Abstract
Adenosine 5′-triphosphate (ATP) is the major energy currency of cells and is involved in many cellular processes. However, there is no method for real-time monitoring of ATP levels inside individual living cells. To visualize ATP levels, we generated a series of fluorescence resonance energy transfer (FRET)-based indicators for ATP that were composed of the ε subunit of the bacterial FoF1-ATP synthase sandwiched by the cyan- and yellow-fluorescent proteins. The indicators, named ATeams, had apparent dissociation constants for ATP ranging from 7.4 μM to 3.3 mM. By targeting ATeams to different subcellular compartments, we unexpectedly found that ATP levels in the mitochondrial matrix of HeLa cells are significantly lower than those of cytoplasm and nucleus. We also succeeded in measuring changes in the ATP level inside single HeLa cells after treatment with inhibitors of glycolysis and/or oxidative phosphorylation, revealing that glycolysis is the major ATP-generating pathway of the cells grown in glucose-rich medium. This was also confirmed by an experiment using oligomycin A, an inhibitor of FoF1-ATP synthase. In addition, it was demonstrated that HeLa cells change ATP-generating pathway in response to changes of nutrition in the environment.
Keywords: fluorescent indicator, FRET, live imaging, oxidative phosphorylation
Adenosine 5′-triphosphate (ATP) is the ubiquitous energy currency of all living organisms. The high phosphate-transfer potential of ATP is used for many biological processes, including muscle contraction, synthesis and degradation of biological molecules, and membrane transport. In addition, it has been suggested that ATP acts as an intracellular or extracellular signaling molecule in cellular processes, such as insulin secretion (1), neurotransmission (2), cell motility (3), and organ development (4). However, it has been difficult to precisely understand how ATP controls cellular processes and how the intracellular ATP level is regulated at the single cell level, because the conventional ATP quantification methods can only provide the averaged ATP level of an ensemble of cells based on cell extract analysis. Moreover, the distribution pattern of ATP between different intracellular compartments is unclear. Several attempts have been made to monitor ATP levels real-time in individual cells; however, these methods present several problems. For example, in chemiluminescence imaging from cells expressing firefly luciferase (5), chemiluminescence by luciferase depends not only on the intracellular ATP level but also on the luciferase concentration, as well as the other substrates, oxygen, and luciferin. Moreover, pH also affects luciferase activity. Another drawback of this method is that the intracellular ATP level could be perturbed because of ATP consumption. Furthermore, the dim luminescence of luciferase requires longer exposure time for image acquisition, making real-time observation cumbersome. Other approaches include measurement of the ion channel activity (6) or conformational change (7) of the ATP-sensitive potassium channel (KATP channel). These methods are limited to measurements of regions around the plasma membrane. In addition, KATP channel expression may also affect the characteristics of cells. Recently, a fluorescent indicator of the ATP:ADP ratio was developed (8). Although this probe is suitable for monitoring the cellular energy level, it cannot be used to directly measure the ATP level.
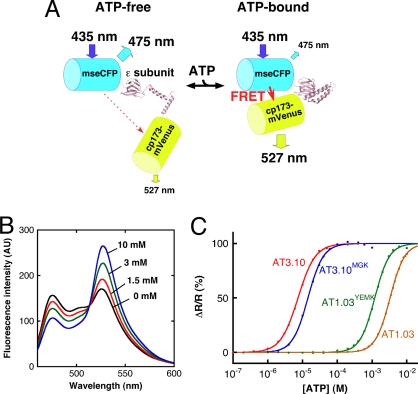
Here, we report on genetically-encoded fluorescence resonance energy transfer (FRET)-based ATP indicators that employ the ε subunit of the bacterial FoF1-ATP synthase. By using these indicators, the ATP levels in different cellular compartments and the dynamics of ATP in real-time can be monitored at the single cell level. The bacterial ε subunit is one of the smallest ATP-binding proteins (14 kDa), which is composed of one N-terminal β-barrel domain and two C-terminal α-helices (Fig. 1). The physiological role of the ε subunit is thought to be the regulation of the FoF1-ATP synthase activity depending on the intracellular ATP level (9–11). The ε subunit confers several advantages for a FRET-based ATP sensor. One is that the ε subunit binds to ATP, but does not hydrolyze it. Another advantageous feature of the ε subunit is the very high specificity for ATP over the other nucleotides; ADP, GTP, CTP, and UTP (12). Furthermore, the ε subunit undergoes a large conformational change into a folded form upon ATP binding, by bundling the two α-helices, which are relaxed in the absence of ATP (13, 14) (Fig. 2A). This large conformational change upon ATP binding may ensures a large dynamic range of the FRET signal.
