Abstract
Organophosphorus esters (OP) bind covalently to the active site serine of enzymes in the serine hydrolase family. Recently, mass spectrometry identified covalent binding of OP to tyrosine in a wide variety of proteins when purified proteins were incubated with OP. In the present work, manual inspection of MSMS data led to the realization that lysines also make a covalent bond with OP. OP-labeled lysine residues were found in 7 proteins that had been treated with either chlorpyrifos oxon or diisopropylfluorophosphate: human serum albumin (K212, K414, K199, and K351), human keratin 1 (K211 and K355), human keratin 10 (K163), bovine tubulin alpha (K60, K336, K163, K394, and K401), bovine tubulin beta (K58), bovine actin (K113, K291, K326, K 315 and K328), and mouse transferrin (K296 and K626). These results suggest that OP binding to lysine is a general phenomenon. Characteristic fragments specific for chlorpyrifos-oxon labeled lysine appeared at 237.1, 220.0, 192.0, 163.9, 128.9 and 83.9 amu. Characteristic fragments specific for diisopropylfluorophosphate labeled lysine appeared at 164.0, 181.2 and 83.8 amu. This new OP-binding motif to lysine suggests new directions to search for mechanisms of long-term effects of OP exposure and in the search for biomarkers of organophosphorus agent exposure.
Keywords: Mass spectrometry, organophosphorus esters, chlorpyrifos oxon, diisopropylfluorophosphate, serum albumin, keratin, tubulin, actin transferrin
Introduction
Organophosphorus agents (OP)1 include pesticides and chemical warfare agents [1, 2]. The intended target of these agents is the active site serine of acetylcholinesterase [AChE, EC 3.1.1.7]. Inhibition of acetylcholinesterase explains the acute symptoms that are observed upon exposure to high doses of OP [3, 4]. However, chronic exposure to OP at doses too low to generate cholinergic symptoms has been implicated in a variety of adverse effects including memory loss, learning disability, fatigue, depression, and Parkinson’s disease [5, 6, 7, 8, 9]. These symptoms may appear after exposures too low to significantly inhibit acetylcholinesterase [10]. These observations suggest that there are clinically relevant targets for OP in addition to acetylcholinesterase.
Reactions of OP with a variety of serine hydrolases in vitro have long been known. Targets include, but are not limited to fatty acid amide hydrolase, acyl peptide hydrolase, carboxylesterase, phosphoglucomutase and trypsin [2, 11, 12, 13]. Enzymes without a serine active site also react with OP, such as lysyl oxidase and the M2 muscarinic receptor [2, 14]. Results from electrospray ionization mass spectrometry have demonstrated that OP can react with tyrosines on proteins such as transferrin [15], serum albumin [16, 17, 18, 19, 20, 21] and tubulin [22]. Though there has been a renewed interest in the reaction of tyrosine with OP recently, reaction of tyrosines from human serum albumin and bovine serum albumin with the organophosphorus agent diisopropylfluorophosphate (DFP) was reported by Sanger in 1963 [23]. Between 1965 and 1971, DFP was shown to react with tyrosine residues on bromelain [24], papain [25], and lysozyme [26]. We have enlarged the group of proteins for which tyrosine serves as an OP target to include: kinesin 3C, alpha 2-glycoprotein 1 zinc, pro-apolipoprotein AI, keratin, actin, ATP synthase, adenine nucleotide Translocase I, chymotrypsinogen and pepsin [27]. Reaction of free tyrosine with OP was demonstrated by Ashbolt and Rydon [28]. Taken together, these observations firmly establish tyrosine as a target for OP in proteins.
In the course of our investigations into OP labeling of tyrosine, we found that the ε-amine of lysine was also labeled. This finding was quite unexpected because it has long been known that the phosphoamidate bond, such as that in ε-N-phospholysine, is sensitive to hydrolysis, especially at pH values below 8 [29]. The pH sensitivity is of particular importance for our experiments because, as is the general practice when preparing peptide samples for electrospray ionization mass spectrometry, we used solvents containing 0.1% formic acid. Despite the unexpected nature of the observations, we have found OP-labeled lysine in 7 proteins. To our knowledge, there are no previous reports of proteins containing an OP-labeled lysine.
Mass spectrometry is an excellent tool for identifying markers of protein modification. Three mass spectral features can be used for the identification of protein modification. First, is the mass of the parent ion, which must be consistent with the mass of a known peptide plus the mass of the modification. Second, is the presence of a gap in the amino acid sequence from an MSMS spectrum that is consistent with the mass of a modified amino acid. And, third is the presence of fragments in the MSMS spectrum that are characteristic of the modification. We have employed tandem quadrupole electrospray ionization mass spectrometry in conjunction with collision induced dissociation (CID) to study peptides containing OP-labeled lysine. All three mass spectral features have been found.
This presentation has four goals. The first is to document the existence of OP-labeled lysine in proteins. The second is to identify the OP-labeled peptides and the specific OP-labeled lysine residues. The third is to describe the characteristic fragment ions for the OP-labeled lysine, along with the frequency at which each ion appears and the relative intensity of the signals for each characteristic fragment. The fourth goal is to establish that OP reacts with lysine in a number of proteins.
Here we report on the reaction of seven proteins from three species (human serum albumin, human keratin 1, human keratin 10, bovine tubulin alpha, bovine tubulin beta, bovine actin, and mouse transferrin) with two OP (chlorpyrifos-oxon and diisopropylfluorophosphate). In all cases, the mass of the parent ion in the MS spectrum was consistent with the presence of the OP label. Manual analysis of the MSMS spectra from the labeled peptides typically revealed gaps in the b- and/or y-ion series that were consistent with the mass of OP-modified lysine. These gaps confirmed the presence of the OP and yielded the location of the labeled residue in the peptide sequence. Finally, characteristic fragments that are diagnostic for the presence of OP-labeled lysine were identified for each OP.
Methods and Materials
Materials
Purified human serum albumin (essentially fatty acid free, cat # 05418), bovine actin (cat # 3653), human epidermal keratin (cat # K0253), diisopropylfluorophosphate (DFP, cat # D0879), iodoacetamide (cat # I6125), Glu-fibrinopeptide B (cat # F3261), and mouse transferrin (cat # T0523) were obtained from Sigma/Aldrich/Fluka (St. Louis, MO). Dithiothreitol (electrophoresis grade, cat # BP172-25) was from Fisher Biotech (Fair Lawn, NJ). Modified porcine trypsin (TPCK treated, reductively methylated, sequencing grade, cat # V5113) was purchased from Promega (Madison, WI). Bovine brain tubulin (>99% pure, cat # TL238) was from Cytoskeleton Inc (Denver, CO). Chlorpyrifos-oxon (CPO, cat # MET-674B) was from Chem Service Inc (West Chester, PA).
