregation and thermotolerance, and in that case, ssd1-d2 fully complemented the ssd1 deletion in disaggregation and zymolyase sensitivity and partially restored thermotolerance (Mir et al., 2009). These observations make it clear that the common C-terminal truncation of Ssd1 retains some of its functions.
Early studies showed that overexpression of SSD1-V could suppress conditional mutations in RNA polymerase I, II, and III (Stettler et al., 1993) and in splicing factors (Luukkonen and Seraphin, 1999). Ssd1 has also been identified through its interaction with the phosphorylated C-terminal repeat of RNA polymerase II (Phatnani and Greenleaf, 2006). These observations are all consistent with Ssd1 having a general effect on RNA levels.
Ssd1 binds RNA in vitro and has some homology to ribonucleases, but it has no discernible ribonuclease activity (Uesono et al., 1997). Recent work has shown that Ssd1-v binds a discrete group of mRNAs in vivo. These mRNAs are enriched for those that encode cell wall components and proteins that localize to the bud (Hogan et al., 2008). Ssd1 does not contain a recognizable RNA-binding domain, and hence its binding to these mRNAs may be indirect.
SSD1-V was initially characterized for its ability to suppress the G1 arrest of a Sit4 phosphatase mutant (Sutton et al., 1991). The role of Sit4 in the cell cycle remains mysterious, but SSD1-V has also been shown to restore the viability of swi4 and cln1cln2 mutants (Cvrckova and Nasmyth, 1993), all of which are important for the G1-to-S transition. SSD1-V also enhances replicative life span (RLS; Kaeberlein et al., 2004), which is defined as the number of cell divisions a mother cell completes in her lifetime (Mortimer and Johnston, 1959). These phenotypes are consistent with a possible role for Ssd1 in the initiation of the cell cycle. We have explored this possibility by characterizing genome-wide transcript levels across the cell cycle in isogenic SSD1-V and ssd1-d cells. We find that cell cycle regulated transcripts have sharper oscillations in SSD1-V, but no specific class is affected. The largest class of affected transcripts are involved in ribosome biogenesis and other pathways that affect longevity. Using a new assay of longevity in purified quiescent cells, we find that SSD1-V cells also have a significantly longer chronological life span (CLS) than ssd1-d cells.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
Strains used are all in the W303 background and are listed in Table 1. The SSD1-V allele in BY4260 was introduced by four backcrosses to W303Va (Wijnen and Futcher, 1999). The W303 ssd1-d prototrophs were made by homologous gene replacement of auxotrophic markers with wild-type DNA in BY2125 and/or BY2124 (W303 MATα). These replacements were verified by sequencing, and then crosses were performed to generate the prototrophic W303 (BY6500 and BY6501) and other auxotrophic combinations, including BY6504 (W303 MATa his3-11,15). The entire SSD1 gene was deleted in BY2125 and replaced by the HIS3MX6 gene (Longtine et al., 1998) to produce BY6464. The isogenic ssd1Δ::HIS3 prototroph (BY6562) was generated from crossing BY6464 to BY6504. To introduce the SSD1-V allele to BY2125, the SSD1-V allele was amplified by PCR from W303Va (Wijnen and Futcher, 1999) and inserted into the URA3 integrating vector pRS306 (Sikorski and Hieter, 1989) to produce pBD3000. pBD3000 was integrated at the ssd1-d locus of BY2125, and the ssd1-d and plasmid sequences were evicted by selection against URA3 (Scherer and Davis, 1979; Boeke et al., 1984) and verified by sequencing to produce BY6580. This W303 SSD1-V auxotroph was crossed to BY6562 (MATα his3-11,15 ssd1::HIS3) to generate W303 his3-11,15 SSD1-V, and then the his3-11,15 was replaced as above to generate the prototroph BY6564 (MATα SSD1-V). The isogenic MATa prototrophs were generated by appropriate crosses (Table 1). All studies described in this manuscript were carried out with the MATa isolates.
Table 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotypea | Source |
|---|---|---|
| BY2125 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 ssd1-d2 | Breeden lab W303a |
| BY2124 | MATα ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 ssd1-d2 | Breeden lab W303α |
| BY4260 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 SSD1-V | Breeden lab |
| BY6464 | MATα ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 ssd1Δ::HIS3 | This study |
| BY6580 | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 SSD1-V | This study |
| BY6500 | MATa ssd1-d2 | This study |
| BY6501 | MATα ssd1-d2 | This study |
| BY6504 | MATa his3-11,15 ssd1-d2 | This study |
| BY6562 | MATα his3-11,15 ssd1Δ::HIS3 | 6504 × 6464 |
| BY6563 | MATa his3-11,15 ssd1Δ::HIS3 | 6562 × 6580 |
| BY6564 | MATα SSD1-V | This study |
| BY6641 | MATa SSD1-V | 6564 × 6563 |
| W303Va | MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 SSD1-V | Wijnen and Futcher (1999) |
a All strains are W303 background and include can1-100 rad5 ho in addition to the genotype as listed.
Strains were cultured at 30°C with aeration in yeast rich medium (YPD) with 2% glucose and a 55 mg/L adenine supplement (YSC; Ausubel et al., 2003) buffered to pH 5.8 with succinic acid (10 g/l) and NaOH (6 g/l) and supplemented with 2% glucose. Other nutritional supplements were added as specified (Ausubel et al., 2003) except in the case where fourfold excess supplementation was used for auxotrophic requirements: adenine (final concentration 160 mg/l), uracil (80 mg/l), leucine (240 mg/l), tryptophan (160 mg/l), and histidine (80 mg/l).
Budding and DNA Analysis
Cells were examined by microscopy for the presence of new buds. Two hundred cells were examined every time point. The budding percentage was calculated as the number of budded cells divided by the number of total cells at each time point. To monitor the DNA synthesis through the cell cycle, cells stained with Sytox-Green (Invitrogen, Carlsbad, CA) were analyzed on a FACScan cytometer (BD Biosciences, San Jose, CA) as described in Pramila et al. (2002).
Viability Assays of Stationary Phase Cultures
Assays were performed as previously published (Fabrizio and Longo, 2003) with additional supplementation of required compounds as previously described (Fabrizio et al., 2004; Powers et al., 2006), but our YSC media is buffered (see above). Cells were grown to stationary phase (SP) in 25 ml YSC synthetic medium with a fourfold excess of required nutrients with aeration at 30°C in 250-ml flasks. Cultures were sampled at times indicated, diluted, and plated in duplicate on YPD plates. Plates were incubated at 30°C for 3 d before colonies forming units (CFUs) were counted. The percentage of surviving cells was calculated with the plateau CFUs attained by the culture set as 100%.
Fractionation and Purification of Quiescent Cells
Density gradient fractionation of quiescent cells (Q cells; Allen et al., 2006) was scaled down to 2-ml total volume. Cells are grown in 25 ml YPD medium in 250-ml flask for 7 d at 30°C. Separation of Q cells from these SP cultures was done by using RediGrad (GE Healthcare, Waukesha, WI) density gradients. To generate the gradient, RediGrad is diluted 9:1 (vol/vol) with 1.5 M NaCl to a final NaCl concentration of 167 mM. Then 1.75 ml gradient solution was put into a 2-ml sterile microfuge tube and centrifuged for 15 min at 13,400 rpm (19,100 × g) at room temperature. About 30 OD600 of SP cells in 100 μl Tris buffer was layered onto the preformed gradient¤, and centrifuged at 1370 rpm (400 × g) for 1 h. Q cells were collected from the bottom 0.5 ml of the gradient. Q cell yield was calculated as the percentage of the total loaded OD600 units harvested from this bottom fraction.
Chronological Life Span Analysis with Purified Quiescent Cells
Purified Q cells from 1-wk-old SP cultures were inoculated into water at an optical density (A600) of ∼1 and incubated at 30°C with aeration. Samples were diluted into distilled water before plating onto YPD plates in duplicate. Plates were incubated at 30°C for 3 d before colony counting. Survival was determined by CFU. CFU at the first week was set to be the initial survival (100%).
Viability Assay
To determine viability, cells were stained using the LIVE/DEAD FungaLight yeast viability kit (Agilent Technologies, Wilmington, DE). The negative viability control was prepared by incubating cells at 75°C for 10 min before the staining. Exponentially growing cells were used for the positive viability control. Cells with no dye added were used as an autofluorescence control. All cell samples were diluted in 1 ml of 50 mM Tris buffer (pH 7.5) with 1 μl of SYTO9 dye and 1 μl of propidium iodide added,¤ and were incubated at 37°C for 15 min before analysis. For flow cytometry, 30,000 cells per sample were analyzed by using 488-nm wavelength excitation and collecting fluorescent emission with filters at 530/30 nm for the FL-1 parameter and 585/42 nm for the FL-2 parameter. To avoid coincidence error, flow rate was maintained at ≤1000 events/s. Cell Quest software (BD Biosciences, San Jose, CA) was used for data collection and analysis.
Microarray Hybridization and Data Processing
Alpha factor synchronizations were carried out as described in Breeden (1997), and the cells were sampled every 10 min for about two cell cycles. Total RNA, 30 μg, was extracted from both ssd1-d and SSD1-V cells and used for cDNA synthesis. Labeling was performed using the amino-allyl reverse transcription labeling protocol, the labeled cDNAs were hybridized to yeast cDNA arrays (Pramila et al., 2002). Array analysis was performed using the GenePix Prosoftware from Axon Instruments (Foster City, CA). Expression data analysis, normalization, and clustering were done by Genespring GX 7.3.1. The SSD1-V dataset is available from the GEO Database (Accession number GSE16911).
