Abstract
The Drosophila HIFα homologue, Sima, is localized mainly in the cytoplasm in normoxia and accumulates in the nucleus upon hypoxic exposure. We have characterized the mechanism governing Sima oxygen-dependent subcellular localization and found that Sima shuttles continuously between the nucleus and the cytoplasm. We have previously shown that nuclear import depends on an atypical bipartite nuclear localization signal mapping next to the C-terminus of the protein. We show here that nuclear export is mediated in part by a CRM1-dependent nuclear export signal localized in the oxygen-dependent degradation domain (ODDD). CRM1-dependent nuclear export requires both oxygen-dependent hydroxylation of a specific prolyl residue (Pro850) in the ODDD, and the activity of the von Hippel Lindau tumor suppressor factor. At high oxygen tension rapid nuclear export of Sima occurs, whereas in hypoxia, Sima nuclear export is largely inhibited. HIFα/Sima nucleo-cytoplasmic localization is the result of a dynamic equilibrium between nuclear import and nuclear export, and nuclear export is modulated by oxygen tension.
INTRODUCTION
Hypoxia-inducible factors (HIFs) are a family of mammalian transcription factors that regulate the expression of a wide array of hypoxia-inducible genes. They have been characterized as α/β heterodimers of basic-helix-loop-helix-PAS (bHLH-PAS) proteins in which the β-subunit is constitutive and the α-subunit is regulated by oxygen levels (Wang et al., 1995). Oxygen-dependent regulation of the α-subunit is exerted through different mechanisms, including control of protein half-life (Maxwell et al., 1999), recruitment of transcriptional coactivators (Sang et al., 2002), and regulation of subcellular localization (Kallio et al., 1998; Tanimoto et al., 2000). In normoxia, α-subunits undergo polyubiquitination and rapid degradation at the 26S proteasome, and in hypoxia proteolysis is largely prevented (Ivan et al., 2001; Jaakkola et al., 2001). The molecular basis of this regulation is now well understood; in normoxia two key prolyl residues that reside in the so-called oxygen-dependent degradation domain (ODDD) are hydroxylated in a reaction catalyzed by 2-oxoglutarate and iron (Fe2+)-dependent dioxygenases that utilize molecular oxygen as a cosubstrate (Ivan et al., 2001; Jaakkola et al., 2001; Masson et al., 2001; Yu et al., 2001). Prolyl hydroxylation enables interaction between the HIFα ODDD and the von Hippel Lindau (VHL) tumor suppressor protein, which is the substrate recognition subunit of the VCB (VHL; Cullin2; Elongin B-C) E3 ubiquitin ligase complex (Maxwell et al., 1999; Ohh et al., 2000). Thus, at low oxygen levels hydroxylation is prevented, ubiquitination does not occur, and hence HIFα is stabilized. Therefore, the enzymes that catalyze prolyl hydroxylation, termed PHD1, PHD2. and PHD3, are considered bona fide molecular oxygen sensors that ultimately control the half-life of HIFα subunits (Bruick and McKnight, 2001; Epstein et al., 2001).
Recruitment of HIF transcriptional coactivators is another oxygen-regulated step of HIF activity. It is governed by a second oxygen-sensing system that involves hydroxylation of a specific asparagine residue located in the transactivation domain, next to ΗΙFα C-terminal end (Lando et al., 2002b). This hydroxylation reaction is catalyzed by another member of the 2-oxoglutarate and iron-dependent family of dioxygenases, called factor-inhibiting HIF (FIH; Lando et al., 2002a). This regulatory mechanism is also well characterized: FIH-mediated hydroxylation of the asparagine residue prevents physical interaction with the transcriptional coactivator P-300, which was previously shown to be required for maximal HIF transcriptional activity. Thus, in hypoxia asparaginyl hydroxylation is prevented and interaction with P-300 can take place, enabling full transcriptional activation of the system.
Regulation of HIF oxygen-dependent subcellular localization is less well understood. Studies in cell culture have shown that HIFα is mostly cytoplasmic in normoxia and accumulates in the nucleus in hypoxia and that hypoxia-dependent nuclear import accounts for this differential localization (Kallio et al., 1998; Chilov et al., 1999). Although structure-function studies have led to the identification of a relevant nuclear localization signal (NLS) and other protein domains required for this regulation (Kallio et al., 1998; Luo and Shibuya, 2001), the molecular mechanism governing HIFα subcellular localization is so far unclear.
We and others have demonstrated that an oxygen-inducible transcriptional response occurs in Drosophila (Lavista-Llanos et al., 2002; Douglas and Haddad, 2003; Gorr et al., 2004; Centanin et al., 2005; Dekanty et al., 2005), and that Similar (Sima; Nambu et al., 1996) and Tango (Tgo; Sonnenfeld et al., 1997) bHLH-PAS proteins are the HIF-α and HIF-β homologues, respectively (Lavista-Llanos et al., 2002; Gorr et al., 2004; Romero et al., 2007). Although Tango is expressed constitutively regardless of oxygen levels, Sima protein is rapidly degraded in normoxia and stabilized in hypoxia; stability of the protein depends on a central protein domain encompassing amino acids 692-863, functionally homologous to HIFα ODDD (Lavista-Llanos et al., 2002). More recently, we have identified Drosophila mutants for a HIF prolyl hydroxylase homologue that we have named Fatiga, which exhibited strong constitutive upregulation of both Sima protein and HRE reporter gene expression (Centanin et al., 2005, 2008). Thus the whole basic machinery controlling the transcriptional response to hypoxia is apparently conserved between Drosophila and mammalian cells.
By overexpressing Sima in transgenic embryos under control of an engrailed-gal4 driver, we overrode the rapid rate of protein degradation and found that Sima was mostly cytoplasmic in normoxia and nuclear in hypoxia (Lavista-Llanos et al., 2002). This phenomenon is not an all-or-none response but rather, Sima localization depends on oxygen levels in a graded manner and interestingly, it is influenced by developmental parameters (Dekanty et al., 2005). More recently, we have found that Sima nuclear import depends on an atypical bipartite NLS, localized next to the C-terminal end of the protein and that nuclear export is mediated in part by two conserved nuclear export signals (NESs) mapping at the bHLH domain of the protein (Romero et al., 2008).
In this study we have further explored the mechanism of oxygen-dependent regulation of HIFα/Sima subcellular localization. We show that Sima localization emerges from a dynamic equilibrium between nuclear import and nuclear export and that oxygen levels can modify the ratio between these two processes. A cryptic CRM1-dependent NES occurs in the ODDD, contributing to the regulation of Sima nuclear export. Consistent with the occurrence of this NES in the ODDD, prolyl hydroxylation and VHL activity are absolutely required for CRM1-dependent nuclear export of Sima and moreover, graded activation of the prolyl hydroxylase/VHL pathway modulates the rate of Sima nuclear export. This regulatory mechanism has not been so far reported in mammalian HIFα proteins.