Labeling
Reaction of human serum albumin with DFP was performed as follows: 8.8 mg of serum albumin were dissolved in 5 ml of 10 mM Tris/HCl, pH 8.0 (to yield 1.76 mg protein/ml or 2.6×10−5 M protein) and treated with 26.5 μl of 0.1 M DFP in isopropanol (final concentration 5.3×10−4 M) for 2 h at room temperature. The protein was denatured in 8 M urea, reduced with 10 mM dithiothreitol (with boiling, for 10 min in a water bath), alkylated with 90 mM iodoacetamide (for 1 h at 37°C in the dark) and dialyzed against 10 mM ammonium bicarbonate at 4°C overnight, using Spectrapor dialysis membrane (MWCO = 12,000 to 14,000 from Spectrum Medical Industries, Los Angeles CA. cat # 132700). 500 μl of the mixture was digested with trypsin (1:50 ratio w/w) overnight at 37 °C. The tryptic digest was dried in a Jouan SpeedVac (model RC10-10 from Thermo Fisher Scientific, Waltham, MA) and redissolved in 5% acetonitrile/95% water/0.1% formic acid to make a final solution of 7 pmol albumin peptides per μl, which was used for electrospray ionization mass spectrometry.
Reaction of human albumin with chlorpyrifos-oxon was performed in an analogous manner, using 15 μM albumin and 150 μM CPO [19].
Reaction of tubulin with chlorpyrifos-oxon was performed as described by Grigoryan et al. [30]. Bovine tubulin was dissolved in 200 μl of 15 mM ammonium bicarbonate, pH 8.3, to give a concentration of 0.6 mg tubulin/ml or 12 μM. This was treated with 500 μM CPO (10 μl of a 10 mM CPO stock solution in dimethyl sulfoxide) for 24 hours at 37°C. The tubulin was denatured by heating for 10 minutes in boiling water then the denatured protein was dialyzed against 4 liters of ammonium bicarbonate, pH 8.3 using a Slide-A-Lyzer dialysis cassette (MWCO = 7000 from Pierce, Madison, WI cat #66370). Sixty μg of dialyzed tubulin was digested with 1.5 μg of sequencing grade trypsin for 16 hours at 37°C. The tryptic peptides were dried in a SpeedVac and redissolved in 5% acetonitrile/water plus 0.1% formic acid to a final concentration of about 2 pmole/μl, which was used for mass spectral analysis.
Reaction of actin with chlorpyrifos-oxon was performed as described by Schopfer et al. [31]. Bovine actin was dissolved in 130 μl of 10 mM ammonium bicarbonate, pH 8.3, to give a final concentration of 2.5 mg actin/ml or 48 μM. This was reacted with 240 μM CPO (3 μl of a 10 mM CPO stock solution in dimethyl sulfoxide) at 37°C for 24 hours. The actin was denatured, reduced, alkylated, dialyzed, digested with trypsin, dried and resuspended in 5% acetonitrile/0.1% formic acid for mass spectral analysis as described for tubulin.
Reaction of keratin with chlorpyrifos-oxon was performed as described by Schopfer et al. [31]. One hundred microliters of a mixture of denatured human keratins at 1 mg protein/ml (in 5 mM Tris, 8 M urea, 1 mM beta-mercaptoethanol and 0.1% azide, pH 8.4) was renatured by dialysis against 25 mM Tris/Cl, pH 7.5, then treated with 2 mM CPO (25 μl of a 10 mM CPO stock solution in dimethyl sulfoxide) at 37°C for 24 hours. The keratin was then processed as described for actin.
Reaction of transferrin with chlorpyrifos-oxon was performed as described by Li et al. [15]. One hundred microliters of mouse transferrin at 1 mg transferrin/ml was treated with 0.5 mM CPO (5 μl of a 10 mM CPO stock solution in ethanol) at 37°C for 16 hours. The transferrin was denatured in 8 M urea, then reduced, alkylated, dialyzed, digested with trypsin (at a transferrin to trypsin ratio of 50:1, by weight), dried and resuspended in 5% acetonitrile/0.1% formic acid for mass spectral analysis.
Quadrupole mass spectrometry
Five to ten microliters of a tryptic digest (30–50 pmole) were injected onto an HPLC nanocolumn (218MS3.07515 Vydac C18 polymeric reverse phase, 75 micron I.D. × 150 mm long; P.J. Cobert Assoc, St. Louis, MO). Peptides were separated with a 90 minute linear gradient from 5 to 60% acetonitrile at a flow rate of 0.3 μl/min and electrosprayed through a fused silica emitter (360 micron O.D., 75 micron I.D., 15 micron taper, New Objective, Woburn, MA) directly into the QTRAP 2000 (a hybrid quadrupole linear ion trap mass spectrometer, Applied Biosystems, Foster City, CA). An ion-spray voltage of 1900 V was maintained between the emitter and the orifice. Information dependent acquisition was used to collect MS, high resolution MS, and MSMS spectra. All spectra were collected in the enhanced mode, using the trap function. The three most intense MS peaks in each cycle having masses between 200 and 1700 m/z, charge of +1 to +4, and intensities greater than 10,000 cps were selected for high resolution MS and MSMS analysis. Precursor ions were excluded for 30 s after one MSMS spectrum had been collected. MSMS fragmentation was obtained by low energy collision induced dissociation (CID). The collision cell was pressurized to 40 μTorr with pure nitrogen. Collision energies between 20 and 40 eV were determined automatically by the software based on the mass and charge of the precursor ion. The mass spectrometer was calibrated on selected fragments from the MSMS spectrum of human Glu-fibrinopeptide B.
Results
OP labeled peptides
A lysine from each of twenty-two peptides was found to have reacted with either chlorpyrifos-oxon or diisopropylfluorophosphate. Those 22 peptides came from seven different proteins: 5 from human serum albumin, 2 from human keratin 1, 1 from human keratin 10, 6 from bovine tubulin alpha, 1 from bovine tubulin beta, 5 from bovine actin, and 2 from mouse transferrin (see Table 1).
Table 1.
P-Labeled Lysine-Containing Peptides Observed in Tryptic Digests
| # | Species | Protein | Peptidea | Residueb number | gi: numberc | Peptided mass | OPe |
|---|---|---|---|---|---|---|---|
| 1 | Human | Serum Albumin | AFK*AWAVAR | K212 | 28592 | 1155.4 | CPO |
| 2 | Human | Serum Albumin | K*VPQVSTPTLVEVSR | K414 | 28592 | 1775.5 | CPO |
| 3 | Human | Serum Albumin | Y*TKK*VPQVSTPTLVEVSR | K414 Y411 | 28592 | 2307.4 | CPO |
| 4 | Human | Serum Albumin | LK*CASLQK | K199 | 28592 | 1083.4 | CPO |
| 5 | Human | Serum Albumin | LAK*TYETTLEK | K351 | 28592 | 1461.0 | DFP |
| 6 | Human | Keratin 1 | FLEQQNQVLQTK*WELLQQVDTSTR | K211 | 119395750 | 3069.1 | CPO |
| 7 | Human | Keratin 1 | SLDLDSIIAEVK*AQYEDIQK | K355 | 119395750 | 2486.5 | CPO |
| 8 | Human | Keratin 10 | LASYLDK*VR | K163 | 47744568 | 1200.7 | CPO |
| 9 | Bovine | Tubulin alpha | DVNAAIATIK*TK | K336 | 73586894 | 1380.8 | CPO |
| 10 | Bovine | Tubulin alpha | LSVDY*GK*K | K163 Y161 | 73586894 | 1181.6 | CPO |
| 11 | Bovine | Tubulin alpha | LDHK*FDLMYAK | K394 | 73586894 | 1517.0 | CPO |
| 12 | Bovine | Tubulin alpha | FDLMY*AK*R | K401 Y399 | 73586894 | 1315.6 | CPO |
| 13 | Bovine | Tubulin alpha | Y*AK*R | K401 Y399 | 73586894 | 809.4 | CPO |
| 14 | Bovine | Tubulin alpha | TIGGGDDSFNTFFSETGAGK*HVPR | K60 | 73586894 | 2633.4 | CPO |
| 15 | Bovine | Tubulin beta | K*Y*VPR | K58 Y59 | 75773583 | 934.0 | CPO |
| 16 | Bovine | Actin | VAPEEHPTLLTEAPLNPK*ANR | K113 | 62287933 | 2433.7 | CPO |
| 17 | Bovine | Actin | DLTDYLMK*ILTER | K291 | 62287933 | 1746.4 | CPO |
| 18 | Bovine | Actin | EITALAPSTMK*IK | K326 | 62287933 | 1538.4 | CPO |
| 19 | Bovine | Actin | MQK*EITALAPSTMK | K315 | 62287933 | 1684.3 | CPO |
| 20 | Bovine | Actin | IK*IIAPPER | K328 | 62287933 | 1172.5 | CPO |
| 21 | Mouse | Transferrin | DLLFK*DSAFGLLR | K296 | 21363012 | 1631.4 | CPO |
| 22 | Mouse | Transferrin | STTK*DLLFR | K626 | 21363012 | 1217.0 | CPO |
K* indicates the labeled lysine. Y* indicates a labeled tyrosine in the same peptide.
