Abstract
The DNA replication machinery plays additional roles in S phase checkpoint control, although the identities of the replication proteins involved in checkpoint activation remain elusive. Here, we report that depletion of the prereplicative complex (pre-RC) protein Cdc6 causes human nontransformed diploid cells to arrest nonlethally in G1-G1/S and S phase, whereas multiple cancer cell lines undergo G1-G1/S arrest and cell death. These divergent phenotypes are dependent on the activation, or lack thereof, of an ataxia telangiectasia and Rad3-related (ATR)-dependent S phase checkpoint that inhibits replication fork progression. Although pre-RC deficiency induces chromatin structural alterations in both nontransformed and cancer cells that normally lead to ATR checkpoint activation, the sensor mechanisms in cancer cells seem to be compromised such that higher levels of DNA replication stress/damage are required to trigger checkpoint response. Our results suggest that therapy-induced disruption of pre-RC function might exert selective cytotoxic effects on tumor cells in human patients.
INTRODUCTION
In all eukaryotic cells, DNA replication initiates from multiple replication origins throughout the genome, and each segment of DNA replicates only once per cell cycle. Primary regulation of DNA replication is exerted at the initiation of DNA synthesis, when replication origins are licensed by assembly of prereplication complexes (pre-RCs) in early G1 (Bell and Dutta, 2002; Blow and Dutta, 2005). This process begins with origin recognition complex (ORC) binding to a nascent replication origin, resulting in recruitment of the loading factors Cdc6, and Cdt1, which function together to load the putative DNA replicative helicase minichromosomal maintenance (MCM) complex. Subsequently, phosphorylation of pre-RC and other DNA replication factors by the S phase-promoting kinases, cyclin-dependent kinases and Dbf4-dependent kinase Cdc7, in G1/S and S promotes loading of Cdc45, MCM10, GINS, RPA, and DNA polymerases to the pre-RC/origin, triggering the initiation of DNA replication.
DNA replication fidelity is ensured by S phase checkpoint mechanisms that monitor aberrant DNA replication, replication stress, DNA damage, and chromatin structure alterations in S phase. The S phase checkpoints are mainly governed by the phosphoinositide 3-kinase–related kinases (PIKKs) ataxia telangiectasia mutated- (ATM) or ataxia telangiectasia and Rad3-related (ATR)–dependent signaling pathways. On activation, the ATM/ATR checkpoint pathways immediately suppress late origin firing to prevent further DNA replication and stabilize stalled replication forks to ensure proper replication restart once the replication block/DNA damage has been repaired or removed. Although the ATM-dependent checkpoint responds to DNA double-strand breaks (DSBs), the ATR-dependent checkpoint responds to a broad spectrum of DNA lesions, such as single-strand breaks, fork stalling, or chromatin structural alterations (Abraham, 2001; Yang and Zou, 2006; Cimprich and Cortez, 2008). These checkpoint pathways function interdependently; for example, DSBs trigger an initial response from the ATM checkpoint machinery that leads to processing of the damaged DNA to structures recognized by the ATR checkpoint apparatus (Yang and Zou, 2006; Paulsen and Cimprich, 2007; Cimprich and Cortez, 2008). In response to replication stress or DNA damage, sensor proteins, such as RPA or NBS1, are recruited to the damage sites, and these proteins, in turn, provoke the recruitment of a complex array of DNA damage response proteins, including ATM, the ATRIP-ATR complex, TopBP1, MRE11, Rad50, Rad17, and 9-1-1 complex. Depending on the type of DNA damage, either ATR or ATM functions as the initiating protein kinase that engages a complex network of downstream proteins through phosphorylation of these proteins at Ser/Thr-Gln (S/T-Q) sites (Cimprich and Cortez, 2008). Prominent substrates for ATR and ATM are the protein serine-threonine kinases Chk1 and Chk2, respectively, which act as signal amplifiers in these checkpoint pathways. Ultimately, the signals emanating from the active site impinge on the cell cycle machinery to block DNA replication, stabilize stalled forks or broken DNA, arrest the cell cycle, and initiate DNA repair.
Because DNA replication and the S phase checkpoint are intimately linked, pre-RC proteins have long been proposed to play roles in checkpoint signaling/response. Direct interaction between pre-RC components and checkpoint proteins, including the binding of Cdc6/Cdc18 or MCM proteins to Rad3/ATR, Rad17, or Cds2/Chk2, and phosphorylation of MCM subunits by ATR/ATM have been reported previously (Cortez et al., 2004; Tsao et al., 2004; Hermand and Nurse, 2007; Bailis et al., 2008). Depletion of ORC subunits, Cdc6, Cdt1, MCM proteins, or Cdc7 kinase from a variety of organisms and cell types results in DNA replication inhibition, cell cycle arrest, and/or cell death (Murakami et al., 2002; Shimada et al., 2002; Feng et al., 2003; Montagnoli et al., 2004; Oehlmann et al., 2004; Lau et al., 2006; Teer et al., 2006; Kim et al., 2008; Ogi et al., 2008). These cellular consequences are probably attributed to pre-RC depletion-induced replication inhibition and S phase checkpoint responses (for review, see Lau and Jiang, 2006). Overexpression studies of pre-RC proteins also demonstrate a linkage between pre-RC proteins and checkpoint signaling, because overexpression of Cdc6 or Cdt1 activates the ATR-Chk1 or ATM-Chk2 checkpoint (Clay-Farrace et al., 2003; Tatsumi et al., 2006; Fersht et al., 2007; Hermand and Nurse, 2007; Liu et al., 2007). Together, these results indicate that interplay between pre-RC and S phase checkpoint proteins is essential for proper DNA replication, cell cycle progression, and cell viability, although the exact mechanistic relationship between pre-RC and checkpoint activation/signaling remains unclear.
We investigated previously the role of the pre-RC protein Cdc6 in maintaining proper origin firing and temporal replication dynamics in HeLa cells (Lau et al., 2006). We showed that the S phase depletion of Cdc6 in transformed HeLa cells resulted in aberrant DNA replication, and, ultimately, cell death in mitosis. Consistent with our findings, cell death induction was observed in several different transformed cancer cell lines by depletion of Orc2, Cdc6, and Cdt1 proteins or by Cdc7 kinase (Wohlschlegel et al., 2000; Shreeram et al., 2002; Feng et al., 2003; Montagnoli et al., 2004; Prasanth et al., 2004a). Recent studies, however, revealed that nontransformed mammalian cell lines are resistant to killing by manipulations that induce pre-RC insufficiency, suggesting that these cells, unlike their transformed counterparts, are able to mount protective cellular responses that prevent inappropriate DNA replication (Feng et al., 2003; Montagnoli et al., 2004; Lau and Jiang, 2006). To better understand the mechanistic basis for the differential responses of nontransformed versus transformed cells to pre-RC functional disruption, we comparatively examined the effects of pre-RC perturbation, especially Cdc6 depletion, on DNA replication dynamics, checkpoint activation, cell cycle progression, and cell death in both types of cells.
MATERIALS AND METHODS
Cell Culture, Synchronization, Transfection, and Drug Treatment
14F, A549, HCT116, HeLa, and MDA-MB-231 cells were cultured in DMEM containing 10% fetal calf serum (FCS). RPE1 cells were cultured in DMEM:F-12 (1:1) containing 10% FCS and 1.2g/l sodium bicarbonate. All cells were cultured at 37°C in 5% CO2. RPE1 cells were synchronized to G0 by incubation in serum-free media for 72–96 h and released into the cell cycle by addition of media containing 10% FCS. Small interfering RNA (siRNA) transfection was conducted using Oligofectamine for A549, HCT116, HeLa, and MDA-MB-231 cells or with Lipofectamine 2000 for 14F and RPE1 cells, according to manufacturer's protocol (Invitrogen, Carlsbad, CA). Caffeine treatment consisted of addition of 2.5 mM caffeine to cell culture media for indicated incubation times. Aphidicolin treatment consisted of addition of 1 μM to cell culture media for indicated times.
siRNA Synthesis and Antibodies
Heterogeneous, pooled endonuclease-prepared siRNAs specifically targeting luciferase (siLuc: coding region 538–983 bp), Cdc6 (siCdc6: coding region 842–1252 bp), or Orc2 (siOrc2: coding region 100–501 bp) were synthesized as described previously (Lau et al., 2006). ATR-targeted siRNA, anti-ATR, anti-MCM2, anti-Cdc6, anti-cyclin D1, anti-cyclin E antibodies were described previously (Jiang et al., 1999; Lau et al., 2006; Tsuji et al., 2006). Anti-Orc2, anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH), anti-pS317-Chk1, anti-5-bromo-2′-deoxyuridine (BrdU), anti-5′-chloro-2′-deoxyuridine (CldU), anti-5′-iodo-2′-deoxyuridine (IdU), and anti-pS33 RPA32 antibodies were purchased from EMD Biosciences (San Diego, CA), Abcam (Cambridge, MA), Cell Signaling Technology (Danvers, MA), Sigma-Aldrich (St. Louis, MO), Invitrogen (Carlsbad, CA), CellTech (UCB, Brussels, Belgium), and Bethyl Laboratories (Montgomery, TX), respectively. All secondary antibodies were purchased from Southern Biotechnology Associates (Birmingham, AL) and Jackson ImmunoResearch Laboratories (West Grove, PA).
