Abstract
The miR-17 family of microRNAs has recently been recognized for its importance during lung development. The transgenic overexpression of the entire miR-17-92 cluster in the lung epithelium led to elevated cellular proliferation and inhibition of differentiation, while targeted deletion of miR-17-92 and miR-106b-25 clusters showed embryonic or early post-natal lethality. Herein we demonstrate that miR-17 and its paralogs, miR-20a, and miR-106b, are highly expressed during the pseudoglandular stage and identify their critical functional role during embryonic lung development. Simultaneous downregulation of these three miRNAs in explants of isolated lung epithelium altered FGF10 induced budding morphogenesis, an effect that was rescued by synthetic miR-17. E-Cadherin levels were reduced, and its distribution was altered by miR-17, miR-20a and miR-106b downregulation, while conversely, beta-catenin activity was augmented, and expression of its downstream targets, including Bmp4 as well as Fgfr2b, increased. Finally, we identified Stat3 and Mapk14 as key direct targets of miR-17, miR-20a, and miR-106b and showed that simultaneous overexpression of Stat3 and Mapk14 mimics the alteration of E-Cadherin distribution observed after miR-17, miR-20a, and miR-106b downregulation. We conclude that the mir-17 family of miRNA modulates FGF10-FGFR2b downstream signaling by specifically targeting Stat3 and Mapk14, hence regulating E-Cadherin expression, which in turn modulates epithelial bud morphogenesis in response to FGF10 signaling.
Keywords: Lung, miRNAs, E-Cadherin, structural homeostasis
Introduction
Lung development is under the control of a complex network of regulatory proteins including those that participate in the FGF, TGFbeta, SHH, and WNT pathways (Cardoso and Lu, 2006; Warburton et al., 2005). Branching and growth of the epithelium are principally controlled by the FGF10-FGFR2b signaling pathway, wherein a mesenchymal FGF10 gradient acts as a chemoattractant for the endoderm and promotes its proliferation (Bellusci et al., 1997; Park et al., 1998) Fgf10 expression is strictly controlled by endodermal SHH pathways that function to finely tune Fgf10 expression, assuring the correct size and shape of the growing buds (Pepicelli et al., 1998; Warburton, 2008). The growth of the developing lung must be continuously calibrated. In the mesenchyme, for example, fibronectin deposition is strictly controlled by WNT signaling during the early stages of branching morphogenesis, and alterations of such regulation lead to impaired branching with enlarged terminal buds (De Langhe et al., 2005).
Cadherins are a large family of glycoproteins that mediate specific cell-cell adhesion in a Ca2+-dependent manner (Van Roy F. and Berx G., 2008). E-Cadherin (CDH1) is an important cadherin that is highly expressed in the epithelium throughout lung development. Structurally, CDH1 is comprised of a single transmembrane domain, an ectodomain for binding to other adjacent cells (Leckband and Prakasam, 2006), and a cytoplasmic domain that interacts with a variety of molecules that link CDH1 to the actin cytoskeleton, and hence to cell signaling and trafficking mechanisms (Bryant and Stow, 2004; Mege et al., 2006; Yap and Kovacs, 2003). Cdh1 expression is promoted by transcription factors that bind cis-regulatory elements present in introns 1 and 2 (Goomer et al., 1994; Hennig et al., 1996; Liu et al., 2005), and can also be downregulated by specific transcription factors and corepressors (Peinado et al., 2007; Shi et al., 2003). CDH1 can undergo endocytosis and recycling through an endosomal pathway that modulates its expression at the cell surface during dynamic morphogenetic processes (Bryant and Stow, 2004). CDH1 endocytosis, ubiquitination, and transport to the cell surface are finely regulated, adding another level of control over its activity (Davis et al., 2003; Palacios et al., 2002; Palacios et al., 2005). The regulation of Cdh1 and its network of regulatory molecules has been widely investigated at the transcriptional and post-translational levels, but the effect of translational regulation is poorly understood.
An emerging understanding of the functional roles of microRNAs (miRNAs) in the post-transcriptional regulation of gene expression has shed light on the growing importance of these sub-cellular interactions. MiRNAs are small non-coding RNAs that can bind the 3’-UTR of target mRNAs and thus specifically inhibit their translation. The function of miRNAs can vary from acting as a molecular switch for specific events, to finely tuning or maintaining the stability and robustness of specific states. MiRNAs can also function as a control to eliminate specific gene expression on a translational level in specific locations (Brennecke et al., 2003; Cohen et al., 2006; Hornstein et al., 2005; Johnston and Hobert, 2003; Kwon et al., 2005; Sokol and Ambros, 2005).
In the lung, tissue-specific deletion of Dicer, the enzyme responsible for producing active miRNAs, results in branching arrest, revealing the importance of miRNAs for epithelial morphogenesis (Harris et al., 2006). The miR-17 family of miRNA, is comprised of three paralog clusters (miR-17–92, miR-106a-363, and miR-106b-25). The miR17–92 cluster has recently been described for its importance during lung development. In mice the miR-17–92 cluster is located on chromosome 14 and contains six miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92-1). The miR-106a-363 cluster is located on mouse chromosome X and contains six miRNAs (miR-106a, miR-18b, miR-20b, miR-19b-2, miR-92-2, miR-363), while miR-106b-25 is located on mouse chromosome 5 and contains three miRNAs (miR-106b, miR-93, miR-25). Transgenic overexpression of the miR-17–92 cluster has been produced, showing its importance in controlling differentiation and proliferation of lung progenitor cells (Lu et al., 2007). Targeted deletion of the miR-17–92 cluster has been shown to produce smaller embryos with postnatal lethality, characterized by severely hypoplastic lungs. Furthermore during combined deletion of miR-17–92 cluster and its paralog miR-106b-25, mice showed earlier lethality before embryonic day 15 (Ventura et al., 2008). Herein we report a specific functional role for miR-17, miR-20a, and miR-106b, in maintaining the structural homeostasis of developing lung epithelium. We show that miR-17, miR-20a, and miR-106b, targets Mapk14 and Stat3 and thereby contributes to correctly modulating CDH1 expression in the epithelium in response to FGF10-FGFR2b signaling.
Materials and methods
RT-PCR and Real-time PCR
Total RNA was extracted using miRNesasy Mini Kit (Qiagen) following the manufacturer’s recommendations. For RNA extraction from lung tissue a pool of three samples was used. RT-PCR for mRNA was carried out using SuperScript II reverse transcriptase (Invitrogen) with random primers, while in RT-PCR for miRNA the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) was used. In both cases, reactions were assembled following the manufacturer’s recommendations.
Real-time PCR was performed on a LightCycler 480 system (Roche). The universal probe library (Roche) was used for the analysis of mRNA expression. The miRNA Early Access Kit or Taqman microRNA assays (Applied Biosystems) were used for the screening of miRNAs differentially expressed during different stages of lung development. Primers and probes were used at a final concentration of 250nM and 125nM respectively. Following denaturation for 10min at 95°C, 45 cycles were performed with 10s at 95°C and 30s at 60°C. A relative quantification was performed using LightCycler 480 Software, and data were normalized with Actb for mRNA and U6 RNA for miRNA.
Immunoblotting
Samples were lysed in RIPA buffer (50mM Tris–HCI, pH 7.4, 150mM NaCI, 1mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) containing 1:100 dilution of protease inhibitor cocktail set III (Calbiochem). Immunoblotting was performed as previously described (Ding et al., 2007). The following primary antibodies were used: CDH1 (BD bioscences) was used at 1:1000, STAT3 (Cell Signaling) was used at 1:2000, and MAPK14 (Cell Signaling) was used at 1:2000. ACTB (Chemicon) was used as a loading control and was used at 1:2000. Protein quantification was produced by densitometry analysis with the software Image J (Rasband W.S., U. S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij).
