Abstract
We designed a high-density mouse genotyping array containing 623,124 SNPs that capture the known genetic variation present in the laboratory mouse. The array also contains 916,269 invariant genomic probes that are targeted to functional elements and regions known to harbor segmental duplications. The array opens the door to the characterization of genetic diversity, copy number variation, allele specific gene expression and DNA methylation and will extend the successes of human genome-wide association studies to the mouse.
Array based hybridization platforms allow simultaneous genotyping of many single nucleotide polymorphisms (SNPs). Among these platforms, whole genome sampling analysis1, which reduces genomic complexity by selective amplification of restriction fragments, has been used to genotype large human cohorts to conduct genome wide association studies for a variety of human diseases. These studies have been highly successful at identifying loci associated with certain diseases.
The laboratory mouse with its fully sequenced and annotated genome, targeted germline modification, and many inbred strains, is a popular tool in biomedical research that complements the strengths of human studies. Mice and humans are eutherian mammals sharing genomes of similar size, content and organization. The mouse has been widely used as a model of human disease and to characterize basic biological processes. However, current microarray-based genotyping platforms are limited to a few thousand markers2 or have not been available for wide use3. To overcome these limitations and to enable the potential for genome wide association (GWA) studies in the mouse, we have developed a high-density SNP array.
The Mouse Diversity array was designed to capture the full spectrum of genetic diversity present in current stocks of laboratory mice, including classical and wild derived inbred strains. Classical strains have been the most popular tools in mouse genetics and have contributed disproportionately to the genotypes stored in databases. Genotyping platforms based on SNPs discovered by resequencing of classical strains, however, have shortcomings. They lack SNPs that discriminate among subspecies and SNPs that tag variation within subspecies other than M. m. domesticus, because, contrary to a previous hypothesis4 the genome of classical strains includes only limited representation of subspecies other that M. m. domesticus5. Furthermore, a significant fraction of the genome is identical by descent among the classical laboratory strains5.
Biases due to SNP ascertainment plague all existing microarrays and databases6. The methods used for SNP discovery and selection determine the type of variation that can be observed in subsequent genotyping experiments. This can result in distorted allele frequencies and inaccurate phylogenies if ascertainment is not taken into account. Thus, future analysis and interpretation of array data requires documentation of ascertainment strategies used to select SNPs. Previous SNP discovery projects that included wild-derived strains, such as a project from NIEHS7, are critical to the design of a genotyping array that will be useful for studies that include wild derived strains or wild-caught mice including the Collaborative Cross (CC)8.
We employed several complementary strategies to select SNPs, giving rise to distinct categories of SNP probes (Supplementary Fig.1 and Online Methods). Our aim was to capture maximum diversity among the phylogenetic clades represented in laboratory mouse stocks while retaining the ability to discriminate among commonly used classical inbred strains. SNPs within each category have a uniform spatial distribution and most selection strategies were iterated to ensure depth and redundancy. Only SNPs that are “chippable” according to our criteria (see Online Methods) were selected and SNPs supported by multiple public data sets were preferred. We initially selected 725,086 SNPs that were printed on test arrays. From these, we selected 623,124 well performing SNPs for the final array. The numbers of SNPs selected within each category are summarized in Table 1 and further information is provided in Supplementary Table 1.
Table 1.
Final SNP and invariant genomic sequence selection.