Fluorescence-activated Cell Sorting (FACS) Analysis
Cells were fixed in 70% ethanol/30% 1× phosphate-buffered saline (PBS) for 1 h at −20°C. After fixation, cells were washed once in 1× PBS, resuspended, and incubated in propidium iodide (PI) buffer (60 μg/ml PI and 0.1 mg/ml RNase A) for 45 min at room temperature. Flow cytometry was conducted on at least 10,000 cells per condition using an FACSort and CellQuest version 3.3 (BD Biosciences, San Jose, CA). Cell cycle profiles were processed and analyzed for cell cycle phase distribution using FlowJo version 6.4.7 (Tree Star, Ashland, OR).
Cell Lysates, Subcellular Fractionation, Immunoblotting, Immunofluorescence, and DNA Fiber Analyses
Cell lysates and subcellular/chromatin fractionation were made as described previously (Jiang et al., 1999; Cook et al., 2002; Lau et al., 2006; Anantha et al., 2007). For immunoblotting analysis, whole-cell lysates or chromatin fractions were resolved on 6–15% SDS polyacrylamide gels, transferred onto polyvinylidene difluoride membranes, and immunoblotted with antibodies. For immunofluorescence analysis, after indicated treatment(s), coverslip-grown cells were cytoskeleton (CSK) extracted and immunostained with antibodies as described previously (Zhu and Jiang, 2005; Lau et al., 2006; Tsuji et al., 2006). Imaging for coverslips was carried out with a 63× oil objective on a DMIRE2 fluorescent microscope (Leica Microsystems, Deerfield, IL) by using Simple PCI software (Hamamatsu, Sewickley, PA).
DNA fiber analysis was performed as described previously (Li et al., 2003; Merrick et al., 2004; Lau et al., 2006). Imaging for labeled DNA fibers was also carried out with a 63× oil objective on a DMIRE2 fluorescent microscope (Leica Microsystems) by using Simple PCI software. Different replication structures were quantitated by manual counting of labeling patterns. Replication fork lengths were quantitated using the Line Measurement tool of ImageJ (National Institutes of Health, Bethesda, MD). ImageJ values were imported into Excel (Microsoft, Redmond, WA) and converted to micrometers by using the 63× micrometer-calibrated equation derived for a DMIRE2 fluorescent microscope (Leica Microsystems): ([ImageJ value] + 0.0095)/1.3477) × 10. Replication fork lengths were manually sorted into micrometer-defined bins, and histograms were generated. Three independent experiments were conducted per condition, with a final count of ≥950 labeled fibers per condition.
RESULTS
Pre-RC Deficiency Induces Nonlethal Cell Cycle Block in Human Nontransformed Cells but Cell Cycle Block and Cell Death in Cancerous Cells
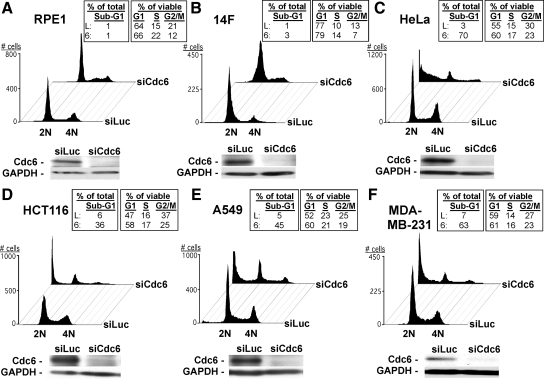
A panel of human nontransformed and cancer cell lines was transfected with a pool of endonuclease-prepared siLuc (a.k.a. control siRNA) or for Cdc6 (siCdc6). The siCdc6 pool has been shown to specifically and effectively deplete Cdc6 protein in human cells (Figure 1; Lau et al., 2006). FACS analysis showed that in response to Cdc6 depletion, asynchronously growing nontransformed cells (hTERT-immortalized retinal pigment epithelial-1, RPE1, or normal dermal fibroblasts, 14F) exhibited cell cycle arrest in G1-G1/S and in S compared with siLuc-treated cells (Figure 1, A and B). In contrast, HeLa (cervical), HCT116 (colon), A549 (lung), or MDA-MB-231 (breast) cancer cells exhibited a G1-G1/S block and significant cell death induction after Cdc6 depletion compared with siLuc-treated cells (Figure 1, C–F). Similar results were also obtained from the nontransformed and cancer cells treated with Orc2 siRNA (Supplemental Figure S1). Thus, consistent with our and others' previous observations (Wohlschlegel et al., 2000; Shreeram et al., 2002; Feng et al., 2003; Montagnoli et al., 2004; Prasanth et al., 2004a; Lau and Jiang, 2006; Lau et al., 2006), these results indicate that pre-RC deficiency induces G1-G1/S and S arrest in nontransformed cells and G1-G1/S arrest and cell death in cancer cells. The G1-G1/S arrests observed in nontransformed and cancer cells by Cdc6 or Orc2 depletion are probably due to insufficient DNA replication origin licensing in G1 that inhibits overall DNA replication, and/or to the activation of a G1-G1/S checkpoint as reported previously (Lau et al., 2006; Teer et al., 2006; Nevis et al., 2009). However, the discrepancies of S phase block and cell death between nontransformed cells and cancer cells suggest that, unlike cancer cells, nontransformed cells might exert a critical S phase checkpoint(s) to mount a protective response to disruption of pre-RC function during S phase.
Figure 1.
Depletion of Cdc6 results in nonlethal cell cycle block in nontransformed cells, but cell cycle arrest and cell death in cancer cells. (A–F) Indicated cell types were transfected with 100 nM siLuc or siCdc6 for 72 h. Transfected cells were fixed and stained with PI, followed by FACS analysis of >10,000 cells per condition. Sub-G1 populations were calculated based upon all counts per sample; G1/S/G2/M populations were calculated from non–sub-G1 counts. Below each FACS profile, siRNA-treated cells were lysed and subjected to immunoblotting analysis with anti-Cdc6 or anti-GAPDH (loading control) antibody. Note: It is not possible to strictly distinguish G1 and early S phase cells or late S phase cells and G2/M cells by FACS analysis. Thus, G1 contains G1 and G1/S cells and G2/M contains late S and G2/M cells.
Checkpoint Response to Cdc6 Deficiency in Nontransformed Cells Requires the ATR-dependent S Phase Checkpoint
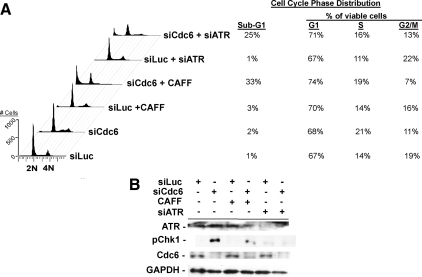
Previously, we showed that S phase deficiency of Cdc6 in cancerous HeLa cells inhibited new origin firing and prolonged DNA synthesis but failed to activate the ATR-Chk1 checkpoint, ultimately causing cell death in mitosis (Lau et al., 2006). Because the ATR-Chk1 checkpoint is intimately linked to DNA replication and responds to a variety of replication stress and damage (Paulsen and Cimprich, 2007), we hypothesized that this checkpoint might prevent nontransformed cells with pre-RC deficiency from pursuing a lethal program of DNA replication, and this protective checkpoint was perturbed in transformed cells. To test this possibility, we treated siLuc- or siCdc6-transfected RPE1 cells with caffeine, a nonspecific inhibitor of PIKK catalytic activities, to abrogate ATR-Chk1 signaling (Sarkaria et al., 1999). FACS analysis indicated that although caffeine cotreatment did not detectably affect cell cycle progression in siLuc-treated cells, it abolished the S phase-arrested population in siCdc6-treated cells, recapitulating the cell death detected in Cdc6-depleted cancer cells (Figure 2A). Immunoblotting analysis showed that depletion of Cdc6 in RPE1 cells induced phosphorylation of Chk1, an ATR-mediated event during checkpoint activation (Zhao et al., 2001; Smits 2006), which was abrogated by caffeine treatment (Figure 2B). Treatment of siLuc- or siCdc6-treated cells with specific ATR-targeted siRNA (siATR) produced similar results as observed in caffeine-treated cells, indicating that ATR function was required for the checkpoint response to pre-RC deficiency (Figure 2A). Together, these results indicate that nontransformed cells cope with Cdc6 deficiency in S phase through activation of the ATR-dependent checkpoint pathway, whereas cancer cells with Cdc6 deficiency fail to activate this checkpoint pathway as we reported previously (Lau et al., 2006).