In situ hybridization
Embryonic lung and epithelial lung explants were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) with rocking at 4°C for 2h and 5min respectively, washed twice for 5min in PBS at 4°C, dehydrated in several washes with 70% ethanol (EtOH), and stored in absolute EtOH at −20°C. For the in situ hybridization of miR-17 we used a modified protocol (Winnier et al., 1995) after taking the short size of mature miRNA into consideration. Samples were washed in Tris-Buffered Satoe (TBS)/Tween-20 0.1% (TBST), digested with Proteinase K, fixed again in 4% PFA and washed in TBST. The samples were then transferred to hybridization buffer (Formamide 50%, SSC 5X, tRNA 0.05mg/mL, sodium dodecyl sulfate 1%, Heparin 0.05mg/mL) for 1 hour at 50°C and then hybridized with the probe at 55°C over night. Locked Nucleic Acid (LNA) probes (Exiqon) were used to increase the stability of the hybridization. The next day the lung was washed with standard washing solutions. All washes were performed at 50°C. After the washes the lungs were added to blocking solution made with sheep serum for 1 hour and then incubated over night a 4°C with anti-DIG antibody. The color reaction was performed with BM Purple solution after washes with TBST and NTMT (NaCI 100mM, Tris pH9.5 100mM, Tween-20 0.1%).
Isolated epithelial explant culture
Lungs from E11.5 embryos were treated with dispase (BD Bioscences) for 5min at 4°C and then transferred to Dulbecco’s Modified Eagle’s Medium:Nutrient Mixture F-12 (DMEM:F12) with 10% fetal bovine serum (FBS) (Invitrogen) to block the enzymatic reaction. The epithelial buds were separated from mesenchyme using tungsten needles (Fine Science Tools), embedded in 200µl of Matrigel (BD Biosciences) diluted 1:1 with DMEM:F12 serum-free media containing 250ng/ml FGF10 (R&D Systems), and left to polymerize by incubation at 37°C for 20 minutes. The same media used for the matrigel mix was then added to the surface of the polymerized gel, and the samples were incubated at 37°C (modified from Bellusci et al, 1997). Locked nucleic acid against miR-17 (LNA17) or scrambled (SCRA) (Exiqon), or miR-17 mimic molecules (MIM17) (Thermo Scientific), were added at 1.5µM to the isolated epithelium in presence of 0.3% lipofectamine RNAiMAX (Invitrogen). Image analysis
Phase-contrast images of the samples were recorded using a digital camera (Diagnostic Instruments) connected to a reversed phase-contrast microscope (Leica). The public domain software ImageJ was broadened by routines specifically developed by us and used to process and analyze the images. The main processing steps applied to the images are shown in Fig. 2D. Briefly, after correction for uneven illumination, the image underwent grey level threshold and contour smoothing to obtain the binary image of the tissue area. By using binary thinning procedures (Seul et al., 2000), the skeleton of this image was also derived and pruned to remove small artifactual branches. The number of terminal points and the number of branching points of the binary skeleton were then estimated. The former corresponds to the number of recognizable tissue branches; the latter provides an index of structural complexity of the sample (Guidolin et al., 2004). Finally, by tracing a poly-line with vertices located at the middle-point of each terminal branch of the binary skeleton, the area of tissue forming branches was measured and expressed as a percent of the total tissue area. The number of formed branches, percentage of tissue area involved in branching, and branching complexity (i.e. the number of branching points of the binary skeleton) were the morphometric parameters estimated from each of 15 samples per group. Data were processed by using statistical analysis software (GraphPad Prism 3.03, GraphPad Software Inc., San Diego, USA). P ≤ 0.05 was considered statistically significant. Comparisons of morphometric parameters between LNA17 and SCRA treated tissue cultures were performed by Student’s two-sample t-test.
Fig 2.
miR-17 downregulation disrupted progression of embryonic lung epithelium branching. (A) Real-time PCR using total RNA extracted from isolated epithelial explants grown for 48h in the presence of locked nucleic acid, scrambled (SCRA) or miR-17 specific (LNA17) was performed to determine the specificity of the LNA17 knockout probe. miR-17, miR-20a and miR-106b (three paralogs of the miR-17 family with high sequence conservation) showed significantly reduced expression after LNA17 administration, while miR-19b, a control member of the miR-17-92 cluster, was not affected. Data are reported as means ± SEM. (B, 40X) Fluorescein-labeled LNA17 probe was transfected into the epithelial lung explants and was visible after 36h. (C, 12X and 4X) Epithelial lung explants were grown for 72h in a matrigel-based three-dimensional scaffold in the presence of 250ng/ml FGF10. SCRA administration did not alter the normal pattern of branching progression (left two images), while LNA17 interfered with branching in the presence of FGF10 (right two images). (D) Morphometric parameters of LNA17- and SCRA-treated epithelial lung explants were collected and analyzed to determine their statistical significance. To the left, a description of the processing steps is depicted: the image of the tissue was corrected for uneven illumination and thresholded to obtain a binary image of the tissue area. The binary skeleton of this image was estimated and terminal branches (yellow) and branching points (blue) were identified. They are shown superimposed onto the image of the tissue area. The fraction of tissue involved in branching was then objectively defined as the area external to the polyline with vertices located at the midpoint of each terminal branch of the binary skeleton (red). To the right, the morphometry from fifteen samples per group was analyzed. Data are reported as means ± SEM. *=P ≤ 0.05.
Cell culture
Mouse lung epithelial cells (MLE12) (ATCC) were grown in DMEM/F12 (Invitrogen) in the presence of transferrin at 5µg/ml, hydrocortisone at 10µM, β-estradiol at 10µM, HEPES at 10mM, and L-glutamine 2mM (SIGMA). Cells were grown in presence of Penicillin/ streptomycin and 2% FBS. Mouse mammary gland epithelial cells (NMuMG) (ATCC) were grown in DMEM in the presence of glucose at 4.5g/L, sodium bicarbonate at 1.5 g/L, bovine insulin at 0.01 mg/ml (SIGMA) and 10% FBS.
Cell transfection and transduction
Modulation of miR-17 was obtained by use of LNA17 or SCRA, or MIM17. For Stat3 or Mapk14 silencing pools of specifically optimized small interfering RNA (siMapk14 and siStat3) and scrambled siRNA (siScra) were used (On-Target plus siRNA, Thermo Scientific). Molecules were added at 5nM to the cells in presence of 0.1% lipofectamine RNAiMAX (Invitrogen) in media without antibiotics. After 5h media was replaced with fresh complete media and cells grown for 36h. Cell were then starved for 12h with minimum media without serum in presence of 250ng/ml FGF10 and then harvested for the analysis or fixed for staining. Adenoviruses for Mapk14, Stat3 or GFP were added 24h after plating of the cells at 50% confluence and then the same procedure used after transfection was followed.
Experiments were repeated in triplicate. Adenoviruses having comparable titers were used. Effectiveness of siRNAs and adenoviruses were tested by real-time PCR on MLE12 cells.
Immunohistochemistry for whole mount and sectioned tissue
Epithelial lung explants were fixed in 4% PFA, and hydrated by washing several times in Wash PBS (PBS containing 0.05% Tween-20, 0.1% BSA and 0.02% Sodium Azide). Tissues were then pre-incubated with 0.1% Triton X-100 in PBS for 5 minutes for membrane permeabilization, and washed 5x with Wash PBS. Tissues were then incubated in FBS diluted 1/20 in PBS for 45 minutes. Then, tissues were drained and incubated overnight at 4°C with CDH1 antibody at 1:50 diluted in Wash PBS. The following day, samples were rinsed 5 times in Wash PBS. For the analysis on slides, tissues were fixed in 4% PFA, gradually dehydrated in ethanol, impregnated with toluene, embedded in paraffin, and sectioned into 5um slices on poly-L-lysine coated slides. Antigen retrieval was performed by boiling the sample for 12 minutes in Na-citrate buffer (10mM, pH 6.0) (Vector). CDH1 antibody (Ab) (BD bioscences) was used at 1:100, and MAPK14 antibody (Cell Signaling) was used at 1:20. Incubation at 4° was performed in TBST with 3% bovine serum albumin. Secondary fluorescence antibodies (Jackson Immunoresearch) were used for protein detection. DAPI (a nuclear dye of 4,6-diamidino-2- phenylindole; Vector Labs) was used to counterstain the DNA. Photomicrographs were taken using a Leica DMRA fluorescence microscope with a Hamamatsu digital camera. Digital analysis of CDH1 intensity and distribution was performed with the software MetaMorph (Molecular Devices). Cells analyzed in four different epithelial lung explants for each condition were analyzed. Data were reported as means ± SEM and P ≤ 0.05 was considered statistically significant.