Numbering is for the mature sequence, except for the keratins which are numbered from the gene sequence.
The gi number is the NCBI accession number for the protein from PubMed.
These are the measured, singly-charged, average masses [M+H]1+ for the labeled peptides. Peptides containing Cys are carbamidomethylated.
CPO= chlorpyrifos oxon; DFP=diisopropyl fluorophosphate
When chlorpyrifos-oxon reacts with lysine, the mass added to the peptide is 136 amu (for diethoxyphosphate). When diisopropylfluorophosphate reacts with lysine, the mass added to the peptide is 164 amu (for diisopropoxyphosphate). Most of the peptides in Table 1 exhibited a parent ion mass equal to the mass of the amino acid sequence plus the mass added from the OP-adduct (to within 0.2 amu). In 5 instances, the mass of the peptide was equal to the mass of the amino acid sequence plus twice the mass of the OP-adduct. In the latter instances, a label was found on both a tyrosine and a lysine. The sequence of each peptide was confirmed by manual analysis of the CID MSMS spectrum. The “MS Product” algorithm (a component of Protein Prospector, v 5.2.2, on the http//prospector.ucsf.edu website from the University of California at San Francisco) was used to help identify fragments from the MSMS spectra. The Mascot database algorithm (Matrix Science, London, UK, http://www.matrixscience.com) was also used in the identification of labeled peptides [32]. The masses for OP-labeled lysine have been added to the Unimod protein modification database (http//www.unimod.org) to facilitate the use of Mascot in searches for OP-modified residues. They are listed as O-diethylphosphate (for chlorpyrifos-oxon) and diisopropylphosphate (for diisopropylfluorophosphate). The precise location of the labeled amino acid could generally be established from the observed sequence. Finally, characteristic, non-sequence masses were identified that supported the proposed labeling (see Table 2).
Table 2.
OP-Lysine Characteristic Ionsa
| Mass amub | Name | Structure | Per Centc | Rel Intensed |
|---|---|---|---|---|
| Chlorpyrifos-oxon | ||||
| 21 tryptic peptides analyzed | ||||
| 220.0 | diethoxyphospho Lys immonium minus NH3 | 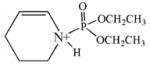 |
95 | 6–100 |
| 192.0 | monoethoxyphospho Lys immonium minus NH3 | 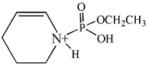 |
85 | 4–38 |
| 163.9 | phospho Lys immonium minus NH3 | 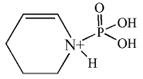 |
75 | 5–26 |
| 128.9 | α-amino caprolactam | 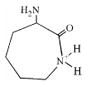 |
30 | 4–13 |
| 129.9 | Pipecolic acid | 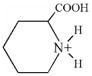 |
10 | 10–15 |
| 83.9 | Lys immonium minus NH3 | 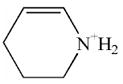 |
50 | 4–27 |
| 237.1 | diethoxyphospho Lys immonium | 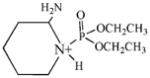 |
50 | 3–18 |
| 237 | monoethoxyphospho α-amino caprolactam | 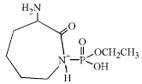 |
-- | -- |
| 209.1 | monoethoxyphospho Lys immonium | 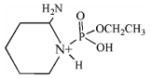 |
10 | 2–5 |
| Diisopropylfluorophosphate | ||||
| 1 tryptic peptide analyzed | ||||
| 164.0 | phospho Lys immonium minus NH3 | 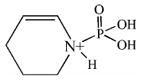 |
100 | 42 |
| 181.2 | phospho Lys immonium | 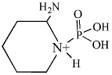 |
100 | 8 |
| 8\ | Lys immonium minus NH3 | 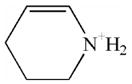 |
100 | 8 |
All masses are for the protonated, dehydro form of the amino acid.
Masses are given as the average of all measurements.
“Per Cent” refers to the fraction of the tryptic peptides that exhibited this mass.
“Rel Intense” refers to the intensity of the mass relative to the most intense peak in the MSMS spectrum. This value is generally given as a range, in percentage. Entries without a range represent masses which appeared only once.
Reaction of OP with lysine
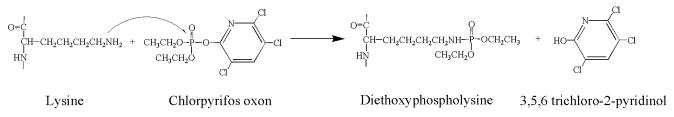
Reaction of an OP with a lysine (in a protein) would be expected to proceed via nucleophilic attack of the ε-amino group from the lysine on the phosphorus of the OP. This reaction would result in addition of a single organophosphorous moiety to the ε-amino group of the lysine (Figure 1).
Figure 1.
Illustration of the reaction of lysine with OP, using chlorpyrifos oxon as an example for the OP.
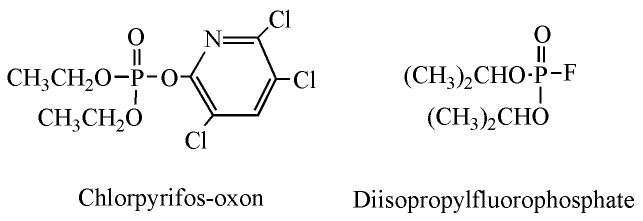
The OP used in the following work were chlorpyrifos-oxon and diisopropylfluorophosphate (see Figure 2 for structures). The groups that are displaced from the OP as a consequence of nucleophilic attack are the O-(3,5,6-trichloro-2-pyridinyl) of chlorpyrifos-oxon and the fluoride of diisopropylfluorophosphate. The resulting adducts are diethoxyphospho-lysine and diisopropoxyphospho-lysine, respectively.
Figure 2.