Figure 2.
Nonlethal cell cycle block induced by Cdc6 deficiency in RPE1 cells requires ATR. (A) Asynchronous RPE1 cells were treated with siLuc or siCdc6 for 60 h; with siLuc or siCdc6 for 48 h and then 2.5 mM caffeine (CAFF) for 12 h; or with siLuc or siCdc6 for 12 h and then with siATR for 48 h, as indicated. Treated cells were harvested and subjected to FACS. Sub-G1 populations were calculated based upon all counts per sample; G1/S/G2/M populations were calculated from non–sub-G1 counts. As mentioned in Figure 1, G1 contains G1 and G1/S cells and G2/M contains late S and G2/M cells. (B) Cells treated as described in A were subjected to immunoblotting analysis with indicated antibodies.
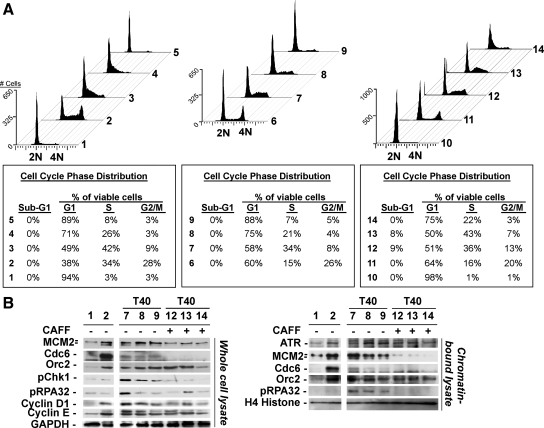
To investigate the temporal nature of ATR-checkpoint response to Cdc6 deficiency in nontransformed cells, we examined ATR-checkpoint activation in siLuc- or siCdc6-treated synchronously proliferating RPE1 cells. To avoid the confounding effects of replication checkpoint activation induced by cell cycle synchronization compounds, such as thymidine or aphidicolin, we synchronized RPE1 cells at G0 stage by serum deprivation and then stimulated cells to re-enter the cell cycle by addition of serum (Supplemental Figure S2). After serum addition, cells were immediately treated with siLuc or increasing concentrations of siCdc6, and the effects of partial or complete Cdc6 depletion at subsequent time points were determined by FACS and immunoblotting analyses. We postulated that complete depletion of Cdc6 in synchronous G1 cells should lead to complete replication licensing deficiency and G1-G1/S-phase arrest. Incomplete depletion of Cdc6 in synchronous G1 cells might support partial pre-RC function to allow abnormal S phase entry/DNA replication, resulting in activation of the protective ATR-dependent S phase checkpoint, similar to that of pre-RC deficiency in asynchronous S phase cells. Consistent with this notion, 20 h after serum stimulation and siRNA treatment, RPE1 cells exposed to 200 nM siLuc (Figure 3A, 2) had advanced into S-G2, whereas siCdc6-treated cells (Figure 3A, 3–5) exhibited extremely delayed/blocked cell cycle progression (Figure 3A, 1–5). As predicted, the effect was dependent upon the extent of Cdc6 knockdown as low-dose siCdc6-treatment (20 nM) yielded G1/S and S blocks, whereas higher dose siCdc6 treatment (100 nM) also resulted in G1/S and S arrests, but with less S phase cells (Figure 3A, 3 and 4). Transfection with the highest dose (200 nM) of siCdc6 resulted in a G1-G1/S block with few S phase cells (Figure 3A, 5). We also attempted to determine the effects of DNA replication and S phase progression in siLuc- or siCdc6-transfected synchronous RPE1 cells in detail by pulse labeling cells with BrdU followed by FACS analysis, but these efforts were unsuccessful because depletion of Cdc6 strongly inhibited DNA replication (see Figures 4 and 5).
Figure 3.
Cdc6 deficiency induces distinct G1-G1/S arrest and activation of ATR-dependent S phase checkpoint signaling required for S phase arrest in RPE1 cells. (A) G0 serum deprivation-synchronized RPE1 cells (1) were stimulated with 10% fetal bovine serum (FBS) and immediately transfected with 200 nM siLuc (2), 20 nM siCdc6 (3), 100 nM siCdc6 (4), or 200 nM siCdc6 (5). Twenty hours after transfection, cells were harvested and subjected to FACS. G0-synchronized RPE1 cells were stimulated with 10% FBS and transfected with 200 nM siLuc (6), 20 nM siCdc6 (7), 100 nM siCdc6 (8), or 200 nM siCdc6 (9). Forty hours after transfection, cells were harvested and subjected to FACS. G0-synchronized cells as in 1 or 6–9 were subjected either to additional 40 h of serum deprivation (10) or to additional 20 h of transfection + 2.5 mM caffeine (11–14) and were harvested and analyzed by FACS. Sub-G1 populations were calculated based upon all counts per sample; G1/S/G2/M populations were calculated from non–sub-G1 counts. As mentioned in Figure 1, G1 contains G1 and G1/S cells and G2/M contains late S and G2/M cells. (B) Whole-cell lysates (left) or chromatin-bound lysates (right) from the G0-synchronized RPE1 cells subjected to indicated conditions as described in A were immunoblotted with indicated antibodies. Note: Faster migrating band observed in anti-MCM2 blots comprises phosphorylated MCM2 species (Tsuji et al., 2006).
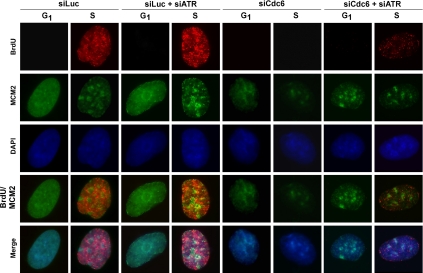
Figure 4.
S phase arrest induced by Cdc6 deficiency in nontransformed cells results from ATR-dependent DNA replication inhibition. Asynchronously growing RPE1 cells were transfected with indicated siRNA for 72 h. Transfected cells were pulse labeled with 20 μM BrdU for 15 min, fixed, CSK extracted and immunostained with indicated antibodies and DAPI (DNA). Representative nuclei from each siRNA treatment condition are shown.
Figure 5.
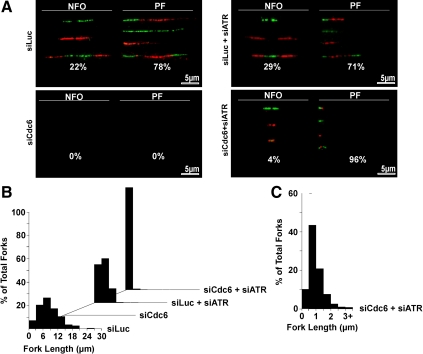
Cdc6 deficiency in nontransformed cells results in ATR-mediated checkpoint suppression of progressing forks. (A) Dual-labeled (green, CldU and red, IdU) DNA fibers were generated from asynchronously growing RPE1 cells treated with indicated siRNA(s). Representative labeled DNA fibers and percentage of total DNA fibers for new firing origins (NFO) or progressing forks (PF) for each condition are shown. We counted ≥950 labeled DNA fibers from three independent experiments for each condition. (B) Lengths of all counted replication forks were measured by ImageJ (National Institutes of Health), sorted into increasing 3-μm bins, and plotted as percentage of total fork histograms on Excel (Microsoft). (C) Histogram of increased bin resolution (0.5-μm bins) showing a more detailed replication fork length distribution of siCdc6- and siATR-cotreated RPE1 cells.
We next analyzed siRNA-treated RPE1 cells after an additional 20 h (a total of 40 h from addition of serum and siRNA transfection) in the absence or presence of caffeine. Regardless of caffeine cotreatment, the percentage of siLuc-treated cells in G1 increased over time, suggesting that these cells completed the cell cycle and reentered G1 again (compare Figure 3A, 2 with 6 and 11). In contrast, cells treated with 20 or 100 nM siCdc6 exhibited partial cell cycle progression, with G1 cells remaining in G1 phase, and the remainder undergoing an abnormally protracted transit through S-G2 phases (Figure 3A, 7 and 8). Cells treated with 200 nM siCdc6-treated cells remained arrested in G1-G1/S (Figure 3A, 9). In the presence of caffeine, cell cycle progression was restored to the G1/S-S phase populations in 20 and 100 nM siCdc6-treated cells (Figure 3A, 12 and 13) and to a minor extent to the G1/S population in 200 nM siCdc6-treated cells (Figure 3A, 14) but at extremely compromised rates compared with controls.
