Abstract
A systematic study is reported on the effect of linker size and its chemical composition toward ligand binding to a surface-immobilized aptamer, measured using surface plasmon resonance. The results, using thrombin as the model system, showed that as the number of thymidine (T) units in the linker increases from 0 to 20 in four separate increments (T0, T5, T10, T20), the surface density of the aptamer decreased linearly from approximately 25 to 12 pmol•cm-2. The decrease in aptamer surface density occurred due to the increased size of the linker molecules. In addition, thrombin binding capacity was shown to increase as the linker length increased from 0 to 5 thymidine nucleotides; and then, decreased as the number of thymidine residues increased to 20 due to a balance between two different effects. The initial increase was due to increased access of thrombin to the aptamer as the aptamer was moved away from the surface. For linkers greater in length than T5, the overall decrease in binding capacity was primarily due to a decrease in the surface density. Incorporation of a hexa(ethylene glycol) moiety into the linker did not affect the surface density, but increased the amount of thrombin bound. In addition, the attachment of the linker at the 3′ versus the 5′-end of the aptamer resulted in increased aptamer surface density. However, monolayers formed with equal surface densities showed similar amounts of thrombin binding irrespective of the point of attachment.
Introduction
The use of aptamers as the affinity component in assays and sensors is beginning to rival antibodies for these applications.1,2 A primary reason for this is that once the DNA or RNA sequence of an aptamer is known for a protein or other target molecule, it can be produced using automated synthetic approaches which may reduce cost and reliance on animal sources.3 This has facilitated research on sensors, separations, and binding assay applications utilizing aptamers in a number of readout formats including electrochemical,4,5 surface plasmon resonance (SPR),6,7 quartz crystal microbalance (QCM)8,9 etc. For most of these applications, aptamers must be immobilized onto a solid surface.10 In general, the surface density of receptors and their ligand capture efficiency are two of the most important factors that determine the sensitivity of the receptor-immobilized surface.7 For aptamer-based self-assembled monolayers (SAMs), both of these factors are controlled primarily by the choice of linker that connects the aptamer to the surface.11
Linkers are used to provide a “chemical” spacer between the solid surface and the receptor that is anchored to the surface by an appropriate functional group; in the case of SAMs on gold, this functional group is a thiol. The role of the linker is to spatially extend the receptor from the surface to increase its accessibility by the solution ligand and to remove any non-specific adsorption. Although there are a few reports of random oligonucleotide linkers,10 the most commonly used linker for aptamer immobilization on gold incorporates oligothymidine groups10 that can be easily added to the aptamer sequence during solid phase synthesis. Thymidine is the nucleotide of choice for linker design because it adsorbs less strongly on gold surfaces compared to the other three nucleotides, providing the most inert linker of all possible oligonucleotides.12 There are several literature reports of thymidine oligonucleotides in the range of T1 - T20 that have been used as linkers for aptamer immobilization on gold.10
There have not been any systematic studies on the effects of linker length on aptamer performance. Thus, to understand the effect of linker on surface immobilized aptamer performance we choose thrombin binding aptamer, referred to as HD-1,13 as a model.6,7,9, 14-27 This aptamer, which binds to exosite-I on thrombin, was selected because its structure is well-known28,29 (G-quartet) and it is relatively short (15-base sequence, GGTTGGTGTGGTTGG) facilitating synthesis and purification. We prepared monolayers of this aptamer containing different length of oligothymidine moieties, T1-T20, to do a systematic investigation of length of oligothymidine linkers on the binding performance of surface immobilized HD-1 to thrombin. Further, we have recently reported that the use of oligoethylene glycol units incorporated into the linker can improve aptamer binding performance; thus a parallel study was carried out using different oligothymidine linkers with the hexaethylene glycol linker group (EG6).7 Further, studies in this paper were extended to determine if attachment of the linker to either the 3′ or the 5′ terminus of the aptamer imparted a significant factor in thrombin binding performance to surface immobilized aptamers.