Fig. 1.
Three-dimensional structure of Bacillus sp. PS3 ε subunit complexed with ATP (14). The N-terminal β-sandwich domain (residues 1–84) and C-terminal α-helical domain (residues 85–133) are colored green and magenta, respectively. ATP is represented as a cyan sphere model.
Fig. 2.
FRET-based ATP probes, ATeam. (A) Schematic drawing of AT1.03 probe. Variants of CFP (mseCFP) and YFP (cp173-mVenus) were connected by the ε subunit of Bacillus subtilis FoF1-ATP synthase. In the ATP-free form (left), extended and flexible conformations of the ε subunit separate the two fluorescent proteins, resulting in low FRET efficiency. In the ATP-bound form, the ε subunit retracts to draw the two fluorescent proteins close to each other, which increases FRET efficiency. (B) ATP-dependent fluorescence spectral change of AT1.03 in vitro. Fluorescence emission of AT1.03 proteins at various ATP concentrations ([ATP]) and at 37 °C in 50 mM Mops-KOH (pH 7.3), 50 mM KCl, 0.5 mM MgCl2, and 0.05% Triton X-100 was measured by exciting with 435 nm light. (C) ATP-dependent fluorescence emission ratio (R) changes of four ATeam proteins in vitro. The fraction of emission ratio (527/475 nm) change was plotted against [ATP]. Fluorescence of ATeam proteins was measured as in (B). Plots were fitted using a Hill equation: ΔR/R = ΔR x [ATP]n/([ATP]n + Kdn). The apparent dissociation constant (Kd) and Hill coefficient (n) were calculated to be 7.4 μM and 1.7 (AT3.10), 14 μM and 2.0 (AT3.10MGK), 1.2 mM and 2.1 (AT1.03YEMK), and 3.3 mM and 2.1 (AT1.03), respectively.
Results
Development of a Series of ATP Indicators.
We invented a FRET-based ATP indicator by genetically linking mseCFP (15)—a variant of cyan fluorescent protein (CFP)—with monomeric (A206K) Venus (mVenus) (16, 17)—a variant of yellow fluorescent protein (YFP)—each at either the N- or C terminus of the ε subunit, which was derived from Bacillus subtilis FoF1-ATP synthase. The generated indicator was termed ATeam (Adenosine 5′-Triphosphate indicator based on Epsilon subunit for Analytical Measurements). Four hydrophobic amino acid residues (Val-9, Leu-42, Phe-67, and Leu-78) forming a hydrophobic surface to interact with the FoF1 complex were replaced by hydrophilic residues, to retard incorporation into the FoF1 and reduce nonspecific hydrophobic interaction with other proteins. The dynamic range of FRET signal of the initial construct was not considerably large (data not shown). It has been demonstrated that by altering the relative orientations between donor and acceptor fluorescent proteins of a cameleon calcium indicator using a circularly permuted fluorescent protein, dynamic range of FRET signal can be efficiently enhanced (18). To improve the dynamic range of the ATeam, we substituted the mVenus part of the ATeam with several circularly permuted mVenus (18). A circularly permuted mVenus having the 173rd amino acid as its N terminus (cp173-mVenus) yielded the largest dynamic range; the construct is termed AT1.03 (Fig. 2A and Fig. S1). The FRET signal increased by ≈2.3-fold (as assessed by the emission ratio of 527/475 nm) upon the addition of ATP in vitro (Fig. 2B). The dissociation constant (Kd) of AT1.03 was determined as 3.3 mM at 37 °C from plots of the emission ratio against ATP concentration (here after [ATP]) (Fig. 2C). Thus, AT1.03 can be used for measuring intracellular ATP levels in the millimolar range. To develop an indicator with higher affinity, we constructed an ATeam employing the ε subunit of Bacillus sp. PS3, termed AT3.10 (Fig. S1), on the basis of the findings of previous studies, wherein the isolated ε subunit derived from the thermophilic Bacillus sp. PS3 was shown to exhibit ≈500-fold higher affinity to ATP than that derived from mesophilic B. subtilis (19, 20). As expected, the fluorescence emission ratio of AT3.10 changed at significantly lower [ATP] than AT1.03, and the apparent Kd of AT3.10 was determined to be 7.4 μM at 37 °C (Fig. 2C). It is unclear why the affinity between the two ε subunits differs markedly, despite the high similarity of amino acid sequences (≈70% identity) and high