Consistent with the FACS results, immunoblotting analysis of whole-cell lysates and chromatin-bound proteins showed that Cdc6 depletion resulted in a siRNA dose-dependent reduction of chromatin association and phosphorylation of MCM2 compared with controls (Figure 3B, 1 and 2, 7–9, left and right). Cdc6-depletion also induced phosphorylation of Chk1 and RPA p32 subunit (RPA32), consistent with active ATR signaling (Figure 3B, 1 and 2, 7–9, left and right). The levels of phosphorylated Chk1 and RPA32 were dramatically elevated after treatment with the lower concentration (20 nM) of siCdc6, consistent with the ability of partially Cdc6-depleted cells to undergo G1-to-S phase progression, leading to effective engagement of ATR-dependent checkpoint response (Figure 3B, 7). In contrast, cells challenged with the higher concentration (200 nM) of siCdc6 displayed a strong G1-G1/S block and hence lower levels of ATR-dependent Chk1 and RPA32 phosphorylation (Figure 3B, 9). Caffeine treatment abolished phosphorylation of Chk1 and RPA32 and reduced chromatin-bound levels of MCM2 and residual Cdc6 but not Orc2 (Figure 3B, 12–14). Although cyclin D1 levels were marginally decreased in siCdc6-treated cells compared with controls as reported recently (Liu et al., 2009), cyclin E levels did not exhibit appreciable alterations in these cells (Figure 3B, left). Together, these results indicate that Cdc6 knockdown in nontransformed cells triggers an ATR-dependent arrest that blocks pre-RC-deficient cells in S phase.
Cdc6 Deficiency-induced S Phase Arrest Is the Result of ATR-dependent Inhibition of DNA Replication
To determine how DNA replication was affected in pre-RC deficiency in RPE1 cells, we performed detailed immunocytological analyses. We pulse-labeled siLuc- and siCdc6-treated cells with BrdU and monitored BrdU incorporation, chromatin association of MCM2, and DNA content (4′,6-diamidino-2-phenylindole [DAPI] staining) by immunofluorescence staining. Previously, we and others showed that the intensities of DAPI staining together with the combined localization patterns of chromatin-bound MCM2 and BrdU staining revealed the cell cycle phase of individual cells in an asynchronous population and also allowed identification of early versus late S phase cells (Dimitrova et al., 1999; Tsuji et al., 2006). G1 cells were characterized by lower levels of DAPI staining, a uniform pattern of chromatin-bound MCM2 staining and lack of BrdU staining. Early/mid-S phase cells were characterized by higher levels of DAPI staining, a speckled pattern of chromatin-bound MCM2, and BrdU staining. Late S phase cells were characterized by higher levels of DAPI staining, a diminished, dappled pattern of chromatin-bound MCM2, and BrdU staining. Finally, G2/M cells were characterized by highest levels of DAPI staining and the absence of chromatin-bound MCM2 and BrdU staining (for details, see Supplemental Figure S3A). Our immunostaining results revealed that siLuc-treated cells exhibited a cell cycle distribution with 42% of cells in G1, 34% in early/mid-S phase, 5% in late S, and 19% in G2/M (Figure 4 and Supplemental Figure S3A), similar to that recorded by FACS analysis (Figures 1A and 2A). In contrast, siCdc6-treated RPE1 cells displayed a marked reduction of chromatin-bound MCM2 staining and lack of detectable BrdU staining. Of the cells analyzed, half (50%) exhibited lower DAPI staining and extremely reduced but diffuse chromatin-bound MCM2 staining, indicating that these cells were in G1 (Figure 4 and Supplemental Figure S3B). The other half displayed higher DAPI staining with reduced levels of chromatin-bound MCM2 staining pattern in early/mid-S phase (24%) and in late S phase 26% (Figure 4 and Supplemental Figure S3B). Thus, consistent with the FACS and immunoblotting results (Figures 1–3), these results indicate that depletion of Cdc6 in nontransformed RPE1 cells perturbs pre-RC formation, blocks G1-G1/S and S cell cycle progression, and inhibits DNA replication.
To determine whether inhibition of ATR in Cdc6-depleted cells restored DNA replication, thereby promoting cell cycle progression, we subsequently transfected siLuc- or siCdc6-transfected RPE1 cells with siATR and examined BrdU incorporation, chromatin association of MCM2, and DAPI staining. siLuc- and siATR-cotreated cells exhibited similar cell cycle distribution profiles but with increased BrdU staining intensity in S phase cells compared with cells treated with siLuc alone, consistent with recent reports that inhibited ATR-Chk1 signaling activates dormant replication origin firing and increases active replication origin density (Figure 4 and Supplemental Figure S3C) (Woodward et al., 2006). In contrast, knockdown of ATR in siCdc6-treated cells restored BrdU incorporation but at significantly reduced levels, indicating resumption of DNA replication and S phase progression in these cells (Figure 4 and Supplemental Figure S3D). Similar results were also obtained in siLuc- and siCdc6-treated cells treated with caffeine (Supplemental Figure S3, E and F). Together, these results indicate that the caffeine-sensitive inhibition of S phase progression and DNA replication observed in nontransformed cells in response to loss of Cdc6 function is attributable to the activation of an ATR-dependent DNA replication checkpoint.
Inhibition of ATR Checkpoint Response to Cdc6 Deficiency Restores Replication Fork Progression with Extremely Reduced Rates
To investigate Cdc6 deficiency-induced ATR-dependent checkpoint inhibition of DNA replication in more detail, we analyzed DNA replication in higher resolution by examination of origin firing and fork progression by using DNA fiber analysis. After siRNA treatment, cells were sequentially pulse labeled (10 min/pulse) with differentially halogenated nucleoside precursors, 5-Chloro-2′-Deoxyuridine (CldU) or 5-Iodo-2′-Deoxyuridine (IdU), which incorporate into actively replicating DNA. DNA fibers were generated, allowing visualization of DNA replicating structures by immunofluorescent microscopy, as described previously (Lau et al., 2006). Changes in global replication dynamics induced by siRNA treatment were determined by analysis of labeled DNA fibers for two particular characteristics (Supplemental Figure S4A): 1) types of active replication structures (i.e., newly fired origins or progressing/terminating forks) by quantitating nucleoside incorporation patterns (Lau et al., 2006) and 2) rate of DNA replication fork progression by measurement of labeled fiber lengths (Conti et al., 2007a). Figure 5 and Supplemental Figure S4B show representative labeled DNA fibers and the summarized quantitative results. Analysis of >950 labeled DNA fibers in three independent experiments indicated that 22% of total DNA replication structures in asynchronous siLuc-treated RPE1 cells were newly fired origins and 78% of DNA replication structures were progressing forks. In contrast, siCdc6-treated RPE1 cells did not display any detectable DNA fiber labeling, confirming our immunocytological data (Figure 4) that Cdc6 depletion resulted in inhibition of global replication activity. Cotreatment of RPE1 cells with siLuc and siATR caused a slight, but statistically significant, increase in newly fired origins (29%) and a decrease in progressing forks (71%), consistent with our immunofluorescence data (Figure 4) and the previously described role of ATR in dormant origin suppression (Woodward et al., 2006). Combined treatment of siCdc6-treated RPE1 cells with siATR restored DNA replication predominantly as progressing forks (>95%) but not new origin firing because depletion of Cdc6 blocked pre-RC assembly and inhibited the initiation of DNA replication (origin firing). However, the lengths of restored replication forks in siCdc6- and siATR-cotreated cells were significantly shorter than those of siLuc alone or siLuc- and siATR-cotreated cells (Figure 5, B and C). siLuc-treated cells exhibited median replication fork lengths between 6 and 9 μm (∼15.6–23.4 kb; 1-μm fiber, ∼2.6 kb; Jackson and Pombo, 1998; Li and Stern, 2005), whereas siLuc- and siATR-cotreated cells exhibited median replication fork lengths of 3–6 μm (∼7.8–15.6 Kb). The slight shortening of fork lengths in siLuc- and siATR-cotreated cells is consistent with reports that inhibition of ATR increases global origin firing and decreases origin–origin distance and replication fork progression rates (Conti et al., 2007b). In sharp contrast, siCdc6- and siATR-cotreated cells only exhibited median replication fork lengths between 0.5 and 1 μm (1.3–2.6 kb) (Figure 5C). The significantly shorter fork lengths in siCdc6- and siATR-cotreated cells indicated that resumption of DNA replication in Cdc6-depleted cells by ATR inhibition resulted from restoration of stalled fork progression, albeit in a greatly perturbed manner.