Identifying Differentially Expressed Genes
DiffExp (Bar-Joseph et al., 2003) was used to identify genes differentially expressed across the cell cycle between ssd1-d and SSD1-V. Repeat datasets of ssd1-d were used to determine the noise model. The significance threshold for selecting differentially expressed genes was set at 0.005. DiffExp first represents each expression time series as continuous curves so that expression patterns of the same gene are comparable. Then, for each gene, the algorithm computes the global difference between its continuous representation in ssd1-d and SSD1-V and the corresponding p-value. Finally, it ranks the genes according to their p-values, and outputs those genes with a p-value lower than the given threshold.
Clustering Differentially Expressed Genes
The differentially expressed genes were analyzed by the Clustering of Regression Models (CORM) method (Qin and Self, 2006) to identify clusters of genes whose transcript profiles are similar under two different conditions (SSD1-V and ssd1-d). The number of clusters was set at 25.
Identifying and Ranking Cycling Genes
To identify the periodic transcripts in ssd1-d and SSD1-V, we used a method that combines a score for the periodicity of the expression profile with a score for the magnitude of the expression. Previous work (de Lichtenberg et al., 2005; Lu et al., 2007) has shown that such a score performs best among all proposed methods for identifying cycling genes. To compute these scores, we first performed Fast Fourier Transform on the gene expression time series. We then combined the fraction of spectral power around the cell cycle period, and the height of the peak spectrum to obtain the score. Intuitively, the method assigns high scores to genes whose expression level fluctuates strongly with the cell cycle.
Computing the p-value for Differences in Cycling Genes
To evaluate the effects of the SSD1-V versus ssd1-d on cycling genes, we identified genes that received a high cycling score in one cell type but low in the other. To evaluate the significance of these differences, we used randomization analysis as follows. We assume that the cyclic score assigned to a gene is the sum of its true cyclic score and a random error term that follows the Gaussian distribution N (0, σ2). We estimated σ to be 0.85 based on the zero time point technical replicates. We randomly generated the score for each gene by drawing from a Gaussian distribution N (0, 1). This score was assigned to the gene in both strains. Next, we added a random error term sampled from the error distribution to each gene (independently for each strain). We then counted the number of genes that had a high score (rank <1000) in one cell type and a low score (rank >5000) in the other. By repeating this process multiple times, we empirically estimated the probability of observing more than a certain number of genes that are highly periodically expressed in one and not in the other cell type.
RESULTS
SSD1-V Affects Transcript Levels Globally
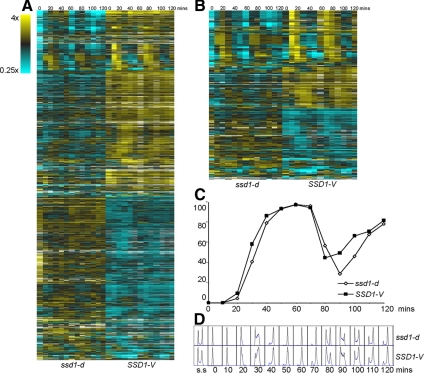
We have generated three microarray data sets through the cell cycle with W303 (Pramila et al., 2006), which carries the ssd1-d2 allele,¤ and another data set with an isogenic strain carrying SSD1-V. All four data sets were obtained from cells synchronized with alpha factor and followed through two consecutive cell cycles. We have compared the global transcript profiles across the cell cycle using DiffExp (Bar-Joseph et al., 2003) and find that there are 889 genes (15%) that show significant differences between cells carrying SSD1-V versus ssd1-d. One-half of these transcripts are up-regulated, and the other half are significantly down-regulated by SSD1-V (Figure 1A). SSD1-V also affects ∼20% of the cell cycle–regulated (ccr) transcripts (Figure 1B), but we find no evidence that any specific class of ccr transcripts are enriched among the differentially expressed genes (see Supplemental Figures S1 and S2).
Figure 1.
Differentially expressed genes between cells carrying ssd1-d versus SSD1-V. (A) Heat map of BY2125 (ssd1-d) and BY4260 (SSD1-V) microarray data through two cell cycles showing the 889 differentially expressed genes identified by DiffExp. Each column represents one time point. Each row represents a gene. (B) Heat map of BY2125 and BY4260 microarray data showing the 227 cell cycle regulated genes that are differentially expressed in cells carrying ssd1-d versus SSD1-V. (C) Progress through the cell cycle was monitored by budding index and (D) DNA synthesis from 0 min to 120 min after alpha factor release. ssd1-d, ◇; SSD1-V, ■.
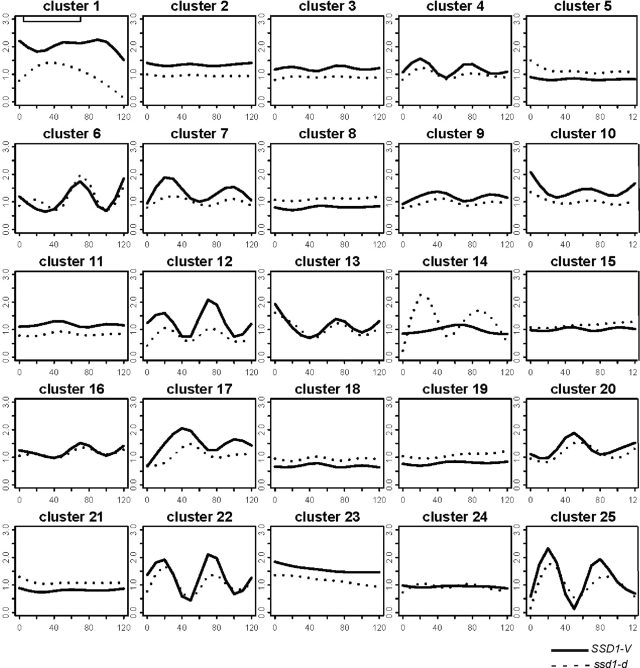
To identify genes whose transcripts might be coordinately regulated by Ssd1, the 889 transcripts that are differentially expressed in SSD1-V versus ssd1-d were analyzed by CORM (Qin and Self, 2006; Tanaka et al., 2007). This algorithm groups genes that share similar relationships to the covariate, which is SSD1. Figure 2 shows 25 clusters of genes whose mean transcript profiles across the cell cycle follow one pattern in SSD1-V cells (solid line) and a second, different pattern in ssd1-d cells (dotted line). Inspection of these profiles shows that about half the affected clusters are periodic. In nearly every case, the ccr transcripts in the SSD1-V profiles have sharper peaks and troughs than those of the ssd1-d cells. These effects can also be seen in the heat maps (Figure 1B). We note that budding and DNA synthesis occur slightly earlier in the SSD1-V strain than the ssd1-d strain (Figure 1, C and D). This slightly accelerated and more synchronous cycle may contribute to the sharper peaks observed in the SSD1-V ccr transcripts. It is also possible that SSD1-V reinforces these oscillations in a more direct manner. (Cluster membership can be found in Supplemental Table S1).
Figure 2.
Clustering analysis of the 899 differentially expressed genes in ssd1-d versus SSD1-V. Twenty-five groups of similarly affected transcripts were found by Clustering of Regression Models (CORM) analysis. ssd1-d (BY2125), dashed line; SSD1-V (BY4260), solid line.
We have used GO analysis to identify enrichment for functionally related gene products within these clusters. We find that a subset of the Ty1 retro-transposons (in clusters 1 and 11) and subtelomeric helicases (cluster 12) are among the most up-regulated transcripts in SSD1-V. However, the sequence homologies within these families make it difficult to know if all or just a fraction of them are up-regulated. We also find that a family of genes involved in iron transport is up-regulated in SSD1-V (in clusters 9, 17, and 20). This is likely to be an indirect effect, because SSD1-V cells also show a very significant up-regulation of both AFT1 and YAP5 mRNA levels (Table 2A). Aft1 and Yap5 are transcription factors that activate transcription of genes involved in iron transport (Yamaguchi-Iwai et al., 1995) and iron storage (Li et al., 2008). Increased expression of these factors may explain why at least 24 of the genes that are bound in vivo by Aft1 (Harbison et al., 2004) are also up-regulated in the SSD1-V cells. A subset of these iron transport gene profiles are shown in Supplemental Figure S3.
Table 2.