β-galactosidase staining
TOPGAL mice were obtained from The Jackson Laboratory. TOPGAL epithelial explants were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) at 4 °C for 5min with rocking, washed twice for 5min in PBS at 4°C, then transferred into freshly prepared X-gal solution and stained at 37°C until a clear precipitate formed, modified from (Hogan et al., 1994). After rinsing with PBS, epithelial explants were post-fixed in 4% PFA in PBS. For vibratome sections, samples were embedded in a mixture of 300mg/ml albumin, 5mg/ml gelatin, and 0.6% glutaraldehyde, and sectioned at 30µm.
Proliferation analysis
Ki67 antibody (Labvision) was used at 1:200, and signal was developed with the Histostatin Rabbit Primary Kit (Zymed-lnvitrogen) following the manufacturer’s instructions. Slides were counterstained with Hematoxilyn. The total number of cells and the number of Ki67 positive cells in the epithelial lung explants were scored in 6 photomicrographs at the tip portions of 3 different sections in each of 3 MIM17-, 3 LNA17- and 3 SCRA-treated epithelial lung explants. The significance of the difference in proliferation between SCRA-treated and MIM17- or LNA17-treated samples was evaluated by Student’s two-sample t-test.
Migration assay
A colorimetric chemotaxis cell migration assay (Millipore) was used for this analysis according with the manufacturer protocol. Briefly, MLE12 cells were transfected with SCRA, LNA17, and MIM17. After 48h cells were harvested and plated at 30% confluence into 8µm pore size membrane and grown in DMEM:F12 without FBS. After 24h fresh media containing 250ng/ml FGF10 was added, and after 14h cells were stained and spectrophotometer reading at 560nm (ND-1000, Nanodrop) was performed. Three experiments were performed for each group.
Adenoviral constructs
Adenoviral constructs for Stat3 and Mapk14 were produced using an adenoviral vector carrying a CMV promoter (ViraPower Adenoviral Expression System, Invitrogen). Coding sequences were NM_011951 for Mapk14 and NM_213659 for Stat3 and were cloned into an entry vector (pENTR-11, Invitrogen) before being transferred to the final vector. Adenoviruses were produced with HEK293A cells carrying E1a and E1b proteins necessary to generate the adenovirus. After repeated cycles of amplification, the adenoviruses were concentrated using a spin column based system (Cell Biolabs) and titered using an immunoassay with an antibody against hexon (Cell Biolabs). The titer obtained was between 107 and 108 transducing U/ml. Adenovirus containing GFP was used as a control. Experiments were repeated in triplicate with adenoviruses having comparable titers.
Computational target prediction
Stat3 and Mapk14 were identified as principal miR17 targets based on the on-line available prediction databases Target-Scan (http://www.targetscan.org/) and Miranda (http://www.microrna.org/microrna/). Seventy base pairs at the 5’UTR and 3’UTR of the miR-17 binding position were further analyzed with the software mfold (Mathews et al., 1999; Zuker, 2003) (http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi) to determine the secondary structure and free energy. As a threshold for mouse sequences we used the previously reported value dG=-13.4 Kcal mol−1 (Zhao et al., 2005). Stems ≥10bp were considered stabilizing elements, while hairpin loops ≥11bp, interior loops ≥9bp, bulge loops ≥7bp, multiple branching loops ≥11bp, and joint sequences and free ends ≥11bp were considered destabilizing elements (Zhao et al., 2005).
Luciferase reporter assay
The predicted binding sequences on the 3’UTR of Mapk14 and Stat3 were synthetically produced by PCR and transferred into a luciferase reporter vector. For each binding position, reporter vectors with one, two or three copies of the sequence were created. To determine the specificity of the interaction, reporter vectors containing “seed” mutated sequences were generated using specific primers. The specific primer sequences were the following: for Mapk14 wild type, Forward (F) 5’-GGCTAGTCTAGATTTTCTGA-3’ and Reverse (R) 5’-GGCTAGTCTAGACAAAGTGC-3’, were used with the single copy template 5’-GGCTAGTCTAGATTTTCTGAATTTTGAGCACTTTGTCTAGACTAGCC-3’, the double copy template 5’-GGCTAGTCTAGATTTTCTGAATTTTGAGCACTTTGTTTTCTGAATTTTGAGCA CTTTGTCTAGACTAGCC-3’, and the triple copy template 5’-GGCTAGTCTAGATTTTCTGAA TTTTGAGCACTTTGTTTTCTGAATTTTGAGCACTTTGTTTTCTGAATTTTGAGCACTTTGTC TAGACTAGCC-3’. For Mapk14 mutant, R 5’- GGCTAGTCTAGAGTCGTCTG-3’ was used with the same forward used for the wild type. Templates for Mapk14 mutant were: GGCTAGTCTAGATTTTCTGAATTTTGTCAGACGACTCTAGACTAGCC for the single copy, GGCTAGTCTAGATTTTCTGAATTTTGTCAGACGACTTTTCTGAATTTTGTCAGACGACTCT AGACTAGCC for the double copy, and GGCTAGTCTAGATTTTCTGAATTTTGTCAGACGA CTTTTCTGAATTTTGTCAGACGACTTTTCTGAATTTTGTCAGACGACTCTAGACTAGCC for the triple copy For Stat3 wild type, Forward (F) 5’ GGCTAGTCTAGAGCTATCT-3’ and Reverse (R) 5’- GGCTAGTCTAGAAAAGTGCC-3’, were used with the single copy template 5’- GGCTAGTCTAGAGCTATCTTTGGGCAATCTGGGCACTTTTCTAGACTAGCC-3’ and the double copy template 5’GGCTAGTCTAGAGCTATCTTTGGGCAATCTGGGCACTTTGCTATC TTTGGGCAATCTGGGCACTTTTCTAGACTAGCC-3’, while using 5’- GGCTAGTCTAGATAT CTTTGGGCAATCTGGGCACTTTTATCTTTGGGCAATCTGGGCACTTTTATCTTTGGGCAAT CTGGGCACTTTTCTAGACTAGCC-3’ for the triple copy template, the same R was used with the F 5’-GGCTAGTCTAGATATCTTTG-3’. For Stat3 mutant, R 5’-GGCTAGTCTAGATCGTAC TC-3’ was used with the same forward used for the wild type. Templates for Stat3 mutant were: GGCTAGTCTAGAGCTATCTTTGGGCAATCTGGAGTACGATCTAGACTAGCC for the single copy, GGCTAGTCTAGAGCTATCTTTGGGCAATCTGGAGTACGAGCTATCTTTGGGC AATCTGGAGTACGATCTAGACTAGCC for the double copy, and GGCTAGTCTAGATATCTT TGGGCAATCTGGAGTACGATATCTTTGGGCAATCTGGAGTACGATATCTTTGGGCAATCTG GAGTACGATCTAGACTAGCC for the triple copy. The vector backbone was a pGL4 luciferase reporter (Promega) carrying a synthetic firefly luciferase (luc2), and our constructs were inserted downstream of the luciferase gene into a unique Xba-l restriction site. A vector carrying Renilla luciferase (Promega) was used as a control to normalize luminescence levels. Experiments were carried out in triplicate using a Dual-luciferase reporter assay system (Promega) with a Victor3 Luminometer (PerkinElmer).