OP structures
CID fragmentation of ε-N-modified lysine from peptides
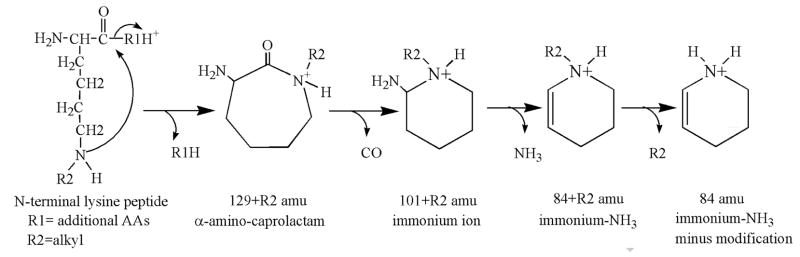
When peptides containing ε-N-modified lysine are subjected to low energy CID, a sequence of reactions ensues that yields prominent characteristic fragment ions corresponding to 1) a modified α-amino-caprolactam (at 129 amu plus the mass of the modification), 2) a modified-lysine immonium ion (at 101 amu plus the mass of the modification), 3) a modified-lysine (at 84 amu plus the added mass of the modification), and 4) a lysine immonium ion, minus NH3 (at 84 amu). Figure 3 illustrates the immonium ion, minus the modification and minus NH3 sequence of structures involved in this fragmentation process. Details of the process were worked-out by Fenaille et al. using MSMS and pseudo-MS3 analysis of peptides containing lysine modified on the ε-amine with hexanal [33]. Their proposed sequence of steps is consistent with the process described by Yalcin and Harrison for CID fragmentation of unmodified lysine-containing peptides [34]. CID fragmentation of OP-labeled lysine would be expected to follow the pathway described in Figure 3. This is essentially what is observed. We have found fragment masses that are consistent with these structures from peptides containing OP-modified lysine (where the OP is either chlorpyrifos-oxon or diisopropylfluorophosphate) (see Table 2).
Figure 3.
CID Fragmentation of ε-N-modified lysine. The mass of R2 for DFP can be +164 for diisopropoxyphosphate, or +80 for DFP that has lost both isopropyl groups. The mass of R2 for CPO can be +136 for diethoxyphosphate, or +108 for monoethoxyphosphate, or +80 for is indicated as CPO that has lost both ethoxy groups. The immonium ion minus NH3 immonium-NH3.
An alternative fragmentation pathway has also been reported that leads to pipecolic acid (130 amu) [33, 34]. This mass was occasionally observed in the CID of peptides containing OP-labeled lysine.
Characteristic fragments from OP labeled lysine
Chlorpyrifos-oxon
When chlorpyrifos-oxon reacts with lysine, the added mass is 136 amu (for diethoxyphosphate). Under CID conditions, there is a relatively facile gas-phase elimination of one or both ethylene side-chains (28 amu each) from the diethoxyphosphate [31] resulting in a monoethoxyphosphate adduct (added mass 108 amu) or a phosphate adduct (added mass 80 amu). This side-chain elimination explains the lysine related masses observed during CID fragmentation of chlorpyrifos-oxon modified lysine-containing peptides (see Table 2).
CID fragmentation of peptides containing diethoxyphosphate-modified lysine yields non-sequence, characteristic ions at 237, 220, 209, 192, 164, 130, 129, and 84 amu. These masses correspond to the diethoxyphosphate adduct of the lysine immonium ion (136 + 101 = 237 amu), the diethoxyphosphate adduct of the lysine immonium ion minus NH3 (136 + 84 =220 amu), the monoethoxyphosphate adduct of the lysine immonium ion (108 + 101 = 209 amu) the monoethoxyphosphate adduct of the lysine immonium ion minus NH3 (108 + 84 = 192), the phosphate adduct of the lysine immonium ion minus NH3 (80 + 84 = 164), pipecolic acid (130 amu), α-amino-caprolactam (129 amu), and the lysine immonium ion minus NH3 and minus all vestiges of the diethoxyphosphate label (84 amu). Table 2 lists these fragments, shows their structures, indicates the frequency with which they appeared in the 21 MSMS spectra of chlorpyrifos-oxon modified peptides, and indicates their intensities relative to the most intense mass in each spectrum.
As indicated in Table 2, the 237 amu mass could be interpreted as either the diethoxyphospho-lysine immonium ion or the monoethoxyphospho α-amino-caprolactam. If the latter interpretation were correct, one might expect to see ions consistent with the diethoxyphospho α-amino-caprolactam (at 265 amu). No peak at 265 amu appeared in any of the 21 MSMS spectra, suggesting that the α-amino-caprolactam interpretation is incorrect. Conversely, masses for both the diethoxyphospho-lysine immonium ion (237 amu) and the monoethoxyphospho-lysine immonium ion (209 amu) were detected (see Table 2), suggesting that the lysine immonium ion interpretation is correct. Modified immonium ions are most often the source of characteristic fragments [35].
The most prevalent characteristic ion was the diethoxyphospho-lysine immonium ion minus (220 amu), appearing in 95% of the MSMS spectra (20 out of 21). This is consistent with NH3 to be a the results of Fenaille et al. who found the modified-lysine immonium ion minus NH3 prominent mass in MSMS spectra of hexanal-modified lysine containing peptides [33]. Though the intensity of the 220 amu ion varied from 6% to 100%, of the most intense ion in the spectrum, its intensity was generally around 30%. On three occasions it was the most intense ion in the spectrum. The two next most common characteristic ions were also modified-derivatives of the lysine immonium ion minus NH3: monoethoxyphospho-lysine immonium ion minus NH3 (163.9 amu). The lysine immonium (192.0 amu) and phospho-lysine immonium ion minus NH3 ion (without modification, 83.9 amu) and the diethoxyphospho-lysine immonium ion (237.1 amu) also were frequently present. Occasionally, masses for the α-amino caprolactam (128.9 amu), pipecolic acid (129.9 amu), and monoethoxyphospho-lysine immonium ion (209.1 amu) appeared.
Once identified, the characteristic ions for the diethoxyphosphate-adduct were particularly useful in that they provided a convenient way to identify diethoxyphosphate-labeled lysine in other peptides.
Diisopropylfluorophosphate
Only human serum albumin was treated with diisopropylfluorophosphate, and only one labeled peptide was found, LAK*TYETTLEK. Since no other proteins were treated with diisopropylfluorophosphate, it is entirely possible that other diisopropylfluorophosphate-labeled peptides may be found in the future.
When diisopropylfluorophosphate reacts with lysine, the added mass is 164 amu (for diisopropoxyphosphate). Under CID conditions, gas-phase elimination of the isopropylene side-chains (42 amu) is even more facile than elimination of ethylene is for the diethoxyphospho-adducts [31]. Isopropylene elimination is so easy that there is no evidence for characteristic ions that retain a side chain.
CID fragmentation of the peptide containing the diisopropoxyphosphate-modified lysine yielded non-sequence, characteristic ions at 181, 164 and 84 amu. These masses correspond to the phospho-lysine immonium ion (80 + 101 = 181 amu), the phospho-lysine immonium ion minus NH3 (80 + 84 = 164 amu), and the lysine immonium ion minus NH3 and minus all vestiges of the diisopropoxyphosphate label (84 amu). Table 2 lists these fragments, shows their structures, and indicates their intensities relative to the most intense mass in the spectrum. Since only one diisopropoxyphospho-peptide was identified, the frequency with which they appear in the MSMS spectra is set at 100%.