Differentially transcribed transcription factors
| Gene | Function | p value |
|---|---|---|
| A. Transcription factors up-regulated by SSD1-V | ||
| YAP5 | bZIP transcription factor; interacts with AFT1 | 0 |
| MET4 | Transcriptional activator responsible for the regulation of the sulfur amino acid pathway; requires different combinations of the auxiliary factors Cbf1p, Met28p, Met31p and Met32p | 1.98E-14 |
| MAL33 | MAL-activator protein | 1.03E-10 |
| HAL7 | Leucine zipper transcription factor; overexpression increases sodium and lithium tolerance | 1.52E-09 |
| RDS1 | Zinc cluster transcription factor involved in conferring resistance to cycloheximide | 3.03E-07 |
| HCM1* | Transcription factor (fork head domain) regulating genes involved in chromosome dynamics | 6.28E-07 |
| RDR1* | Transcriptional repressor involved in the control of multidrug resistance; negatively regulates expression of the PDR5 gene | 2.35E-06 |
| MSN2 | Transcriptional activator related to Msn4p; activated in stress conditions | 4.11E-06 |
| AFT1 | Transcription factor binds the consensus site PyPuCACCCPu and activates the expression of target genes in response to changes in iron availability | 5.22E-06 |
| HAL9* | Putative transcription factor containing a zinc finger; overexpression increases salt tolerance | 5.34E-05 |
| MET18 | DNA repair and TFIIH regulator; involved in telomere maintenance | 2.33E-04 |
| MET32 | Zinc-finger DNA-binding protein, involved in transcriptional regulation of the methionine biosynthetic genes, similar to Met31p | 4.38E-04 |
| MBF1* | Transcriptional coactivator that bridges the DNA-binding region of Gcn4p and TATA-binding protein Spt15p | 5.08E-04 |
| INO2 | Helix-loop-helix protein | 7.75E-04 |
| NDD1* | Nuclear Division Defective 1 | 0.002 |
| OPI1 | Negative regulator of phospholipid biosynthesis; involved in telomere maintenance | 0.002 |
| YHP1* | One of two homeobox transcriptional repressors (see also Yox1p); restrict ECB-mediated transcription to the M/G1 interval | 0.003 |
| RRN3 | Protein required for transcription of rDNA by RNA polymerase I | 0.003 |
| GAT1 | Transcriptional activator of genes involved in nitrogen catabolite repression | 0.01 |
| B. Transcription factors down regulated by SSD1-V | ||
| ROX1 | HMG-domain site-specific DNA binding protein; Heme-dependent repressor of hypoxic genes | 4.44E-16 |
| YER130C* | Transcription factor activity; contains two tandem C2H2-type zinc fingers | 1.67E-15 |
| FAP1 | FKBP-associated protein that conferring rapamycin resistance | 2.04E-08 |
| HAP5 | Transcriptional activator of respiratory gene expression | 2.80E-05 |
| ARG80 | Transcription factor, involved in arginine metabolic process | 6.18E-05 |
| FAP7* | Transcription cofactor activity; Essential NTPase required for small ribosome subunit synthesis | 7.18E-05 |
| KRE33 | Transcription factor activity; required for 40S ribosomal subunit biogenesis | 2.1E-04 |
| RTS2 | Similar to mouse KIN7 protein | 2.6E-04 |
| STB1* | Interacts with the putative transcription factor Sin3p | 8.4E-04 |
| FKH2* | Fork Head homolog two | 8.6E-04 |
| MSS1 | Polypeptide with poly-glutamine and poly-asparagine domains | 9.3E-04 |
| MDS3 | Negative regulator of early meiotic genes | 0.001 |
| YRR1* | Zn2-Cys6 zinc-finger transcription factor that activates genes involved in multidrug resistance | 0.003 |
| RSF2* | Transcription factor that involved in glycerol utilization, respiration | 0.005 |
*, cell cycle regulated genes
In addition of AFT1 and YAP5, there are 31 other transcription factors that are either up- or down-regulated by SSD1-V (Table 2B). We also see increased mRNA levels for transcription factors involved in methionine biosynthesis (MET4, MET18 and MET32), salt tolerance (HAL7 and HAL9) and the stress response (MSN2). Efflux pumps involved in multidrug resistance may also be down-regulated indirectly in SSD1-V cells, because the Rdr1 repressor of pleiotropic drug response genes (Hellauer et al., 2002) is up-regulated and the Yrr1 activator of drug response genes (Onda et al., 2004) is down-regulated.
Structural Components of Cytoplasmic Ribosomes and Many Ribosome Biogenesis Genes Are Down-Regulated by SSD1-V
The most striking enrichment we observe is for the down-regulation of genes involved in translation in SSD1-V cells. Cluster 18 (Figure 2) includes 23 genes involved in ribosome biogenesis (p = 10−11). The promoters of ribosome biogenesis genes contain PAC elements (Dequard-Chablat et al., 1991) and at least one copy of the RRPE element (Hughes et al., 2000). To see if either of these elements could be influenced by SSD1's status, we compared our list of 889 differentially expressed genes with the genes found to contain one or both of these elements in their promoters. Only ∼10% of all genes carrying RRPE or PAC elements were differentially expressed, and of those, about half were up- and half were down-regulated (data not shown). We conclude that direct influences on the activities of these promoter elements are unlikely.
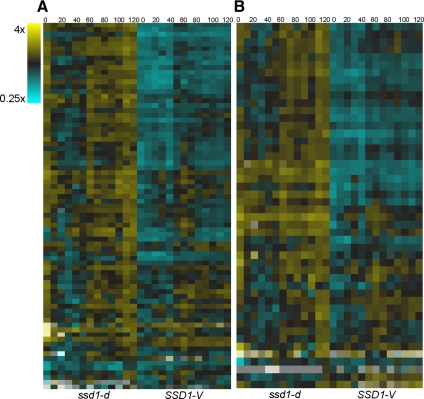
In addition, cluster 19 includes 24 genes that encode structural components of cytoplasmic ribosomes (p = 10−20). These genes also display reduced expression in SSD1-V cells. Figure 3 shows the transcript levels for all proteins of the large and small subunits of cytoplasmic ribosomes across two cell cycles after release from alpha factor arrest in ssd1-d and SSD1-V cells. These heat maps suggest that the impact of SSD1-V on ribosomal (r) protein genes is global. All the r protein transcript levels are lower in the alpha factor arrest (0 time point) and most remain low across two cell cycles in the SSD1-V time course. R protein genes are transcriptionally regulated by Rap1, Fhl1, Ifh1¤ and Crf1 (Moehle and Hinnebusch, 1991; Martin et al., 2004; Rudra et al., 2005). None of these factors are affected by SSD1-V at the RNA level. Moreover, genome-wide chromatin immunoprecipitation studies have identified 207 promoters that are bound in vivo by Fhl1 (Harbison et al., 2004), but only 38 of those are significantly down-regulated in the presence of SSD1-V based on our DiffExp analysis. This, in addition to the fact that the r protein transcripts are affected to differing extents, suggests that another form of regulation is involved.
Figure 3.
The global impact on the transcript levels of cytoplasmic ribosomal proteins by SSD1-V. Data are displayed as in Figure 1. (A) Large subunit protein transcripts. (B) Small subunit protein transcripts.
Differentially Regulated Transcripts of Genes That Interact Physically or Genetically with SSD1
The recent identification of mRNAs that copurify with Ssd1-v (Hogan et al., 2008) led us to first ask if these bound mRNAs are expressed differentially in SSD1-V and ssd1-d cells. Surprisingly, we found no correlation. Of the 60 mRNAs that SSD1-V binds, only eight were differentially expressed and six of these were expressed at a higher level in SSD1-V cells. These results do not support the hypothesis that Ssd1 binding to mRNA influences their degradation rate¤, or that Ssd1 is a ribonuclease in vivo.
Table 3 identifies the 20 genes whose mRNA levels are significantly affected by SSD1 (p < 0.01) and that interact, either genetically or physically with Ssd1 based on BIOGRID (ver. 2.0.49). We find that SSD1-V down-regulates mRNA levels for two subunits of the prefoldin complex (Gim3 and Gim1). This may explain why ssd1 genetically interacts with five of the six prefoldin subunit mutants (Tong et al., 2004; Collins et al., 2007). Among the most affected transcripts are two that encode other RNA-binding proteins (Dhh1 and Mpt5), which are up-regulated by SSD1-V. Both of these proteins promote RNA decay via the Ccr4 complex (Garneau et al., 2007; Goldstrohm et al., 2007). DHH1 and SSD1 functionally complement each other¤ and double mutants are synthetically lethal (Moriya and Isono, 1999). Mpt5 physically interacts with Dhh1 (Goldstrohm et al., 2006) and genetically interacts with Ssd1 (Kikuchi et al., 1994). Interestingly, MPT5, like SSD1, is a polymorphic locus in budding yeast (Kennedy et al., 1995). In addition, loss of any one of these four genes (ssd1, ccr4, mpt5¤ or dhh1) is synthetically lethal with swi4 (Kaeberlein and Guarente, 2002), which encodes a transcription factor that is rate-limiting for the G1-to-S transition (McInerny et al., 1997).
Table 3.