Results
Developmental expression of miR-17, miR-20a and miR-106b in the embryonic lung
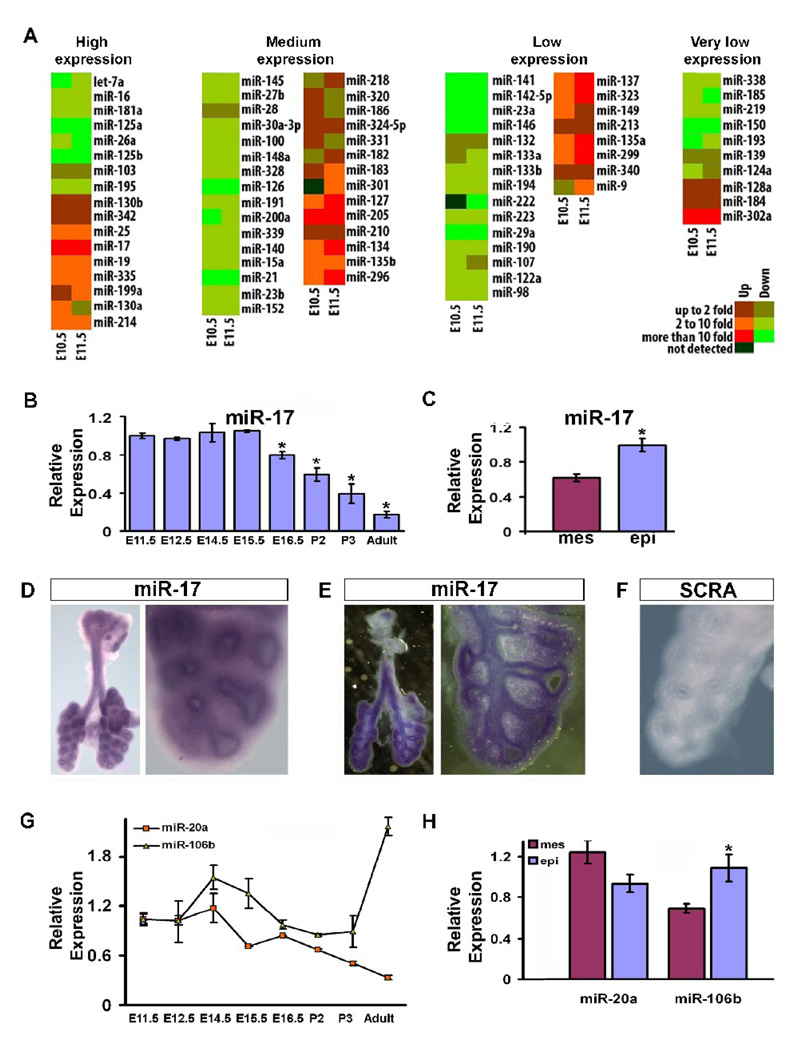
We used real-time PCR to study the expression of miRNAs in embryonic lungs as compared to the adult lung (summarized as a heat map in Figure 1A). Among the 80 miRNAs analyzed, 17 were highly expressed, 30 at medium levels, 23 at low levels, and 10 at very low levels. Furthermore, 39 miRNAs were highly expressed in the adult lung as compared with the embryonic lung. Among them are miRNAs that may participate in maintaining adult lung homeostasis: miR-126 has been noted for its importance in regulating the response of endothelial cells to VEGF, controlling vascular integrity and angiogenesis (Fish et al., 2008); and let-7a can regulate cellular proliferation, targeting Ras in human colon cancer cells (Akao et al., 2006) potentially contributing to the maintenance of correct cellular growth in the adult lung through this mechanism. Conversely, 19 miRNAs had maximum expression during early embryonic stages, suggesting they may play a prominent role during lung development. Our miRNA expression data generally agreed with a previous report on microarray analysis of miRNA expression in embryonic lung (Lu et al., 2007). Among the miRNAs more highly expressed during lung development, miR-17 stood out because if its high and stable level of expression during the pseudoglandular stage. Its expression then progressively declined, reaching a minimum level in the adult lung (Fig. 1B). Analysis of miR-17 distribution in the embryonic lung by real-time PCR and in situ hybridization at E12.5 showed expression in both mesenchyme and epithelium with relatively higher expression in the epithelium (Fig. 1C–F). Three miRNA clusters exist within the miR-17 family. miR-106a-363 expression is not detectable (Ventura et al., 2008) and therefore miR-106a and miR-20b were not taken under consideration. Within miR-17–92 and miR-106b-25 clusters, miR-20a and miR-106b have high sequence similarity with miR-17 (Fig. 2A) and therefore were also analyzed. miR-20a temporal expression during lung development is similar to miR-17 (Fig. 1G), but it is ubiquitously expressed in E12.5 embryonic lung (Fig. 1H). miR-106b during embryonic stages is highly expressed starting from E12.5 and has an even higher level of expression in adult lung (Fig. 1G). The spatial expression of miR-106b correlated with miR-17, being higher in the epithelium compared with the mesenchyme of E12.5 embryonic lung (Fig. 1H). Taken together these data suggest a possible functional role of miR-17 and its paralogs miR-20a and miR-106b during embryonic lung development.
Fig 1.
MicroRNA expression profiles in embryonic versus adult lung and miR-17, miR-20a, and miR-106b spatial and temporal expression in embryonic lung. (A) Real-time PCR was done using primers that can identify 80 individual miRNAs, as listed in the panel, on total RNA extracted from embryonic (E10.5 and E11.5) versus adult lung to identify differentially expressed miRNAs. Results are shown as a heat map wherein green represents and red represent higher expression in the adult and embryonic lung respectively, mir-17 was clearly the most highly expressed in the early embryonic lung as compared to that of the adult. (B) Real-time PCR specifically tracking miR-17 in the embryonic, post-natal, and adult lung was carried out to determine its temporal expression. (C) Real-time PCR of miR-17 in the epithelium versus the mesenchyme of E12.5 embryonic lung was performed to determine its spatial localization within the embryonic lung (D-F), and the results were confirmed by in situ hybridization (ISH) in E12.5 embryonic lung. Whole mount ISH (D, 3.2 and 14X) was further analyzed with vibratome sectioning (E, 3.2 and 14X). Control ISH was carried out using a probe composed of a scrambled sequence (F, 14X). (G) Real-time PCR specifically comparing miR-20a and miR-106b expression in the embryonic, post-natal, and adult lung is shown. (H) Real-time PCR of miR-20a and miR-106b in the epithelium versus the mesenchyme of E12.5 embryonic lung showing their relative spatial localization within the embryonic lung. Data in (B), (C), (G), and (H) are reported as means ± SEM.*=P ≤ 0.05. mes, mesenchyme; epi, epithelium; SCRA, scrambled probe.
Abnormal branching in embryonic lung epithelial explants after miR-17, miR-20a and miR-106b downregulation
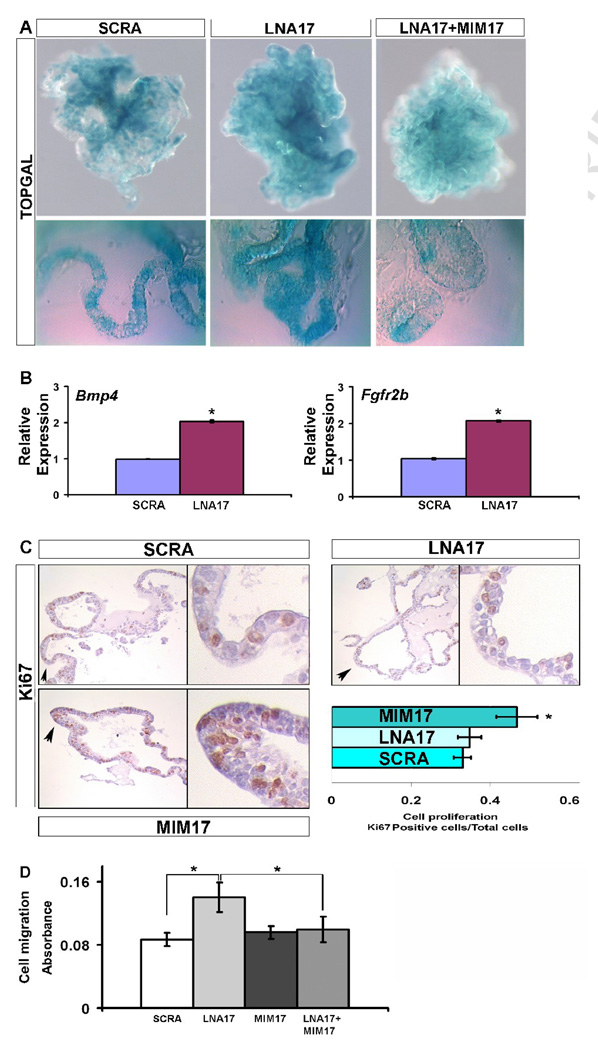
To determine the specific effect of miR-17 paralogs during lung morphogenesis, we downregulated miR-17, miR-20a and miR-106b expression in epithelial lung explants using a miR-17 specific LNA probe (LNA17) and a scrambled LNA (SCRA) as control. In total, 22 LNA17 and 20 SCRA transfections were carried out. Within the LNA17 treated tissues, 15 samples that showed altered phenotype were analyzed. Positive transfection was observed as fluorescence of FlTC labeled LNA17 (Fig. 2B) or by real-time PCR miR-17 expression (Fig. 2A). miR-17, miR-20a and miR-106b have high sequence homology (Fig. 2A top) and LNA17 was able to simultaneously downregulate expression of all three of them (Fig. 2A bottom). A control miRNA member of the miR-17 family, miR-19b, with low sequence similarity to miR-17 was used as a specificity control (Fig 2A bottom). For the 7 samples that did not show changes of phenotype, real-time PCR revealed miR-17 downregulation ≤ 20% compared with the 70% of samples showing altered phenotype. At 72h after transfection, epithelial explants showed significant branching defects compared with lung grown with FGF10 and SCRA (Fig. 2C). Morphometric analysis showed increases in both the number of terminal branches and the area of the branches. Nevertheless, the number of primary branching points did not significantly change, suggesting that when miR-17 paralogs expression is inhibited, the ordered and sequential progression and extension of early isolated epithelial branching is disrupted rather than the initiation of individual early branching events (Fig. 2D).