| Selection Criteria | Type of Variant | Training Array | Final Array | Probes in + strand | Probes in − strand | Total number of probes | Unmapped Probes | Total number of probes (Build 37) |
|---|---|---|---|---|---|---|---|---|
| Paigen Set | SNP | 187,256 | 157,653 | 4 | 4 | 1,261,224 | 0 | 1,261,224 |
| B6 Singletons | SNP | 3,376 | 2,773 | 4 | 4 | 22,184 | 3 | 22,160 |
| MSM Singletons | SNP | 49,930 | 37,597 | 4 | 4 | 300,776 | 4 | 300,744 |
| NIEHS/Perlegen | SNP | 482,862 | 423,893 | 4 | 4 | 3,391,144 | 14 | 3,391,032 |
| extra | SNP | 569 | 339 | 4 | 4 | 2,712 | 7 | 2,656 |
| Wild-derived | SNP | 990 | 799 | 4 | 4 | 6,392 | 0 | 6,392 |
| chrY | SNP | 83 | 51 | 4 | 4 | 408 | 0 | 408 |
| chrM | SNP | 20 | 19 | 4 | 4 | 152 | 0 | 152 |
| SubTotal | SNP | 725,086 | 623,124 | 4,984,992 | 28 | 4,984,768 | ||
| Exon 1 | CNV | na | 373,667 | 1 | 1 | 747,334 | 14 | 747,306 |
| Exon 2 | CNV | na | 224,091 | 1 | 1 | 448,182 | 6 | 448,170 |
| UCEs | CNV | na | 34,739 | 1 | 1 | 69,478 | 0 | 69,478 |
| MSM Zero Hit | CNV | na | 94,471 | 1 | 1 | 188,942 | 0 | 188,942 |
| MSM Single Hit | CNV | na | 19,399 | 1 | 1 | 38,798 | 0 | 38,798 |
| Gap Filling Nsp | CNV | na | 87,145 | 1 | 1 | 174,290 | 471 | 173,348 |
| Gap Filling Sty | CNV | na | 82,757 | 1 | 1 | 165,514 | 355 | 164,804 |
| SubTotal | CNV | na | 916,269 | 1,832,538 | 846 | 1,830,846 | ||
| Total | na | 725,086 | 1,539,393 | na | na | 6,817,530 | na | 6,815,614 |
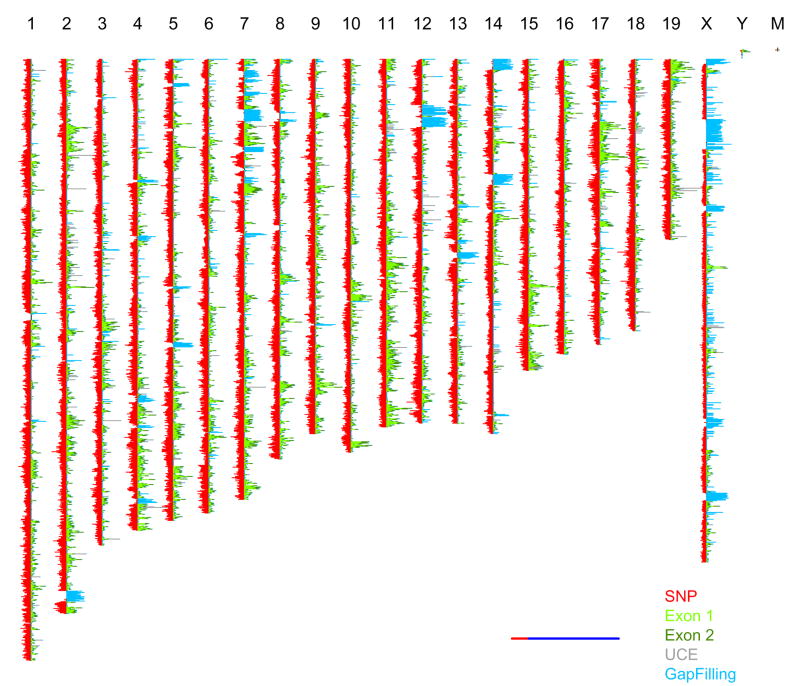
For the final array we also selected 916,269unique invariant genomic probes (IGPs)to tag functional elements of the genome and putative structural differences (Supplementary Fig.1, Table 1). These probes are devoid of known SNPs. IGPs were selected to capture 93.4% of the 214,869 exons defined in Ensemble (version 49). For 125,390 exons (58.4%), we identified three chippable probes (Exon 1). For 75,204 exons (35.0%), we selected three probes regardless of chippable status (Exon 2). We tiled probes across 238 chippable ultra-conserved elements. In regions of the mouse genome known to harbor segmental duplications, most selected probes failed to meet the uniqueness criteria. We relaxed this constraint and selected three probes on each chippable NspI and StyI fragment in these regions (Gap Filling probes in Supplementary Fig.1). Lastly, we attempted to identify probes for sequences that are not present in the C57BL/6J reference genome using BAC-end sequences from the MSM/Ms strain. IGPs were printed as complementary pairs, one on each strand of DNA. Spatial distributions of SNPs and IGPs are shown in Figure 1, and the distances between consecutive probes are shown in Supplementary Fig.2 (online).