Illustration of CID fragmentation for peptides containing OP-lysine adducts
Figures 4 through 7 show representative MSMS fragmentation spectra for peptides containing OP-labeled lysine. There is a spectrum for the diisopropoxyphosphate adduct (Figure 4), a spectrum for a peptide carrying a single diethoxyphosphate adduct on lysine which yielded characteristic fragments (Figure 5), a spectrum for a diethoxyphosphate adduct on lysine which did not yield characteristic fragments (Figure 6), and a spectrum for a peptide carrying a diethoxyphosphate adduct on lysine and another diethoxyphosphate adduct on tyrosine which yielded fragments characteristic of both labeled amino acids (Figure 7).
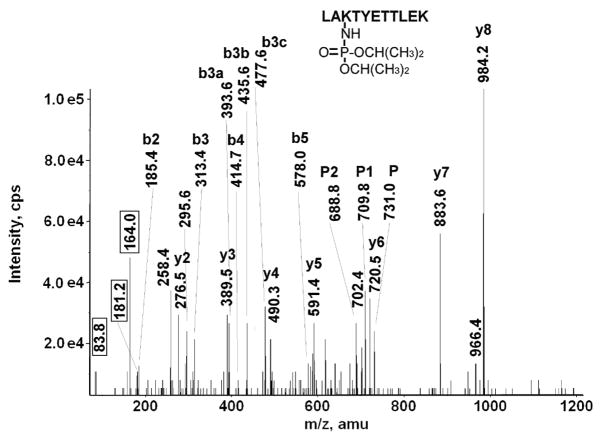
Figure 4.
A CID mass spectrum of the diisopropoxyphosphate-labeled, human serum albumin, tryptic peptide LAK*TYETTLEK. The doubly-charged parent ion at 731.0 amu is designated by the letter P. Sequential neutral loss of isopropylene (42 amu) from the diisopropoxyphosphate parent ion is designated by P1 (709.8 amu) and P2 (688.3 amu). Sequential neutral loss of isopropylene from the b3 ion is indicated by b3c (diisopropoxyphosphate form at 477.6 amu), b3b (monoisopropoxyphosphate form at 435.6 amu), and b3a (phosphate form at 363.6 amu). The values enclosed in the boxes are the masses of the characteristic fragments for diisopropoxyphosphate-labeled lysine. Lysine 351 is covalently modified by diisopropylfluorophosphate.
Figure 7.
A CID mass spectrum of the diethoxyphosphate-labeled, bovine tubulin beta, tryptic peptide K*Y*VPR. The values enclosed in the boxes are the masses of the characteristic fragments for diethoxyphosphate-labeled lysine and diethoxyphosphate-labeled tyrosine. The parent ion is marked by [M+2H]+2. Lysine 58 and tyrosine 59 are labeled by chlorpyrifos oxon.
Figure 5.
A CID mass spectrum of the diethoxyphosphate-labeled, mouse transferrin, tryptic peptide STTK*DLLFR. The values enclosed in the boxes are the masses of the characteristic fragments for diethoxyphosphate-labeled lysine. The parent ion is marked by [M+2H]+2. Lysine 626 is labeled by chlorpyrifos oxon.
Figure 6.
A CID mass spectrum of the diethoxyphosphate-labeled, bovine actin, tryptic peptide VAPEEHPTLLTEAPLNPK*ANR. The parent ion is marked by the letter P. There are no characteristic fragments for diethoxyphosphate-labeled lysine. Lysine 113 is covalently modified by chlorpyrifos oxon.
Figure 4 shows the MSMS spectrum of the diisopropoxyphosphate-labeled peptide LAK*TYETTLEK from human serum albumin. It is labeled on the lysine that is three residues from the N-terminus. That this lysine was not cleaved during the tryptic digestion supports the proposal that it is labeled. The parent ion is doubly-charged with an m/z of 731.0, which includes the mass of the amino acid sequence plus an added mass of 164 amu for the diisopropoxyphosphate.
The spectrum is complicated by the facile loss of isopropylene (42 amu) from the diisopropoxyphosphate [31]. This neutral loss fragmentation accounts for the doubly-charged peaks at 709.8, and 688.3 amu, designated P1 and P2. These are associated with the parent ion (at 731.0 amu, designated P), and are consistent with the loss of first one and then both isopropylene groups from the diisopropoxyphosphate adduct of the parent ion.
A similar phenomenon occurs for the b3 ion. This ion appears in four forms, designated b3, b3a, b3b, and b3c. The mass of the b3c fragment (477.6 amu) is consistent with the N-terminal three amino acids, LAK, plus the mass of diisopropoxyphosphate. Fragment b3b is 42 amu smaller (at 435.6 amu) consistent with the loss of one isopropylene group. Fragment b3a (at 393.6 amu) is smaller by yet another 42 amu. Finally, the b3 fragment (at 313.4 amu) is 80 amu smaller than b3a indicative of the loss of the phospho-moiety. The masses of these b3 fragments are all consistent with diisopropoxyphosphate labeling of the lysine at position 3.
The b-series, without any added mass, continues from b3 to b5. In addition, there is a y-series that extends from y2 to y8. None of the residues in this series show any indication of being labeled. The y-series includes residues y7 and y8, which carry the two other potentially reactive groups, threonine and tyrosine. The fact that their masses do not include the adduct mass indicates that they are not labeled. Most of the other major peaks in the spectrum are consistent with loss of water from the sequence ions or with characteristic fragments. The foregoing observations: the parent ion mass, the neutral losses from the parent ion, the sequence data, and the neutral loss from the b3 fragment clearly identify this peptide as being labeled by diisopropoxyphosphate on lysine.
Having established the identity of the label, we can turn to the non-sequence fragments in the CID spectrum to identify masses that are characteristic of this particular label. Three such masses appear in Figure 4. They are enclosed in boxes for emphasis. The most intense of these masses is at 164.0 amu. It is consistent with phospho-lysine immonium ion minus NH3. The phospho-lysine immonium ion is also present (at 181.2 amu), as is the lysine immonium ion at 83.6 amu. Structures for these compounds are given in Table 2.
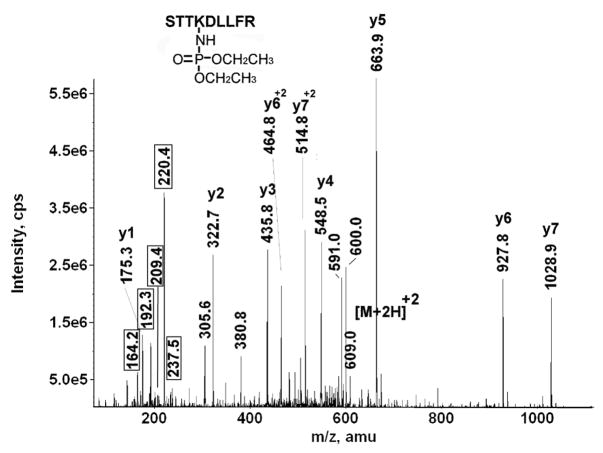
Figure 5 shows the MSMS spectrum of the diethoxyphosphate-labeled peptide STTK*DLLFR from mouse transferrin. It is labeled on the lysine that is four residues from the N-terminus. That this lysine was not cleaved during the tryptic digestion supports the proposal that it is labeled. The parent ion is doubly-charged with an m/z of 609.0, which includes the mass of the amino acid sequence plus an added mass of 136 amu for the diethoxyphosphate.