conservation of the amino acid residues that directly interact with ATP (Fig. S2A). We found that the 60th residue in the N-terminal β-sandwich domain and the 132nd residue in the C-terminal α-helical domain, which closely interact in the crystal structure of the ATP-bound form of Bacillus sp. PS3 ε subunit, are not conserved between the two ε subunits (Fig. S2). Residues adjacent to the 132nd residue that do not have direct interaction with ATP are also not conserved. Expecting that these residues might contribute to the affinity of the ε subunit, these residues were exchanged between AT1.03 and AT3.10 to construct AT1.03YEMK and AT3.10MGK (Fig. S1). The apparent Kd values of AT1.03YEMK and AT3.10MGK were 1.2 mM and 14 μM at 37 °C (Fig. 2C), which is approximately three-fold less and two-fold more than that of the original constructs. These results indicate that the affinity of the ε subunit is partly determined by the stability of the folded form, and that the affinity of the ATeam to ATP can be adjusted to conduct individual experiments, by simply substituting the ε subunit with that of a different bacteria and/or by introducing point mutations into the interface between the N- and C-terminal domains of the ε subunit.
Characterization of ATeam Indicators In Vitro.
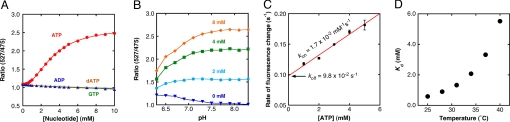
We further examined the characteristics of the purified ATeam indicators in vitro. In contrast to the effect observed on ATP addition, addition of up to 10 mM GTP or ADP had little effect on the YFP/CFP emission ratio of AT1.03 (Fig. 3A), which was consistent with previous findings (19). We also found that addition of even dATP did not increase the emission ratio of AT1.03 (Fig. 3A); rather, the emission ratio gradually decreased for unknown reasons (≈10% decrease at 10 mM ADP). AT3.10 also exhibited high selectivity for ATP: it showed 30-fold and 300-fold lower affinity to ADP (Kd = 0.23 mM) and GTP (Kd = 2.6 mM), respectively (Fig. S3), also consistent with previous studies (12, 13). The pH dependence of the FRET signal of AT1.03 was also examined. The fluorescence emission ratio was almost invariant from pH 7.1 to 8.5 (Fig. 3B), suggesting that the fluorescence signal from AT1.03 will not be affected by small pH fluctuations around normal cytoplasmic pH, which is near 7.3 (21). The rates of ATP binding (kon) and dissociation (koff) were determined to be 1.7 × 10−2 mM−1 s−1 and 9.8 × 10−2 s−1, respectively (Fig. 3C). Thus, AT1.03 can be used to monitor the dynamics of ATP changes with rates up to 0.1 s−1. Further, the temperature dependence of the affinity was investigated (Fig. 3D): the dissociation constant of AT1.03 was elevated approximately five-fold by an increase of 10 °C, close to the temperature-sensitivity of the isolated Bacillus sp. PS3 ε subunit (13).
Fig. 3.
Characterization of purified AT1.03 in vitro. (A) Nucleotide selectivity of AT1.03. The fluorescence emission ratio (527/475 nm) at 37 °C was plotted against nucleotide concentrations. Red filled circle, ATP; blue filled circle, ADP; green open square, GTP; orange open circle, dATP. Plots were fitted with Hill equations; R = (Rmax − Rmin) x [S]n/([S]n + Kdn) + Rmin, where Rmax and Rmin are the maximum and minimum fluorescence ratios, respectively, Kd is the apparent dissociation constant, and n is a Hill coefficient. (B) pH dependence of AT1.03. The fluorescence ratios (527/475 nm) at 37 °C at 0, 2, 4, and 8 mM ATP in the pH range of 6.3–8.3 are shown. The buffer contained 50 mM Mops-KOH (pH 6.3–7.5) or Hepes-KOH (pH 7.7–8.3), 50 mM potassium chloride, 0.5 mM magnesium chloride, and 0.05% Triton X-100. (C) Reaction rate constants of AT1.03. Apparent rate constants (kapp = kon[ATP] + koff) at 37 °C, which were determined by fitting the CFP fluorescence decrease after ATP addition with a single exponential equation, were plotted against ATP concentrations ([ATP]). From a linear fit to the plot, kon and koff were calculated as 1.7 × 10−2 mM−1s−1 and 9.8 × 10−2 s−1, respectively. (D) Dependence of Kd on temperature. Kd values for ATP were measured as in (A) at 25, 28, 31, 34, 37, and 40 °C.