Figure 1. Spatial distribution of SNP and invariant genomic probes.
Number of SNP and IGPs in each 200Kb interval. IGPs are color coded by type (Exon1, Exon2, UCE and Gap Filling). Invariant MSM probes are not shown. The left and right histograms have different scales as indicated by the relative proportions of red and blue in the horizontal bar (lower right).
To train the SNP calling algorithm and to establish the performance of the Mouse Diversity array we analyzed 136 DNA samples. A complete list of strains and additional resources can be found in Online Methods. We have removed 41,452 SNPs (6.65%) with unreliable genotype calls. (Supplementary Fig. 3). Performance among the remaining 581,672 SNPs was assessed based on call rate, concordance with known genotypes, concordance rate with test array, concordance between biological replicates and heterozygosity rate(Supplementary Table 2). Among classical inbred strains there is good concordance with genotypes reported in databases (99.6% of genotypes called in both resources) with little variation between strains. There is also good concordance between the training and final arrays (mean 99.9%, interquartile range(IQR),99.8%–99.9%). In addition, concordance across biological replicates is good (mean 99.2%, IQR 97.6%–100%). The F1 hybrid genotypes provide an additional consistency check, as these are predictable from the parental strain genotypes (mean 99.1%, IQR 98.8%–99.3%). Finally, the array performed extremely well with 99.6% average call rate (IQR 99.4%–99.7%) and 1.3% heterozygosity call rate (IQR 0.9%–1.6%).
Heterozygous calls are a useful indicator of performance in inbred strains, since these samples should not have any heterozygosity. Heterozygous genotype calls can reflect true heterozygosity at the relevant SNP due to incomplete inbreeding or homoplasy. We have confirmed the ability of the Mouse Diversity array to detect and fine map these regions in an inbred strain, SSL/LeJ, known to be segregating two historical mutations of biomedical interest, piebald and piebald-lethal, at the Ednrb locus (Supplementary Fig. 4). However, given the fact that we have filtered poorly performing SNPs and that heterozygosity rate increases with phylogenetic distance from the C57BL/6J genome (i.e, M.m. domesticus) we suspect that most of the apparent heterozygote calls are due to the presence of additional off-target variation within the probes.
The NIEHS strain7 set includes four wild derived strains WSB/EiJ, PWD/PhJ, CAST/EiJ, and MOLF/EiJ representing M. m. domesticus, M. m. musculus, M. m. castaneus and M. m. molossinus subspecies, respectively. Among these strains WSB/EiJ has the highest call and concordance rates and the lowest heterozygosity rate. Genotyping performance decreases for PWD/PhJ, MOLF/EiJ and CAST/EiJ as the genetic divergence from the C57BL/6J strain increases. The C57BL/6J genome is 92% of M. m. domesticus origin, and classical laboratory mice have a similar degree of M. m. domesticus 5, explaining the performance difference between M. m. domesticus and other subspecies. We also genotyped SPRET/EiJ and PANCEVO/EiJ, strains from M. spretus and M. spicilegus species, respectively, for which call rates are substantially lower (92.4% and 92.1%), and heterozygosity rates are higher (14.8% and 12.4%, respectively). Most of the observed heterozygosity is likely due to off-target variation within probe sequences. Concordance rate with known genotypes remains high (98.6%) indicating that the array works well in other Mus species. The array performs best for classical laboratory or wild derived strains from M. m. domesticus subspecies followed by strains from other Mus subspecies reflecting the degree of genetic divergence from C57BL/6J.