Experimental Section
Materials
The gold sensor slides were obtained from Biacore (SIA kit Au, Biacore). Thrombin was purchased from Haematologic Technologies, Vermont, USA; Bovine serum albumin (BSA) was obtained from Sigma. All of the modified oligonucleotides were obtained from Eurogentec North America, Inc. (San Diego, CA). These include 3′ thiol-modified oligonucleotide HS(CH2)6-T-GGTTGGTGTGGTTGG-3′ (C6-T1-A1-3′); and the 5-thiol-modified oligonucleotides: 5′-HS(CH2)6-GGTTGGTGTGGTTGG (5′-C6-T0-A1), 5′-HS(CH2)6-T-GGTTGGTGTGGTTGG (5′-C6-T1-A1), 5′-HS(CH2)6-TTTTT-GGTTGGTGTGGTTGG (5′-C6-T5-A1), 5′-HS(CH2)6-TTTTTTTTTT-GGTTGGTGTGGTTGG (5′-C6-T10-A1), 5′-HS(CH2)6- TTTTTTTTTTTTTTTTTTTT-GGTTGGTGTGGTTGG (5′-C6-T20-A1). In addition, the following 5′-thiol-modified oligonucleotides containing an additional hexaethylene glycol unit were used: 5′-HS(CH2)6-OPO2-(CH2CH2O)6-GGTTGGTGTGGTTGG-3′ (5′-C6EG6-T0-A1), 5′-HS(CH2)6-OPO2-(CH2CH2O)6-TTTTT-GGTTGGTGTGGTTGG-3′ (5′-C6EG6-T5-A1), 5′-HS(CH2)6-OPO2-(CH2CH2O)6-TTTTTTTTTT-GGTTGGTGTGGTTGG-3′ (5′-C6EG6-T10-A1) and 5′-HS(CH2)6-OPO2-(CH2CH2O)6-TTTTTTTTTTTTTTTTTTTT-GGTTGGTGTGGTTGG-3′ (5′-C6EG6-T20-A1) for the formation of the SAMs. 2-Mercapto-1-ethanol (MCE) was used as received from Fluka. HBS-EP buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% v/v Surfactant P20) was purchased from Biacore. GR ACS grade potassium phosphate monobasic (KH2PO4) was obtained from Fischer Chemicals. All the buffer solutions were prepared in DI water with resistivity greater than 18 MΩcm, obtained by passing water through a Barnstead reverse osmosis filter and then through a Nanopure filtration exchange system.
SPR measurements
The SPR measurements were carried out with a Biacore X instrument. The gold sensor surface (SIA kit Au, Biacore) was cleaned by UV-oxidation for 1 h and then rinsed with water, ethanol and finally with water and dried in a stream of dry nitrogen.30 The clean gold cover slips were affixed to a plastic plate and the resultant assembly placed in a cassette, which was subsequently introduced into the SPR instrument.
For aptamer monolayer formation, 100 μL of 2.0 μM solution of aptamer thiol in 1.0 M potassium phosphate buffer (pH 3.5) was injected at a flow rate of 2 μL•min-1 followed by 20 μL of 2.0 μM solution of aptamer thiol. This was followed by 50 μL of a 0.1 M solution of MCE at a flow rate of 5 μL•min-1. For aptamer monolayer formation, 1.0 M potassium phosphate buffer (pH 3.5) was used as a running buffer.6 For protein binding studies, 10 μL of thrombin at different concentrations was injected at a flow rate of 2 μL•min-1 (for 10 min). SAMs were regenerated between experiments using 100 μL of a 2.0 M NaCl solution at a flow rate of 100 μL•min-1.6 The running buffer used was HBS-EP (Biacore).