Differentially transcribed genes reported to interact with SSD1-V
| Gene | Function | Interaction | Reference | p value |
|---|---|---|---|---|
| A. Genes up-regulated by SSD1-V | ||||
| SDS3 | (Putative) transcriptional regulator; involved in transcriptional silencing and required for sporulation | Genetic | Vannier et al. (2001) | 9.50E-11 |
| ATG1 (APG1) | Protein ser/thr kinase required for vesicle formation in autophagy and the cytoplasm-to-vacuole targeting (Cvt) pathway | Physical | Ptacek et al. (2005) | 1.00E-09 |
| COG8 | Required to maintain wild-type vacuolar morphology | Genetic | Collins et al. (2007) | 3.25E-09 |
| IES1 | Subunit of the INO80 chromatin remodeling complex | Genetic | Collins et al. (2007) | 1.35E-07 |
| RPB8 | RNA polymerase subunit(common to polymerases I, II, and III) | Genetic | Briand et al. (2001) | 3.32E-07 |
| DHH1 | (Putative) DEAD box RNA helicase, stimulates mRNA decapping, coordinates distinct steps in mRNA function and decay | Genetic | Moriya and Isono (1999) | 8.94E-07 |
| DOT1 | Involved in meiosis and transcriptional silencing | Genetic | Collins et al. (2007) | 2.09E-06 |
| SRO9 | Cytoplasmic RNA-binding protein that associates with translating ribosomes; involved in heme regulation of Hap1p as a component of the HMC complex | Physical | Tarassov et al. (2008) | 3.45E-06 |
| HYP2 | Translation initiation factor eIF-5A, possible role in translation elongation | Genetic | Zanelli et al. (2005) | 3.70E-06 |
| JNM1 | Coiled-coil domain protein required for nuclear migration | Genetic | Wilson et al. (1991) | 1.10E-05 |
| SRB2 | Subunit of the RNA polymerase II mediator complex | Genetic | Collins et al. (2007) | 0.0004 |
| SIT4 | Type 2A–related protein phosphatase that functions in the G1/S transition of the mitotic cycle | Genetic | Stettler et al. (1993) | 0.001 |
| CKB2 | Beta' regulatory subunit of casein kinase 2, a Ser/Thr protein kinase with roles in cell growth and proliferation | Genetic | Collins et al. (2007) | 0.0029 |
| MPT5 | Member of the Puf family of RNA-binding proteins; involved in longevity, maintenance of cell wall integrity, and sensitivity to and recovery from pheromone arrest | Genetic | Kikuchi et al. (1994) | 0.0036 |
| B. Genes down-regulated by SSD1-V | ||||
| SAP30 | Subunit of a histone deacetylase complex that is involved in silencing at telomeres, rDNA, and silent mating-type loci | Genetic | Collins et al. (2007) | 3.20E-12 |
| GIM3 | Subunit of the heterohexameric cochaperone prefoldin complex, which binds specifically to cytosolic chaperonin and transfers target proteins to it | Genetic | Tong et al. (2004) | 7.50E-06 |
| YKE2 (GIM1) | Subunit of the heterohexameric Gim/prefoldin protein complex involved in the folding of α -tubulin, β -tubulin, and actin | Genetic | Collins et al. (2007) | 1.11E-05 |
| SDC1 | Required for proper transcriptional silencing of rDNA, telomeric silencing | Genetic | Collins et al. (2007) | 3.25E-05 |
| NBP2 | Interacts with Nap1, which is involved in histone assembly | Genetic | Ohkuni et al. (2003) | 0.00052 |
| NHP10 | Protein related to mammalian high mobility group proteins; likely component of the INO80 complex | Genetic | Collins et al. (2007) | 0.0006 |
| PCL1 | G1 cyclin that associates with PHO85 | Physical | Ptacek et al. (2005) | 0.00094 |
| YPT6 | Highly homologous to the human GTPase, Rab6 | Genetic | Li and Warner (1996) | 0.04226 |
Transcripts That Influence Replicative Life Span Are Affected by SSD1-V
One-quarter of the genes listed in Table 3, A and B, are involved in chromatin modification. Topping these lists are two proteins that associate with Rpd3 (SDS3 and SAP30). Rpd3 is a histone deacetylase that influences transcription and silencing across the genome (Rundlett et al., 1996). Both rpd3 and ssd1 mutations have been identified as antisilencing factors (Raisner and Madhani, 2008), and both influence replicative aging, but rpd3 extends (Jiang et al., 2002) and ssd1 shortens RLS (Kaeberlein et al., 2004). Interestingly, SDS3 is up-regulated (p = 10−11), and SAP30 is down-regulated (p = 10−12) by SSD1-V. This suggests that the number or nature of Rpd3 complexes formed in SSD1-V cells may differ from that of ssd1-d cells. BIOGRID indicates that SSD1 interacts with seven of the 12 subunits of the large Rpd3 complex.
SSD1-V is known to increase mean RLS in at least three genetic backgrounds (Kaeberlein and Guarente, 2002; Longo, 2003; Kaeberlein et al., 2004). Early transcript microarrays identified 97 transcripts that differed by 1.5-fold or greater in exponentially growing SSD1-V versus ssd1-d cells (Kaeberlein et al., 2004). None of the transcripts identified in that study have been implicated in life span determination. Only a few of those transcripts meet our criteria for differential expression in our SSD1-V/ssd1-d comparison (Table 4). These include four of the iron transport genes.
Table 4.
Overlap in gene expression profiles with SSD1-V/ssd1-d
| Life-span–extending intervention | No. of genes differentially transcribed | Expression pattern | Overlap with SSD1-V/ssd1-d and the expression patterna | |
|---|---|---|---|---|
| SSD1-V/ssd1-db | 95 | 49 up | 8 | 7 up1; 1 dn |
| 46 dn | 11 | 5 dn2; 6 up | ||
| High osmolarityc | 117 | 50 up | 11 | 7 up3; 4 dn |
| 50 dn | 11 | 7 dn4; 4 up | ||
| Calorie restriction | 124 | 114 up | 19 | 15 up5; 4 dn |
| 10 dn | 0 | 0 | ||
| HAP4 overexpression | 255 | 255 up | 39 | 15 up6; 22 dn |
a Genes affected: 1. YBR131W (CCZ1), YDR534C(FIT1), YHL040C(ARN1), YKR075C, YLL053C, YLR108C, YOR383C; 2. YDL043C(PRP11), YKL096W(CWP1), YLR200W(YKE2), YMR193C-A, YPR078C; 3. YLR108C, YLR338W(OPI9), YMR169C(ALD3), YMR170C(ALD2), YMR250W(GAD1), YNR076W(PAU6), YPL223C(GRE1); 4. YBL028C, YDR152W(GIR2), YDR469W(SDC1), YGR030C(POP6), YHR055C(CUP1–2), YMR193C-A, YNL114C; 5. YDL105W(QRI2), YDR290W, YDR440W(DOT1), YDR534C(FIT1), YEL065W(SIT1), YHL040C(ARN1), YIL084C(SDS3), YIL177C, YLL011W(SOF1), YLL062C(MHT1), YNL088W(TOP2), YOR306C(MCH5), YOR374W(ALD4), YOR383C(FIT3), YPL058C(PDR12); and 6. YBL099W(ATP1), YGL149W, YGL261C(PAU11), YIL084C(SDS3), YJL102W(MEF2), YJL182C, YJR157W, YLR338W(OPI9), YLR422W, YMR007W, YOL131W, YOL166C, YPL061W(ALD6),YPL172C(COX10), YPL223C(GRE1).
b See Kaeberlein et al. (2004).
c This extends the life span of ssd1-d cells.
High osmolarity can extend life span (Kaeberlein et al., 2002) and suppress the short RLS observed with ssd1-d cells (Kaeberlein et al., 2004). Caloric restriction (CR) and Hap4 overexpression also increase RLS (Lin et al., 2002). Thus we have looked for transcripts whose levels change twofold or greater in microarray data collected after these treatments (Kaeberlein et al., 2002) and are differentially expressed in our study. Table 4 shows that there is about a 10% overlap between our results and these other studies, but none of these genes have been implicated in longevity.
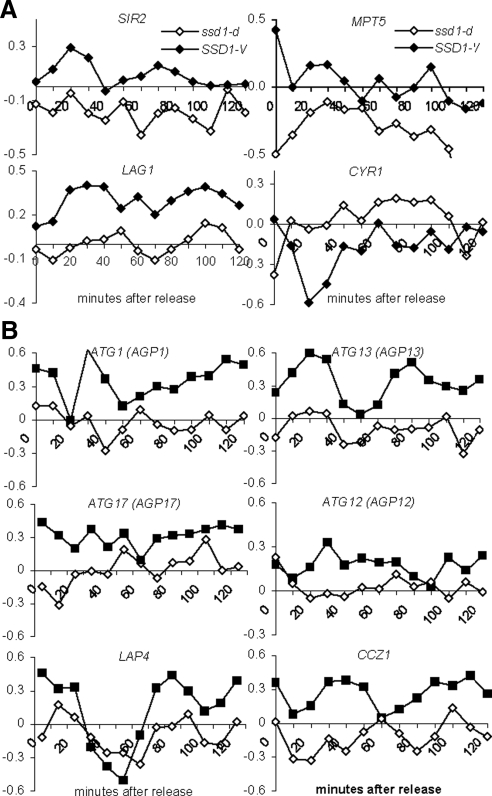
To see if gene products that are known to influence RLS are affected by SSD1-V in W303, we looked for differentially expressed genes whose deletion or overexpression has been reported to increase RLS in yeast (Supplemental Table S2). Figure 4A shows the profiles for the four differentially expressed genes from our analysis (p < 0.01) that are known to influence RLS. The SIR2 (Kaeberlein et al., 1999), MPT5 (Kennedy et al., 1997) and LAG1 (Jiang et al., 2004) gene products promote longevity, and their mRNA levels are higher in SSD1-V cells. Loss of Cyr1 activity promotes longevity (Longo, 2003), and this mRNA is expressed at a lower level in SSD1-V cells. Any or all of these transcriptional changes could contribute to increased longevity of SSD1-V cells.
Figure 4.
Differentially expressed transcripts that affect the life span extension of yeast cells are affected by SSD1-V (closed symbols) versus ssd1-d (open symbols). Log2 ratios for each time point plotted using the transcript level in asynchronous populations as the denominator. (A) Microarray expression profiles of four transcripts that influence the RLS are affected in SSD1-V versus ssd1-d. (B) Microarray expression profiles of the six transcripts that influence autophagy that are up-regulated in SSD1-V versus ssd1-d.