To assess the performance of IGPs we compared the distribution of the hybridization intensities of each type of IGP with the distribution of the perfect match SNP probes within a given DNA sample (Supplementary Fig.5a). The latter distribution provides a standard intensity that should be matched by the IGP. This expectation is fulfilled by the Exon 1 probes. Exon 2 probes show a distribution shifted towards lower intensities due to the larger length of the PCR amplicons (Exon1 median 854bp, Exon2 median 1,955bp). Lower intensity observed for UCE probes can be explained by their low G+C content. The gap-filling probes show a much wider range of intensity as expected for probes in segmental duplications. The performance of both SNP and IGPs benefits from normalization that accounts for G+C content of the probe and the lengths of the NspI and StyI fragments on which probes reside(Supplementary Fig.5b). Gap filling probes should be excluded from estimation of normalization parameters as they are highly enriched for repeated sequences, and thus present a broad distribution of intensity values. Finally, the majority of MSM/Ms IGPs have very low intensity (similar to background noise) because these probes were selected to be absent from the C57BL/6J reference genome. MSM/Ms IGPs cannot be normalized because we lack information on NspI and StyI fragments on which each probelies.
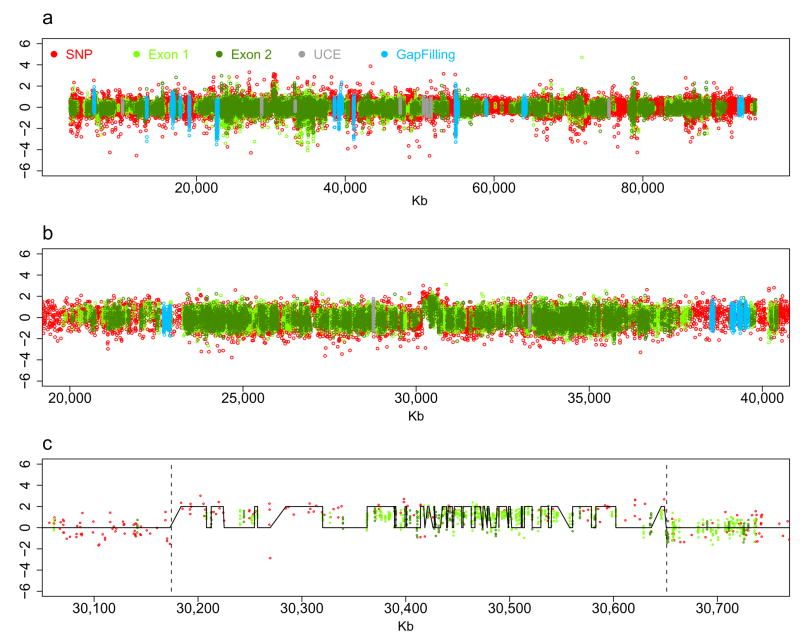
To assess the performance of the array to detect copy number variation we compared the intensity of SNP and IGP probes on chromosome 17 between C57BL/6J and BALB/cByJ (Fig.2). The Mouse Diversity array detects the presence of a 475kb duplication described previously in the BALB/cBy genome9. We further analyzed 80 previously reported CNVs using our array10. Among them 53 intervals can be mapped to Build 36, and we confirmed 83% (44) of these CNV using our array.
Figure 2. Detection of CNV using the Mouse Diversity array.
Open circles of different colors represent different types of probesets. (a) Difference in probeset intensity along the entire chromosome 17 between BALB/cByJ and C57BL/6J strains. (b) Z-score of the intensity of each probe set in BALB/cBy in a 23 Mb window of chromosome 17. (c) Z-score of the intensity of each probe set in BALB/cBy in a 700kb window that span a know duplication on BALB/cBy. Red lines are from HMM fit and black vertical lines denote the boundaries of the duplication described previously9.