A y-ion series from y1 to y7 is present. The delta mass for the diethoxyphosphate-lysine adduct appears in the sequence between y5 and y6. This delta mass is 263.9 amu, consistent with the expected value for lysine (128 amu) plus diethoxyphosphate (136 amu). Unlike the diisopropoxyphosphate adduct, neutral loss is not evident from either the parent ion or the sequence ions. This is similar to our experience with modified tyrosine [31]. The masses at 464.8 amu and 514.8 amu are consistent with doubly-charged forms of y6 and y7. Most other significant masses could be assigned as fragments due to loss of ammonia from sequence ions, to loss of water from the parent ion, to internal fragments or characteristic ions. The parent ion mass and the sequence data, including the y5–y6 interval for diethoxyphospho-lysine, clearly identify this peptide as being labeled on lysine by diethoxyphosphate.
Non-sequence, characteristic fragments appeared at 237.5 amu (diethoxyphospho-lysine immonium ion), 220.4 amu (diethoxyphospho-lysine immonium ion minus NH3), 209.4 amu (monoethoxyphospho-lysine immonium ion), 192.3 amu (monoethoxyphospho-lysine immonium ion minus NH3) and 164.2 amu (phospho-lysine immonium ion minus NH3). Though neutral loss of ethylene (28 amu) from neither the parent ion nor the sequence ions was observed, neutral loss from the immonium ion seems to be more facile. A similar observation was made for diethoxyphospho-tyrosine [31]. Structures of the characteristic ions are given in Table 2.
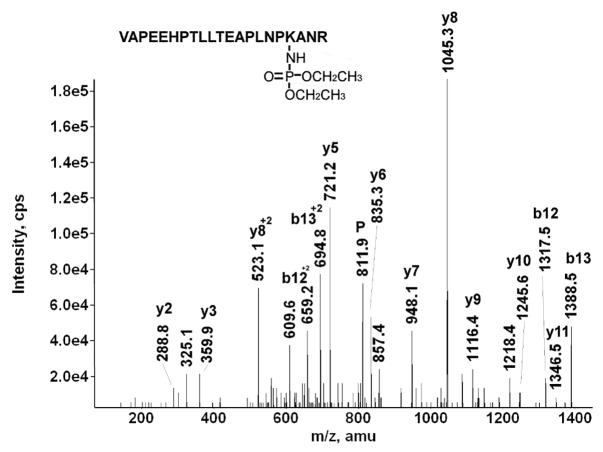
Figure 6 shows the MSMS spectrum of the diethoxyphosphate-labeled peptide VAPEEHPTLLTEAPLNPK*ANR from bovine actin. It is labeled on the lysine that is four residues from the C-terminus. That this lysine was not cleaved during the tryptic digestion supports the proposal that it is labeled. The parent ion is triply-charged with an m/z of 811.9, which includes the mass of the amino acid sequence plus an added mass of 136 amu for the diethoxyphosphate.
A y-ion series from y2 to y11 is present. There is a gap in the sequence at y4, which is the position of the proposed lysine adduct. The delta mass for this gap is too large for simply the lysine adduct. However, it is consistent with the interval between y3 and y5 (361.3 amu) which would include lysine plus diethoxyphosphate plus proline (128 + 136 + 97 = 361 amu). This places the diethoxyphospho-adduct on either lysine or proline. Since proline is not a viable candidate for labeling, it may be concluded that the label resides on lysine. In addition to the y-series, there are strong peaks for the b12 and b13 fragments, both the singly-charged forms (at 1317.5 and 1388.5 amu) and the doubly-charged forms (at 659.2 and 694.8 m/z). Most other major peaks can be assigned to internal fragments.
The parent mass and the sequence data, including the y3–y5 interval for diethoxyphospho-lysine and proline, clearly identify this peptide as being labeled by diethoxyphosphate on lysine. However, there is no evidence for characteristic ions in the MSMS spectrum. This was the only peptide of the 21 CPO-labeled peptides we analyzed that did not show any characteristic fragments.
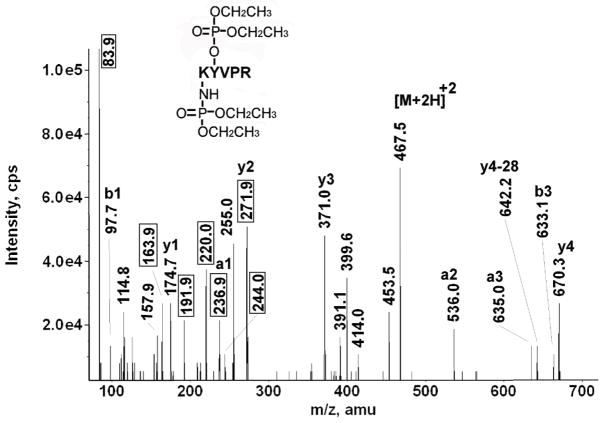
Figure 7 shows the MSMS spectrum of the diethoxyphosphate-labeled peptide K*Y*VPR from bovine tubulin beta. This peptide is doubly-labeled, once on the N-terminal lysine and again on the neighboring tyrosine. In support of the foregoing statement, the parent ion mass is 467.5 m/z (doubly-charged), which includes the mass of the amino acid sequence plus two-times the added mass of 136 amu, for the two diethoxyphosphate labels. The N-terminal lysine and the neighboring tyrosine are the only reasonable candidates for labeling in this peptide. Simultaneous labeling of both lysine and tyrosine occurred in four other peptides (Table 1): Y*AK*R, LSVDY*GK*K and FDLMY*AK*R (all three from bovine tubulin alpha) and Y*TKK*VPQVSTPTLVEVSR (from human serum albumin). Thus, double-labeling is not an isolated phenomenon. Labeling of two tyrosines in a single peptide was observed for two peptides from mouse transferrin [31].
A complete y-ion series for K*Y*VPR is present in Figure 7. The interval between y3 and y4 (299.3 amu), is consistent with a diethoxyphosphate-labeled tyrosine (163 + 136 = 299 amu). Addition of 264 amu to y4 (670.3 amu) yields the singly-charged parent ion mass of 934 amu. The mass for diethoxyphosphate-labeled lysine is 264 amu (128 + 136 amu). There is a neutral loss from y4 of 28 amu (to give the peak at 642.2 amu) which is most likely due to dissociation of ethylene from the diethoxyphosphate adduct on tyrosine. A similar loss of 28 amu from b3 (to give the peak at 635.0 amu) is more difficult to interpret. B-ions readily lose CO (28 amu) to yield a-ions. Alternatively, the 28 amu loss could again reflect loss of ethylene. The 635.0 amu mass has been designated a3 in Figure 7.