Autophagy genes are required for life span extension in worms and flies, and this conserved pathway may play a similar role in yeast (Powers et al., 2006; Alvers et al., 2009). Autophagy plays an important role in the removal of damaged organelles and proteins, which accumulate in all ageing cells (Vellai, 2009). This process counteracts stresses (e.g., nutrient availability, oxidative damage) by engulfing cytoplasmic organelles and other constituents and transporting them to the vacuole for degradation and recycling (Vellai, 2009). We find that ATG1 (Table 3) as well as five other autophagy-promoting genes are up-regulated by SSD1-V (Figure 4B).
By far the most striking global effect of SSD1-V is its down-regulation of transcripts involved in ribosomal structure and biogenesis. Deletion of a single ribosomal protein gene has been shown to extend life span (Kaeberlein et al., 2005b; Chiocchetti et al., 2007), so this global down-regulation would be expected to contribute to the longevity of this strain.
A Specific Assay for Chronological Life Span in Quiescent Budding Yeast
SSD1-V has a significant positive effect on RLS (Kaeberlein and Guarente, 2002; Longo, 2003; Kaeberlein et al., 2004), and we have identified many expression changes that are likely to contribute to this increase in RLS. A possible role for SSD1-V in CLS, that is, the length of time that cells can survive in a nongrowing state, has not been tested. Moreover, SP cultures of the W303 ssd1-d strain survive longer in this nondividing state than many other strains, including BY4743, which carries the SSD1-V allele (Qin and Lu, 2006). This led us to wonder if SSD1-V was a significant variable in CLS.
The long-term survival of SP cultures that have exhausted their nutrients and stopped dividing has been used as a means to approximate the CLS of nondividing cells (Fabrizio et al., 2001, 2003). However, a recent study has traced mortality in SP cultures to the accumulation of acetic acid in the media (Burtner et al., 2009), so it is unclear how relevant this assay is as a model of aging in yeast. Moreover, these assays of SP cultures are typically carried out with auxotrophic strains. Starved auxotrophs have short but variable life spans, depending on the limiting nutrient (Henry, 1973; Boer et al., 2008). They also have a nonuniform arrest (Saldanha et al., 2004), accumulate more reactive oxygen species and DNA fragmentation (Gomes et al., 2007)¤ and exhaust their glucose supply faster than prototrophs (Brauer et al., 2006). In SP cultures, auxotrophs have been shown to be genetically unstable and rely on the DNA damage checkpoint to retain viability (Weinberger et al., 2007). One potential solution to this problem is to add fourfold more of the required compounds than are typically added (Powers et al., 2006; Wei et al., 2008).
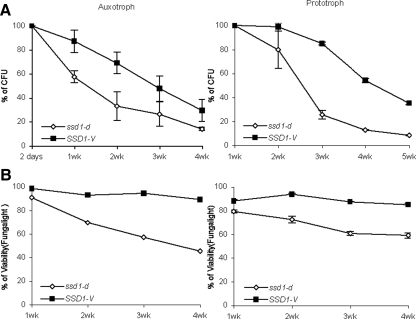
For comparison to this recent work, we assayed the long-term survival of SP cultures of SSD1-V and ssd1-d strains, both as auxotrophs and prototrophs, using fourfold oversupplementation of the compounds required by the auxotroph. Figure 5 shows the survival curves for SSD1-V and ssd1-d SP cultures in an auxotrophic (left) and prototrophic (right) background. The top panels plot the fraction of the population that can resume cell division and form colonies or CFUs. The bottom plots show the FungaLight viability assay (Molecular Probes, Invitrogen), which quantifies the fraction of cells with sufficient metabolic activity to maintain intact membranes and exclude propidium iodide. By both assays, in auxotrophs and prototrophs, the SSD1-V allele enhances long-term viability compared with ssd1-d.
Figure 5.
Comparison of the long-term survival of SP cultures of ssd1-d and SSD1-V in an auxotrophic and a prototrophic background. (A) Survival curves of ssd1-d versus SSD1-V were measured by colony forming units (CFU). Maximum CFUs for the culture was set as 100% survival. BY6500, prototrophic ssd1-d strains; BY6641, prototrophic SSD1-V strain; BY2125, auxotrophic ssd1-d strains; BY4260, auxotrophic SSD1-V strains. (B) The viabilities of both ssd1-d and SSD1-V in culture were determined by using Fungalight Yeast Viability Kit and plotted from 1 to 4 wk.
However, these studies also reveal a large and unexpected source of variability in these assays (Table 5). The auxotrophic cultures survive poorly compared with their prototrophic, but otherwise isogenic counterparts. The ssd1-d auxotroph retains 50% CFUs for 10 d, but the ssd1-d prototroph retains 50% CFUs until day 18. The same is true with the SSD1-V pair, but to a lesser extent. The auxotrophic SSD1-V culture retains 50% CFUs for 21 d, and the prototroph lasts 30 d. Hence, it is clear that colony-forming ability declines more rapidly in auxotrophs than prototrophs, despite the provision of fourfold excess of required compounds, and this decline is more rapid in auxotrophs carrying ssd1-d. In summary, this direct comparison shows that SSD1-V promotes long-term survival of SP cells, but prototrophy is also important for survival under these conditions. Auxotrophy has a significant, but variable, negative impact on the reproductive capacity of both strains, and this deleterious effect is not mitigated by oversupplementation of required compounds. We conclude that in order to assess the effect of Ssd1 or any gene product on survival after nutrient exhaustion, the assays must be performed in prototrophs.
Table 5.
Summary of chronological life span results
| SP survival |
Q cell CLS Protd | Q cell yield (%)a |
||||
|---|---|---|---|---|---|---|
| Auxb | Protc | 1 wk | 2 wk | 3 wk | ||
| SSD1-V | 21 ± 3 | 30 ± 0.5 | 57 ± 5 | 77 ± 3 | 66 ± 5 | 48 ± 1 |
| ssd1-d | 10 ± 2 | 18 ± 0.5 | 33 ± 1 | 45 ± 9 | 20 ± 1 | 7 ± 5 |
| ssd1Δ | n.d. | 19 ± 5 | 41 ± 1 | 44 ± 2 | 23 ± 6 | 9 ± 4 |
n.d., not determined.
a Q cell yield from SP cultures grown for 1, 2, or 3 wk before purification.
b Days to 50% CFU in SP culture with auxotrophs.
c As in A, but with prototrophs.
d Days to 50% CFU with Q cells purified from 1-wk-old SP culture and resuspended in water at 30°C.
Another variable that affects the life span of a SP culture is the fact that these cultures contain cells that are already dead and cells that are viable but cannot resume cell division (Herker et al., 2004; Allen et al., 2006). These cells obscure the properties of the quiescent cells in the population. To specifically assay the longevity of the Q cells, we have used a new purification procedure that separates Q cells from all others by virtue of their increased density (Allen et al., 2006). The cause of this increased density is not understood, but it serves as a convenient property by which to purify a population of cells that retain all the features associated with quiescence. These Q cells arrest with G1 DNA content and maintain high levels of storage carbohydrates. They also show little or no indication of reactive oxygen species or apoptosis (Allen et al., 2006). By assaying the long-term survival of purified, prototrophic Q cells, we can obtain a direct measure of CLS and assess the influence of mutations on the longevity of quiescent cells.
Repeated purifications show that the yield of Q cells is reproducible. However, Q cell yield is dramatically affected by the amount of time the cells are in SP culture before purification and by the genotype of the strain (Table 5). Q cell yield is characteristically higher and drops more slowly in SP cultures of SSD1-V cells than for ssd1-d. We have also purified Q cells from an SSD1 deletion (ssd1Δ). The Q cell yield over time with the null allele was comparable to that of ssd1-d.
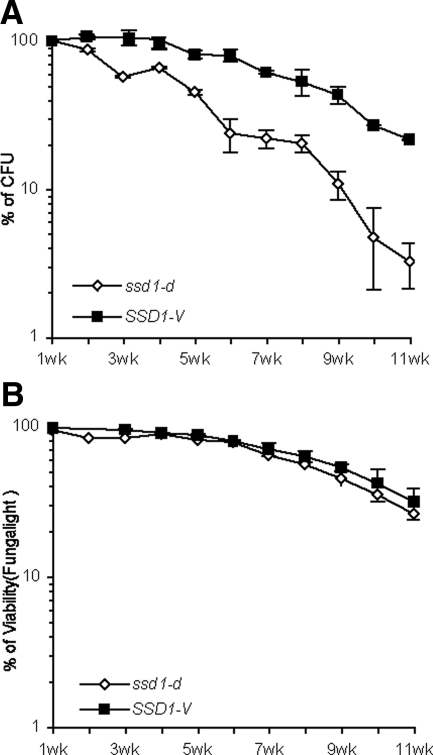
SSD1-V Increases the CLS of Q Cells
Q cells were purified from 1-wk-old SP cultures of prototrophic SSD1-V and ssd1-d cells. These cells were transferred to water and assayed for CFU and membrane integrity over 11 wk (Figure 6). ssd1-d Q cells lose reproductive capacity much more rapidly than SSD1-V cells, but they retain sufficient metabolic activity to exclude propidium idodide almost as well as SSD1-V cells. This indicates that ssd1-d is specifically defective in maintaining the long-term replicative capacity of Q cells, but it does not affect their ability to survive in a nondividing, senescent state. We have also assayed the longevity ssd1Δ and found little or no difference between the CLS of ssd1-d and the null mutant. We conclude that ssd1-d displays a phenotype similar to the null mutant in this assay.