In our analyses, we have used the BRLMM-P algorithm, originally developed for human genotyping arrays11. It assumes that genotypes will form three clusters, whereas inbred strains will generally have two distinct genotypes. We circumvented this problem by including multipleF1 samples in our training set to ensure the presence of sufficient numbers of heterozygous genotypes. Sex chromosomes presented additional challenges. For this study we constructed specialized R scripts to analyze Y chromosome. The X chromosome requires that male and female samples be distinguished. With this information BRLMM-P calls are reasonably good, but modifications will be needed to achieve the same performance levels as for autosomal loci. Performance statistics reported here do not include X, Y or mitochondrial SNPs.
The Mouse Diversity array will allow the comprehensive analysis of the origin of all mouse resources, detection of residual heterozygosity, contamination and drift in strains and cell lines, and de novo copy number variation arising in mouse strain resources and in somatic tissues such as tumor samples.
We have previously shown that reliable imputation is possible provided that density of typed SNPs is sufficiently high12. Genotyping of most inbred stocks with the Mouse Diversity array will immediately allow imputation of these strains and increase the reliability of imputed genotypes. Projects are underway to obtain complete sequences of 17 inbred mouse strains including several wild derived strains. This should substantially extend the range of strains for which reliable imputation can be achieved. We envision that Mouse Diversity array genotyping of the CC recombinant inbred strains together with the genome sequences of the parental strains will enable us to impute the complete genome sequences of the CC strains with high accuracy.
The high density of SNP information will enable genome-wide association studies in both advanced crosses and in wild mouse populations. Given the nature of the probes included in the array and their extensive annotation, we expect that novel uses such as allele specific gene expression and DNA methylation13–15 will become routine applications for this array. The Mouse Diversity Array is distributed by Affymetrix (Santa Clara, CA).
Online METHODS
We implemented 11 selection strategies for SNP and IGP probes(Supplementary Fig. 1)
We first selected SNPs to represent variation among 25 widely used classical laboratory strains. The genome was divided into non-overlapping 40 kb intervals and, in each interval, three SNPs with high minor allele frequency and low missing rate were selected.
Next we selected SNPs based on local phylogenetic trees constructed on 100 kb non-overlapping intervals using the 15 NIEHS strains7 plus C57BL/6J. The procedure was repeated after shifting the window 50kb. This set represents the largest class of SNPs, encompassing 67% of the total. Three substrategies were used to select SNPs. First, for intervals with >20 SNPs in which 30% or more have complete genotypes, we constructed local phylogenetic trees and corrected branch lengths to account for sampling bias in the SNP discovery data6(Supplementary Fig.6, online). In each window, we selected one SNP corresponding to each branch in the local tree until either 98% of the variation was represented or a maximum of 22 SNPs were selected. The threshold of 22 SNPs allowed us to capture over 98% of the variation in 90% of the intervals, and 95% of the variation in the remaining 10% of intervals (Supplementary Fig.7, online). These SNPs represent 95% (402,719) of the total SNPs in this strategy. Next, for intervals having >20 NIEHS SNPs7 but with less than 30% having complete genotypes, we used the same procedure but used imputed SNP Genotypes12. These SNPs represent 4.9% (21,138) of the total SNPs in this strategy. Finally, for windows having fewer than 20 NIEHS SNPs7, we selected SNPs from other sources. These SNPs represent 0.008% (36) of the total SNPs in this strategy.
C57BL/6J singleton SNPs are completely absent from the NIEHS data7. Therefore, we identified private C57BL/6J SNPs by comparing the genotypes of six strains used in other SNP discovery experiments,27 C57BL/6J, A/J, DBA/2J,129X1/SvJ, 129S1/SvImJ, and MSM/Ms. We selected three C57BL/6J singletons in every 1Mb interval.