Two possibilities could account for the unique fiber patterns in siCdc6 and siATR cotreated RPE1 cells. Inhibition of ATR in Cdc6-depleted cells might restore stalled fork progression either 1) at significantly reduced progression rates or 2) with increased fork instability, leading to fork stalling and/or collapse. To distinguish between these two possibilities, we repeated the above-described experiments, with longer CldU/IdU pulse times (1 h each) to gain a clearer view of fork progression rates. We anticipated that short replication forks resulting from reduced fork progression rates would be manifested by labeling patterns that would be qualitatively similar to those seen with cells subjected to shorter periods of CldU/IdU labeling. Conversely, short replication forks resulting from unstable/collapsing replicating structures would be manifested by erratic, asymmetrical nucleotide incorporation, i.e., no or reduced labeling with the second IdU pulse. As shown in Supplemental Figure S4C, prolonged nucleotide labeling times yielded longer labeled tracks resembling those produced by shorter pulses (Supplemental Figure S4B) in both siLuc- and siATR- or siCdc6- and siATR-cotreated cells. Thus, knockdown of ATR in cells, especially siCdc6-treated cells, did not lead to rampant fork instability but rather permitted replication fork progression at a greatly reduced rate.
Together, these DNA fiber analyses indicate that inhibition of all forms of replication activity in Cdc6-deficient nontransformed cells is due to 1) inhibition of new origin firing by pre-RC insufficiency and 2) inhibition of existing fork progression by the ATR-dependent checkpoint.
Cdc6-deficient Cancer Cells Exhibit a Defect in ATR-dependent Checkpoint Activation
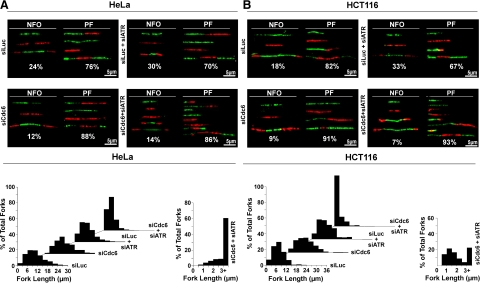
Although DNA fiber analysis revealed that the ATR checkpoint was required for inhibition of existing fork progression in nontransformed Cdc6-deficient S phase cells, it was unclear how pre-RC deficiency triggered ATR checkpoint activation. We reasoned that examination of the differences in replication and checkpoint responses between nontransformed and transformed cells might clarify the issue because transformed cancer cells continued replication activity and S phase progression during Cdc6 deficiency (Lau et al., 2006). Therefore, we analyzed labeled DNA fibers from similarly siRNA-treated HeLa and HCT116 cancer cells. As shown in Figure 6 and Supplemental Figure S5A, siLuc-treated HeLa or HCT116 cells exhibited a DNA replication distribution (∼20% new firing origins and ∼80% progressing forks) and median replication fork lengths (∼7–10 μm; ∼18.2–26 kb) similar to those of siLuc-treated RPE1 cells. However, unlike RPE1 cells, HeLa or HCT116 cells treated with siCdc6 did not completely inhibit DNA replication, displaying fewer labeled DNA fibers, which were predominantly progressing forks (∼90%), consistent with our previously published results (Lau et al., 2006). Moreover, we observed a slight increase in overall replication fork length, indicating that replication fork speed was increased in these cells, consistent with the notion of increased distance between firing origins due to inhibition of late origin firing by pre-RC deficiency and lack of sufficient checkpoint activation to suppress this altered replication activity (Lau et al., 2006; Conti et al., 2007a). siLuc and siATR cotreatment of HeLa or HCT116 cells increased new origin firing and decreased progressing forks (∼30 vs. ∼70%) with shorter replication fork lengths, similar to RPE1 cells (Figure 5). Cotreatment with siCdc6 and siATR in HeLa or HCT116 cells increased labeled DNA fibers, with distribution similar to siCdc6-treated cells (predominantly progressing forks ∼90%). Furthermore, siCdc6 and siATR cotreatment in these cells induced shortening of replication fork lengths (median fork length, ∼3 μm; ∼7.8 kb). Although these forks were not as short as those observed in the RPE1 cells, long-labeling experiments indicated that these short fibers in HeLa cells also resulted from a reduced fork progression rate (Supplemental Figure S5B). Although siCdc6- and siATR-cotreated HeLa or HCT116 cells exhibited short replication forks, unlike RPE1 cells, these cells also exhibited longer replication forks, similar to those observed during siCdc6 treatment alone (Figure 6 and Supplemental S5A). The appearance of the shorter replication forks amid the longer replication forks in siCdc6- and siATR-cotreated HeLa or HCT116 cells indicated that these pre-RC–deficient cancer cells did partially inhibit DNA replication. Given our previous results that depletion of pre-RC proteins in human cancer cells did not result in detectable activation of S phase checkpoint response, such as Chk1 phosphorylation (Lau et al., 2006; see below Figure 7C), our data indicate that cancer cells, unlike nontransformed RPE1 cells, do not fully mount S phase checkpoint activation in response to pre-RC deficiency and do not completely inhibit DNA replication.
Figure 6.
HeLa and HCT116 cancer cells do not completely inhibit replication fork progression in response to Cdc6 deficiency. Dual-labeled (green, CldU and red, IdU) DNA fibers were generated for asynchronously growing HeLa (A) or HCT116 (B) cells treated with indicated siRNA(s). Representative labeled DNA fibers and percentage of total labeled DNA fibers for new firing origins (NFO) or progressing forks (PF) for each condition are shown. We counted ≥950 labeled DNA fibers from three independent experiments for each condition. Lengths of all counted replication forks were measured as described in Figure 4, B and C.
Figure 7.
Cdc6 deficiency alters HP1 localization and increases γ-H2AX staining in HeLa and HCT116 cancer cells in the presence of aphidicolin. Asynchronously growing RPE1, HeLa, or HCT116 cells were treated with indicated siRNA for 48 h (A) or with indicated siRNA and then an additional 4 h in the presence or absence of 1 μM APH (B). Cells were fixed and immunostained with indicated antibodies and DAPI (DNA). (C) Whole-cell lysates from cells treated as described in B were immunoblotted with indicated antibodies.
Effects on Localization of Heterochromatin Protein HP1α and Low-Dose Treatment of DNA Replication Inhibitor Aphidicolin (APH) in Cdc6-deficient RPE1 and Cancer Cells
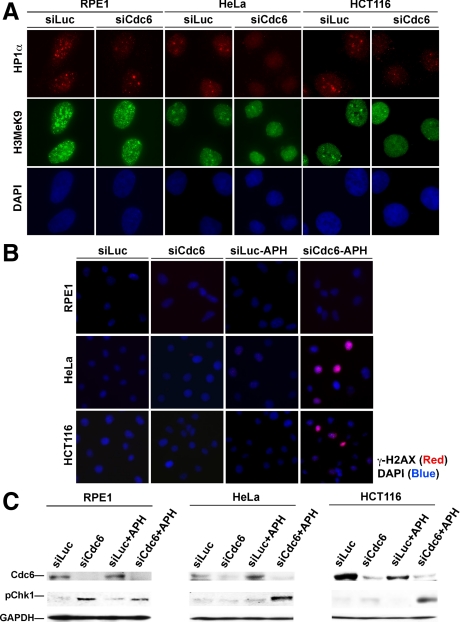
To understand the mechanism(s) by which nontransformed cells, but not cancer cells, activated an ATR-checkpoint response to pre-RC deficiency to completely inhibit DNA replication, we first examined possible causes of reduced fork progression rates after ATR inhibition in siCdc6-treated cells. Although ATR might directly promote fork progression and depletion of ATR would reduce fork speed, this possibility was unlikely because siLuc- and siATR-cotreated RPE1, HeLa, and HCT116 cells did not uniformly display dramatically reduced fork progression speeds compared with siCdc6- and siATR-cotreated cells. We speculated that chromatin structural changes induced by abnormal replication during pre-RC depletion might impede fork progression, reducing fork speeds. In support of this notion, altered chromatin structure resulting from perturbation of Orc2, has been reported previously (Prasanth et al., 2004b). To test whether Cdc6 depletion altered global chromatin structure, we examined the localization and levels of the chromatin structural marker heterochromatin protein 1 α-subunit (HP1α) and the methylation status of histone H3 at lysine 9 (H3MeK9), which is required for HP1α binding to heterochromatin, in siLuc- or siCdc6-treated RPE1, HeLa, or HCT116 cells. As shown in Figure 7A and Supplemental Figure S6, siLuc-treated RPE1 cells exhibited prominent, bright HP1α foci and speckled H3MeK9 staining patterns, whereas siCdc6-treated RPE1 cells displayed a marked reduction of HP1α foci staining and marginal reduction of speckled H3MeK9 staining. Unexpectedly, similar results were also observed in HeLa or HCT116 cells (Figure 7A and Supplemental S6). Thus, Cdc6 depletion in nontransformed and transformed cells resulted in chromatin structural alterations, which abrogated HP1α localization and to a lesser extent the H3MeK9 required for HP1 heterochromatin association.