Comparison of ATP Levels in Different Subcellular Compartments.
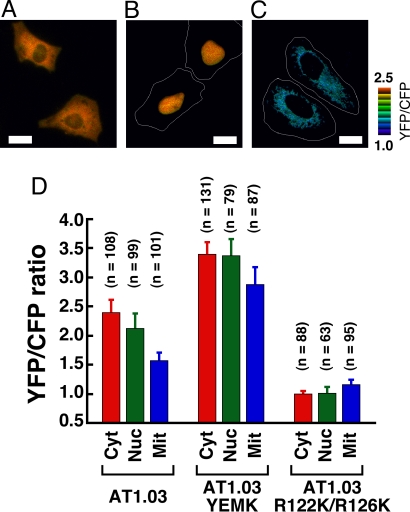
Next, we visualized the ATP levels inside subcellular compartments of single living HeLa cells expressing AT1.03 for the investigation of ATP distribution among cellular compartments. When AT1.03 without a targeting sequence was expressed in HeLa cells, it was mainly distributed in the cytoplasm, and slightly in the nucleus (Fig. 4A). For the targeting ATeam into the nucleus or mitochondrial matrix, a triplet of the nuclear localization signal of SV40 large T-antigen or a duplex of the mitochondrial targeting signal of cytochrome c oxidase subunit VIII were each fused to the N terminus of AT1.03 (nucAT1.03 and mitAT1.03; Fig. S1). These indicators were properly localized to the desired cellular compartments (Fig. 4 B and C). The YFP/CFP emission ratio was calculated from CFP and YFP fluorescence images of the individual cells, which were recorded using a wide-field fluorescence microscope. We found that ATeam localized in the mitochondrial matrix showed a significantly lower YFP/CFP emission ratio than that in the cytoplasm or nucleus (Fig. 4). The nucleus also showed a lower YFP/CFP emission ratio than the cytoplasm, but to a smaller extent (Fig. 4D), suggesting that ATP can pass through nuclear pores almost freely. The mitochondrial targeting sequence will not affect the fluorescence ratio, because as demonstrated for the cameleon calcium indicator, the targeting sequence is cleaved after transport into mitochondria (22). Higher mitochondrial pH does not account for the lower YFP/CFP emission ratio, because the fluorescence of AT1.03 is almost invariant at pH conditions >7.3 (Fig. 3B). To confirm that the low YFP/CFP emission ratio in mitochondria is not due to some environmental effects but ATP level, HeLa cells expressing derivatives of AT1.03 with different affinity to ATP were also investigated. First, the same measurements were carried out using AT1.03YEMK, which has a higher affinity as described above. Although the YFP/CFP emission ratio of mitochondria was lower than that of cytoplasm, their difference (16%) was much smaller than that (34%) observed with AT1.03 (Fig. 4D). This means that the FRET signal of cytoplasm is almost saturated. The nucleus showed the same ratio as cytoplasm also due to the saturation of the FRET signal. Then, we used another type of ATeam, which was designed not to response to ATP. Structural studies revealed that two arginine residues in the position of 122 and 126 of the ε subunit have a direct contact with the α and β phosphates of ATP (Fig. S4A), suggesting their crucial roles on the ATP recognition. A recent biochemical study showed that the substitution of these residues with other ones leads drastic decrement of the affinity to ATP (20). We constructed the AT1.03R122K/R126K mutant by substituting the arginine residues with lysine residues. This mutant ATeam did not respond even to 10 mM ATP (Fig. S4B). Then, HeLa cells were observed with AT1.03R122K/R126K. As expected, the cytoplasm, nucleus and mitochondria showed essentially the same level of the YFP/CFP emission ratio (Fig. 4D). Thus, it was confirmed that the ATP level in mitochondria is lower than that of cytoplasm or nucleus. However, the lower [ATP] level in mitochondria may seem paradoxical when we consider that mitochondria are the major ATP-producing organelles of cells. One possible explanation is that adenine nucleotide translocator pumps ATP from the mitochondria, thereby maintaining a high ADP/ATP ratio; this seems preferable for FoF1-ATP synthase to produce ATP from ADP and phosphate. One might point out that the low dependence on oxidative phosphorylation (OXPHOS) for ATP production of cancer cells like HeLa cells (23) (also see below) causes the low [ATP] in mitochondria. However, this possibility is unlikely, because we did not observe clear changes of both cytoplasmic and mitochondrial ATP level in HeLa cells grown in galactose medium (data not shown), whose major ATP-generating pathway is OXPHOS (see below). A lower [ATP] in mitochondria was also observed in noncancer NIH 3T3 cells (data not shown). For the generality and the mechanism of this phenomenon, further investigations remain to be done.