Results and Discussion
Effects of linker length on aptamer surface loading density and thrombin binding
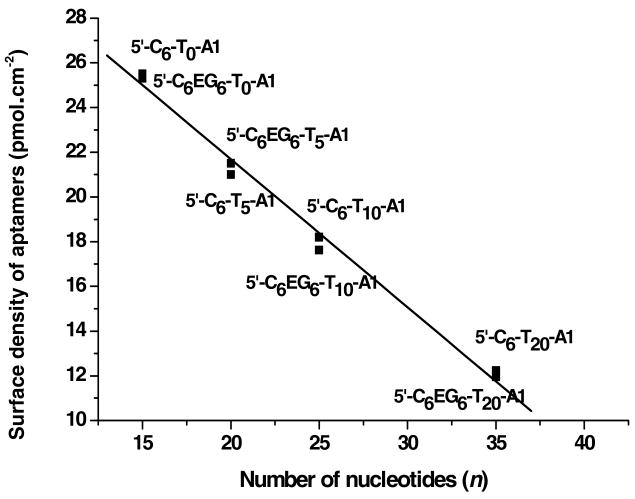
Aptamer monolayers were formed in-situ in a Biacore X SPR instrument, which enabled real-time monitoring of monolayer formation. From the SPR response (RU) for each of the aptamer monolayers, the aptamer surface density was determined using the published conversion factor of 10 RU = 1.0 ng•cm-2 for DNA.31 Although a number of conversion factors (from 0.73 to 1.25 ng•cm-2) have been reported in the literature, the value of 1.0 ng•cm-2 was used because the refractive index of single stranded DNA is reported to be same as that for proteins, 1.45. The conversion factor for proteins32 is known to be 10 RU = 1.0 ng•cm-2, and because the SPR response is directly related to the refractive index, this value should be used for single stranded DNA as well. The surface density of aptamer was calculated by methods described by Georgiadis and co-workers, which takes into account desorption of aptamer during subsequent blocking step by addition of mercaptoethanol.33 The surface density values for each aptamer monolayer is reported in the third column of Table 1; moles are reported in the table after conversion using the molecular weight of each thiol-linked aptamer. A plot of surface density of the aptamer versus the number of bases, including both the linker and the aptamer sequence, is shown in Figure 1. A linear dependence on surface density as a function of the total number of nucleotides (including aptamer and linker) is observed, yielding a slope of -0.663 pmol•cm-2 per DNA base; therefore, each nucleotide in the linker decreased the surface density of aptamer by 0.663 pmol/cm2. Steel et al. reported earlier that the surface density of oligonucleotides decreased as the length of the oligonucleotide increases; however, the degree of linearity in our case is much more pronounced.34 The high degree of linearity observed may be useful for prediction of aptamer surface density for a given linker length. Moreover, the aptamer surface density for linkers containing the same number of thymidine residues was similar irrespective of whether or not the EG6 group was included in the linker structure. This indicated that the low-molecular-weight EG6 group did not change the packing density of the aptamer on the surface. The overall decreasing trend in Figure 1 is most likely due to the increasing size of the molecule with increasing number of oligothymidine groups, which caused each molecule to occupy more volume and therefore more surface area and thus, lowering the surface concentration.35
Table 1. Aptamer surface density as a function of thrombin binding and efficiency for SAMs formed with different thymidine linkers.
| Entry | Thiol-linked aptamer | Surface density of aptamer in the mixed SAM (pmol•cm-2)a | Maximum thrombin bound to the mixed SAM (pmol•cm-2) | Thrombin/Aptamer ratio at saturation (%) |
|---|---|---|---|---|
| 1 | 5′-C6-T0-A1 | 25.3 | 2.96 | 11.7 |
| 2 | 5′-C6-T5-A1 | 21.0 | 4.10 | 19.5 |
| 3 | 5′-C6-T10-A1 | 18.2 | 3.89 | 21.4 |
| 4 | 5′-C6-T20-A1 | 12.23 | 3.39 | 27.7 |
| 5 | 5′-C6EG6-T0-A1 | 25.5 | 4.70 | 18.6 |
| 6 | 5′-C6EG6-T5-A1 | 21.5 | 6.49 | 30.2 |
| 7 | 5′-C6EG6-T10-A1 | 17.62 | 5.36 | 30.4 |
| 8 | 5′-C6EG6-T20-A1 | 11.95 | 4.82 | 40.4 |
Maximum standard deviation from duplicate values was ± 0.03 pmol•cm-2.
Figure 1.
Plot of aptamer surface density versus the total number of bases of the aptamer and the oligo-thymidine linker. (Slope = -0.663 ± 0.020 and R = - 0.996). Maximum standard deviation from duplicate values was ± 0.03 pmol•cm-2.