Despite global chromatin structure alterations (abnormal HP1 localization) in both nontransformed and cancer cells upon pre-RC deficiency, cancer cells were nonetheless insufficiently responsive to pre-RC deficiency. Previous studies showed that cancer cells required a higher level of DNA replication stress or damage to activate cell cycle checkpoints compared with nontransformed cells (Bartkova et al., 2005). We showed that the ATR checkpoint could be trigged by additional genotoxic stress in Cdc6-deficient HeLa cells (Lau et al., 2006). To determine whether a relative insensitivity to replication stress underlies the defective ATR checkpoint response to pre-RC deficiency in cancer cells, we artificially raised replication stress levels by exposing siLuc- or siCdc6-treated RPE1, HeLa, or HCT116 cells to aphidicolin at a concentration (1 μM) shown previously to slow fork progression without triggering overt replication checkpoint activation (Luciani et al., 2004). Immunofluorescence analysis of checkpoint response and the replication-dependent DNA damage marker γ-H2AX showed that neither siLuc nor siCdc6 treatments, alone, or in the presence of aphidicolin, up-regulated γ-H2AX, because DNA replication was already inhibited in siCdc6-treated RPE1 cells (Figure 7B). Furthermore, addition of aphidicolin to siLuc-treated HeLa or HCT116 did not induce γ-H2AX (Figure 7B). However, cotreatment with siCdc6 and aphidicolin induced robust H2AX phosphorylation in HeLa and HCT116 cells. These results were further substantiated by immunoblotting analysis Chk1 phosphorylation (Figure 7C). Consistent with our previous report (Lau et al., 2006), neither siCdc6 nor low-dose aphidicolin treatment triggered significant Chk1 phosphorylation in HeLa or HCT116 cells. In contrast, Cdc6 depletion alone resulted in Chk1 phosphorylation in RPE1 cells. However, upon cotreatment of low-dose aphidicolin and siCdc6 in HeLa or HCT116 cells, Chk1 phosphorylation was observed. Together, these results suggest that the additional replication stress imposed by low-dose aphidicolin exceeds the threshold for activation of the replication checkpoint response in Cdc6-depleted cancer cells.
DISCUSSION
We have investigated pre-RC deficiency and checkpoint response in human cells. Our results show that depletion of pre-RC component Cdc6 yields two distinct outcomes. G1-G1/S arrest is an outcome observed in both nontransformed and cancer cells and is triggered by profound abrogation of origin licensing and/or a G1-checkpoint that blocks S phase entry (Lau et al., 2006; Teer et al., 2006; Nevis et al., 2009). The second outcome is S phase specific; cell cycle arrest is observed in nontransformed cells, whereas abnormal DNA replication and ultimately cell death are observed in cancer cells. We show that in S phase-nontransformed cells, deficiency of Cdc6 results in activation of the ATR-dependent S phase checkpoint that halts replication fork progression. Codepletion of Cdc6 and ATR in these cells abrogates checkpoint responses; restores fork progression at extremely reduced rates; and ultimately causes cell death, recapitulating the phenotype of cancer cells.
Pre-RC Deficiency and ATR-dependent Checkpoint Activation
We determined the underlying reason(s) for disparate activation of the ATR-dependent S phase checkpoint between nontransformed and cancer cells arising from pre-RC deficiency. Our results suggest that Cdc6 deficiency, which decreases new origin firing and presumably causes increased interorigin distance for progressing forks, can lead to altered chromatin structure and subsequent activation of the ATR checkpoint, resulting in the suppression of replication fork progression and S phase arrest in nontransformed cells. Consistently, abrogation of the ATR checkpoint in Cdc6-depleted cells restores fork progression at an extremely reduced rate, suggesting that chromatin structural alterations induced by pre-RC deficiency slow DNA replication rate in Cdc6 and ATR codepleted cells. Although Cdc6 deficiency also causes similar chromatin structural changes in S phase cancer cells, these changes fail to sufficiently activate ATR-dependent checkpoint response due to an elevated DNA damage/stress threshold. Supporting evidence for this comes from the finding that the ATR-dependent checkpoint response can be triggered in cancer cells if basal replication stress levels are increased by a low concentration of the DNA polymerase inhibitor aphidicolin. Although an elevated replication checkpoint activation threshold may confer a proliferative advantage to cancer cells exposed to a stressful tumor microenvironment, this alteration in checkpoint function seems to render cancer cells more prone to attempt a catastrophic S-to-M phase progression in the setting of abnormal DNA replication (Lau et al., 2006).
Pre-RC proteins have long been proposed to play critical roles in checkpoint responses. Overexpression of Cdt1 and/or Cdc6, for example, induces checkpoint activation, whereas pre-RC deficiency compromises checkpoint response (Murakami et al., 2002; Clay-Farrace et al., 2003; Oehlmann et al., 2004). Recent studies demonstrated direct interactions between pre-RC proteins and checkpoint proteins, suggesting that pre-RC proteins may function as chromatin anchors and/or crucial downstream targets for S phase checkpoint initiation and maintenance (Cortez et al., 2004; Tsao et al., 2004; Hermand and Nurse, 2007; Bailis et al., 2008). However, our observations of ATR checkpoint activation, as indicated by phosphorylation of Chk1 and RPA32, in pre-RC–deficient, nontransformed human cells indicate that the interaction of checkpoint proteins with specific individual pre-RC proteins may be dispensable in regulating ATR-checkpoint activation and signaling. Instead, our results suggest that abnormal chromatin structure resulting from aberrant DNA replication due to pre-RC deficiency might be a trigger for S phase checkpoint activation. Recently, chromatin alterations have been shown to play an important role for ATR-checkpoint activation, because deregulation of DNA licensing and alteration of chromatin structure indicated by changes of HP1α localization, activate S phase checkpoint signaling cascades (Davidson et al., 2006; Lin and Dutta, 2007; Ayoub et al., 2008). Similar chromatin structural alterations also were observed in Orc2 depletion or carcinogen/replication inhibitor studies, where inhibition of new origin firing, replication/chromatin structural alterations, checkpoint activation, and reduced replication rates were reported (Prasanth et al., 2004b; Conti et al., 2007a). It will be of interest to determine how abnormal replication/chromatin structure induced by perturbed pre-RC function in S phase initiates the S phase checkpoint response that protects pre-RC–deficient cells from inappropriate S-to-M phase progression in the future.
Deregulation of pre-RC/S Phase Checkpoint Control and Cancer
Despite inhibited new origin firing, perturbed DNA replication and altered chromatin structure induced by pre-RC deficiency, several human cancer cell lines failed to activate the ATR-dependent S phase checkpoint under this condition. Our results suggest that the deficient S phase checkpoint response in cancer cells might be due to elevated DNA replication stress/damage response thresholds. Previous studies demonstrated that the cellular transformation process perturbs cell growth and genome stability, which result in DNA damage and replication stress/errors. These problems initially alarm the checkpoint machineries to trigger S phase checkpoint response that leads to cell cycle arrest, damage repair, senescence, and/or cell death to prevent or delay tumorigenesis. However, the nature of checkpoint response also creates a selective pressure that favors the outgrowth of malignant clones with genetic or epigenetic defects in the checkpoint machineries (Bartkova et al., 2005; Bartek et al., 2007a,b). Thus far, the majority of cancer cells examined exhibit some extent of genetic or functional defect(s) in checkpoint pathways. Because many checkpoint proteins are essential for embryonic development, cell homeostasis, and survival, homozygous depletion of the checkpoint genes or complete elimination of the functions of their proteins in tumor cells is rarely observed. Instead, tumor cells harboring heterozygous deletion of checkpoint genes (haploinsufficiency) or reduction of checkpoint protein function (partial deficiency) are commonly observed. Hence, haploinsufficient and/or partially defective checkpoint control render tumor cells less sensitive/more tolerant to genotoxic insults and aberrations, including replication stress, than normal cells. Bartkova et al. (2005) showed that early cancerous lesions exhibit activated DNA replication checkpoint proteins, which comprise an anticancer barrier that induces growth arrest or cell death, constraining tumor progression. However, during tumorigenesis, malignant cells within the lesions overcome checkpoint control through acquisition of defects in DNA damage checkpoint response components (such as ATR, ATM, Chk1, Chk2, or p53) that either raises the threshold for, or qualitatively alters, the DNA damage-induced checkpoint response. Our results are consistent with these findings, suggesting that elevated DNA damage/stress thresholds in cancer cells account for the lack of ATR-dependent S phase checkpoint activation and cell cycle block in response to the aberrant replication structures generated during pre-RC deficiency.