Fig. 4.
Comparison of ATP concentrations ([ATP]) between different cellular compartments. (A–C) Expression of ATeam in different cellular compartments. Ratiometric pseudocolor images of AT1.03 expressed in cytoplasm (A), nucleus (B), and mitochondria (C) of HeLa cells. White lines represent cell outline. (Scale bar, 20 μm.) (D) Comparison of YFP/CFP emission ratio of ATeams in different cellular compartments. Three ATeams (AT1.03, AT1.03YEMK, AT1.03R122K/R126K), which have different affinity to ATP, were expressed in cytoplasm, nucleus, or mitochondria of HeLa cells. The ratio was calculated from fluorescent images. The numbers of cells used for calculating the ratio are indicated. Error bars are standard deviation of ratios.
Real-Time Monitoring of Intracellular ATP Depletion.
We next monitored the dynamics of ATP levels in living HeLa cells after the addition of 2-deoxyglucose (2DG) and potassium cyanide (KCN), which are inhibitors of glycolysis and OXPHOS, respectively. Fig. 5A shows sequential images of the CFP and YFP channels. Addition of the inhibitors induced an increase in CFP intensity and a decrease in YFP intensity, which rapidly decreased the YFP/CFP emission ratio (i.e., decrease of [ATP]; Fig. 5 A and C). Interestingly, the cytoplasmic [ATP] partially recovered after the transient decay. This partial [ATP] recovery would be the result of a cellular response to a large decrease in intracellular [ATP]. Addition of 2DG alone also induced a decrease of [ATP], although the drop rate was much slower than when the two inhibitors were added. In addition, recovery of [ATP] was hardly observed. In contrast to 2DG, KCN itself had almost no effect on the intracellular ATP level. The result is consistent with the fact that ATP synthesis in actively proliferating cancer cells is mainly dependent on glycolysis, rather than OXPHOS in mitochondria, even in the aerobic environment (23). Our result showed that 2DG and KCN have a synergistic rather than additive effect on the intracellular ATP level, thereby suggesting that the intracellular ATP level is synergistically regulated by both glycolysis and OXPHOS. It has been shown that >80% of uptaken glucose is metabolized through glycolysis, when HeLa cells are grown in medium containing glucose (24). In contrast, when galactose is used in medium instead of glucose, most of galactose is metabolized through pentose phosphate pathway, and contribution of glutamine to generation of ATP through citrate cycle drastically increases (24). In addition, HeLa cells grown in galactose show elevated mitochondrial respiration rate and increased synthesis of mitochondrial proteins, as compared to those grown in glucose (25). Therefore, it is thought that ATP generation mainly depends on OXPHOS in the cells grown in galacotose medium. To confirm this, we compared changes of cytoplasmic [ATP] between cells grown in glucose and galactose in response to inhibition of FoF1-ATP synthase by oligomycin A. When oligomycin A was added to the cells grown in glucose medium, apparent change in cytoplasmic ATP level was not observed (Fig. 6A). This is consistent with the above result that OXPHOS inhibition by KCN did not affect cytoplasmic [ATP] (Fig. 5D). On the other hand, cytoplasmic [ATP] of HeLa cells grown in galactose was rapidly depleted by the addition of oligomycin A. Cells did not show partial recovery of [ATP] in this experiment, unlike those grown in glucose medium and treated with 2DG and KCN (Fig. 5 C and D). These data clearly show that HeLa cells growing in galactose medium generate most of cellular ATP using OXPHOS, whereas those growing in glucose use glycolysis for ATP production. This flexible alternation of energy metabolism seems to be advantageous for the cells to propagate in glucose-limiting conditions.
Fig. 5.