MATERIALS AND METHODS
DNA Constructs
Enhanced green fluorescent protein (EGFP)-NES and EGFP in pMT/Bip/V5-His for S2 cell transfections were previously described (Roth et al., 2003). Sima constructs for transgenic lines generation were all engineered in a pNB40 plasmid and then subcloned into the pCaSpeR-UAS vector as EcoRI-NotI fragments. EGFP-ODDD1-3 chimeras were generated by in-frame cloning of sequences amplified by PCR into XbaI-NotI sites of the pMT/Bip/V5-His-EGFP plasmid (Roth et al., 2003). The EGFP-ODDD fusion was generated by cloning in-frame the EGFP sequence in a SacII site upstream of the sequence encoding Sima 665-1507 (Bacon et al., 1998) in a pcDNA3.1 vector (Invitrogen, Carlsbad, CA). The plasmid encoding the EGFP-Sima 665-1507 fusion was used as a template to amplify the EGFP-ODDD sequence (Sima amino acids 665-871) using the primers: 5′-GCTTCGCCGCGGTCTAGATGGTGAGCAAGGGCGAGGAG-3′and 5′-GCTTCGTCTAGACCACATGAGATCCGTCTCCGTTAGC-3′ that included XbaI restriction sites on their 5′ ends and was subcloned in an XbaI site of a pMT/Bip/V5-His vector.
Mutated EGFP-ODDD was generated by mutagenic divergent PCR in the original pMT/Bip/V5-His-EGFP-ODDD plasmid, with the QuikChange Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA) by using the primers 5′-CTCGGGCAGCGCGCAGGTGCCCATGGCGCCAACCAACATGTC-3′ and 5′-GACATGTTGGTTGGCGCCATGGGCACCTGCGCGCTGCCCGAG-3′. Underlined codons represent the mutations introduced in the sequence.
S2 Cell Transfections and Analysis of Subcellular Localization of EGFP Fusion Constructs
All expression constructs were cloned in a pMT/Bip/V5-His plasmid (Invitrogen). S2 cells were maintained at 25°C in Schneider Drosophila medium (Sigma, St. Louis, MO), with 10% fetal bovine serum (Invitrogen), 50 U/ml penicillin, and 50 μg/ml streptomycin in 25- or 75-cm2 T-flasks (Greiner, Monroe, NC). Transfections (5 μg of total DNA) were performed with a calcium phosphate transfection kit (Invitrogen), and after 24 h expression was induced by addition of 0.4 mM CuSO4 for 5–16 h. Cells were then observed at the confocal microscope (Carl Zeiss, Thornwood, NY; LSM5, Pascal). For hypoxic treatments, cells were exposed to hypoxia (3% O2) during 20 h in a Forma Scientific 3131 incubator (Marietta, OH). Leptomycin B (Sigma-Aldrich) was applied for 2 h, and results were immediately analyzed in the confocal microscope; cell nuclei were visualized by Hoechst 33342 staining. Quantitative analysis of subcellular localization was performed in the confocal microscope by measuring intensity of EGFP fluorescence per μm2 in the nucleus (N) and cytoplasm (C) and assessing the N–C ratio.
Fly Stocks and Analysis of Sima Localization In Vivo
The transgenic lines UAS-embargoed, UAS-VHL, UAS-simaP850A, and other UAS-sima–mutagenized variants were generated by P-element germline transformation using standard procedures. The following fly strains were also used: UAS-sima and UAS-nGFP-LacZ (Lavista-Llanos et al., 2002), embargoed3 (Collier et al., 2000), fatiga1 (Centanin et al., 2005), engrailed-Gal4 and Df(2R)en-A deficiency (Bloomington Stock Center). The UAS-VHL double-stranded RNA line (no. 32163) was obtained from the Vienna Drosophila RNAi Centre (VDRC).
Analysis of Sima subcellular localization in vivo was performed in transgenic embryos overexpressing the Sima wild-type protein or altered variants of Sima under control of an en-Gal4 driver. To obtain synchronized embryos, egg-laying agar plates were replaced every 3 h, and embryos were grown at 25°C in normoxia until the desired stage and then transferred to hypoxia for 4 h. Hypoxic treatments were applied in a Forma Scientific 3131 incubator at 25°C; embryos were then recovered, fixed, and immunostained with anti-Sima and/or anti-βgal antibodies (Bacon et al., 1998; Lavista-Llanos et al., 2002). Observations were performed in an Olympus BX60 microscope (Melville, NY) or in a Carl Zeiss LSM5 Pascal confocal microscope. To analyze the effect of embargoed, fatiga, or VHL loss-of-function, recombinant chromosomes carrying UAS-sima or en-gal4 elements and the mutation of interest were generated by meiotic recombination. Similarly, for emb or VHL gain-of-function experiments UAS-sima/UAS-VHL or UAS-emb recombinant chromosomes were generated and used for crosses with the en-gal4 driver. Quantitative analyses of subcellular localization were carried out as previously described (Dekanty et al., 2005). Briefly, each embryo was classified into one of three categories of Sima subcellular localization: cytoplasmic, where more than 90% of the cells in the engrailed-expressing stripes show cytoplasmic localization; nuclear, where more than 90% of the cells in the engrailed-expressing stripes have nuclear localization; and ubiquitous, where <90% of the cells have cytoplasmatic or nuclear localization. The percentage of embryos assigned to each subcellular localization category was calculated, and category distributions in the different genotypes were compared with a χ2 test.
RESULTS
Sima Is Continuously Imported to the Nucleus and Reexported to the Cytoplasm
We have recently shown in cell culture that Sima-EGFP chimeric proteins become more nuclear upon addition of leptomycin B (LMB), a drug that inhibits the nuclear export receptor CRM1, which is responsible for nuclear export of most proteins in eukaryotic cells (Romero et al., 2008). We therefore, sought to investigate whether Sima steady-state subcellular localization depends on CRM1 in living embryos, by analyzing en-Gal4/UAS-sima individuals carrying a homozygous loss-of-function mutation in the Drosophila CRM1 homologous gene embargoed (Collier et al., 2000; Roth et al., 2003). Sima subcellular localization in these mutants was compared with that in wild-type control embryos. As we reported previously (Dekanty et al., 2005), in wild-type embryos Sima was predominantly cytoplasmic in normoxia and gradually more nuclear as oxygen concentration decreased (Figure 1, A and B). In embargoed mutant embryos (emb3/emb3), Sima localization was clearly more nuclear than in wild-type controls (Figure 1B), supporting the notion that Sima steady-state subcellular localization depends at least in part on active nuclear export mediated by the nuclear export receptor CRM1. To get direct evidence that a nuclear export defect in emb mutants accounts for the shift in Sima subcellular localization, we measured nuclear export directly in a reoxygenation assay as described before (Romero et al., 2008). Embryos expressing transgenic Sima protein were exposed to 1% O2 for 4 h (Sima is fully nuclear in this condition) and then transferred to normoxia and fixed at different times after reoxygenation for Sima subcellular localization analysis. As depicted in Figure 1C, Sima nuclear export is affected in the emb mutant embryos, supporting the notion that increased Sima steady-state nuclear localization in these mutants is due to slower nuclear export that partially depends on Embargoed.