Recent studies have also revealed other features of the S phase replication checkpoint, such as replication fork pausing. In yeast and certain normal mammalian cell types, abundant origin firing in early S phase is followed by replication forks “pausing” before resumption later in S phase (Caldwell et al., 2008; Frum et al., 2008). This pause-and-release mechanism is not evident in transformed cells, and the additional loss of this regulation may account for the lack of S phase checkpoint response to pre-RC deficiency in cancer cells. In certain tumors, deregulation of DNA replication checkpoints in early oncogenesis may be attributed to changes in pre-RC proteins themselves. For example, up-regulation of Cdc6 results in the specific methylation and silencing of tumor suppressors in premalignant lesions (Gonzalez et al., 2006), indicating that deregulation of pre-RC proteins might alter transcription of tumor suppressors or oncogenes. Furthermore, defects in or loss of p16INK4A, or deregulated cyclin D1 or E have been demonstrated to result in abnormally long replication structures and attenuated DNA damage response (Bartkova et al., 2005; Tort et al., 2006), similar to the consequences of pre-RC deficiency that we observed in this study. The notion that pre-RC deregulation directly promotes early pro-oncogenic events, such as decreased DNA damage sensitivity, is consistent with the fact that pre-RC proteins are frequently deregulated in a multitude of tumor types and can recapitulate tumor phenotypes when similarly perturbed in vitro and in vivo (Seo et al., 2005; Gonzalez et al., 2006; Honeycutt et al., 2006; Lau et al., 2007; Shima et al., 2007; Blow and Gillespie, 2008).
Pre-RC Involvement in DNA Replication/S Phase Checkpoint
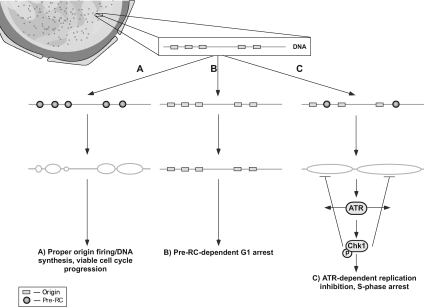
Together, our current findings and a previous report (Lau et al., 2006) demonstrate that nontransformed and cancer cells exhibit distinct checkpoint responses to pre-RC perturbation. During normal DNA replication, optimal assembly of pre-RC complexes and origin licensing ensures an appropriate distribution of newly fired origins and replicon lengths conducive for complete and accurate genome duplication during S phase (Figure 8A). In both nontransformed and cancer cells, extensive pre-RC deficiency in early G1 phase results in abrogation of origin licensing and blocks the G1-to-S phase transition (Figure 8B). However, partial pre-RC insufficiency in G1 or more profound loss of pre-RC in S phase cells reduces pre-RC assembly and/or pre-RC function, thus suppressing S phase origin firing. According to our model, this reduced distribution of S phase origin firing results in abnormal elongation of progressing forks and altered chromatin structure, which is sufficient to activate the ATR-checkpoint response and inhibit DNA replication in nontransformed cells (Figure 8C). In contrast, cancer cells, which exhibit an elevated threshold for ATR-checkpoint activation, are relatively permissive for such aberrant fork progression during pre-RC insufficiency. This relative insensitivity to pre-RC insufficiency and its effects on replication renders such transformed cells more prone to undergo S-to-M phase progression with incompletely/abnormally replicated DNA and its lethal consequences. The selective cytotoxic effect of pre-RC inhibition on cancer cells suggests that the pre-RC might be an attractive target for the development of drugs that kill proliferating malignant cells but spare actively proliferating host cells. Thus, the exploitation of the differences in normal and cancer cells through the selective targeting of pre-RC proteins could lead to anticancer therapies with increased selectivity and efficacy.
Figure 8.
Schematic of cell cycle response to pre-RC depletion. Schematic of cell cycle response to pre-RC depletion is shown (for details, see text).
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Jim Borowiec, Gen-Sheng Feng, Ze'ev Ronai, and Xiaohua Wu for reagents and discussions. E. L. was supported by National Institutes of Health predoctoral training grant 2T32 CA77109-06A2. This study was supported by National Institutes of Health grants CA-97950 (to R.T.A. and W. J.) and GM-67859 (to W. J.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-01-0022) on July 8, 2009.
REFERENCES
- Abraham R. T. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Anantha R. W., Vassin V. M., Borowiec J. A. Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair. J. Biol. Chem. 2007;282:35910–35923. doi: 10.1074/jbc.M704645200. [DOI] [PubMed] [Google Scholar]
- Ayoub N., Jeyasekharan A. D., Bernal J. A., Venkitaraman A. R. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- Bailis J. M., Luche D. D., Hunter T., Forsburg S. L. Minichromosome maintenance proteins interact with checkpoint and recombination proteins to promote S-phase genome stability. Mol. Cell Biol. 2008;28:1724–1738. doi: 10.1128/MCB.01717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J., Bartkova J., Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007a;26:7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- Bartek J., Lukas J., Bartkova J. DNA damage response as an anti-cancer barrier: damage threshold and the concept of ‘conditional haploinsufficiency’. Cell Cycle. 2007b;6:2344–2347. doi: 10.4161/cc.6.19.4754. [DOI] [PubMed] [Google Scholar]
- Bartkova J., et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Blow J. J., Dutta A. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow J. J., Gillespie P. J. Replication licensing and cancer–a fatal entanglement? Nat. Rev. Cancer. 2008;8:799–806. doi: 10.1038/nrc2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell J. M., Chen Y., Schollaert K. L., Theis J. F., Babcock G. F., Newlon C. S., Sanchez Y. Orchestration of the S-phase and DNA damage checkpoint pathways by replication forks from early origins. J. Cell Biol. 2008;180:1073–1086. doi: 10.1083/jcb.200706009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich K. A., Cortez D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay-Farrace L., Pelizon C., Santamaria D., Pines J., Laskey R. A. Human replication protein Cdc6 prevents mitosis through a checkpoint mechanism that implicates Chk1. EMBO J. 2003;22:704–712. doi: 10.1093/emboj/cdg046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti C., Sacca B., Herrick J., Lalou C., Pommier Y., Bensimon A. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Mol. Biol. Cell. 2007a;18:3059–3067. doi: 10.1091/mbc.E06-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti C., Seiler J. A., Pommier Y. The mammalian DNA replication elongation checkpoint: implication of Chk1 and relationship with origin firing as determined by single DNA molecule and single cell analyses. Cell Cycle. 2007b;6:2760–2767. doi: 10.4161/cc.6.22.4932. [DOI] [PubMed] [Google Scholar]
- Cook J. G., Park C. H., Burke T. W., Leone G., DeGregori J., Engel A., Nevins J. R. Analysis of Cdc6 function in the assembly of mammalian prereplication complexes. Proc. Natl. Acad. Sci. USA. 2002;99:1347–1352. doi: 10.1073/pnas.032677499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D., Glick G., Elledge S. J. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. USA. 2004;101:10078–10083. doi: 10.1073/pnas.0403410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson I. F., Li A., Blow J. J. Deregulated replication licensing causes DNA fragmentation consistent with head-to-tail fork collision. Mol. Cell. 2006;24:433–443. doi: 10.1016/j.molcel.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova D. S., Todorov I. T., Melendy T., Gilbert D. M. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J. Cell Biol. 1999;146:709–722. doi: 10.1083/jcb.146.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D., Tu Z., Wu W., Liang C. Inhibiting the expression of DNA replication-initiation proteins induces apoptosis in human cancer cells. Cancer Res. 2003;63:7356–7364. [PubMed] [Google Scholar]
- Fersht N., Hermand D., Hayles J., Nurse P. Cdc18/CDC6 activates the Rad3-dependent checkpoint in the fission yeast. Nucleic Acids Res. 2007;35:5323–5337. doi: 10.1093/nar/gkm527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frum R. A., Chastain P. D., 2nd, Qu P., Cohen S. M., Kaufman D. G. DNA replication in early S phase pauses near newly activated origins. Cell Cycle. 2008;7:1440–1448. doi: 10.4161/cc.7.10.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez S., Klatt P., Delgado S., Conde E., Lopez-Rios F., Sanchez-Cespedes M., Mendez J., Antequera F., Serrano M. Oncogenic activity of Cdc6 through repression of the INK4/ARF locus. Nature. 2006;440:702–706. doi: 10.1038/nature04585. [DOI] [PubMed] [Google Scholar]