Characteristic ions for diethoxyphospho-lysine appear at 220.0, 191.9, 163.9, and 83.9 amu. In addition, there is a mass at 236.9 amu which could be interpreted as the diethoxyphospho-lysine immonium ion or the a1 ion. There is a significant peak at 244.0 amu, which is consistent with the monoethoxyphospho-tyrosine immonium ion [31]. A more commonly observed characteristic ion for chlorpyrifos oxon labeled tyrosine is the diethoxyphospho-tyrosine immonium ion at 272 amu [31]. Though an intense peak exists at 271.9 amu, its interpretation is complicated by the possibility of a proline-arginine internal fragment (nominal mass of 272 amu). Support for the existence of the internal fragment is the 97.7 amu mass which is consistent with the N-terminal proline expected from such an internal fragment. At a minimum, the 271.9 amu mass is a combination of the proline-arginine internal fragment and the diethoxyphospho-tyrosine immonium characteristic ion.
A final complication in this MSMS spectrum is a group of doubly-charged fragments (at 453.5, 414.0, 399.6 and 391.1 m/z) in the vicinity of the parent ion. The 453.5 m/z fragment is 28 amu smaller than the parent ion, making it tempting to assign 453.5 to a neutral loss of ethylene from the parent ion. Similarly, the 399.6 m/z fragment is 136 amu smaller than the parent, making it reasonable to assign 399.6 to a neutral loss of the entire diethoxyphosphate group, i.e. HPO(OCH2CH2)2. However, a neutral loss of this sort has not been seen before, either for diethoxyphospho-tyrosine or diethoxyphospho-lysine adducts. The m/z 414.0 is still more confusing. Being 108 amu smaller than the parent, it would appear to be due to neutral loss of monoethoxyphosphate where the second ethylene remains attached to the peptide; however, we can conceive of no mechanism to accommodate such an interpretation. The 391.1 m/z is a simple loss of 17 amu (NH3) from 399.6 m/z. One might attribute these masses to contaminants or some other sort of artifact except that an analogous pattern of peaks is seen in the MSMS spectrum of Y*AK*R (data not shown).
The parent ion mass, the sequence data (including the y3 to y4 and y4 to parent ion intervals), and the characteristic fragments strongly argue that this peptide is doubly-labeled by chlorpyrifos-oxon, carrying diethoxyphosphate groups on both lysine and tyrosine.
Discussion
Reaction of OP with tyrosine in a variety of proteins has been thoroughly documented [31]. We have now found a comparable reaction between OP and lysine. Table 1 lists 22 peptides from 7 proteins that are labeled on lysine. Involvement of this many different proteins strongly suggests that the reaction is commonplace. Nonetheless, we are aware of no other reports of OP reacting with lysine on protein.
Analysis of the MSMS spectra for 4 of the 22 peptides is presented. Comparable data exist for the remaining 18. These data firmly establish the existence of this reaction. A useful list of ions that are characteristic of diethoxyphospho-lysine and diisopropoxyphospho-lysine are given in Table 2. We have found the characteristic ions for diethoxyphospho-lysine to be especially helpful in finding unknown peptides that were labeled.
Until recently, investigations into the reaction of OP with biological targets have been confined largely to studies on serine hydrolases where the OP react with the active site serine [2 11, 12]. Early, work did demonstrate that OP could react with selected tyrosines on some proteins [23, 24, 25, 26]. These studies laid the groundwork for the possibility that reaction of OP with proteins other than acetylcholinesterase could be clinically relevant. As a result of increased interest in the hypothesis that there are clinically relevant targets for OP, other than acetylcholinesterase, the reaction of OP with tyrosine in proteins has been revisited [18, 20, 21, 31]. The observation of OP-reactive lysine described in this paper enlarges the field of targets which must be considered when searching for potential, clinically significant OP-targets.
Acknowledgments
This work was supported by U.S. Army Medical Research and Materiel Command [W81XWH-07-2-0034 to OL]; National Institutes of Health [U01 NS058056 to OL, P30CA36727 to Eppley Cancer Center]; Direction Générale de l’Armement of the French Ministry of Defense [DGA grant 03co010-05/PEA01 08 7; DGA/PEA 08co501]; and Agence Nationale pour la Recherche [ANR-06-BLAN-0163]. Mass spectra were obtained with the support of the Mass Spectrometry and Proteomics core facility at the University of Nebraska Medical Center.
Footnotes
The address to which proofs should be mailed is the same as that of the corresponding author.
Abbreviations: OP, organophosphate; CID, collision induced dissociation; AChE, acetylcholinesterase; DFP, diisopropylfluorophosphate, O,O-diisopropyl fluorophosphate; CPO, chlorpyrifos-oxon, O,O-diethyl-(3,5,6-trichloro-2-pyridyl) phosphate; MS, mass spectrometry; MSMS, tandem mass spectrometry fragmentation; pseudo-MS3, fragmentation of a fragment from MSMS; MWCO, molecular weight cut off
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pope CN. Organophosphorus pesticides: do they all have the same mechanism of toxicity? J Toxicol Environ Health B Crit Rev. 1999;2:161–181. doi: 10.1080/109374099281205. [DOI] [PubMed] [Google Scholar]
- 2.Casida JE, Quistad GB. Organophosphate toxicology: safety aspects of nonacetylcholinesterase secondary targets. Chem Res Toxicol. 2004;17:983–998. doi: 10.1021/tx0499259. [DOI] [PubMed] [Google Scholar]
- 3.Maxwell DM, Brecht KM, Koplovitz I, Sweeney RE. Acetylcholinesterase inhibition: does it explain the toxicity of organophosphorus compounds? Arch Toxicol. 2006;80:756–760. doi: 10.1007/s00204-006-0120-2. [DOI] [PubMed] [Google Scholar]
- 4.Brown MA, Brix KA. Review of health consequences from high-, intermediate- and low-level exposure to organophosphorus nerve agents. J Appl Toxicol. 1998;18:393–408. doi: 10.1002/(sici)1099-1263(199811/12)18:6<393::aid-jat528>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 5.Abou-Donia MB. Organophosphorus ester-induced chronic neurotoxicity. Arch Environ Health. 2003;58:484–497. doi: 10.3200/AEOH.58.8.484-497. [DOI] [PubMed] [Google Scholar]
- 6.Salvi RM, Lara DR, Ghisolfi ES, Portela LV, Dias RD, Souza DO. Neuropsychiatric evaluation in subjects chronically exposed to organophosphate pesticides. Toxicol Sci. 2003;72:267–271. doi: 10.1093/toxsci/kfg034. [DOI] [PubMed] [Google Scholar]
- 7.Beseler C, Stallones L, Hoppin JA, Alavanja MC, Blair A, Keefe T, Kamel F. Depression and pesticide exposures in female spouses of licensed pesticide applicators in the agricultural health study cohort. J Occup Environ Med. 2006;48:1005–1013. doi: 10.1097/01.jom.0000235938.70212.dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamel F, Engel LS, Gladen BC, Hoppin JA, Alavanja MC, Sandler DP. Neurologic symptoms in licensed pesticide applicators in the Agricultural Health Study. Hum Exp Toxicol. 2007;26:243–250. doi: 10.1177/0960327107070582. [DOI] [PubMed] [Google Scholar]