Figure 1.
Sima nuclear export depends on the nuclear export receptor CRM1/Embargoed. (A) Categories of Sima subcellular localization in en-Gal4/UAS-Sima/UAS-nGFP.LacZ transgenic embryos: nuclear, ubiquitous, and cytoplasmic sample photographs of groups of cells overexpressing Sima in embryonic ectodermal cells (for quantitative criteria see Materials and Methods; see also Dekanty et al., 2005). (B) Sima becomes more nuclear in embargoed/CRM1 mutant embryos. □, cytoplasmic localization of Sima; ■, nuclear localization; and ▩, ubiquitous localization. Sima subcellular localization in wild-type embryos (WT) was progressively more nuclear as embryogenesis proceeds and gradually more nuclear as oxygen levels decrease (χ2 test; p < 10−4; n > 50). Nuclear localization increased in embargoed (emb3) homozygous mutant embryos (p < 10−3; n > 50) at 21, 5, and 3% O2 concentrations. At 1% O2 Sima was already totally nuclear in wild-type embryos throughout development. (C) Nuclear export assay of Sima protein upon reoxygenation. Stage 16 embryos expressing Sima protein were exposed to 1% O2 during 4 h and then transferred to normoxia and stained for Sima at different time points after reoxygenation. In WT embryos Sima is totally exported from the nucleus in only 10 min, whereas in emb3 mutants, Sima is exported at a slower rate (Kaplan-Meier; p < 10−4; n > 30). (D) Cytoplasmic localization of Sima was enhanced in embryos overexpressing the Emb protein (p < 10−4; n > 40) at all tested oxygen concentrations throughout embryogenesis, suggesting that increased levels of CRM1 promote nuclear export of Sima protein. No differences occurred at embryonic stages 11–12 in normoxia or 5% O2, as in wild-type embryos (y w), Sima was already fully cytoplasmic in these conditions. (E) Sima nuclear export is faster in embryos overexpressing CRM1/Embargored. Nuclear export assays were carried out as in C, revealing that upon expression of the Embargoed protein in transgenic embryos, Sima is exported more efficiently (Kaplan-Meier; p < 10−4; n > 30). (F) Sima transcriptional activity depends on the rate of nuclear export. A Sima-responsive LDH-LacZ Sima-responsive transcriptional reporter (Lavista-Llanos et al., 2002) was crossed into the UAS lines expressing Sima in embryos that were otherwise wild-type (WT), mutant for CRM1/embargoed (emb3), or overexpressing the Emb protein (UAS.emb), and β-galactosidase activity was determined through a colorimetric assay (Materials and Methods). Activity of the Sima-dependent LacZ reporter was significantly increased in emb3 mutant embryos and reduced in embryos overexpressing the Emb protein (Student's t test; * p < 10−3; n = 3).
To gather additional evidence that CRM1/Emb can control Sima subcellular localization, we overexpressed the Emb protein in transgenic embryos at different oxygen levels throughout embryogenesis. Overexpression of Emb caused an increase of Sima steady-state cytoplasmic localization and this phenomenon could be verified at all tested oxygen concentrations (Figure 1D). Consistent with this, the rate of Sima nuclear export was significantly higher in the embryos overexpressing Emb, as verified in the reoxygenation assay (Figure 1E). Altogether, these experiments show that by varying the levels of CRM1, it is possible to modulate the rate of Sima nuclear export, thereby altering its steady-state subcellular localization at different oxygen concentrations.
Next, we sought to investigate whether the Emb-dependent shift in Sima subcellular localization can modify Sima transcriptional activity, and for this purpose, we analyzed induction of a Sima-inducible LacZ reporter (Lavista-Llanos et al., 2002) in emb mutants and in embryos overexpressing Emb, in comparison with wild-type controls. As shown in Figure 1F, β-galactosidase activity derived from a Sima-inducible LacZ reporter was significantly increased in emb mutant embryos, and reduced in embryos overexpressing emb. These results provide direct functional evidence that alteration of Sima subcellular localization impinges on its transcriptional activity.
Occurrence of a Functional NES in the Sima Oxygen-dependent Degradation Domain
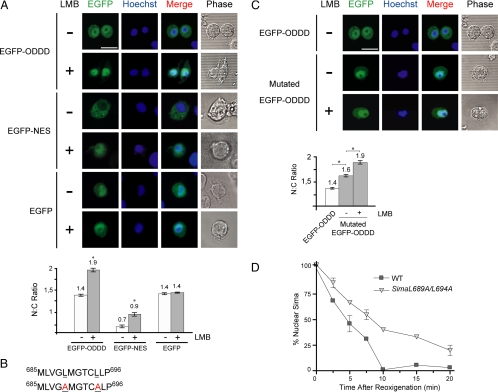
Because deletion of the entire ODDD provokes constitutive nuclear localization of Sima in normoxia, resulting in increased transcriptional activity of the protein (Lavista-Llanos et al., 2002), we explored whether CRM1-dependent NESs occur within this domain. Close inspection of the entire amino acid sequence of the ODDD did not reveal the presence of a canonical NES consensus (L-x(2,3)-[LIVFM]-x(2,3)-L-x[LI] (Bogerd et al., 1996) within this domain. However, because CRM1-dependent NESs often diverge from this canonical consensus, we tested the ability of the ODDD (amino acids 665-871) to promote nuclear export of an EGFP reporter in Drosophila S2 cultured cells. Analysis of subcellular localization of EGFP-ODDD revealed an ubiquitous (slightly nuclear) localization of the chimera, with an N–C fluorescence ratio of 1.4 (Figure 2A). To answer if the steady-state subcellular localization of the fusion protein is influenced by CRM1-dependent nuclear export, we investigated whether subcellular localization is sensitive to LMB. LMB provoked a significant increase of nuclear localization, shifting the N–C fluorescence ratio to 1.9 (Figure 2A). As expected, subcellular localization of an EGFP-NES positive control construct, bearing a NES derived from PKI (Roth et al., 2003), also became more nuclear upon addition of LMB (Figure 2A), and an EGFP construct lacking an export signal (negative control), was insensitive to LMB (Figure 2A). The above results strongly suggest that the Sima ODDD includes a functional CRM1-dependent NES.