Figure 6.
The CLS of Q cells purified from one week old prototrophic cultures of SSD1-V and ssd1-d cells. Survival of colony forming units plotted as a function of weeks after purification and resuspended in water. (A) Survival curves of Q cells from ssd1-d versus SSD1-V measured by CFUs. CFUs of Q cells after the first week in water was set as 100% survival. (B) The viabilities of Q cells from ssd1-d versus SSD1-V in water were determined using Fungalight Yeast Viability Kit and plotted.
Our results are summarized in Table 5. We find that purified Q cells live significantly longer than their respective SP cultures. Instead of dropping to 50% CFUs in 18 d, the ssd1-d Q cells remain above 50% CFUs for ∼33 d. The purified SSD1-V Q cells also double the life span measured for the SP culture, and again they surpass the longevity of ssd1-d cells by many weeks. We conclude that CLS assays carried out with prototrophic Q cells provide the most direct measure of longevity in the nondividing state. With this assay, we observe a significant enhancement of longevity by SSD1-V.
DISCUSSION
Ribosomal Genes Are Globally Down-Regulated by SSD1-V
By far the most striking effect SSD1-V has on transcript levels is the global down-regulation of those that are involved in translation. We see significant enrichment of the ribosome biogenesis genes among transcripts expressed at a lower level in SSD1-V cells, but only a minority of these transcripts is affected. In addition we find that all of the transcripts that encode cytoplasmic ribosomal (r) proteins are affected to some extent by SSD1-V. Interestingly, SSD1 transcript levels are increased by oxidative and heat stress, and during SP (Gasch et al., 2000). All these conditions also lead to drops in r protein mRNA levels (Gasch et al., 2000). This is consistent with the possibility that Ssd1 responds to a shift to adverse conditions and globally regulates translational capacity. However, the mechanism of this control is unknown. The promoter elements known to regulate translational machinery are not likely to be the targets, and there is no correlation with the mRNAs bound by Ssd1.
Ribosomal protein mRNAs are highly abundant and account for about one-third of the cell's mRNA (Holstege et al., 1998) and 50% of transcription initiation events (Warner, 1999). In addition, most r protein mRNAs contain introns (Spingola et al., 1999). The abundance of these intron-containing mRNAs has led to estimates that 90% of all splicing events occur on r protein transcripts (Warner, 1999). It follows that a global reduction of r protein mRNAs would significantly reduce the demand for splicing machinery. This probably explains why mutations in Fhl1, a positive regulator of r protein transcription, suppress splicing defects (Maddock et al., 1994). We note that SSD1-V shares both the global reduction of r protein mRNAs and the ability to suppress a variety of splicing defects (Luukkonen and Seraphin, 1999).
Expression Changes in Transcripts That Influence Longevity
We have identified more than 100 transcript changes that could be contributing to the increased RLS that has been seen in SSD1-V cells (Kaeberlein et al., 2004). Among the best studied are Sir2 and Mpt5, which are both expressed at higher levels in SSD1-V cells. However, both sir2 and mpt5 mutants have been tested and found not to be required for the RLS increase observed in SSD1-V cells (Kaeberlein and Guarente, 2002; Kaeberlein et al., 2004).
Autophagy is a starvation-induced pathway for degrading and recycling cytoplasmic components that is highly conserved and important for longevity in many systems (Diaz-Troya et al., 2008). We find that six of the 31 autophagy-related (ATG) yeast genes are up-regulated in SSD1-V cells. Among these are all three subunits (ATG1, ATG17, and ATG13) of a highly conserved kinase complex that is essential for the initial induction of autophagy in response to starvation (Diaz-Troya et al., 2008). Atg1/17/13 complexes recruit other Atg proteins to a perivacuolar site where phagosomes are formed (Cheong et al., 2008)¤ and its kinase activity is required for mobilization of these phagosomes (Cheong and Klionsky, 2008). The fact that all three subunits of this kinase are coordinately up-regulated by SSD1-V suggests that cells carrying SSD1-V may be more efficient at triggering the autophagic response and hence more protected from the stresses that evoke it.
As indicated above, the coordinate reduction in transcripts involved in translation is the most striking signature in the SSD1-V microarrays. Loss of even a single ribosomal protein gene can significantly increase RLS (Kaeberlein et al., 2005a). We have found that nearly all the r protein transcripts and 10% of the ribosome biogenesis transcripts are down-regulated by SSD1-V in our strain background. While these effects may not be as dramatic at the protein level, this global reduction would certainly be expected to contribute to the increase in longevity of SSD1-V cells.
It has been shown that mutations in several proteins involved in mRNA degradation show all the hallmarks of death by apoptosis in exponentially growing cells (Mazzoni et al., 2003). It is not known whether this apoptosis is elicited by an excess of total mRNA, or by an excess of specific mRNAs that encode apoptosis-triggering gene products. We see no evidence of changes in mRNA levels for specific apoptosis regulators, but it is possible that the total load of mRNA is decreased in SSD1-V cells and this contributes to their increased longevity.
SSD1-V Is Important for the Long-Term Survival of Q Cells
Until recently, monitoring the long-term survival of SP cultures has been the only means to estimate the CLS of budding yeast. However, a surprising density shift of the quiescent population within an SP culture enables us to purify these Q cells and study their CLS and other properties in isolation (Allen et al., 2006). A direct comparison of the Q cell CLS assay with the previous assay using SP cultures showed that the SP assay significantly underestimates the CLS of the quiescent population. Moreover, we find that the SP assay is further compromised by the use of auxotrophs. It is not surprising that starvation for amino acids, or other auxotrophic requirements would be deleterious. These compounds are normally produced by wild, prototrophic yeast. As such, wild yeast would not encounter such limitations and would not have evolved a protective response to them. However, the negative effects of auxotrophy that we observe do not appear to do be due to starvation for required compounds. Despite excess supplementation, the prototroph survives SP much longer than its parent auxotroph.
Even with prototrophs, only a fraction of the cells in a SP culture attain the quiescent state (Allen et al., 2006). We also find that the yield of Q cells in these SP cultures diminishes with time in culture and varies with the genetic background. The gradual loss of Q cells from these dense SP cultures is probably not primarily due to the aging process, because there is an extrinsic factor, namely, the acidification of the medium, that appears to drive the loss of viability in SP cultures (Burtner et al., 2009). However, understanding the genetic components that influence Q cell yield could be very important for understanding the mechanism by which cells attain the quiescent state. In addition, our new CLS assay, which follows longevity in a dilute suspension of purified quiescent cells, does not suffer from the same complexities seen with SP cultures and, as such, it offers a new tool for characterizing the chronological aging process.
With this assay, we have shown that SSD1-V promotes longevity in quiescent cells. We find that prototrophic SSD1-V Q cells have twice the CLS as prototrophic ssd1-d Q cells. Because SSD1-V also increases RLS, we conclude that SSD1-V plays a role in longevity in both dividing and nondividing cells. Consistent with this, we find that loss of SSD1-V does not disrupt the metabolic activities required to keep the membranes of nondividing cells intact. Rather, it specifically impairs a Q cell's ability to return to the cell division cycle and form a colony. This inability to resume cell division from quiescence may not be so different from the inability of an old mother cell to divide and give rise to one more viable daughter cell at the end of her RLS. Hence, it is possible that Ssd1-v is one of a set of core activities that are required for both CLS and RLS. That function of Ssd1 may be difficult to identify because of the large number of transcript differences that are evident in the comparison of SSD1-V and ssd1-d cells, many of which have known effects on longevity. In this case it may be that the range of processes affected by SSD1-V will incrementally or synergistically affect the overall fitness and long-term viability of these cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Braun for scaling down the Q cell purification, A. Prasad for useful comments on the manuscript, and all members of the Breeden lab for helpful discussions. This work was supported by National Institutes of Health Grant GM41073 (L.B.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-04-0347) on July 1, 2009.