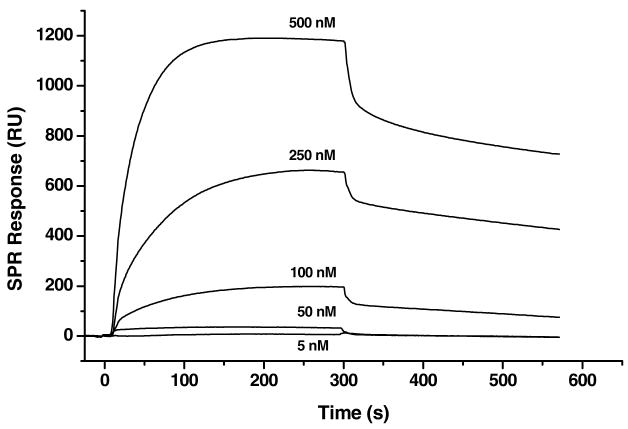
Following aptamer monolayer formation, the films were washed with buffer, and binding isotherms were obtained from SPR measurements of thrombin at different concentrations. Figure 2 illustrates typical SPR responses of the aptamer monolayer (in this case, the linker was 5′-C6-T0-A1) measuring thrombin binding at different concentrations that were used to generate the binding isotherms. Procedurally, sequentially higher concentrations of thrombin were successively measured with regeneration (i.e. removal of thrombin) of aptamer films between each measurement by using 2 M NaCl.6 The SPR response for thrombin binding was converted into pmol•cm-2 using the published conversion factor (10 RU = 1.0 ng•cm-2 for proteins).36 Using this value, the maximum amount of thrombin bound to each film incorporating different linkers was determined at a solution concentration of 1.0 μM thrombin (this has generally been found to be above the binding isotherm saturation level)7; this data are shown in the fourth column of Table 1.37 A correlation between linker length and thrombin binding capacity (column 4 of Table 1) exists for each class of linkers (i.e., linkers with and without the EG6 spacer groups). For each class of linker, as the number of thymidine nucleotides increased to 5, there was an increase in the total amount of thrombin bound; greater than 5 thymidine residues in the linker resulted in a decrease in the amount of thrombin bound to the aptamer surface.
Figure 2.
Representative SPR sensograms for binding of different concentrations of thrombin onto a 5′-C6-T0-A1 aptamer monolayer. Effective regeneration of the aptamer surface between analyses was achieved by removal of bound thrombin with 2 M NaCl.
There are two opposing effects responsible for these trends. First, as the number of thymidine nucleotides in the linker increased, the surface density of aptamer decreased. This decrease in the number of binding sites (per unit area) would be anticipated to result in a proportional lowering of the amount of bound thrombin. However, an increase in the linker length moved the aptamer away from the surface and into a more solvated environment. This resulted in reduced steric hindrance for the protein to approach and bind to the aptamer, causing an increased binding efficiency (i.e., less aptamer was required to bind a larger fraction of thrombin). A balance between these two factors determined the maximum amount of thrombin that could bind (per unit area) to the aptamer monolayer. For both classes of linkers, steric hindrance to thrombin binding was the controlling factor for linkers with 0-5 thymidine nucleotides. For linkers with greater than 5 thymidine residues, reduction of the surface density was the controlling factor for determining the amount of thrombin bound to the aptamer monolayer. Thus, it appeared that the critical point for these opposing effects occurred around 5 thymidine units in the linker, which afforded the highest capacity of thrombin binding to the SAMs. This observation was true for both the linkers with or without the intervening EG6 group.
A consistent observation from the results displayed in Table 1 is that incorporation of the EG6 spacer in the linker increased the amount of thrombin bound to the aptamer monolayer without any changes of the surface density of the aptamer (see column 3 in Table 1). A possible reason for this may have been that the EG6 spacer increased the overall linker length; however, spatial compactness of this spacer (compared to oligothymidine spacers) did not reduce the surface density of aptamer. This ultimately resulted in an increase in the binding efficiency of the aptamer toward thrombin, measured by the ratio of thrombin bound divided by the amount of aptamer on the surface as shown in the last column of Table 1. This data also showed that for both classes of linkers, the binding efficiency increased with increasing number of thymidine nucleotides. Similar aptamer monolayers prepared under static conditions by immersing the gold slide in an aptamer-thiol solution, rather than under flow conditions in the SPR instrument, also showed that inclusion of the EG6 spacer improved thrombin binding efficiency.7 Control experiments to determine non-specific binding were carried out using BSA (a representative sensogram is provided in the supplementary information, Figure S-3). A low level of non-specific BSA adsorption to the thrombin aptamer monolayers was detected; the typical values were 0.2 pmol•cm-2 or less, which corresponds to the range of 3.0 – 6.7% depending on the amount of thrombin bound.