Altered expression and distribution of CDH1 after miR-17, miR-20a and miR-106b downregulation
In order to determine whether the observed phenotype was related to any alteration in epithelial cell-cell adhesion, we analyzed CDH1 expression. Epithelial cells transfected with LNA17 showed a sharp loss of CDH1 distribution at their interacting edges compared with SCRA transfected epithelial cells. The resulting cell shape was rounded with diffuse CDH1 expression within the cell. In experiments in which MIM17 was co-transfected with LNA17, a rescue of the phenotype was observed (Fig. 3A). Similar results and rescue were obtained with epithelial lung explants after transfection with LNA17 alone or co-transfection of LNA17 with MIM17 (Fig. 3B). Cdh1 changes at transcriptional level were not observed after LNA17 treatment (3C), while differences of protein levels were observed after both LNA17 and MIM17 (Fig. 3D). Analysis of sections from epithelial lung explants confirmed these alterations in epithelial cell shape and CDH1 distribution (Fig. 3E). Quantitative analysis on single cells showed that LNA17 treatment was characterized by lower expression and increased lateral diffusion of CDH1 at the cell edges, the points of cell-to-cell interaction (Fig. 3F).
Fig 3.
CDH1 distribution is disrupted after miR-17 downregulation. (A, 20X and 60X) NMuMG cells were transfected with LNA17, SCRA, or LNA17 and MIMWCDH1 IHC was performed to determine the impact of miR-17 downregulation on CDH1 distribution (green). Nuclei were stained with DAPI (blue). (B, 63X) Whole mount IHC was performed using epithelial lung explants transfected with LNA17, SCRA, or LNA17 and MIM17, and grown for 72h in presence of 250ng/ml FGF10 to monitor changes in CDH1 distribution. (C) Real-time PCR using epithelial lung explants from the treatment described in (B) was carried out to determine variations in Cdh1 mRNA levels. (D) Immunoblotting analysis was performed using whole-cell extract from NMuMG cells treated as described in (A) to monitor Cdh1 changes at the translational level. (E, 63X and 330X) IHC was performed for CDH1 (red) using 5um sections from epithelial lung explants treated as described in (B). On the left, images of terminal buds from scrambled LNA (top) or LNA17 (bottom) treated epithelial explants. On the right, electronic magnification of single cells showing the selected area (dashed lines) used to analyze CDH1 signal intensity and distribution (F top). Data from cells analyzed in four different epithelial lung explants for each condition were analyzed (F bottom). Data are reported as means ± SEM. *=P ≤ 0.05.
Downstream effects of miR-17, miR-20a and miR-106b downregulation on epithelial beta-catenin activity, migration, and proliferation
We used epithelial explants obtained from TOPGAL transgenic mice to determine if the alteration of Cdh1 expression observed after miR-17, miR-20a, and miR-106b downregulation affected beta-catenin activity. Transfection with LNA17 produced increased beta-catenin activity as shown by augmented TOPGAL expression in the branching epithelium. This phenotype was rescued after co-transfection of LNA17 and MIM17 (Fig. 4A). Furthermore, direct beta-catenin targets such as Bmp4 and Fgfr2b were positively regulated after miR-17, miR-20a, and miR-106b downregulation, supporting the functional importance of these changes in beta-catenin activity (Fig. 4B). Bmp4 overexpression in embryonic lung epithelium driven by SPC promoter has bee reported and resulted in diminished branching with dilated terminal buds (Bellusci et Al’ 1996). Further, in vitro administration of BMP4 to isolated endoderm migrating toward FGF10-loaded bids has been shown to inhibit bud outgrowth (Weaver et al., 2000). Our observed increase in the area of the branches and disruption of budding shape may be partially produced by the increase in Bmp4, while the increase in the number of secondary branches must be related to different regulators that we speculate may be in turn related to CDH1 modulation.
Figure 4.
miR-17 downregulation affects beta-catenin activity as well as cell migration and proliferation. (A, 10X and 40X) Beta-catenin activity was analyzed using epithelial lung explants obtained from TOPGAL mice. Tissues were treated with 250ng/ml FGF10 and SCRA, LNA17, or MlM17, and harvested after 72h for analysis. Whole-mount (top) and 30um vibratome sections (bottom) are shown. (B) The effect on Beta-catenin activity was confirmed by an analysis of downstream targets with real-time PCR for Bmp4 and Fgfr2b. (C, 20X and 90X) Changes in cell proliferation were monitored by IHC for Ki67 produced on 5um sections from epithelial lung explants treated as described in (A). (D) MLE12 cells treated as described in (A), were harvested after 48h, plated into an 8um pore size membrane and stimulated with 250ng/ml FGF10 after starvation for 24h, to determine changes in migration capability. Data are reported as means ± SEM. *=P ≤ 0.05.
Since CDH1 plays a key role in controlling the ability of epithelial cells to dissociate and migrate, we examined changes in the migratory capability of epithelial cells after miR-17 alteration. Treatment of MLE12 cells with LNA17 showed increased (60%) migration during FGF10 stimulation, compared with cells treated with SCRA. Meanwhile, co-transfection with both LNA17 and MIM17 completely rescued the ability of MLE12 cells to migrate towards FGF10, while transfection with MIM17 alone did not produce any appreciable effect (Fig. 4D).
The branching alteration observed during LNA17 treatment may depend on changes in epithelial cell proliferation. To address this possibility, we analyzed cell proliferation in epithelial lung explants after LNA17 treatment as compared to proliferation after MIM17 administration. Treatment with LNA17 did not show significant changes in epithelial proliferation compared with samples treated with SCRA. However MIM17 transfection produced a significant (40%) increase of epithelial proliferation (Fig. 4C). Increased cellular proliferation was previously reported in transgenic mice misexpressing the entire miR-17–92 cluster (Lu et al., 2007). Both MIM17 and LNA17 treated samples, analyzed with TUNEL assay, did not show signs of apoptosis (data not shown).
Mapk14 and Stat3 are direct targets of miR-17 paralogs at the translational level, and their simultaneous overexpression mimics miR-17 paralogs downregulation
In order to identify transcripts directly regulated by miR-17 paralogs we analyzed available miRNA target prediction software for putative miR-17, miR-20a and miR-106b target genes (TargetScan and Miranda). Candidate genes were analyzed at the target position flanking regions to determine their level of accessibility with the software mfold. Two genes, Mapk14 and Stat3, were shown to posses a free energy higher than the characteristic threshold level for mouse models (dG=-13.4 Kcal mor−1, Zhao et al. 2005) (Fig. 5C) and to have destabilizing elements in their secondary structure that may facilitate the accessibility of miR-17, miR-20a, and miR-106b to the binding site (Table 1). The levels of expression of both Stat3 and Mapk14 in embryonic lung development are stable during the pseudoglandular stage (Fig. 5A), suggesting that their stabilization may be important for correct branching morphogenesis. To determine the ability of miR-17 paralogs to directly bind to the 3’-UTR of these two genes (Fig. 5B), we produced luciferase-expressing vectors containing respectively one, two, or three copies of their target position. Vectors containing one, two or three copies of the target position mutated inside the “seed” sequence were used as reference for the quantification of luciferase expression. Transfection of the luciferase vectors into MLE12 epithelial cells showed a progressive reduction in luminescence. For Stat3 in the presence of one, two, or three copies of the target sequence, 29%, 32%, or 50% luminescence reduction was observed, while for Mapk14 the decrease was respectively of 20%, 37% or 46%. This result demonstrates the specificity of the binding of miR-17, miR-20a and miR-106b to the 3’-UTR of Stat3 and Mapk14 (Fig. 5D).