REFERENCES
- Allen C., et al. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J. Cell Biol. 2006;174:89–100. doi: 10.1083/jcb.200604072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvers A. L., Fishwick L. K., Wood M. S., Hu D., Chung H. S., Dunn W. A., Jr, Aris J. P. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell. 2009;8:353–369. doi: 10.1111/j.1474-9726.2009.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. Current Protocols in Molecular Biology. New York: Wiley Interscience; 2003. 13.13.13. [Google Scholar]
- Bar-Joseph Z., Gerber G., Simon I., Gifford D. K., Jaakkola T. S. Comparing the continuous representation of time-series expression profiles to identify differentially expressed genes. Proc. Natl. Acad. Sci. USA. 2003;100:10146–10151. doi: 10.1073/pnas.1732547100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J., Lacroute F., Fink G. A positive selection for mutants lacking orotidine-5′ phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Boer V. M., Amini S., Botstein D. Influence of genotype and nutrition on survival and metabolism of starving yeast. Proc. Natl. Acad. Sci. USA. 2008;105:6930–6935. doi: 10.1073/pnas.0802601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer M. J., Yuan J., Bennett B. D., Lu W., Kimball E., Botstein D., Rabinowitz J. D. Conservation of the metabolomic response to starvation across two divergent microbes. Proc. Natl. Acad. Sci. USA. 2006;103:19302–19307. doi: 10.1073/pnas.0609508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden L. L. Alpha factor synchronization of budding yeast. Methods Enzymol. 1997;283:332–341. doi: 10.1016/s0076-6879(97)83027-3. [DOI] [PubMed] [Google Scholar]
- Briand J. F., Navarro F., Rematier P., Boschiero C., Labarre S., Werner M., Shpakovski G. V., Thuriaux P. Partners of Rpb8p, a small subunit shared by yeast RNA polymerases I, II and III. Mol. Cell. Biol. 2001;21:6056–6065. doi: 10.1128/MCB.21.17.6056-6065.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner C. R., Murakami C. J., Kennedy B. K., Kaeberlein M. A molecular mechanism of chronological aging in yeast. Cell Cycle. 2009;8:1256–1270. doi: 10.4161/cc.8.8.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Klionsky D. J. Dual role of Atg1 in regulation of autophagy-specific PAS assembly in Saccharomyces cerevisiae. Autophagy. 2008;4:724–726. doi: 10.4161/auto.6375. [DOI] [PubMed] [Google Scholar]
- Cheong H., Nair U., Geng J., Klionsky D. J. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 2008;19:668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiocchetti A., et al. Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Exp. Gerontol. 2007;42:275–286. doi: 10.1016/j.exger.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Collins S. R., et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Cvrckova F., Nasmyth K. Yeast G1 cyclins CLN1 and CLN2 and a GAP-like protein have a role in bud formation. EMBO J. 1993;12:5277–5286. doi: 10.1002/j.1460-2075.1993.tb06223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lichtenberg U., Jensen L. J., Fausboll A., Jensen T. S., Bork P., Brunak S. Comparison of computational methods for the identification of cell cycle-regulated genes. Bioinformatics. 2005;21:1164–1171. doi: 10.1093/bioinformatics/bti093. [DOI] [PubMed] [Google Scholar]
- Dequard-Chablat M., Riva M., Carles C., Sentenac A. RPC19, the gene for a subunit common to yeast RNA polymerases A (I) and C (III) J. Biol. Chem. 1991;266:15300–15307. [PubMed] [Google Scholar]
- Diaz-Troya S., Perez-Perez M. E., Florencio F. J., Crespo J. L. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy. 2008;4:851–865. doi: 10.4161/auto.6555. [DOI] [PubMed] [Google Scholar]
- Du L. L., Novick P. Pag1p, a novel protein associated with protein kinase Cbk1p, is required for cell morphogenesis and proliferation in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:503–514. doi: 10.1091/mbc.01-07-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P., Liou L. L., Moy V. N., Diaspro A., Valentine J. S., Gralla E. B., Longo V. D. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics. 2003;163:35–46. doi: 10.1093/genetics/163.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P., Longo V. D. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- Fabrizio P., Pletcher S. D., Minois N., Vaupel J. W., Longo V. D. Chronological aging-independent replicative life span regulation by Msn2/Msn4 and Sod2 in Saccharomyces cerevisiae. FEBS Lett. 2004;557:136–142. doi: 10.1016/s0014-5793(03)01462-5. [DOI] [PubMed] [Google Scholar]
- Fabrizio P., Pozza F., Pletcher S. D., Gendron C. M., Longo V. D. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- Gank K. D., Yeaman M. R., Kojima S., Yount N. Y., Park H., Edwards J. E., Jr, Filler S. G., Fu Y. SSD1 is integral to host defense peptide resistance in Candida albicans. Eukaryot. Cell. 2008;7:1318–1327. doi: 10.1128/EC.00402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau N. L., Wilusz J., Wilusz C. J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., Brown P. O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm A. C., Hook B. A., Seay D. J., Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- Goldstrohm A. C., Seay D. J., Hook B. A., Wickens M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J. Biol. Chem. 2007;282:109–114. doi: 10.1074/jbc.M609413200. [DOI] [PubMed] [Google Scholar]
- Gomes P., Sampaio-Marques B., Ludovico P., Rodrigues F., Leao C. Low auxotrophy-complementing amino acid concentrations reduce yeast chronological life span. Mech. Ageing Dev. 2007;128:383–391. doi: 10.1016/j.mad.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Harbison C. T., et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellauer K., Akache B., MacPherson S., Sirard E., Turcotte B. Zinc cluster protein Rdr1p is a transcriptional repressor of the PDR5 gene encoding a multidrug transporter. J. Biol. Chem. 2002;277:17671–17676. doi: 10.1074/jbc.M201637200. [DOI] [PubMed] [Google Scholar]
- Henry S. A. Death resulting from fatty acid starvation in yeast. J. Bacteriol. 1973;116:1293–1303. doi: 10.1128/jb.116.3.1293-1303.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herker E., Jungwirth H., Lehmann K. A., Maldener C., Frohlich K. U., Wissing S., Buttner S., Fehr M., Sigrist S., Madeo F. Chronological aging leads to apoptosis in yeast. J. Cell Biol. 2004;164:501–507. doi: 10.1083/jcb.200310014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan D. J., Riordan D. P., Gerber A. P., Herschlag D., Brown P. O. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege F. C., Jennings E. G., Wyrick J. J., Lee T. I., Hengartner C. J., Green M. R., Golub T. R., Lander E. S., Young R. A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Hong E. L., et al. Gene Ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 2008;36:D577–D581. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. D., Estep P. W., Tavazoie S., Church G. M. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- Ibeas J. I., Yun D. J., Damsz B., Narasimhan M. L., Uesono Y., Ribas J. C., Lee H., Hasegawa P. M., Bressan R. A., Pardo J. M. Resistance to the plant PR-5 protein osmotin in the model fungus Saccharomyces cerevisiae is mediated by the regulatory effects of SSD1 on cell wall composition. Plant J. 2001;25:271–280. doi: 10.1046/j.1365-313x.2001.00967.x. [DOI] [PubMed] [Google Scholar]
- Jiang J. C., Kirchman P. A., Allen M., Jazwinski S. M. Suppressor analysis points to the subtle role of the LAG1 ceramide synthase gene in determining yeast longevity. Exp. Gerontol. 2004;39:999–1009. doi: 10.1016/j.exger.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Jiang J. C., Wawryn J., Shantha Kumara H. M., Jazwinski S. M. Distinct roles of processes modulated by histone deacetylases Rpd3p, Hda1p, and Sir2p in life extension by caloric restriction in yeast. Exp. Gerontol. 2002;37:1023–1030. doi: 10.1016/s0531-5565(02)00064-5. [DOI] [PubMed] [Google Scholar]
- Jorgensen P., Nelson B., Robinson M. D., Chen Y., Andrews B., Tyers M., Boone C. High-resolution genetic mapping with ordered arrays of Saccharomyces cerevisiae deletion mutants. Genetics. 2002;162:1091–1099. doi: 10.1093/genetics/162.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., Andalis A. A., Fink G. R., Guarente L. High osmolarity extends life span in Saccharomyces cerevisiae by a mechanism related to calorie restriction. Mol. Cell. Biol. 2002;22:8056–8066. doi: 10.1128/MCB.22.22.8056-8066.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., Andalis A. A., Liszt G. B., Fink G. R., Guarente L. Saccharomyces cerevisiae SSD1-V confers longevity by a Sir2p-independent mechanism. Genetics. 2004;166:1661–1672. doi: 10.1534/genetics.166.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., Guarente L. Saccharomyces cerevisiae MPT5 and SSD1 function in parallel pathways to promote cell wall integrity. Genetics. 2002;160:83–95. doi: 10.1093/genetics/160.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., Kirkland K. T., Fields S., Kennedy B. K. Genes determining yeast replicative life span in a long-lived genetic background. Mech. Ageing Dev. 2005a;126:491–504. doi: 10.1016/j.mad.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M., McVey M., Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., Powers R. W., 3rd, Steffen K. K., Westman E. A., Hu D., Dang N., Kerr E. O., Kirkland K. T., Fields S., Kennedy B. K. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005b;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kennedy B. K., Austriaco N. R., Jr, Zhang J., Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., Oka Y., Kobayashi M., Uesono Y., Toh-e A., Kikuchi A. A new yeast gene, HTR1, required for growth at high temperature, is needed for recovery from mating pheromone-induced G1 arrest. Mol. Gen. Genet. 1994;245:107–116. doi: 10.1007/BF00279756. [DOI] [PubMed] [Google Scholar]
- Kosodo Y., Imai K., Hirata A., Noda Y., Takatsuki A., Adachi H., Yoda K. Multicopy suppressors of the sly1 temperature-sensitive mutation in the ER-Golgi vesicular transport in Saccharomyces cerevisiae. Yeast. 2001;18:1003–1014. doi: 10.1002/yea.747. [DOI] [PubMed] [Google Scholar]
- Li B., Warner J. R. Mutation of the Rab6 homologue of Saccharomyces cerevisiae, YPT6, inhibits both early Golgi function and ribosome biosynthesis. J. Biol. Chem. 1996;271:16813–16819. doi: 10.1074/jbc.271.28.16813. [DOI] [PubMed] [Google Scholar]