Effect of point of attachment of the linker: 5′ vs 3′
Comparison of the sensograms in Figure 2 to an earlier report on thrombin binding by aptamers immobilized at the 3′ end (3′-C6-T1-A1), revealed a significantly higher binding response for the 5′ attached aptamer (even in the absence of thymidine or EG6 spacers).6 To address this difference, it was of interest to determine whether attachment to either the 5′ or 3′-end of the aptamer had an effect on binding efficiency. Thus SAMs were formed using C6-T1-A1 linked to thrombin aptamers at either the 5′ or the 3′ end (abbreviated 5′-C6-T1-A1 and 3′-C6-T1-A1 respectively), to match the conditions of the earlier report.6 Entries 1 and 2 in Table 2 show the comparison of aptamer surface density for both SAMs with the unexpected result that the linker attached to the 3′-end of the aptamer created a significantly more densely packed monolayer of DNA strands per unit area (41.0 pmol•cm-2) versus the aptamer thiol having the same linker with point of attachment at the 5′-end (24.7 pmol•cm-2). To our knowledge, such a significant effect by the point of linker attachment on the oligonucleotide monolayer formation has not been previously reported.
Table 2. The amount of thrombin bound to C6-T1-A-3′ and 5′-C6-T1-A monolayers of comparable surface density.
| Entry | Aptamer | Surface density of aptamer (pmol•cm-2)a | Maximum specific thrombin bound to the mixed SAM (pmol•cm-2) |
|---|---|---|---|
| 1 | 5′-HS(CH2)6-T-A1-3′ | 24.7 | 3.2 |
| 2 | 3′-HS(CH2)6-T-A1-5′ | 41.0 | 1.3 |
| 3 | 3′-HS(CH2)6-T-A1-5′ | 21.3 | 3.4 |
Maximum standard deviation from duplicate values was ± 0.03 pmol•cm-2.
The underlying reasons for differences in the surface loading of aptamers using either 5′ or 3′-end attachment may be due to differences in aptamer structure. In the absence of thrombin, the aptamer may exist in the folded G-quartet structure or the unfolded structure; however, the presence of thrombin causes the aptamer to fold into the G-quartet structure via an “induced-fit” mechanism.38 The higher surface density of aptamer attached at the 3′-position suggests the equilibrium may be more shifted toward the folded G-quartet structure compared to 5′ attachment, enabling the aptamer film to pack more densely. While the surface density of the aptamer was greater for the 3′ attachment, the data in Table 2 show that thrombin binding decreased nearly 60% compared to the SAM with 5′ attachment. It should be noted that the amount of thrombin bound here matched the value for the same SAM prepared using 3′-attached aptamer reported by Baldrich et al.6 The difference in thrombin binding between the 5′- and the 3′-attached aptamers is much higher than the same comparison previously reported for other surface immobilized aptamers.39 Other studies have shown that as the surface density of ligands increases, an optimum value is reached after which protein binding decreases or levels off.40,41 Thus, a second set of binding experiments were performed for the 3′ attached aptamer with the aptamer surface density lowered to equal that of the 5′-attached aptamer. Treating the gold surface with 70 μL of 2 μM 3′-attached aptamer-thiol solution at 2 μL•min-1 flow rate results in a monolayer with the surface density of 21.3 pmol•cm-2, which is comparable to the 5′-attached aptamer (Table 2). Under these conditions, the 3′-C6-T1-A1 monolayer showed the maximum amount thrombin binding (6.1 pmol•cm-2); however, control experiments using BSA showed a high level of non-specific BSA adsorption (2.7 pmol•cm-2). Therefore, after subtraction of non-specific binding from the maximum amount of thrombin bound, the specific thrombin bound is 3.4 pmol•cm-2, which is nearly the same as the 5′-C6-T1-A1 monolayer (Table 2). Thus, an optimum surface density is important for maximizing protein binding by the aptamer, which can be affected by 5′ or 3′ attachment, as well as linker length. Therefore, it can be concluded that differences between the current studies using 5′ attached aptamers, and the previous report using the 3′ attached aptamer are most likely due to differences in surface density.