The wild derived strains PWD/PhJ and MOLF/EiJ used in the NIEHS study7 each carry substantial genomic regions of M. m. domesticus origin6. In order to identify non-domesticus SNPs in these regions and to include additional M. m. molossinus variants, we aligned the BAC end sequences from MSM/Ms27 with strains C57BL/6J, A/J, DBA/2J, 129X1/SvJ, and 129S1/SvImJ. We selected three MSM/Ms private SNPs in every 100kb window. We note that the representative M. m castaneus strain in the NIEHS panel7, CAST/EiJ, also has substantial introgression regions but there are no comparable resources to help make up for this deficiency.
Additional SNPs were identified by small scale resequencing of other Mus species (M. spretus, M. spicilegus and M. macedonicus) and M. m. musculus subspecies.
SNPs identified in the NIEHS study7 that represent the main branches of the Chromosome Y phylogenetic tree6 were selected. In addition we searched the NCBI database for Y chromosome sequences from M. spretus, M. spicilegus and M. macedonicus, SNPs present in the NIEHS data7 were eliminated.
SNPs identified in resequencing studies that represent the main branches of the mitochondrial phylogenetic tree were selected.
Ultraconserved elements (UCE) are short non-genic sequences with exceptionally high degree of sequence conservation between species. Among the 481 UCEs identified between human and mouse16 we tiled the entire sequence of 238 that were chippable in the C57BL/6J genome.
For every annotated exon we selected three probes spanning the proximal, intermediate and distal regions. The majority of exons (58%) (denoted Exon 1 probes) have probes that are chippable according to Affymetrix specifications. We selected probes in most of the remaining exons with unique sequences (35%), regardless of whether they were chippable using the same procedure (denoted as Exon 2 probes).
We included probes from sequence reads of MSM/Ms BAC ends that had no corresponding sequence in the C57BL/6J genome. We selected three probes per BAC end from the proximal, middle and distal regions. The chippable status of most of these probes is unknown.
At this stage, there were950 100 kb intervals with fewer than 10probes (SNP or IGP). The majorities of these were contiguous and fell into genome regions known to contain segmental duplications17. This is consistent with the absence of NIEHS data7 and the failure of probes to satisfy the selection criteria for unique sequences. For these intervals, we identified all chippable probes, either unique or nonspecific, and selected one or two probes per NspI or StyI fragment.
Chippable probes
To identify chippable probes we used the following criteria: 1) SNP should be on NspI or StyI fragments with sizes of 50bp to 1kb, computed on C57BL/6J reference genome; 2) SNP should be at least 10bp away from cut site; 3) There should be no other known SNPs in ± 12bp flanking sequences around target SNP; and 4) The 33mer centered on the SNP must BLAT as unique with no alignments which have >28bp matches against C57BL/6J genome.
SNP calling
We used the BRLMM-P algorithm11 implemented in Affymetrix Power Tools to obtain genotype calls. We used quantile normalization of probe intensities and median polish to summarize probe sets. We then applied a transformation that converts intensities of the A-allele (Sa) and B-allele (Sb) into contrast (as in(K(Sa−Sb)/(Sa+Sb)) and strength (log (Sa + Sb)) values. The parameter K in this transformation can be adjusted to optimize the contrast between A and B allele intensities. We used K=2 for the test arrays and K=4 for the final Mouse Diversity array. Genotype calls are based on clustering of contrast values. For genotyping we used only the contrast value, although there is clearly additional information in the strength value. We used the silhouette score18 to assess genotype class separation and we used variance of intensity as a measure of within cluster consistency to guide probe selection. We used 116 samples, including 76 F1 hybrids, many with known genotypes, to train the BRLMM-P algorithm. We have observed that performance continues to improve as more samples are added to the training set.
Annotation of the Mouse Diversity array
We have extensively annotated the Mouse Diversity array to help users interpret results, integrate array data with other genomic resources, and develop new uses for the array. Annotation and information regarding probe performance can be obtained from the Center for Genome Dynamics website (http://genomedynamics.org/tools/diversityarray.shtml). Probe contrast, silhouette scores and heterozygous genotype calls in inbred strains are important indicators of individual SNP performance. We have flagged poorly performing SNPs in the annotation files on the basis of their performance. These SNPs should be ignored or used with caution. In the future, we will annotate SNP and IGP for performance in additional samples, including wild derived strains from different M. musculus subspecies and other Mus species. Annotation files will be updated regularly.