Figure 5.
miR-17 paralogs directly targets the 3’-UTR of Stat3 and Mapk14. (A) Stat3 and Mapk14 temporal differential expression were determined by real-time PCR performed on total RNA extracted from embryonic, post-natal, and adult lungs. (B) miR-17, miR-20a, and miR-106b binding to the 3’UTR of Stat3 and Mapk14 are shown. (C) 70bp miR-17 binding position flanking regions secondary structure and free energy (dG) are shown for Stat3 and Mapk14 based on mfold analysis. (D) MLE12 cells were transfected with expression vectors containing the luciferase open reading frame coupled with one, two, or three copies of the mutated (MU) or wild type (WT) miR-17 paralogs binding sequence of Stat3 or Mapk14. Co-transfection with Renilla luciferase-expressing vector was used as a system to normalize luminescence levels. Data are reported as means ± SEM. *=P ≤ 0.05. (E) Immunoblotting from MLE12 cells whole extract after treatment with SCRA or LNA17 was performed to determine miR-17 paralogs translational regulation of Stat3 and Mapk14.
Table 1.
Analysis of predicted miR-17 binding sites flanking UTRs of Mapk14 and Stat3
| 3 ‘-UTR sequence | mFold dG (Kcal/mole) | Structure | ||
|---|---|---|---|---|
| 5’ 70bp | 3’ 70bp | SE | DSE | |
| Mapk14 | −9.9 | −11.4 | – | HL, FE, MBL |
| Stat3 | −5.4 | −8.9 | – | HL, J |
Free energy (dG) of the 5’-UTR and 3’UTR flanking the miR-17 predicted binding position for Mapk14 and Stat3. No Stabilizing Elements (SE) were present. Destabilizing Elements (DSE) are indicated: HL, Hairpin Loop; FE, Free End; MBL, multi-branching loop; J, Joint.
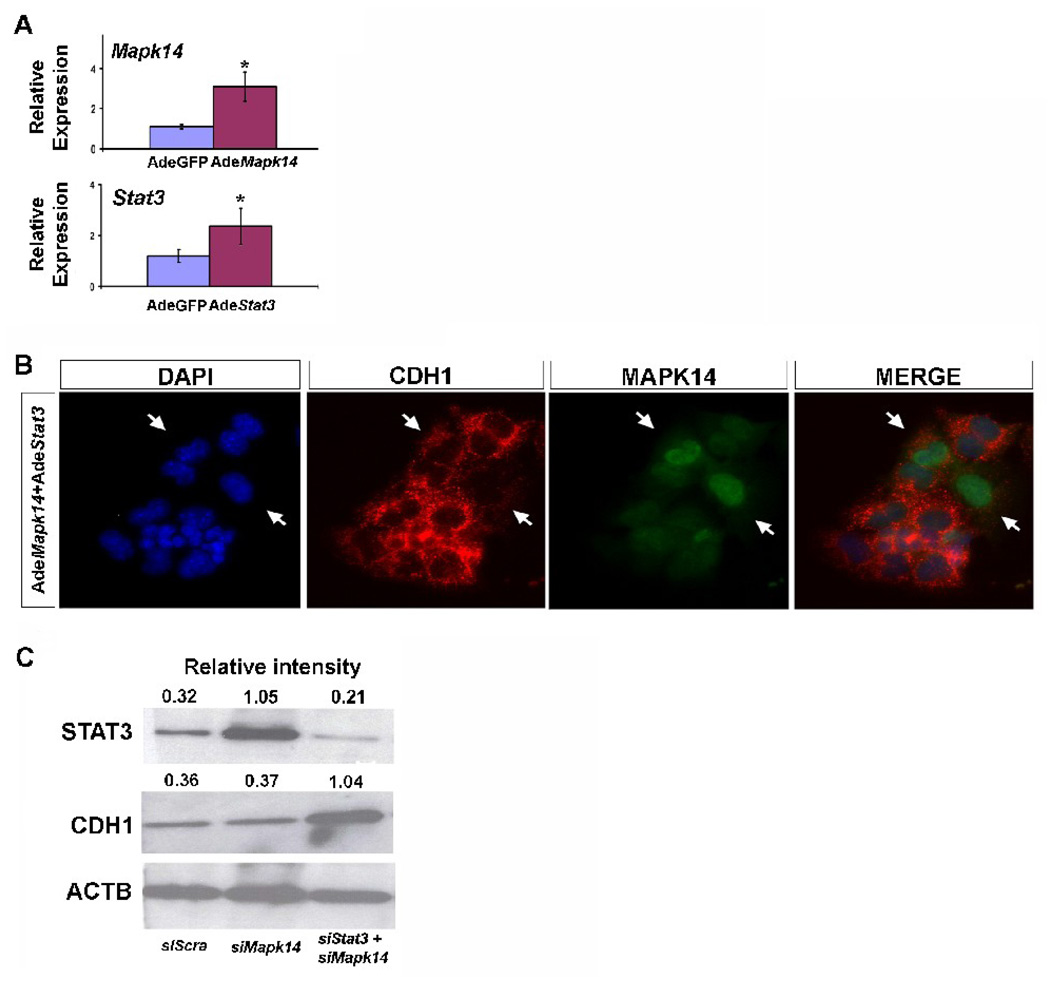
Regulation of Stat3 and Mapk14 expression at the translational level was then analyzed by western blotting after miR-17, miR-20a, and miR-106b downregulation, confirming the expected changes in protein levels (Fig. 5E)
To determine the ability of Stat3 and Mapk14 to control Cdh1 expression, we used adenoviral overexpression approaches in MLE12 epithelial cells. The effectiveness of the adenoviral construct was determined by real-time PCR (Fig. 6A). While the individual overexpression of Stat3 or Mapk14 did not produce evident changes in CDH1 distribution (data not shown), co-overexpression of both molecules strongly replicated the effect observed on CDH1 after LNA17 administration, and co-staining of epithelial cells with CDH1 and MAPK14 showed an inverse correlation between CDH1 levels and MAPK14 expression (Fig. 6B). Furthermore while downregulation of Stat3 alone (data not shown) or Mapk14 alone (Fig. 6C) with specific siRNAs did not modify CDH1 expression, downregulation of both Stat3 and Mapk14 showed specific overexpression of CDH1 (Fig. 6C).
Figure 6.
Simultaneous modulation of Stat3 and Mapk14 mimics miR-17 paralogs effect. (A) Real-time PCR was performed on total RNA extracted from MLE12 cells after transduction with Stat3 and Mapk14 adenovirus to determine the effectiveness of the adenoviral constructs. Data were compared to results from MLE12 cells transduced with a control adenovirus. Data are reported as means ± SEM. *=P ≤ 0.05. (B, 20X) IHC for CDH1 (red) and MAPK14 (green) was performed in MLE12 cells after the overexpression of Stat3 and Mapk14 by transduction with their specific adenovirus. (C) Immunoblotting from MLE12 cells’ whole extract after inhibition of Stat3 and Mapk14 by siRNA transfection was used to determine changes in CDH1.
We therefore conclude that mir-17 family of miRNA, specifically miR-17, miR-20a and miR-106b, negatively modulate FGF10-FGFR2b downstream signaling by specifically targeting Stat3 and Mapk14, hence regulating activation of Cdh1 and beta-catenin, which in turn modulate epithelial bud morphogenesis in response to FGF10 signaling.