Monitoring of cytoplasmic ATP levels of HeLa cells. (A) Sequential wide-field images of YFP (top), CFP (middle) and YFP/CFP emission ratio (bottom, pseudocolored) of a HeLa cell expressing AT1.03. Inhibitors of glycolysis (10 mM 2-deoxyglucose [2DG]) and OXPHOS (1 mM potassium cyanide [KCN]) were added at time = 0 (min). Elapsed time (in minutes) after addition of the inhibitors is shown to the top left of the cells. Images were obtained at 37 °C. (Scale bar, 20 μm). (B) Time course of fluorescence intensity of CFP (blue) and YFP (green) inside ROI1 (solid line) and ROI2 (dashed line) depicted in the top-left image of (A). (C) Time course of YFP/CFP emission ratio inside ROI1 (red solid line) and ROI2 (blue dashed line) depicted in the top-left image of (A). (D) Time course of averaged YFP/CFP emission ratio of HeLa cells expressing AT1.03. 2DG and/or potassium azide was added at time = 0 (min). Red, 2DG and KCN (n = 5); green, 2DG (n = 6); blue, KCN (n = 4). Error bars are standard deviations between measurements.
Fig. 6.
Nutrient dependent alternation of ATP-generating pathway of HeLa cells. Sensitivity of intracellular ATP level to an OXPHOS inhibitor, oligomycin A, was examined for HeLa cells grown in glucose (A) or galactose (B) medium. Time courses of YFP/CFP emission ratio of cells expressing AT1.03, which were grown in DMEM without pyruvate containing 10 mM glucose (A) or 10 mM galactose (B), were monitored as in Fig. 5. Oligomycin A (10 μg/ml) was added to the medium at a time indicated by an arrow head.
Discussion
In this study, we report a method to measure intracellular ATP levels by using genetically-encoded FRET-based indicators for ATP, ATeams. These probes show high selectivity to ATP over other nucleotides. The affinity of ATeam to ATP is tunable by substituting the ε subunit with that from different species or changing the amino acid residues at the domain interface. In principle, it is possible to measure ATP levels at levels ranging from 2 μM to 8 mM by modulating the affinity of ATeam. Organelle-specific ATP level monitoring was also shown to be possible by targeting the probe to the nucleus and mitochondria. This method showed that the mitochondrial ATP level is unexpectedly lower than that in the cytoplasm. ATeam is potentially able to monitor any desired intracellular compartments by attaching a proper signal sequence or by fusion to a localized protein. ATeams can be used in broad fields of biological research. For example, ATeam might become a strong tool to study energy metabolism and mitochondrial function. In addition, estimation of intracellular ATP levels by ATeams might be a good way to evaluate cell viability or toxicity of chemicals, because dead cells lose intracellular ATP.
Materials and Methods
Chemicals.
DNA polymerase and DNA ligation kits were purchased from TaKaRa; oligonucleotides were from Hokkaido System Science; restriction endonucleases were from Roche Diagnostics; ATP, ADP, GTP, and dATP were from Sigma; and other chemical reagents were from Wako Pure Chemicals, unless otherwise noted. ATP and other nucleotides were complexed with equimolar magnesium chloride (MgCl2) before use in the experiments; therefore, the term ATP in this study means MgATP.
Gene Construction.
B. subtilis ε subunit cDNA with humanized codons was synthesized by Hokkaido System Science. The synthesized cDNA also has V9T, V42K, F67N, and Leu78T mutations to disrupt a hydrophobic patch for interaction with the γ subunit of ATP synthase. The cDNA of mseCFPΔC11, the ε subunit, and cp173-mVenus was amplified by PCR (PCR) and then ligated. Ligated DNA was then ligated to XhoI-HindIII sites of the pRSET-B vector (Invitrogen) to obtain pRSET-AT1.03 for expression of AT1.03 in Escherichia coli, and to XhoI–HindIII sites of pcDNA3.1(−) (Invitrogen) to obtain pcDNA-AT1.03 for mammalian expression. pRSET-AT3.10, which is for expression of AT3.10 in E. coli, was constructed by replacing the mseCFPΔC11 part of pRSET-AT1.03 with mseCFPΔC10, and the B. subtilis ε part with Bacillus sp. PS3 ε, which has V9T, V42K, F67N and L78N mutations. PCR-based mutagenesis was used to construct pRSET-AT1.03YEMK and pRSET-AT3.10MGK from pRSET-AT1.03 and pRSET-AT3.10, respectively.
Purification of ATeam.