Figure 2.
Identification of a functional nuclear export signal (NES) in the Sima oxygen-dependent degradation domain (ODDD). (A) Sensitivity of an EGFP-ODDD construct to leptomycin B (LMB). An EGFP-ODDD (amino acids 665–871) fusion protein, an EGFP-NES–positive control or EGFP alone were transfected to S2 cells, and subcellular localization of the constructs was analyzed by confocal microscopy 2 h after addition or not of 30 mM LMB; the nuclear:cytoplasmic (N–C) fluorescence ratio was calculated and is shown in the bottom panel. Although EGFP-ODDD and the EGFP-NES–positive control became clearly more nuclear upon addition of LMB (Student's t test; significant difference: * p < 10−7; n > 20), EGFP was insensitive to LMB, suggesting that a functional CRM1-dependent NES occurs in Sima ODDD. (B) Amino acid sequence of the NES of the Sima ODDD. Top line, wild-type sequence; bottom line mutagenized sequence in which two leucine residues were replaced by alanines (underlined). (C) The NES of the ODDD promotes nuclear export of an EGFP reporter. EGFP fusions were generated with the wild-type or the mutagenized version of the NES. The chimera including the mutagenized NES was remarkably more nuclear than the one with the wild-type NES; treatment with LMB rendered the construct even more nuclear (Student's t test; significant difference: * p < 10−7; n > 20), suggesting that the mutations provoked reduction but not complete elimination of NES function. The N–C fluorescence ratios of these experiments are shown in the bottom panel. (D) A full-length Sima protein, mutagenized at the NES of the ODDD as in B, is exported at a slower rate than wild-type Sima in transgenic embryos, in a reoxygenation assay similar to the one described in Figure 1.
Next, we sought to map the region in the Sima ODDD that includes the functional NES. To this end, we split this domain into three fragments: ODDD1 (amino acids 669-730), ODDD2 (aa 725-803), and ODDD3 (aa 797-870), and generated EGFP fusion proteins as above. Analysis of subcellular localization of the three chimeras in S2 cells revealed that EGFP-ODDD1 but not the other two fusion proteins became more nuclear upon LMB treatment (Supplemental Figure S1), suggesting that the NES maps in the ODDD1 fragment. Careful analysis of ODDD1 sequence revealed the occurrence in this region of a stretch of 10 predominantly hydrophobic amino acids including four leucines (Figure 2B). Because this sequence has features in common with the canonical NES consensus (la Cour et al., 2003), we decided to analyze this stretch of hydrophobic amino acids as a candidate NES. We generated a mutated version of the complete EGFP-ODDD chimera by replacing Leu689 and Leu694 by alanines (Figure 2B). Subcellular localization of the mutated EGFP-ODDD in transfected S2 cells was clearly more nuclear than the wild-type version (Figure 2C), suggesting that the two leucine residues are part of a functional NES. When the cells transfected with mutated EGFP-ODDD were exposed to LMB, the construct became even more nuclear (Figure 2C), suggesting that the functionality of the ODDD NES was reduced but not completely abrogated by the mutations.
To investigate whether this novel NES plays a role in the regulation of subcellular localization of the Sima full-length protein, we replaced Leu689 and Leu694 by alanines (SimaL689A/L694A), generated a transgenic (fly) line expressing this Sima variant, and analyzed the rate of nuclear export upon reoxygenation of the transgene embryos as above. Nuclear export of SimaL689A/L694A was clearly impaired in comparison with wild-type Sima (Figure 2D), confirming that the sequence under study is a functional NES that contributes to the overall subcellular localization of Sima.
Prolyl Hydroxylation and VHL Mediate Sima Nuclear Export
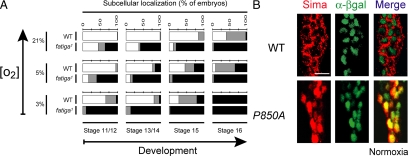
The occurrence of a CRM1-dependent NES in the ODDD prompted us to investigate whether oxygen-dependent hydroxylation of a prolyl residue within this domain is required for Sima nuclear export. We began by performing a quantitative analysis of Sima subcellular localization in embryos carrying a loss-of-function mutation in the fatiga (fga) gene, which encodes the Drosophila HIF prolyl-4-hydroxylase (Centanin et al., 2005). As shown in Figure 3A, embryos homozygous for fga1, the strongest of the fga alleles (Centanin et al., 2005), exhibited a major increase in Sima nuclear localization throughout embryogenesis at different oxygen tensions, indicating that reduced prolyl hydroxylase activity can shift the Sima import/export balance toward nuclear localization.
Figure 3.
Prolyl hydroxylation is essential for Sima nuclear export. (A) In fatiga1 homozygous mutant embryos, Sima was more nuclear than in the wild-type controls (WT; p < 10−4; n > 50) at all tested oxygen levels throughout embryogenesis, indicating that prolyl hydroxylation is necessary for Sima cytoplasmic localization. The experimental design is the same as in Figure 1, B and D. ■, nuclear; ▩, ubiquitous; and □, cytoplasmic. (B) Wild-type Sima protein (red) was predominantly cytoplasmic in normoxia (cell nuclei are marked by anti-βgal staining; green); when Sima proline 850 was replaced by an alanine (P850A), Sima became constitutively nuclear with no signal detected in the cytoplasm. Scale bar, 10 μm.
To analyze further this possibility, we sought to mutagenize proline850, the prolyl residue of Sima ODDD that was reported to undergo oxygen-dependent hydroxylation (Arquier et al., 2006; Centanin et al., 2008), and analyzed subcellular localization in UAS transgenic embryos. After replacing this proline by an alanine (SimaP850A) Sima became totally nuclear throughout embryogenesis, regardless of oxygen tension (Figure 3B). Taken together, these results indicate that prolyl hydroxylation is likely required for Sima nuclear export in normoxia.