Discussion
In this report we have identified a specific role of miR-17 family of miRNAs in maintaining homeostasis of epithelial structure within embryonic lung during branching morphogenesis. We used in vitro gain and loss of function approaches to test our hypothesis that an important function of miR-17, miR-20a, and miR-106b is to maintain correct CDH1 distribution during branching morphogenesis. Changes in branching and epithelial homeostasis were found during miR-17 paralogs loss of function experiments and these were associated with changes in Cdh1 expression. Furthermore, we determined that miR-17, miR-20a, and miR-106b directly target both Mapk14 and Stat3, which in turn regulate CDH1.
Dynamic adhesive mechanisms are important for the maintenance of correct epithelial tissue architecture during development. Cell-cell interaction in the epithelium of the developing lung is subject to dynamic changes during growth and specification of branching patterns (Warburton, 2008). CDH1 is a key adhesion receptor for epithelial cell identification and grouping during development; in its absence, the interaction between extracellular matrix, cytoplasmatic plaques, and other adhesion molecules cannot be maintained. Mouse embryonic lung explants grown in the presence of a CDH1 antibody show an altered pattern of branching (Hirai et al., 1989). The expression of Cdh1, like other cadherins, is regulated at both transcriptional and post-translational levels and is controlled during trafficking and arrangement at the cell surface (Halbleib JM and Nelson WJ, 2006). Therefore, precise control of Cdh1 expression is necessary for cell-cell interaction during lung development. Herein, we provide evidence that miR-17, miR-20a, and miR-106b have a critical function to control Mapk14 and Stat3, thus maintaining correct CDH1 levels and hence avoiding excessive loss of CDH1 during lung branching morphogenesis.
miR-17 and miR-20a expression is higher early in embryonic lung development and decreases with gestational age, miR-106b expression is also high during embryonic stages between E-12.5 and E16.5 and than becomes even higher again in the adult lung. Most importantly these three miRNAs are clearly present during the pseudoglandular stage when the embryonic lung undergoes repetitive budding to develop its characteristic three-dimensional architecture. Mapk14 and Stat3 also both maintain stable expression during the pseudoglandular stage. Taken together with our observation of a synergistic function of Mapk14 and Stat3, this suggests that the stable and correct expression of both Mapk14 and Stat3 are important in maintaining the correct level of CDH1 during the early and critical stages of lung branching morphogenesis.
Mapk14 has been implicated in regulation of developmental processes such as wing development in Drosophila (Adachi-Yamada et al., 1999) and recently its importance in maintaining a correct level of CDH1 degradation has been demonstrated in mice: disruption of MAPK14-interacting protein, a specific MAPK14 activator, can cause defects in neural tube and eye formation as well as gastrulation due to impaired CDH1 degradation (Zohn et al., 2006). Mapk14 also has a functional role in lung branching morphogenesis. Both chemical inhibitors and short hairpin RNAs against Mapk14 produced increased levels of CDH1, leading to impaired branching (Liu et al., 2008). These data are consistent with our results on Mapk14 overexpression following miR-17 downregulation with LNA, in which CDH1 expression was notably altered. On the other hand, overexpression of Mapk14 alone did not affect CDH1 localization (data not shown). Our observations were nevertheless consistent with the data of Liu and colleagues in which Mapk14 downregulation showed changes in Cdh1 expression but not in its localization. Furthermore, our data on Mapk14 and Stat3 co-overexpression and co-downregulation reinforce the concept that both molecules are important in simultaneously controlling CDH1 distribution.
Stat3 has been implicated in the regulation of cell motility during reparative processes in the lung (Kida et al., 2008) and has an important role during branch formation in neural path finding (Conway, 2006). This correlates with our observation of increased migratory ability of MLE12 cells after miR-17 downregulation. Furthermore, Stat3 has been shown to be essential during gastrulation movements in zebrafish because it is responsible for activating snail, a negative regulator of Cdh1 expression (Yamashita et al., 2004). The effect of miR-17, miR-20a, and miR-106b downregulation in our in vitro system can be considered a multiple-target effect. Our results show that co-overexpression of Mapk14 and Stat3 can mimic the effect of miR-17 downregulation. Therefore, Mapk14 and Stat3 are two main molecules that must apparently interact to produce this phenotype.
Other targets of miR-17 have also been identified, but none of them seem to be major candidates for participating in the regulation of epithelial structural homeostasis. E2f1 is an important regulator of apoptosis, but lung development is characterized by the relatively complete absence of apoptosis. In our system, no changes in apoptosis were observed after up or down regulation of miR-17 (data not shown). AIB1 is a proto-oncogene, and cells derived from mice in which this gene was deleted showed decreased proliferation in response to insulin-like growth factor 1 (Wang et al., 2000). Since miR-17 overexpression produced a hyperproliferative effect on epithelial cells, AIB1 cannot be logically implicated in our model. Rbl2 has been proposed as a direct target of miR-17 (Liu et al., 2007), and its deletion produce epithelial cell hyperplasia. Downregulation of Rbl2 in our experiments of miR-17 overexpression could conceivably contribute to the hyper-cellularity that we have observed. However Rbl2 expression becomes important in the lung only during late embryonic stages and is higher in the adult. Therefore, its major involvement during the early stages of lung development is unlikely.
Recently miR-17 has been suggested to have the ability to regulate the G1/S cell cycle checkpoint by targeting both promoters and inhibitors of proliferation in different contexts (Cloonan et al., 2008). While blocking the effects of proliferation inhibitors such as MAPK9 or p21 may account for increased proliferation after miR-17 over expression, in our system inhibition of miR-17 did not produce an arrest of proliferation. This suggests that the changes in Cdh1 expression and localization that we observed are not likely to be caused primarily by changes in cell cycle regulation.
In vivo overexpression of the miR-17–92 cluster has been reported to produce hypercellularity in the lung (Liu et al., 2007). These data are consistent with our results in which specific overexpression of miR-17 produced increased cell proliferation.
Hypoplastic lung at embryonic stage E18 was also noted in the presence of in vivo deletion of the entire miR-17–92 cluster. Furthermore double knockout for miR-17–92 and miR-106b-25 clusters showed a more severe phenotype with lethality before embryonic day 15 (Ventura et al., 2008). In the single deletion of miR-17–92 cluster the authors state that even though hypoplastic lung was observed no branching defects were observed. However, no detailed analysis or discussion of embryonic lung phenotypes in the double knockout were done therein. Our observations on alteration of embryonic lung epithelium structural homeostasis after co-downregulation of miR-17, miR-20a and miR-106b, suggest that the double knock out of miR-17–92 and miR-106b-25 cluster may indeed have embryonic lung developmental abnormalities.
We therefore propose a model in which miR-17, miR-20a, and miR-106b functions to maintain the correct level of expression and distribution of CDH1 in embryonic lung epithelial cells through negative modulation of Mapk14 and Stat3. Thus, if miR-17 paralogs are eliminated or inhibited, the structural integrity of the process of epithelial branching becomes compromised.
Conclusions
In summary we have identified a specific functional role of miR-17, miR-20a, and miR-106b to control the structural homeostasis of the epithelium during early embryonic branching morphogenesis in the lung. The dual targets that we have identified are Mapk14 and Stat3, which can mimic the phenotype elicited by miR-17, miR-20a, and miR-106b co-inhibition, when they are overexpressed together. The detailed molecular links between these two targets and the observed alteration of Cdh1 expression remain to be further defined.