- Hermand D., Nurse P. Cdc18 enforces long-term maintenance of the S phase checkpoint by anchoring the Rad3-Rad26 complex to chromatin. Mol. Cell. 2007;26:553–563. doi: 10.1016/j.molcel.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Honeycutt K. A., Chen Z., Koster M. I., Miers M., Nuchtern J., Hicks J., Roop D. R., Shohet J. M. Deregulated minichromosomal maintenance protein MCM7 contributes to oncogene driven tumorigenesis. Oncogene. 2006;25:4027–4032. doi: 10.1038/sj.onc.1209435. [DOI] [PubMed] [Google Scholar]
- Jackson D. A., Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Wells N. J., Hunter T. Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc. Natl. Acad. Sci. USA. 1999;96:6193–6198. doi: 10.1073/pnas.96.11.6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. M., Kakusho N., Yamada M., Kanoh Y., Takemoto N., Masai H. Cdc7 kinase mediates Claspin phosphorylation in DNA replication checkpoint. Oncogene. 2008;27:3475–3482. doi: 10.1038/sj.onc.1210994. [DOI] [PubMed] [Google Scholar]
- Lau E., Jiang W. Is there a pre-RC checkpoint that cancer cells lack? Cell Cycle. 2006;5:1602–1606. doi: 10.4161/cc.5.15.3124. [DOI] [PubMed] [Google Scholar]
- Lau E., Tsuji T., Guo L., Lu S. H., Jiang W. The role of pre-replicative complex (pre-RC) components in oncogenesis. FASEB J. 2007;21:3786–3794. doi: 10.1096/fj.07-8900rev. [DOI] [PubMed] [Google Scholar]
- Lau E., Zhu C., Abraham R. T., Jiang W. The functional role of Cdc6 in S-G2/M in mammalian cells. EMBO Rep. 2006;7:425–430. doi: 10.1038/sj.embor.7400624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Chen J., Solessio E., Gilbert D. M. Spatial distribution and specification of mammalian replication origins during G1 phase. J. Cell Biol. 2003;161:257–266. doi: 10.1083/jcb.200211127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Stern D. F. Regulation of CHK2 by DNA-dependent protein kinase. J. Biol. Chem. 2005;280:12041–12050. doi: 10.1074/jbc.M412445200. [DOI] [PubMed] [Google Scholar]
- Lin J. J., Dutta A. ATR pathway is the primary pathway for activating G2/M checkpoint induction after re-replication. J. Biol. Chem. 2007;282:30357–30362. doi: 10.1074/jbc.M705178200. [DOI] [PubMed] [Google Scholar]
- Liu E., Lee A. Y., Chiba T., Olson E., Sun P., Wu X. The ATR-mediated S phase checkpoint prevents rereplication in mammalian cells when licensing control is disrupted. J. Cell Biol. 2007;179:643–657. doi: 10.1083/jcb.200704138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Slater D. M., Lenburg M., Nevis K., Cook J. G., Vaziri C. Replication licensing promotes cyclin D1 expression and G1 progression in untransformed human cells. Cell Cycle. 2009;8:125–136. doi: 10.4161/cc.8.1.7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciani M. G., Oehlmann M., Blow J. J. Characterization of a novel ATR-dependent, Chk1-independent, intra-S-phase checkpoint that suppresses initiation of replication in Xenopus. J. Cell Sci. 2004;117:6019–6030. doi: 10.1242/jcs.01400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick C. J., Jackson D., Diffley J. F. Visualization of altered replication dynamics after DNA damage in human cells. J. Biol. Chem. 2004;279:20067–20075. doi: 10.1074/jbc.M400022200. [DOI] [PubMed] [Google Scholar]
- Montagnoli A., Tenca P., Sola F., Carpani D., Brotherton D., Albanese C., Santocanale C. Cdc7 inhibition reveals a p53-dependent replication checkpoint that is defective in cancer cells. Cancer Res. 2004;64:7110–7116. doi: 10.1158/0008-5472.CAN-04-1547. [DOI] [PubMed] [Google Scholar]
- Murakami H., Yanow S. K., Griffiths D., Nakanishi M., Nurse P. Maintenance of replication forks and the S-phase checkpoint by Cdc18p and Orp1p. Nat. Cell Biol. 2002;4:384–388. doi: 10.1038/ncb789. [DOI] [PubMed] [Google Scholar]
- Nevis K. R., Cordeiro-Stone M., Cook J. G. Origin licensing and p53 status regulate Cdk2 activity during G(1) Cell Cycle. 2009;8:1952–1963. doi: 10.4161/cc.8.12.8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehlmann M., Score A. J., Blow J. J. The role of Cdc6 in ensuring complete genome licensing and S phase checkpoint activation. J. Cell Biol. 2004;165:181–190. doi: 10.1083/jcb.200311044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogi H., Wang C. Z., Nakai W., Kawasaki Y., Masumoto H. The role of the Saccharomyces cerevisiae Cdc7-Dbf4 complex in the replication checkpoint. Gene. 2008;414:32–40. doi: 10.1016/j.gene.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Paulsen R. D., Cimprich K. A. The ATR pathway: fine-tuning the fork. DNA Rep. 2007;6:953–966. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- Prasanth S. G., Mendez J., Prasanth K. V., Stillman B. Dynamics of pre-replication complex proteins during the cell division cycle. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004a;359:7–16. doi: 10.1098/rstb.2003.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth S. G., Prasanth K. V., Siddiqui K., Spector D. L., Stillman B. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 2004b;23:2651–2663. doi: 10.1038/sj.emboj.7600255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkaria J. N., Busby E. C., Tibbetts R. S., Roos P., Taya Y., Karnitz L. M., Abraham R. T. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- Seo J., Chung Y. S., Sharma G. G., Moon E., Burack W. R., Pandita T. K., Choi K. Cdt1 transgenic mice develop lymphoblastic lymphoma in the absence of p53. Oncogene. 2005;24:8176–8186. doi: 10.1038/sj.onc.1208881. [DOI] [PubMed] [Google Scholar]
- Shima N., Alcaraz A., Liachko I., Buske T. R., Andrews C. A., Munroe R. J., Hartford S. A., Tye B. K., Schimenti J. C. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat. Genet. 2007;39:93–98. doi: 10.1038/ng1936. [DOI] [PubMed] [Google Scholar]
- Shimada K., Pasero P., Gasser S. M. ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev. 2002;16:3236–3252. doi: 10.1101/gad.239802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shreeram S., Sparks A., Lane D. P., Blow J. J. Cell type-specific responses of human cells to inhibition of replication licensing. Oncogene. 2002;21:6624–6632. doi: 10.1038/sj.onc.1205910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits V. A., Reaper P. M., Jackson S. P. Rapid PIKK-dependent release of Chk1 from chromatin promotes the DNA checkpoint response. Curr. Biol. 2006;16:150–159. doi: 10.1016/j.cub.2005.11.066. [DOI] [PubMed] [Google Scholar]
- Tatsumi Y., Sugimoto N., Yugawa T., Narisawa-Saito M., Kiyono T., Fujita M. Deregulation of Cdt1 induces chromosomal damage without rereplication and leads to chromosomal instability. J. Cell Sci. 2006;119:3128–3140. doi: 10.1242/jcs.03031. [DOI] [PubMed] [Google Scholar]
- Teer J. K., Machida Y. J., Labit H., Novac O., Hyrien O., Marheineke K., Zannis-Hadjopoulos M., Dutta A. Proliferating human cells hypomorphic for origin recognition complex 2 and pre-replicative complex formation have a defect in p53 activation and Cdk2 kinase activation. J. Biol. Chem. 2006;281:6253–6260. doi: 10.1074/jbc.M507150200. [DOI] [PubMed] [Google Scholar]
- Tort F., Bartkova J., Sehested M., Orntoft T., Lukas J., Bartek J. Retinoblastoma pathway defects show differential ability to activate the constitutive DNA damage response in human tumorigenesis. Cancer Res. 2006;66:10258–10263. doi: 10.1158/0008-5472.CAN-06-2178. [DOI] [PubMed] [Google Scholar]
- Tsao C. C., Geisen C., Abraham R. T. Interaction between human MCM7 and Rad17 proteins is required for replication checkpoint signaling. EMBO J. 2004;23:4660–4669. doi: 10.1038/sj.emboj.7600463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji T., Ficarro S. B., Jiang W. Essential role of phosphorylation of MCM2 by Cdc7/Dbf4 in the initiation of DNA replication in mammalian cells. Mol. Biol. Cell. 2006;17:4459–4472. doi: 10.1091/mbc.E06-03-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel J. A., Dwyer B. T., Dhar S. K., Cvetic C., Walter J. C., Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- Woodward A. M., Gohler T., Luciani M. G., Oehlmann M., Ge X., Gartner A., Jackson D. A., Blow J. J. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J. Cell Biol. 2006;173:673–683. doi: 10.1083/jcb.200602108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. H., Zou L. Checkpoint and coordinated cellular responses to DNA damage. Results Probl. Cell Differ. 2006;42:65–92. [PubMed] [Google Scholar]
- Zhao H., Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Jiang W. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc. Natl. Acad. Sci. USA. 2005;102:343–348. doi: 10.1073/pnas.0408438102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.