E. coli strain JM109(DE3) carrying ATeam plasmid was cultured in LB medium at 37 °C for 3 h. Protein expression was then induced by adding 10 μM isopropyl-β-thiogalactopyranoside, followed by overnight incubation at 24 °C. Cells were collected by centrifugation, suspended in buffer A (100 mM sodium phosphate [pH 8.0], 200 mM sodium chloride, and 10 mM imidazole), and then disrupted by sonication. Cell-free extract, obtained by centrifugation at 10,000 × g for 60 min, was applied to a Ni-NTA column (Qiagen) preequilibrated with buffer A. After washing the column with buffer A, the protein was eluted by increasing the imidazole concentration to 200 mM. Fractions containing ATeam were concentrated, followed by application to a Superdex200 gel-filtration column (GE Healthcare) preequilibrated with 20 mM Tris·HCl (pH 8.0) and 150 mM sodium chloride. After adding glycerol to a final concentration of 20%, the purified ATeam protein was stored at −20 °C for further use.
Characterization of ATeam In Vitro.
Protein concentration was determined using light absorption at 515 nm and the extinction coefficient of Venus protein (ε = 92 mM−1cm−1)16. The fluorescence of purified ATeam proteins was investigated in buffer B (50 mM Mops-KOH [pH 7.3], 50 mM KCl, 0.5 mM MgCl2, and 0.05% Triton X-100) using an FP-6500 spectrofluorometer (Jasco) at 37 °C. To obtain the fluorescence spectra, CFP was excited with 435 ± 20 nm light, and emission from 460 to 600 nm was scanned. To measure the time course of CFP emission change, CFP was excited using 435 ± 5 nm light and emission at 475 ± 10 nm was monitored by adding ATP at specific time points.
Cell Culture and Microscopy.
HeLa cells were obtained from JCRB, and maintained in Dulbecco's modified Eagle's medium (DMEM, Sigma) supplemented with 10% FBS (Invitrogen). For glucose-free culture, cells were maintained in DMEM, which contains 10 mM galactose instead of glucose and is supplemented with 10% dialyzed FBS (Invitrogen). Cells were plated on a collagen-coated glass-bottom dish (FPI) and transfected with plasmid coding ATeam cDNA by using FuGENE6 transfection reagent (Roche Diagnostics). Between one and four days after transfection, cells cultured in phenol red-free DMEM were subjected to imaging. Wide-field observations of the cells were performed on a Nikon TE2000-PFS inverted microscope (Nikon Instruments) using a PlanApo VC60, 1.40 numerical aperture (NA), oil-immersion objective (Nikon). Filters used for dual-emission ratio imaging of ATeam were purchased from Semrock: an FF01–427/10 excitation filter, an FF458-Di01 dichroic mirror, and two emission filters (FF01–483/32 for CFP and FF01–542/27 for YFP). The two emission filters were alternated by using a filter changer (Ludl Electronic Products Ltd). Cells were illuminated using a 75-W xenon lamp through 12.5% and 25% neutral density filters. Fluorescence emission from ATeam was imaged by using a cooled charge-coupled device (CCD) camera (ORCA-AG; Hamamatsu Photonics); the exposure times were 500 ms for CFP and YFP images. Cells were maintained on a microscope at 37 °C with a continuous supply of a 95% air and 5% carbon dioxide mixture by using a stage-top incubator (Tokai Hit). Image analysis was performed using MetaMorph (Molecular Devices). The YFP/CFP emission ratio was calculated by dividing pixel-by-pixel a YFP image with a CFP image.
Supplementary Material
Acknowledgments.
We thank Tsuyoshi Nishi and Akihito Yamaguchi for providing us with the equipment for cell culture, Makoto Fujikawa for suggestion for culturing cell in galactose medium, Hiromasa Yagi and Hideo Akutsu for sharing the 3D coordinate crystal structure of ε subunit complexed with ATP before publishing, and Shigeko Kawai-Noma and Hideki Taguchi for plasmids for W1B mutants of CFP and YFP, which were used in the very early stage of this study. We are also indebted to all of the members of the Noji Laboratory for valuable discussions and advice. This work was supported by Grant-in-Aid for Scientific Research 18074005 and 18201025 (to H.N.) and Post-Silicon Materials and Devices Research Alliance (to T.N. and H.N.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and Precursory Research for Embryonic Science (to H.I.) from Japan Science and Technology Agency.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904764106/DCSupplemental.