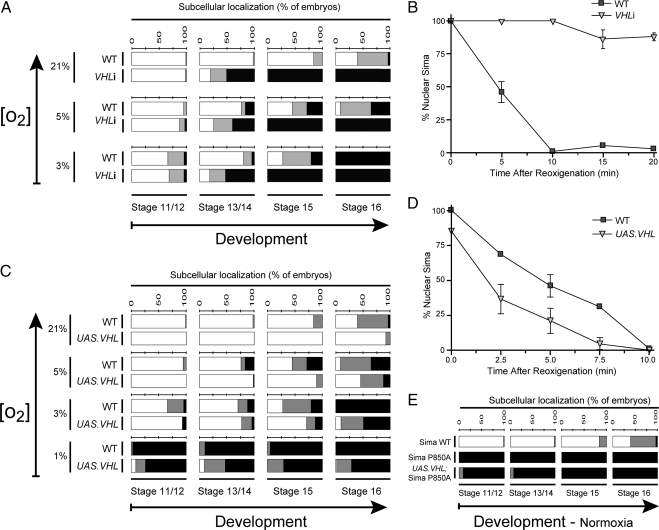
In mammalian VHL-deficient cell lines high levels of stabilized HIFα protein occur, and strong oxygen-independent expression of a battery of HIF target genes was demonstrated (Maxwell et al., 1999), suggesting that VHL loss-of-function leads to HIFα nuclear accumulation (Bonicalzi et al., 2001). To explore the role of Drosophila VHL (Adryan et al., 2000) in Sima subcellular localization, we began by analyzing VHL loss-of-function in embryos expressing VHL double-stranded RNA (VHL RNAi/VHLi) (Dietzl et al., 2007) in UAS transgenic lines. Heat-shock driven expression of VHLi provoked 93% reduction of VHL mRNA expression (data not shown). When VHLi was expressed under an en-Gal4 driver, Sima was remarkably more nuclear than in wild-type controls (Figure 4A), suggesting that VHL is required for cytoplasmic localization in normoxia. Consistent with this, embryos homozygous for the Df(2R)en-A chromosomal deficiency, which includes the VHL gene, exhibited Sima more nuclear than in wild-type siblings (Supplemental Figure S2A). To answer if this effect was indeed due to reduced VHL levels, and not to other genes mapping in the deficiency, we performed a specific rescue by overexpressing the VHL transgenic protein in Df(2R)en-A homozygous embryos. Normal Sima subcellular distribution was largely restored upon reexpressing VHL (Supplemental Figure S2B), confirming that VHL loss-of-function accounts for Sima nuclear localization in Df(2R)en-A homozygous embryos. To get direct evidence for a role of VHL in Sima nuclear export, we subjected the embryos expressing VHLi to a reoxygenation assay as above. We found that Sima nuclear export was dramatically impaired (Figure 4B), supporting the notion that VHL is required in this process. Similar results were obtained upon reoxygenation of embryos homozygous for the Df(2R)en-A deficiency (Supplemental Figure S2C), further supporting a role for VHL in nuclear export.
Figure 4.
VHL promotes cytoplasmic localization of Sima. (A) In embryos expressing a VHL RNAi transgene (VHLi), Sima was more nuclear than in wild-type (WT) controls at all tested oxygen levels after stage 13 (p < 10−4; n > 40). ■, nuclear; ▩, ubiquitous; and □, cytoplasmic. (B) Sima nuclear export assay upon reoxygenation, in embryos expressing VHLi, reveals that Sima export was severely impaired (Kaplan-Meier; p < 10−4; n > 30). (C) Overexpression of Drosophila VHL increased Sima cytoplasmic localization at all tested oxygen levels throughout embryogenesis with the exception of those conditions where in the wild-type controls, Sima was already fully cytoplasmic (p < 10−2; n > 40). (D) Nuclear export of Sima is more efficient in embryos overexpressing VHL. Sima nuclear export assay similar to the one depicted in B (Kaplan-Meier; p < 10−4; n > 30). (E) A mutagenized version of Sima in which proline 850 was replaced by an alanine (SimaP850A) was insensitive to VHL overexpression, remaining totally nuclear even in normoxia (n > 40).
To confirm that VHL levels can modulate the extent of Sima nuclear export, we sought to test if augmented levels of VHL can affect subcellular localization of Sima. To this end, we overexpressed the VHL protein in transgenic embryos and observed that Sima became clearly more cytoplasmic all throughout embryogenesis at different oxygen tensions (Figure 4C). A reoxygenation assay performed in these embryos overexpressing VHL revealed that an augmented rate of nuclear export accounts for cytoplasmic accumulation of Sima when VHL protein levels are higher than normal (Figure 4D). Altogether, the above results support the notion that VHL levels critically determine the rate of Sima nuclear export. The next step was to answer whether VHL requires prolyl hydroxylation to mediate Sima nuclear export, so we studied subcellular localization of SimaP850A in normoxic embryos overexpressing VHL. VHL was unable to alter SimaP850A subcellular localization, which remained totally nuclear. These results indicate that prolyl hydroxylation is absolutely necessary for VHL-dependent nuclear export of Sima (Figure 4E).
CRM1-dependent Nuclear Export of Sima Requires P850 Hydroxylation and VHL Function
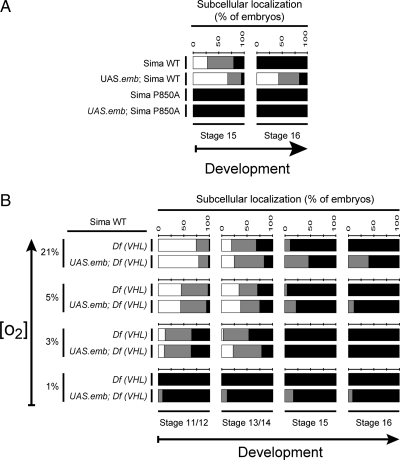
As shown above, overexpression of CRM1/Embargoed can increase the rate of Sima nuclear export (Figure 1D,E). On the basis of this observation, we explored if hydroxylation of Sima is required for this effect, by studying if subcellular localization of SimaP850A is modified upon overexpression of the Emb protein. As depicted in Figure 5A, SimaP850A subcellular localization was not modified upon overexpression of Emb: The protein remained totally nuclear, indicating that Proline 850 is critically required for CRM1-dependent nuclear export of Sima.
Figure 5.
Prolyl hydroxylation and VHL function are required for CRM1-dependent nuclear export of Sima. (A) Overexpression of the nuclear export receptor CRM1/Embargoed enhanced the rate of Sima nuclear export, leading to increased levels of wild-type Sima protein in the cytoplasm in normoxia (p < 10−4; n > 40) but failed to enhance nuclear export of SimaP850A, which remained localized exclusively in the nuclear compartment even in normoxia (n > 40). (B) Sima subcellular localization in VHL loss-of-function embryos [Df (VHL) homozygous for the Df(2R)en-A deficiency] remained largely unchanged upon overexpression of CRM1/Embargoed, revealing that VHL function is required for Sima nuclear export (n > 50). ■, nuclear; ▩, ubiquitous; and □, cytoplasmic.
By following the same rationale, we sought to test whether VHL is also required for CRM1/Emb-dependent nuclear export of Sima, so we analyzed if overexpression of the Emb protein is able to increase Sima cytoplasmic localization in VHL loss-of-function embryos [homozygous for the Df(2R)en-A deficiency]. No significant increase of Sima cytoplasmic localization was observed in these embryos (Figure 5B), indicating that in the absence of VHL, CRM1 is unable to promote nuclear export of the protein. Taken together, the above experiments suggest that both the extent of prolyl hydroxylation and VHL protein levels can determine the rate of CRM1-dependent nuclear export of Sima.