Conclusions
Thymidine nucleotides are most commonly used as linker groups for the formation of aptamer-based SAMs on gold. A survey of the literature indicated that the number of thymidine groups utilized in these studies was arbitrarily chosen. Therefore, a systematic study was undertaken to determine optimum thymidine linker lengths in terms of increasing the receptor surface density and the accessibility of the receptor to the solution ligand. Furthermore, an oligo(ethylene glycol) spacer group was also investigated for improving aptamer binding to its recognition partner. The optimum aptamer binding was found using a linker that incorporated 5 thymidine nucleotides and an oligo(ethylene glycol) spacer group. From a fundamental point of view, the reasons for this lie in a balance between surface density and distance of aptamer from the immobilization surface, making it more accessible to its binding partner; both effects are controlled by the length and chemical nature of the linker. Particularly noteworthy is the incorporation of the oligo(ethylene glycols) into the linker, which did not change the surface packing density of the aptamer, but increased the effective binding of the aptamer ligand. More striking was that surface density is different for thrombin aptamers linked to the surface via the 3′ versus the 5′ attachment point. However, when the surface density was held approximately constant for SAMs with aptamers attached at either end, the overall binding was approximately equal. In conclusion, the data reported here will be useful for the rational selection of linkers that promote enhanced performance of aptasensors and other DNA chip technologies.
Supplementary Material
Representative sensograms showing methods used for monolayer density calculations, thrombin binding on 5′-C6-T1-A1 at different substrate concentrations, and examples of non-specific BSA binding versus thrombin binding. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
We thank the National Institutes of Health (R33CA0099246) and National Science Foundation (EPS-0346411) for financial support of this work. D.A.S. thanks the National Science Foundation for CAREER Program Award CHE-0134290; and NASA grant NNC06AA18A.
References
- 1.Jayasena SD. Clin Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 2.Schlecht U, Malave A, Gronewold T, Tewes M, Lohndorf M. Anal Chim Acta. 2006;573-574:65–68. doi: 10.1016/j.aca.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 3.Klussmann Sven., editor. The Aptamer Handbook. Wiley-VCH Verlag GmbH&Co., KGaA; Weinheim: 2006. [Google Scholar]
- 4.Xu D, Xu D, Yu X, Liu Z, He W, Ma Z. Anal Chem. 2005;77:5107–5113. doi: 10.1021/ac050192m. [DOI] [PubMed] [Google Scholar]
- 5.Xu Y, Yang L, Ye XY, He P, Fang Y. Electroanalysis. 2006;15:1449–1456. [Google Scholar]
- 6.Baldrich E, Restrepo A, O'Sullivan CK. Anal Chem. 2004;76:7053–7063. doi: 10.1021/ac049258o. [DOI] [PubMed] [Google Scholar]
- 7.Balamurugan S, Obubuafo A, Soper SA, McCarley RL, Spivak DA. Langmuir. 2006;22:6446–6453. doi: 10.1021/la060222w. [DOI] [PubMed] [Google Scholar]
- 8.Minunni M, Tombelli S, Gullotto A, Luzi E, Mascini M. Biosens Bioelectron. 2004;20:1149–1156. doi: 10.1016/j.bios.2004.03.037. [DOI] [PubMed] [Google Scholar]
- 9.Liss M, Petersen B, Wolf H, Prohaska E. Anal Chem. 2002;74:4488–4495. doi: 10.1021/ac011294p. [DOI] [PubMed] [Google Scholar]
- 10.Balamurugan S, Obubuafo A, Soper SA, Spivak DA. Anal Bioanal Chem. 2008;390:1009–1021. doi: 10.1007/s00216-007-1587-2. [DOI] [PubMed] [Google Scholar]
- 11.Bini A, Minunni M, Tombelli S, Centi S, Mascini M. Anal Chem. 2007;79:3016–3019. doi: 10.1021/ac070096g. [DOI] [PubMed] [Google Scholar]
- 12.Kimura-Suda H, Petrovykh DY, Tarlov MJ, Whitman LJ. J Am Chem Soc. 2003;125:9014–9015. doi: 10.1021/ja035756n. [DOI] [PubMed] [Google Scholar]