- Li L., Bagley D., Ward D. M., Kaplan J. Yap5 is an iron-responsive transcriptional activator that regulates vacuolar iron storage in yeast. Mol. Cell. Biol. 2008;28:1326–1337. doi: 10.1128/MCB.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. J., Kaeberlein M., Andalis A. A., Sturtz L. A., Defossez P. A., Culotta V. C., Fink G. R., Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Longo V. D. The Ras and Sch9 pathways regulate stress resistance and longevity. Exp. Gerontol. 2003;38:807–811. doi: 10.1016/s0531-5565(03)00113-x. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lu Y., Mahony S., Panayiotis V. B., Rosenfeld R., Simon I., Breeden L. L., Bar-Joseph Z. Combined analysis reveals a core set of cycling transcripts. Genome Biol. 2007;8:R146–R157. doi: 10.1186/gb-2007-8-7-r146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luukkonen B. G., Seraphin B. A conditional U5 snRNA mutation affecting pre-mRNA splicing and nuclear pre-mRNA retention identifies SSD1/SRK1 as a general splicing mutant suppressor. Nucleic Acids Res. 1999;27:3455–3465. doi: 10.1093/nar/27.17.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock J. R., Weidenhammer E. M., Adams C. C., Lunz R. L., Woolford J. L., Jr Extragenic suppressors of Saccharomyces cerevisiae prp4 mutations identify a negative regulator of PRP genes. Genetics. 1994;136:833–847. doi: 10.1093/genetics/136.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. E., Soulard A., Hall M. N. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell. 2004;119:969–979. doi: 10.1016/j.cell.2004.11.047. [DOI] [PubMed] [Google Scholar]
- Mazanka E., Alexander J., Yeh B. J., Charoenpong P., Lowery D. M., Yaffe M., Weiss E. L. The NDR/LATS family kinase Cbk1 directly controls transcriptional asymmetry. PLoS Biol. 2008;6:e203. doi: 10.1371/journal.pbio.0060203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni C., Mancini P., Verdone L., Madeo F., Serafini A., Herker E., Falcone C. A truncated form of KlLsm4p and the absence of factors involved in mRNA decapping trigger apoptosis in yeast. Mol. Biol. Cell. 2003;14:721–729. doi: 10.1091/mbc.E02-05-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerny C. J., Partridge J. F., Mikesell G. E., Creemer D. P., Breeden L. L. A novel Mcm1-dependent element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G1-specific transcription. Genes Dev. 1997;11:1277–1288. doi: 10.1101/gad.11.10.1277. [DOI] [PubMed] [Google Scholar]
- Mir S. S., Fiedler D., Cashikar A. G. Ssd1 is required for thermotolerance and Hsp104-mediated protein disaggregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2009;29:187–200. doi: 10.1128/MCB.02271-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle C. M., Hinnebusch A. G. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991;11:2723–2735. doi: 10.1128/mcb.11.5.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya H., Isono K. Analysis of genetic interactions between DHH1, SSD1 and ELM1 indicates their involvement in cellular morphology determination in Saccharomyces cerevisiae. Yeast. 1999;15:481–496. doi: 10.1002/(SICI)1097-0061(199904)15:6<481::AID-YEA391>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Johnston J. R. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- Ohkuni K., Okuda A., Kikuchi A. Yeast Nap1-binding protein Nbp2p is required for mitotic growth at high temperatures and for cell wall integrity. Genetics. 2003;165:517–529. doi: 10.1093/genetics/165.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onda M., Ota K., Chiba T., Sakaki Y., Ito T. Analysis of gene network regulating yeast multidrug resistance by artificial activation of transcription factors: involvement of Pdr3 in salt tolerance. Gene. 2004;332:51–59. doi: 10.1016/j.gene.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Parsons A. B., Brost R. L., Ding H., Li Z., Zhang C., Sheikh B., Brown G. W., Kane P. M., Hughes T. R., Boone C. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- Phatnani H. P., Greenleaf A. L. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Powers R. W., 3rd, Kaeberlein M., Caldwell S. D., Kennedy B. K., Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramila T., Miles S., GuhaThakurta D., Jemilo D., Breeden L. L. Conserved homeodomain proteins interact with MADS box protein Mcm1 to restrict ECB-dependent transcription to the M/G1 phase of the cell cycle. Genes Dev. 2002;16:3034–3045. doi: 10.1101/gad.1034302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramila T., Wu W., Miles S., Noble W. S., Breeden L. L. The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev. 2006;20:2266–2278. doi: 10.1101/gad.1450606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacek J., et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- Qin H., Lu M. Natural variation in replicative and chronological life spans of Saccharomyces cerevisiae. Exp. Gerontol. 2006;41:448–456. doi: 10.1016/j.exger.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Qin L. X., Self S. G. The clustering of regression models method with applications to gene expression data. Biometrics. 2006;62:526–533. doi: 10.1111/j.1541-0420.2005.00498.x. [DOI] [PubMed] [Google Scholar]
- Racki W. J., Becam A. M., Nasr F., Herbert C. J. Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J. 2000;19:4524–4532. doi: 10.1093/emboj/19.17.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner R. M., Madhani H. D. Genomewide screen for negative regulators of sirtuin activity in Saccharomyces cerevisiae reveals 40 loci and links to metabolism. Genetics. 2008;179:1933–1944. doi: 10.1534/genetics.108.088443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguly T., et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J. Biol. 2006;5:1–28. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke A., Anderson S., McCaffery J. M., Yates J., 3rd, Aronova S., Chu S., Fairclough S., Iverson C., Wedaman K. P., Powers T. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:14752–14762. doi: 10.1074/jbc.M313062200. [DOI] [PubMed] [Google Scholar]
- Rudra D., Zhao Y., Warner J. R. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J. 2005;24:533–542. doi: 10.1038/sj.emboj.7600553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett S. E., Carmen A. A., Kobayashi R., Bavykin S., Turner B. M., Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha A. J., Brauer M. J., Botstein D. Nutritional homeostasis in batch and steady-state culture of yeast. Mol. Biol. Cell. 2004;15:4089–4104. doi: 10.1091/mbc.E04-04-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S., Davis R. W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc. Natl. Acad. Sci. USA. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spingola M., Grate L., Haussler D., Ares M., Jr Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stettler S., Chiannilkulchai N., Hermann-Le Denmat S., Lalo D., Lacroute F., Sentenac A., Thuriaux P. A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol. Gen. Genet. 1993;239:169–176. doi: 10.1007/BF00281615. [DOI] [PubMed] [Google Scholar]
- Sutton A., Immanuel D., Arndt K. T. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol. Cell. Biol. 1991;11:2133–2148. doi: 10.1128/mcb.11.4.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., et al. Saccharomyces cerevisiae SSD1 orthologues are essential for host infection by the ascomycete plant pathogens Colletotrichum lagenarium and Magnaporthe grisea. Mol. Microbiol. 2007;64:1332–1349. doi: 10.1111/j.1365-2958.2007.05742.x. [DOI] [PubMed] [Google Scholar]
- Tarassov K., Messier V., Landry C. R., Radinovic S., Molina M. M., Shames I., Malitskaya Y., Vogel J., Bussey H., Michnick S. W. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- Tong A. H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Tong A. H., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Uesono Y., Fujita A., Toh-e A., Kikuchi Y. The MCS1/SSD1/SRK1/SSL1 gene is involved in stable maintenance of the chromosome in yeast. Gene. 1994;143:135–138. doi: 10.1016/0378-1119(94)90618-1. [DOI] [PubMed] [Google Scholar]
- Uesono Y., Toh-e A., Kikuchi Y. Ssd1p of Saccharomyces cerevisiae associates with RNA. J. Biol. Chem. 1997;272:16103–16109. doi: 10.1074/jbc.272.26.16103. [DOI] [PubMed] [Google Scholar]
- Vannier D., Damay P., Shore D. A role for Sds3p, a component of the Rpd3p/Sin3p deacetylase complex, in maintaining cellular integrity in Saccharomyces cerevisiae. Mol. Genet. Genom. 2001;265:560–568. doi: 10.1007/s004380100447. [DOI] [PubMed] [Google Scholar]
- Vellai T. Autophagy genes and ageing. Cell Death Differ. 2009;16:94–102. doi: 10.1038/cdd.2008.126. [DOI] [PubMed] [Google Scholar]
- Warner J. R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Wei M., Fabrizio P., Hu J., Ge H., Cheng C., Li L., Longo V. D. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 2008;4:e13. doi: 10.1371/journal.pgen.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger M., Feng L., Paul A., Smith D. L., Jr, Hontz R. D., Smith J. S., Vujcic M., Singh K. K., Huberman J. A., Burhans W. C. DNA replication stress is a determinant of chronological lifespan in budding yeast. PLoS ONE. 2007;2:e748. doi: 10.1371/journal.pone.0000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler R. T., Kupiec M., Magnelli P., Abeijon C., Fink G. R. A Saccharomyces cerevisiae mutant with increased virulence. Proc. Natl. Acad. Sci. USA. 2003;100:2766–2770. doi: 10.1073/pnas.0437995100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen H., Futcher B. Genetic analysis of the shared role of CLN3 and BCK2 at the G1-S transition in Saccharomyces cerevisiae. Genetics. 1999;153:1131–1143. doi: 10.1093/genetics/153.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y., Dancis A., Klausner R. D. AFT1, a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 1995;14:1231–1239. doi: 10.1002/j.1460-2075.1995.tb07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. B., Brenner A. A., White T. B., Engler M. J., Gaughran J. P., Tatchell K. The Saccharomyces cerevisiae SRK1 gene, a suppressor of bcy1 and ins1, may be involved in protein phosphatase function. Mol. Cell. Biol. 1991;11:3369–3373. doi: 10.1128/mcb.11.6.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanelli C. F., Valentini S. R. Pkc1 acts through Zds1 and Gic1 to suppress growth and cell polarity defects of a yeast eIF5A mutant. Genetics. 2005;171:1571–1581. doi: 10.1534/genetics.105.048082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.