DISCUSSION
Sima subcellular Localization Is the Result of a Dynamic Equilibrium Between Nuclear Import and Nuclear Export and Depends on the Prolyl Hydroxylase/VHL System
Our work has revealed that in opposition to a static model in which HIFα/Sima is cytoplasmic in normoxia and imported into the nucleus upon hypoxic exposure, Sima localization depends on a dynamic equilibrium between nuclear import and nuclear export in which both processes occur simultaneously (Figure 6). This model is supported by the fact that genetic or pharmacological impairment of either, nuclear import or nuclear export (Romero et al., 2008 and this study) led to an alteration of Sima subcellular localization at different oxygen concentrations. Furthermore, our data suggest that oxygen levels, acting in a dose-dependent manner, can modify the ratio between nuclear import and nuclear export, thereby setting the steady-state subcellular localization of Sima. In this context, the predominant cytoplasmic localization of Sima observed in normoxia is likely due to rapid nuclear export of the protein that overcomes the rate of nuclear import.
Figure 6.
Model for oxygen-dependent regulation of Sima subcellular localization and degradation. Sima is constitutively imported into the nucleus. In hypoxia Sima forms a heterodimer with the β-subunit Tango, and induces transcription; in normoxia the Prolyl 850 residue is hydroxylated by the PHD Drosophila homologue Fatiga, ubiquitinated by VHL and exported to the cytoplasm in a CRM1-dependent manner. Prolyl-hydroxylation, ubiquitination, and degradation at the 26S proteasome can presumably occur both in the nucleus and the cytoplasm (Berra et al., 2001).
This work and previous studies have shown that in both mammalian HIFα, and its Drosophila homologue Sima, nuclear export depends at least in part, on the nuclear export receptor CRM1 (Mylonis et al., 2006; Romero et al., 2008). We have identified a functional NES within the Sima ODDD that contributes to regulated nuclear export of the protein, and hydroxylation of Pro850, the prolyl residue involved in Sima normoxic proteasomal degradation (Arquier et al., 2006; Centanin et al., 2008), is absolutely required for cytoplasmic localization of Sima. Consistent with this, Fatiga, the enzyme responsible for hydroxylation of this prolyl residue (Centanin et al., 2005) is necessary for cytoplasmic localization as well. Thus, our results support the notion that subcellular localization of Sima depends on graded levels of prolyl hydroxylation that can fine tune the ratio between nuclear import and nuclear export in an oxygen-dependent manner, so that high hydroxylation levels favor a cytoplasmic localization, and low hydroxylation levels shift the equilibrium toward Sima nuclear localization (Figure 6). It remains to be determined the molecular mechanism that accounts for oxygen-mediated regulation of nuclear export. It is tempting to speculate that the NES within the Sima ODDD becomes accessible to the export machinery only upon hydroxylation of Pro850.
VHL was shown to be another critical determinant of Sima subcellular localization. Paralleling the requirement of prolyl-hydroxylation for Sima localization in the cytoplasm, high levels of VHL push the equilibrium toward cytoplasmic localization of the protein; conversely, reduced levels of VHL shift the balance in favor of nuclear localization. Thus, our data have shown that, in order for VHL to control Sima subcellular localization, hydroxylation of the Pro850 residue is required. We therefore conclude that the cellular machinery previously reported to be involved in ubiquitination (Maxwell et al., 1999; Ivan et al., 2001; Jaakkola et al., 2001) of HIF-α subunits is also required for cytoplasmic localization of the protein in normoxia.
Sima Nuclear Export Is an Oxygen-regulated Step
Having established that VHL, acting on hydroxylated Pro850 can affect Sima subcellular localization, leading to modification of the import/export ratio of the protein, the next step was to define which of the two processes, nuclear import or nuclear export, is under regulation of the prolyl hydroxylation/VHL pathway. By overexpressing in vivo the Drosophila CRM1 nuclear export receptor homologue, Embargoed, we have been able to increase the rate of Sima nuclear export, thereby enhancing Sima cytoplasmic localization. This strategy led us to conclude that the VHL/hydroxylation pathway is absolutely necessary for Sima Embargoed-dependent nuclear export (Figure 6), because either VHL loss-of-function or mutagenesis of Proline 850 provoked increased nuclear localization of Sima, and, in either case, this localization could not be altered by overexpression of CRM1/Embargoed.
Thus, our data suggest that Sima nuclear export is oxygen-regulated, but do not rule out the possibility that additional mechanisms controlling Sima/HIFα localization, as for example, oxygen-dependent cytoplasmic retention, might occur in parallel. Nuclear import of another transcription factor of the bHLH-PAS family, the aryl hydrocarbon receptor (Ahr), was shown to be elicited by xenobiotic compounds that can bind to its PAS domain, provoking a conformational change that lifts cytoplasmic retention mediated by the heat-shock protein 90 (Hsp90) chaperone complex (Kazlauskas et al., 2001). Interestingly, Hsp90 was shown to bind to HIFα PAS domain (Katschinski et al., 2002), and thus it is possible that the Hsp-90 complex can mediate HIFα cytoplasmic retention and then lifts such retention upon a hypoxia-elicited conformational change. Thus, the possibility that HIFα/Sima is regulated through oxygen-dependent cytoplasmic retention still needs to be investigated.
Compartmentalization of Prolyl Hydroxylation and the Dual Function of VHL
Given that VHL acting on hydroxyprolyl residues is required for Sima nuclear export, hydroxyprolyl Sima molecules are predicted to be present in the nucleus. Several lines of evidence suggest that HIFα/Sima prolyl hydroxylation occurs at least in part in the nuclear compartment. It was reported that the Drosophila HIF-prolyl-hydroxylase/Fatiga is localized in the nucleus in imaginal discs (Frei and Edgar, 2004), and consistent with this, in mammalian cells in culture, prolyl hydroxylase activity occurs in the nuclear compartment (Groulx and Lee, 2002). Subcellular localization of the three mammalian HIF-prolyl hydroxylase isoforms was directly assessed. Although this issue is still matter of some controversy, it is clear that at least one of the PHD isoforms is localized in the nucleus (Metzen et al., 2003).
Nuclear export of p53 is mediated by CRM1 and was shown to be regulated by Mdm2-dependent ubiquitination (Boyd et al., 2000; Geyer et al., 2000). It was demonstrated that Mdm2 can catalyze either p53 mono or polyubiquitination, apparently depending on Mdm2 intracellular levels. Interestingly, monoubiquitination promotes p53 nuclear export and polyubiquitination leads to proteasomal degradation (Li et al., 2003). Evidence from mammalian HIF suggests that this might indeed be the case. Groulx and Lee (2002), working in adenovirus-transfected mammalian cells, showed that HIFα ubiquitination can occur in the nuclear compartment and proposed that HIFα-ubiquitinated species are exported to the cytoplasm previous to degradation at the 26S proteasome. Whether mono- or polyubiquitination of Sima are required for nuclear export remains to be investigated (Figure 6).