- 13.Bock LC, Griffin LC, Latham JA, Vermass EH, Toole JJ. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 14.Potyrailo RA, Conrad RC, Ellington AD, Hieftje GM. Anal Chem. 1998;70:3419–3425. doi: 10.1021/ac9802325. [DOI] [PubMed] [Google Scholar]
- 15.Pavlov V, Xiao Y, Shlyahovsky B, Willner I. J Am Chem Soc. 2004;126:11768–11769. doi: 10.1021/ja046970u. [DOI] [PubMed] [Google Scholar]
- 16.Hansen JA, Wang J, Kawde AN, Xiang Y, Gothelf KV, Collins G. J Am Chem Soc. 2006;128:2228–2229. doi: 10.1021/ja060005h. [DOI] [PubMed] [Google Scholar]
- 17.Radi AE, Acero Sanchez JL, Baldrich E, O'Sullivan CK. J Am Chem Soc. 2006;128:117–124. doi: 10.1021/ja053121d. [DOI] [PubMed] [Google Scholar]
- 18.Ho HA, Leclerc M. J Am Chem Soc. 2004;126:1384–1387. doi: 10.1021/ja037289f. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Wang Z, Li XF, LE XC. Angew Chem, Int Ed. 2006;45:1576–1580. doi: 10.1002/anie.200503345. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida W, Sode K, Ikebukuro K. Biochem Biophys Res Commun. 2006;348:245–252. doi: 10.1016/j.bbrc.2006.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai H, Ming-Hung LT, Hsing IM. Sens Actuators, B: Chem. 2006;114:433–437. [Google Scholar]
- 22.Hianik T, Ostatná Z, Stoikova E, Evtugyn G. Bioorg Med Chem Lett. 2005;15:291–295. doi: 10.1016/j.bmcl.2004.10.083. [DOI] [PubMed] [Google Scholar]
- 23.Kawde AN, Rodriguez MC, Lee TMH, Wang J. Electrochem Commun. 2005;7:537–540. [Google Scholar]
- 24.Maehashi K, Katsura T, Kerman K, Takamura Y, Matsumoto K, Tamiya E. Anal Chem. 2007;79:782–787. doi: 10.1021/ac060830g. [DOI] [PubMed] [Google Scholar]
- 25.Xiao Y, Lubin AA, Heeger AJ, Plaxco KW. Angew Chem, Int Ed. 2005;44:5456–5459. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- 26.Xiao Y, Piorek BD, Plaxco KW, Heeger AJ. J Am Chem Soc. 2005;127:17990–17991. doi: 10.1021/ja056555h. [DOI] [PubMed] [Google Scholar]
- 27.Spiridonova VA, Kopylov AM. Biochem (Moscow) 2002;67:706–709. doi: 10.1023/a:1016110724564. [DOI] [PubMed] [Google Scholar]
- 28.Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Proc Natl Acad Sci USA. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padmanabhan K, Padmanabhan KP, Ferrara JD, Sadler JE, Tulinsky A. J Biol Chem. 1993;24:17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- 30.Ron H, Matlis S, Rubinstein I. Langmuir. 1998;14:1116–1121. [Google Scholar]
- 31.Speck C, Weigel C, Messer W. The EMBO J. 1999;18:6169–6176. doi: 10.1093/emboj/18.21.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson WD. Science. 2002;295:2103–2105. doi: 10.1126/science.295.5562.2103. [DOI] [PubMed] [Google Scholar]
- 33.Georgiadis R, Peterlinz KP, Peterson AW. J Am Chem Soc. 2000;122:3166–3173. [Google Scholar]
- 34.Steel AB, Levicky RL, Herne TM, Tarlov MJ. Biophys J. 2000;79:975–981. doi: 10.1016/S0006-3495(00)76351-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tinland B, Pluen A, Strum J, Weill G. Macromolecules. 1997;30:5763–5765. [Google Scholar]
- 36.Wilson WD. Science. 2002;295:2103–2105. doi: 10.1126/science.295.5562.2103. [DOI] [PubMed] [Google Scholar]
- 37.Stenberg E, Persson B, Roos H, Urbaniczky C. J Colloid Interface Sci. 1991;143:513–526. [Google Scholar]
- 38.Patel DJ, Suri AK, Jiang F, Jiang L, Fan P, Kumar RA, Nonin S. J Mol Biol. 1997;272:645–664. doi: 10.1006/jmbi.1997.1281. [DOI] [PubMed] [Google Scholar]
- 39.Cho EJ, Collett JR, Szafranska AE, Ellington AD. Anal Chim Acta. 2006;564:82–90. doi: 10.1016/j.aca.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 40.Lahiri J, Isaacs L, Grzybowski B, Carbeck JD, Whitesides GM. Langmuir. 1999;15:7186–7198. [Google Scholar]
- 41.Spinke J, Liley M, Schmitt FJ, Guder HJ, Angermaier L, Knoll W. J Chem Phys. 1993;99:7012–7019. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative sensograms showing methods used for monolayer density calculations, thrombin binding on 5′-C6-T1-A1 at different substrate concentrations, and examples of non-specific BSA binding versus thrombin binding. This material is available free of charge via the Internet at http://pubs.acs.org.