- 9.Hancock DB, Martin ER, Mayhew GM, Stajich JM, Jewett R, Stacy MA, Scott BL, Vance JM, Scott WK. Pesticide exposure and risk of Parkinson’s disease: a family-based case-control study. BMC Neurol. 2008;8:6. doi: 10.1186/1471-2377-8-6. (single page) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srivastava AK, Gupta BN, Bihari V, Mathur N, Srivastava LP, Pangtey BS, Bharti RS, Kumar P. Clinical, biochemical and neurobehavioural studies of workers engaged in the manufacture of quinalphos. Food Chem Toxicol. 2000;38:65–69. doi: 10.1016/s0278-6915(99)00123-4. [DOI] [PubMed] [Google Scholar]
- 11.Koshland DE. Correlation of structure and function in enzyme action. Science. 1963;142:1533–1541. doi: 10.1126/science.142.3599.1533. [DOI] [PubMed] [Google Scholar]
- 12.Kidd D, Liu Y, Cravatt BF. Profiling serine hydrolase activities in complex proteomes. Biochemistry. 2001;40:4005–4015. doi: 10.1021/bi002579j. [DOI] [PubMed] [Google Scholar]
- 13.Richards PG, Johnson MK, Ray DE. Identification of acylpeptide hydrolase as a sensitive site for reaction with organophosphorus compounds and a potential target for cognitive enhancing drugs. Mol Pharmacol. 2000;58:577–583. doi: 10.1124/mol.58.3.577. [DOI] [PubMed] [Google Scholar]
- 14.Bomser JA, Casida JE. Diethylphosphorylation of rat cardiac M2 muscarinic receptor by chlorpyrifos oxon in vitro. Toxicol Lett. 2001;119:21–26. doi: 10.1016/s0378-4274(00)00294-0. [DOI] [PubMed] [Google Scholar]
- 15.Li B, Schopfer LM, Grigoryan H, Thompson CM, Hinrichs SH, Masson P, Lockridge O. Tyrosines of human and mouse transferrin covalently labeled by organophosphorus agents: a new motif for binding to proteins that have no active site serine. Toxicol Sci. 2009;107:144–155. doi: 10.1093/toxsci/kfn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li B, Schopfer LM, Hinrichs SH, Masson P, Lockridge O. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry assay for organophosphorus toxicants bound to human albumin at Tyr411. Anal Biochem. 2007;361:263–272. doi: 10.1016/j.ab.2006.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Nachon F, Froment MT, Verdier L, Debouzy JC, Brasme B, Gillon E, Schopfer LM, Lockridge O, Masson P. Binding and hydrolysis of soman by human serum albumin. Chem Res Toxicol. 2008;21:421–431. doi: 10.1021/tx700339m. [DOI] [PubMed] [Google Scholar]
- 18.Peeples ES, Schopfer LM, Duysen EG, Spaulding R, Voelker T, Thompson CM, Lockridge O. Albumin, a new biomarker of organophosphorus toxicant exposure, identified by mass spectrometry. Toxicol Sci. 2005;83:303–312. doi: 10.1093/toxsci/kfi023. [DOI] [PubMed] [Google Scholar]
- 19.Ding SJ, Carr J, Carlson JE, Tong L, Xue W, Li Y, Schopfer LM, Li B, Nachon F, Asojo O, Thompson CM, Hinrichs SH, Masson P, Lockridge O. Five tyrosines and two serines in human albumin are labeled by the organophosphorus agent FP-biotin. Chem Res Toxicol. 2008;21:1787–1794. doi: 10.1021/tx800144z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams NH, Harrison JM, Read RW, Black RM. Phosphylated tyrosine in albumin as a biomarker of exposure to organophosphorus nerve agents. Arch Toxicol. 2007;81:627–639. doi: 10.1007/s00204-007-0191-8. [DOI] [PubMed] [Google Scholar]
- 21.Black RM, Harrison JM, Read RW. The interaction of sarin and soman with plasma proteins: the identification of a novel phosphonylation site. Arch Toxicol. 1999;73:123–126. doi: 10.1007/s002040050596. [DOI] [PubMed] [Google Scholar]
- 22.Grigoryan H, Schopfer LM, Thompson CM, Terry AV, Masson P, Lockridge O. Mass spectrometry identifies covalent binding of soman, sarin, chlorpyrifos oxon, diisopropyl fluorophosphate, and FP-biotin to tyrosines on tubulin: A potential mechanism of long term toxicity by organophosphorus agents. Chem Biol Interact. 2008;175:180–186. doi: 10.1016/j.cbi.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanger F. Amino-acid sequences in the active centers of certain enzymes. Proc Chem Soc. 1963;5:76–83. [Google Scholar]
- 24.Murachi T, Inagami T, Yasui M. Evidence for alkylphosphorylation of tyrosyl residues of stem bromelain by diisopropylphosphorofluoridate. Biochemistry. 1965;4:2815–2825. doi: 10.1021/bi00888a036. [DOI] [PubMed] [Google Scholar]
- 25.Chaiken IM, Smith EL. Reaction of a specific tyrosine residue of papain with diisopropylfluorophosphate. J Biol Chem. 1969;244:4247–4250. [PubMed] [Google Scholar]
- 26.Kato K, Murachi T. Chemical modification of tyrosyl residues of hen egg-white lysozyme by diisopropylphosphorofluoridate. J Biochem. 1971;69:725–737. doi: 10.1093/oxfordjournals.jbchem.a129521. [DOI] [PubMed] [Google Scholar]
- 27.Grigoryan H, Li B, Anderson EK, Xue W, Nachon F, Lockridge O, Schopfer LM. Covalent binding of the organophosphorus agent FP-biotin to tyrosine in eight proteins that have no active site serine. Chem Biol Interact. 2009 doi: 10.1016/j.cbi.2009.03.018. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashbolt RF, Rydon HN. The action of diisopropyl phosphorofluoridate and other anticholinesterases on amino acids. Biochem J. 1957;66:237–242. doi: 10.1042/bj0660237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith RA, Halpern RM, Bruegger BB, Dunlap AK, Fricke O. Chromosomal protein phosphorylation on basic amino acids. Meth Cell Biol. 1978;19:153–159. doi: 10.1016/s0091-679x(08)60020-5. [DOI] [PubMed] [Google Scholar]
- 30.Grigoryan H, Schopfer LM, Peeples E, Duysen E, Lockridge O. Mass spectrometry identifies multiple organophosphorylated sites on tubulin: dynamics of chlorpyrifos oxon binding. 2009 doi: 10.1016/j.taap.2009.07.020. manuscript in preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schopfer LM, Grigoryan H, Li B, Nachon F, Masson P, Lockridge O. Mass spectral characterization of organophosphate-labeled tyrosine-containing peptides: characteristic mass fragments and a new binding motif for organophosphates. submitted to J Chromatog B. 2009 doi: 10.1016/j.jchromb.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 33.Fenaille F, Tabet JC, Guy PA. Study of peptides containing modified lysine residues by tandem mass spectrometry: precursor ion scanning of hexanal-modified peptides. Rapid Commun Mass Spectrom. 2004;18:67–76. doi: 10.1002/rcm.1283. [DOI] [PubMed] [Google Scholar]
- 34.Yalcin T, Harrison AG. Ion chemistry of protonated lysine derivatives. J Mass Spectrom. 1996;31:1237–1243. doi: 10.1002/(SICI)1096-9888(199611)31:11<1237::AID-JMS416>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 35.Hung CW, Schlosser A, Wei J, Lehmann WD. Collision-induced reporter fragmentations for identification of covalently modified peptides. Anal Bioanal Chem. 2007;389:1003–1016. doi: 10.1007/s00216-007-1449-y. [DOI] [PubMed] [Google Scholar]