The Interplay between HIF-α/Sima protein Degradation and Subcellular Localization
Why is it necessary that Sima undergoes rapid nuclear export in normoxia, whereas in parallel, the protein is actively degraded at the 26S proteasome? Regulation of transcription factor activity typically occurs at several different levels through more than one mechanism that operate simultaneously to ensure that regulation is efficiently achieved. Sima regulation does not seem to be an exception to this rule. Degradation and nuclear export of Sima appear to take place simultaneously in an oxygen dose-dependent manner, thereby synergizing in the reduction of Sima nuclear concentration and preventing hypoxia-inducible transcription. According to this model (Figure 6), both degradation and nuclear export are inhibited in hypoxic conditions, provoking an increase of Sima protein levels in the nuclear compartment, where transcription takes place. We therefore propose that Sima regulation involves a dual mechanism (i.e., proteasomal degradation and nuclear export) that relies on a single cellular machinery (i.e., the VHL/prolyl-hydroxylation system) and that this dual mechanism contributes to secure adaptive changes in gene expression to cope with variations of environmental oxygen tension.
Supplementary Material
ACKNOWLEDGMENTS
We thank Christos Samakovlis for discussions and technical help; Jimena Ortega and Herbert Spring for assistance with confocal images, Michael Ashburner (University of Cambridge) for the emb3 line, and the Bloomington and Vienna stock centers for fly strains. We also thank Peter Ratcliffe and Chris Pugh (University of Oxford) for plasmid constructs, Mario Galigniana for critical reading the manuscript, and the whole Wappner's lab for discussions. P.W. is a Howard Hughes Medical Institute International Scholar.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-01-0038) on July 8, 2009.
REFERENCES
- Adryan B., Decker H. J., Papas T. S., Hsu T. Tracheal development and the von Hippel-Lindau tumor suppressor homolog in Drosophila. Oncogene. 2000;19:2803–2811. doi: 10.1038/sj.onc.1203611. [DOI] [PubMed] [Google Scholar]
- Arquier N., Vigne P., Duplan E., Hsu T., Therond P. P., Frelin C., D'Angelo G. Analysis of the hypoxia-sensing pathway in Drosophila melanogaster. Biochem. J. 2006;393:471–480. doi: 10.1042/BJ20050675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon N. C., Wappner P., O'Rourke J. F., Bartlett S. M., Shilo B., Pugh C. W., Ratcliffe P. J. Regulation of the Drosophila bHLH-PAS protein Sima by hypoxia: functional evidence for homology with mammalian HIF-1 alpha. Biochem. Biophys. Res. Commun. 1998;249:811–816. doi: 10.1006/bbrc.1998.9234. [DOI] [PubMed] [Google Scholar]
- Berra E., Roux D., Richard D. E., Pouyssegur J. Hypoxia-inducible factor-1 alpha (HIF-1 alpha) escapes O(2)-driven proteasomal degradation irrespective of its subcellular localization: nucleus or cytoplasm. EMBO Rep. 2001;2:615–620. doi: 10.1093/embo-reports/kve130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd H. P., Fridell R. A., Benson R. E., Hua J., Cullen B. R. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonicalzi M. E., Groulx I., de Paulsen N., Lee S. Role of exon 2-encoded beta -domain of the von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 2001;276:1407–1416. doi: 10.1074/jbc.M008295200. [DOI] [PubMed] [Google Scholar]
- Boyd S. D., Tsai K. Y., Jacks T. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat. Cell Biol. 2000;2:563–568. doi: 10.1038/35023500. [DOI] [PubMed] [Google Scholar]
- Bruick R. K., McKnight S. L. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- Centanin L., Dekanty A., Romero N., Irisarri M., Gorr T. A., Wappner P. Cell autonomy of HIF effects in Drosophila: tracheal cells sense hypoxia and induce terminal branch sprouting. Dev. Cell. 2008;14:547–558. doi: 10.1016/j.devcel.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Centanin L., Ratcliffe P. J., Wappner P. Reversion of lethality and growth defects in Fatiga oxygen-sensor mutant flies by loss of hypoxia-inducible factor-alpha/Sima. EMBO Rep. 2005;6:1070–1075. doi: 10.1038/sj.embor.7400528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier S., Chan H. Y., Toda T., McKimmie C., Johnson G., Adler P. N., O'Kane C., Ashburner M. The Drosophila embargoed gene is required for larval progression and encodes the functional homolog of Schizosaccharomyces Crm1. Genetics. 2000;155:1799–1807. doi: 10.1093/genetics/155.4.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilov D., Camenisch G., Kvietikova I., Ziegler U., Gassmann M., Wenger R. H. Induction and nuclear translocation of hypoxia-inducible factor-1 (HIF-1): heterodimerization with ARNT is not necessary for nuclear accumulation of HIF-1alpha. J. Cell Sci. 1999;112:1203–1212. doi: 10.1242/jcs.112.8.1203. [DOI] [PubMed] [Google Scholar]
- Dekanty A., Lavista-Llanos S., Irisarri M., Oldham S., Wappner P. The insulin-PI3K/TOR pathway induces a HIF-dependent transcriptional response in Drosophila by promoting nuclear localization of HIF-{alpha}/Sima. J. Cell Sci. 2005;118:5431–5441. doi: 10.1242/jcs.02648. [DOI] [PubMed] [Google Scholar]
- Dietzl G., et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Douglas R. M., Haddad G. G. Genetic models in applied physiology: effect of oxygen deprivation on cell cycle activity: a profile of delay and arrest. J. Appl. Physiol. 2003;94:2068–2083. doi: 10.1152/japplphysiol.01029.2002. [DOI] [PubMed] [Google Scholar]
- Epstein A. C., et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- Frei C., Edgar B. A. Drosophila cyclin D/Cdk4 requires Hif-1 prolyl hydroxylase to drive cell growth. Dev. Cell. 2004;6:241–251. doi: 10.1016/s1534-5807(03)00409-x. [DOI] [PubMed] [Google Scholar]
- Geyer R. K., Yu Z. K., Maki C. G. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat. Cell Biol. 2000;2:569–573. doi: 10.1038/35023507. [DOI] [PubMed] [Google Scholar]
- Gorr T. A., Tomita T., Wappner P., Bunn H. F. Regulation of Drosophila hypoxia-inducible factor (HIF) activity in SL2 cells: identification of a hypoxia-induced variant isoform of the HIFalpha homolog gene similar. J. Biol. Chem. 2004;279:36048–36058. doi: 10.1074/jbc.M405077200. [DOI] [PubMed] [Google Scholar]