Supplementary Material
Supplementary Figure 1: Design strategy and content of the mouse diversity array.
Supplementary Figure 2: Distribution of the distance between consecutive probe sets.
Supplementary Figure 3: Poorly performing SNPs.
Supplementary Figure 4: Residual heterozygosity in strain SSL/LeJ.
Supplementary Figure 5: Intensity distributions for different probe types.
Supplementary Figure 6: Selection strategy for SNPs from the NIEHS data.
Supplementary Figure 7: Strain distribution patterns in each 100 kb interval.
Supplementary Table 1: SNP selection criteria in the test array.
Supplementary Table 2: Performance of SNP probes.
Acknowledgments
This work was supported by the National Institute of General Medical Sciences(NIGMS)National Centers of Systems Biology program, grant GM-076468. The authors wish to thank the Array Design and Bioinformatics teams at Affymetrix for their advice and assistance in the design and implementation of the Mouse Diversity array.
References
- 1.Kennedy GC, et al. Nat Biotechnol. 2003;10:1233–7. doi: 10.1038/nbt869. [DOI] [PubMed] [Google Scholar]
- 2.Shifman S, et al. PLOS Biology. 2006;4:2227–2237. [Google Scholar]
- 3.Lindblad-Toh K, et al. Nature Genetics. 2000;24:381–386. doi: 10.1038/74215. [DOI] [PubMed] [Google Scholar]
- 4.Wade CM, et al. Nature. 2002;420:574–578. doi: 10.1038/nature01252. [DOI] [PubMed] [Google Scholar]
- 5.Yang H, Bell TA, Churchill GA, Pardo-Manuel de Villena F. Nature Genetics. 2007;39:1100–1107. doi: 10.1038/ng2087. [DOI] [PubMed] [Google Scholar]
- 6.Clark AG, Hubisz MJ, Bustamante CD, Williamson SH, Nielsen R. Genome Res. 2005;15:1496–502. doi: 10.1101/gr.4107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frazer KA, et al. Nature. 2007;448:1050–3. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- 8.Chesler EJ, et al. Mamm Genome. 2008;19:382–9. doi: 10.1007/s00335-008-9135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams R, IV, et al. PLoS ONE. 2009;4(3):e4649. doi: 10.1371/journal.pone.0004649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graubert TA, et al. PLoS Genet. 2007;3:e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Affymetrix. Technical report. Affymetrix; 2006. [Google Scholar]
- 12.Szatkiewicz JP, et al. Mamm Genome. 2008;19:199–208. doi: 10.1007/s00335-008-9098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Science. 2007;318:1136–40. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 14.Bemmo A, et al. BMC Genomics. 2008;9:529. doi: 10.1186/1471-2164-9-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerkel K, et al. Nature Genetics. 2008;40:904–8. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- 16.She X, Cheng Z, Zöllner S, Church DM, Eichler EE. Nature Genetics. 2008;40:909–14. doi: 10.1038/ng.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bejerano G, et al. Science. 2004;304:1321–5. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 18.Lovmar L, Ahlford A, Jonsson M, Syvanen AC. BMC Genomics. 2005;6:35. doi: 10.1186/1471-2164-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Design strategy and content of the mouse diversity array.
Supplementary Figure 2: Distribution of the distance between consecutive probe sets.
Supplementary Figure 3: Poorly performing SNPs.
Supplementary Figure 4: Residual heterozygosity in strain SSL/LeJ.
Supplementary Figure 5: Intensity distributions for different probe types.
Supplementary Figure 6: Selection strategy for SNPs from the NIEHS data.
Supplementary Figure 7: Strain distribution patterns in each 100 kb interval.
Supplementary Table 1: SNP selection criteria in the test array.
Supplementary Table 2: Performance of SNP probes.