References
- 1.Ashcroft FM. ATP-sensitive potassium channelopathies: Focus on insulin secretion. J Clin Invest. 2005;115:2047–2058. doi: 10.1172/JCI25495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finger TE, et al. ATP signaling is crucial for communication from taste buds to gustatory nerves. Science. 2005;310:1495–1499. doi: 10.1126/science.1118435. [DOI] [PubMed] [Google Scholar]
- 3.Davalos D, et al. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 2005;8:752–758. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- 4.Masse K, Bhamra S, Eason R, Dale N, Jones EA. Purine-mediated signalling triggers eye development. Nature. 2007;449:1058–1062. doi: 10.1038/nature06189. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy HJ, et al. Glucose generates sub-plasma membrane ATP microdomains in single islet β-cells. Potential role for strategically located mitochondria. J Biol Chem. 1999;274:13281–13291. doi: 10.1074/jbc.274.19.13281. [DOI] [PubMed] [Google Scholar]
- 6.Gribble FM, et al. A novel method for measurement of submembrane ATP concentration. J Biol Chem. 2000;275:30046–30049. doi: 10.1074/jbc.M001010200. [DOI] [PubMed] [Google Scholar]
- 7.Tsuboi T, Lippiat JD, Ashcroft FM, Rutter GA. ATP-dependent interaction of the cytosolic domains of the inwardly rectifying K+ channel Kir6.2 revealed by fluorescence resonance energy transfer. Proc Natl Acad Sci USA. 2004;101:76–81. doi: 10.1073/pnas.0306347101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berg J, Hung YP, Yellen G. A genetically encoded fluorescent reporter of ATP:ADP ratio. Nat Methods. 2009;6:161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato-Yamada Y, et al. ε subunit, an endogenous inhibitor of bacterial F1-ATPase, also inhibits FoF1-ATPase. J Biol Chem. 1999;274:33991–33994. doi: 10.1074/jbc.274.48.33991. [DOI] [PubMed] [Google Scholar]
- 10.Rondelez Y, et al. Highly coupled ATP synthesis by F1-ATPase single molecules. Nature. 2005;433:773–777. doi: 10.1038/nature03277. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki T, et al. FoF1-ATPase/synthase is geared to the synthesis mode by conformational rearrangement of ε subunit in response to proton motive force and ADP/ATP balance. J Biol Chem. 2003;278:46840–46846. doi: 10.1074/jbc.M307165200. [DOI] [PubMed] [Google Scholar]
- 12.Kato-Yamada Y, Yoshida M. Isolated ε subunit of thermophilic F1-ATPase binds ATP. J Biol Chem. 2003;278:36013–36016. doi: 10.1074/jbc.M306140200. [DOI] [PubMed] [Google Scholar]
- 13.Iino R, et al. Real-time monitoring of conformational dynamics of the ε subunit in F1-ATPase. J Biol Chem. 2005;280:40130–40134. doi: 10.1074/jbc.M506160200. [DOI] [PubMed] [Google Scholar]
- 14.Yagi H, et al. Structures of the thermophilic F1-ATPase ε subunit suggesting ATP-regulated arm motion of its C-terminal domain in F1. Proc Natl Acad Sci USA. 2007;104:11233–11238. doi: 10.1073/pnas.0701045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuda T, Miyawaki A, Nagai T. Direct measurement of protein dynamics inside cells using a rationally designed photoconvertible protein. Nat Methods. 2008;5:339–345. doi: 10.1038/nmeth.1193. [DOI] [PubMed] [Google Scholar]
- 16.Nagai T, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 17.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 18.Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci USA. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato-Yamada Y. Isolated ε subunit of Bacillus subtilis F1-ATPase binds ATP. FEBS Lett. 2005;579:6875–6878. doi: 10.1016/j.febslet.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 20.Kato S, Yoshida M, Kato-Yamada Y. Role of the ε subunit of thermophilic F1-ATPase as a sensor for ATP. J Biol Chem. 2007;282:37618–37623. doi: 10.1074/jbc.M707509200. [DOI] [PubMed] [Google Scholar]
- 21.Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc Natl Acad Sci USA. 1998;95:6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filippin L, et al. Improved strategies for the delivery of GFP-based Ca2+ sensors into the mitochondrial matrix. Cell Calcium. 2005;37:129–136. doi: 10.1016/j.ceca.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 24.Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem. 1979;254:2669–2676. [PubMed] [Google Scholar]
- 25.Rossignol R, et al. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004;64:985–993. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.