- Groulx I., Lee S. Oxygen-dependent ubiquitination and degradation of hypoxia-inducible factor requires nuclear-cytoplasmic trafficking of the von hippel-lindau tumor suppressor protein. Mol. Cell. Biol. 2002;22:5319–5336. doi: 10.1128/MCB.22.15.5319-5336.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M. K. K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. S., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for oxygen sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Jaakkola P., et al. Targeting of HIF-alpha to the Von Hippel Lindau ubiquitylation complex by O2-regulatred prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Kallio P. J., Okamoto K., O'Brien S., Carrero P., Makino Y., Tanaka H., Poellinger L. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1alpha. EMBO J. 1998;17:6573–6586. doi: 10.1093/emboj/17.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katschinski D. M., Le L., Heinrich D., Wagner K. F., Hofer T., Schindler S. G., Wenger R. H. Heat induction of the unphosphorylated form of hypoxia-inducible factor-1alpha is dependent on heat shock protein-90 activity. J. Biol. Chem. 2002;277:9262–9267. doi: 10.1074/jbc.M110377200. [DOI] [PubMed] [Google Scholar]
- Kazlauskas A., Sundstrom S., Poellinger L., Pongratz I. The hsp90 chaperone complex regulates intracellular localization of the dioxin receptor. Mol. Cell. Biol. 2001;21:2594–2607. doi: 10.1128/MCB.21.7.2594-2607.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Cour T., Gupta R., Rapacki K., Skriver K., Poulsen F. M., Brunak S. NESbase version 1.0, a database of nuclear export signals. Nucleic Acids Res. 2003;31:393–396. doi: 10.1093/nar/gkg101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando D., Peet D. J., Gorman J. J., Whelan D. A., Whitelaw M. L., Bruick R. K. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002a;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando D., Peet D. J., Whelan D. A., Gorman J. J., Whitelaw M. L. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002b;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- Lavista-Llanos S., Centanin L., Irisarri M., Russo D. M., Gleadle J. M., Bocca S. N., Muzzopappa M., Ratcliffe P. J., Wappner P. Control of the hypoxic response in Drosophila melanogaster by the basic helix-loop-helix PAS protein similar. Mol. Cell. Biol. 2002;22:6842–6853. doi: 10.1128/MCB.22.19.6842-6853.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Brooks C. L., Wu-Baer F., Chen D., Baer R., Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- Luo J. C., Shibuya M. A variant of nuclear localization signal of bipartite-type is required for the nuclear translocation of hypoxia inducible factors (1alpha, 2alpha and 3alpha) Oncogene. 2001;20:1435–1444. doi: 10.1038/sj.onc.1204228. [DOI] [PubMed] [Google Scholar]
- Masson N., Willam C., Maxwell P. H., Pugh C. W., Ratcliffe P. J. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 2001;20:5197–5206. doi: 10.1093/emboj/20.18.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell P. H., Wiesener M. S., Chang G. W., Clifford S. C., Vaux E. C., Cockman M. E., Wykoff C. C., Pugh C. W., Maher E. R., Ratcliffe P. J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Metzen E., et al. Intracellular localisation of human HIF-1 alpha hydroxylases: implications for oxygen sensing. J. Cell Sci. 2003;116:1319–1326. doi: 10.1242/jcs.00318. [DOI] [PubMed] [Google Scholar]
- Mylonis I., Chachami G., Samiotaki M., Panayotou G., Paraskeva E., Kalousi A., Georgatsou E., Bonanou S., Simos G. Identification of MAPK phosphorylation sites and their role in the localization and activity of hypoxia-inducible factor-1alpha. J. Biol. Chem. 2006;281:33095–33106. doi: 10.1074/jbc.M605058200. [DOI] [PubMed] [Google Scholar]
- Nambu J. R., Chen W., Hu S., Crews S. T. The Drosophila melanogaster similar bHLH-PAS gene encodes a protein related to human hypoxia-inducible factor 1 alpha and Drosophila single-minded. Gene. 1996;172:249–254. doi: 10.1016/0378-1119(96)00060-1. [DOI] [PubMed] [Google Scholar]
- Ohh M., Park C. W., Ivan M., Hoffman M. A., Kim T., Huang E., Pavlevich N., Chau V., Kaelin W. G. Ubiquitination of hypoxia-inducible factor requires direct binding to the b-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2000;2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- Romero N. M., Dekanty A., Wappner P. Cellular and developmental adaptations to hypoxia: a Drosophila perspective. Methods Enzymol. 2007;435:123–144. doi: 10.1016/S0076-6879(07)35007-6. [DOI] [PubMed] [Google Scholar]
- Romero N. M., Irisarri M., Roth P., Cauerhff A., Samakovlis C., Wappner P. Regulation of the Drosophila hypoxia-inducible factor alpha Sima by CRM1-dependent nuclear export. Mol. Cell. Biol. 2008;28:3410–3423. doi: 10.1128/MCB.01027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth P., Xylourgidis N., Sabri N., Uv A., Fornerod M., Samakovlis C. The Drosophila nucleoporin DNup88 localizes DNup214 and CRM1 on the nuclear envelope and attenuates NES-mediated nuclear export. J. Cell Biol. 2003;163:701–706. doi: 10.1083/jcb.200304046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang N., Fang J., Srinivas V., Leshchinsky I., Caro J. Carboxyl-terminal transactivation activity of hypoxia-inducible factor 1 alpha is governed by a von Hippel-Lindau protein-independent, hydroxylation-regulated association with p300/CBP. Mol. Cell. Biol. 2002;22:2984–2992. doi: 10.1128/MCB.22.9.2984-2992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenfeld M., Ward M., Nystrom G., Mosher J., Stahl S., Crews S. The Drosophila tango gene encodes a bHLH-PAS protein that is orthologous to mammalian Arnt and controls CNS midline and tracheal development. Development. 1997;124:4571–4582. doi: 10.1242/dev.124.22.4571. [DOI] [PubMed] [Google Scholar]
- Tanimoto K., Makino Y., Pereira T., Poellinger L. Mechanism of regulation of the hypoxia-inducible factor-1 alpha by the von Hippel-Lindau tumor suppressor protein. EMBO J. 2000;19:4298–4309. doi: 10.1093/emboj/19.16.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. L., Jiang B. H., Rue E. A., Semenza G. L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., White S. B., Zhao Q., Lee F. S. HIF-1alpha binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl. Acad. Sci. USA. 2001;98:9630–9635. doi: 10.1073/pnas.181341498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.