Acknowledgments
We would like to thank Dr. Yannan Ouyang of cellular imaging core at the Saban Research Institute for the discussion of image analysis. HL60231, HL44977 and HL44060 to DW and American Heart Association Western Affiliates Senior Postdoctoral Fellowship to GC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Bibliography
- Adachi-Yamada T, Nakamura M, Irie K, Tomoyasu Y, Sano Y, Mori E, Goto S, Ueno N, Nishida Y, Matsumoto K. p38 mitogen-activated protein kinase can be involved in transforming growth factor beta superfamily signal transduction in Drosophila wing morphogenesis. Mol Cell Biol. 1999;19:2322–2329. doi: 10.1128/mcb.19.3.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- Bellusci S, Henderson R, Winnier G, Oikawa T, Hogan BL. Evidence from normal expression and targeted misexpression that bone morphogenetic protein (Bmp-4) plays a role in mouse embryonic lung morphogenesis. Development. 1996;122:1693–1702. doi: 10.1242/dev.122.6.1693. [DOI] [PubMed] [Google Scholar]
- Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Bryant DM, Stow JL. The ins and outs of E-cadherin trafficking. Trends Cell Biol. 2004;14:427–434. doi: 10.1016/j.tcb.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Cardoso WV, Lu J. Regulation of early lung morphogenesis: questions, facts and controversies. Development. 2006;133:1611–1624. doi: 10.1242/dev.02310. [DOI] [PubMed] [Google Scholar]
- Cloonan N, Brown MK, Steptoe AL, Wani S, Chan WL, Forrest AR, Kolle G, Gabrielli B, Grimmond SM. The miR-17-5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 2008;9:R127. doi: 10.1186/gb-2008-9-8-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM, Brennecke J, Stark A. Denoising feedback loops by thresholding-a new role for microRNAs. Genes Dev. 2006;20:2769–2772. doi: 10.1101/gad.1484606. [DOI] [PubMed] [Google Scholar]
- Conway G. STAT3-dependent pathfinding and control of axonal branching and target selection. Dev Biol. 2006;296:119–136. doi: 10.1016/j.ydbio.2006.04.444. [DOI] [PubMed] [Google Scholar]
- Davis MA, Ireton RC, Reynolds AB. A core function for p120-catenin in cadherin turnover. J Cell Biol. 2003;163:525–534. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Langhe SP, Sala FG, Del Moral PM, Fairbanks TJ, Yamada KM, Warburton D, Burns RC, Bellusci S. Dickkopf-1 (DKK1) reveals that fibronectin is a major target of Wnt signaling in branching morphogenesis of the mouse embryonic lung. Dev Biol. 2005;277:316–331. doi: 10.1016/j.ydbio.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Ding W, Shi W, Bellusci S, Groffen J, Heisterkamp N, Minoo P, Warburton D. Sprouty2 downregulation plays a pivotal role in mediating crosstalk between TGF- beta 1 signaling and EGF as well as FGF receptor tyrosine kinase-ERK pathways in mesenchymal cells. J Cell Physiol. 2007;212:796–806. doi: 10.1002/jcp.21078. [DOI] [PubMed] [Google Scholar]
- Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goomer RS, Hoist BD, Wood IC, Jones FS, Edelman GM. Regulation in vitro of an L-CAM enhancer by homeobox genes HoxD9 and HNF-1. Proc Natl Acad Sci U S A. 1994;91:7985–7989. doi: 10.1073/pnas.91.17.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidolin D, Vacca A, Nussdorfer GG, Ribatti D. A new image analysis method based on topological and fractal parameters to evaluate the angiostatic activity of docetaxel by using the Matrigel assay in vitro. Microvasc.Res. 2004;67:117–124. doi: 10.1016/j.mvr.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci U S A. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig G, Lowrick O, Birchmeier W, Behrens J. Mechanisms identified in the transcriptional control of epithelial gene expression. J Biol Chem. 1996;271:595–602. doi: 10.1074/jbc.271.1.595. [DOI] [PubMed] [Google Scholar]
- Hirai Y, Nose A, Kobayashi S, Takeichi M. Expression and role of E- and P- cadherin adhesion molecules in embryonic histogenesis. II. Skin morphogenesis. Development. 1989;105:271–277. doi: 10.1242/dev.105.2.271. [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Constantini F, Lacy E. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. Manipulating the Mouse Embryo. [Google Scholar]
- Hornstein E, Mansfield JH, Yekta S, Hu JK, Harfe BD, McManus MT, Baskerville S, Bartel DP, Tabin CJ. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Kida H, Mucenski ML, Thitcftf AR, Le Cras TD, Park KS, Ikegami M, Muller W, Whitsett JA. GP130-STAT3 regulates epithelial cell migration and is required for repair of the bronchiolar epithelium. Am J Pathol. 2008;172:1542–1554. doi: 10.2353/ajpath.2008.071052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci U S A. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leckband D, prakasam A. Mechanism and dynamics of cadherin adhesion. Annu Rev Biomed Eng. 2006;8:259–287. doi: 10.1146/annurev.bioeng.8.061505.095753. [DOI] [PubMed] [Google Scholar]
- Liu Y, Martinez L, Ebine K, Abe MK. Role for mitogen-activated protein kinase p38 alpha in lung epithelial branching morphogenesis. Dev Biol. 2008;314:224–235. doi: 10.1016/j.ydbio.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YN, Lee WW, Wang CY, Chao TH, Chen Y, Chen JH. Regulatory mechanisms controlling human E-cadherin gene expression. Oncogene. 2005;24:8277–8290. doi: 10.1038/sj.onc.1208991. [DOI] [PubMed] [Google Scholar]
- Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17‒92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Mege RM, Gavard J, Lambert M. Regulation of cell-cell junctions by the cytoskeleton. Curr Opin Cell Biol. 2006;18:541–548. doi: 10.1016/j.ceb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Palacios F, Schweitzer JK, Boshans RL, D’Souza-Schorey C. ARF6-GTP recruits Nm23-H1 to facilitate dynamin-mediated endocytosis during adherens junctions disassembly. Nat Cell Biol. 2002;4:929–936. doi: 10.1038/ncb881. [DOI] [PubMed] [Google Scholar]
- Palacios F, Tushir JS, Fujita Y, D’Souza-Schorey C. Lysosomal targeting of E- cadherin: a unique mechanism for the down-regulation of cell-cell adhesion during epithelial to mesenchymal transitions. Mol Cell Biol. 2005;25:389–402. doi: 10.1128/MCB.25.1.389-402.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park WY, Miranda B, Lebeche D, Hashimoto G, Cardoso WV. FGF-10 is a chemotactic factor for distal epithelial buds during lung development. Dev Biol. 1998;201:125–134. doi: 10.1006/dbio.1998.8994. [DOI] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Pepicelli CV, Lewis PM, McMahon AP. Sonic hedgehog regulates branching morphogenesis in the mammalian lung. Curr Biol. 1998;8:1083–1086. doi: 10.1016/s0960-9822(98)70446-4. [DOI] [PubMed] [Google Scholar]
- Seul M, O’Gorman L, Sammon MJ. New York: Cambridge Univ. Press; 2000. Practical Algorithms for Image Analysis. [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y. Coordinated histone modifications mediated by a CtBP corepressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Rose DW, Hermanson O, Liu F, Herman T, Wu W, Szeto D, Gleiberman A, Krones A, Pratt K, Rosenfeld R, Glass CK, Rosenfeld MG. Regulation of somatic growth by the p160 coactivator p/CIP. Proc Natl Acad Sci U S A. 2000;97:13549–13554. doi: 10.1073/pnas.260463097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton D. Developmental biology: order in the lung. Nature. 2008;453:733–735. doi: 10.1038/453733a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton D, Bellusci S, De Langhe S, Del Morals PM, Fleury V, Mailleux A, Tefft D, Unbekandt M, Wang K, Shi W. Molecular mechanisms of early lung specification and branching morphogenesis. Pediatr Res. 2005;57 doi: 10.1203/01.PDR.0000159570.01327.ED. 26R‒37R. [DOI] [PubMed] [Google Scholar]
- Weaver M, Dunn NR, Hogan BL. Bmp4 and Fgf10 play opposing roles during lung bud morphogenesis. Development. 2000;127:2695–2704. doi: 10.1242/dev.127.12.2695. [DOI] [PubMed] [Google Scholar]
- Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- Yamashita S, Miyagi C, Fukada T, Kagara N, Che YS, Hirano T. Zinc transporter LIVI controls epithelial-mesenchymal transition in zebrafish gastrula organizer. Nature. 2004;20:298–302. doi: 10.1038/nature02545. [DOI] [PubMed] [Google Scholar]
- Yap AS, Kovacs EM. Direct cadherin-activated cell signaling: a view from the plasma membrane. J Cell Biol. 2003;160:11–16. doi: 10.1083/jcb.200208156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- Zohn IE, Li Y, Skolnik EY, Anderson KV, Han J, Niswander L. p38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation. Cell. 2006;125:957–969. doi: 10.1016/j.cell.2006.03.048. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]