Abstract
Background
Hyperbilirubinemia is a common complication of sepsis. Elevated bilirubin may induce inflammation and apoptosis. We hypothesized that increased serum bilirubin on ICU admission contributes to sepsis-related ARDS.
Methods
Serum bilirubin on ICU admission was measured in 1006 septic patients. Serial serum bilirubin was analyzed prospectively in septic patients with ARDS for a period of 28 days. The effects of clinical factors and variants of UGT1A1 gene on serum bilirubin levels were determined. Outcomes were ARDS risk and mortality.
Results
During 60-day follow-up, 326 septic patients developed ARDS in whom 144 died from ARDS. The hyperbilirubinemia (≥2.0 mg/dL) rate in patients with ARDS (22.4%) was higher than those without ARDS (14.1%, p = 0.002). For each 1.0 mg/dL increase in admission bilirubin, ARDS risk, 28- and 60-day ARDS mortalities were increased by 7% (OR = 1.07; p = 0.003), 20% (OR = 1.20; p = 0.002), and 18% (OR = 1.18; p = 0.004), respectively. Compared with subjects with bilirubin levels <2.0 mg/dL, patients with hyperbilirubinemia had higher risks of ARDS (OR = 2.12; p = 0.0007), 28-day (OR = 2.24; p = 0.020), and 60-day ARDS mortalities (OR = 2.09; p = 0.020). In sepsis-related ARDS, serial bilirubin levels in non-survivors were consistently higher than in survivors (p<0.0001). Clinical variables explained 29.5% of the inter-individual variation in bilirubin levels, whereas genetic variants of UGT1A1 contributed 7.5%.
Conclusion
In sepsis, higher serum bilirubin level on ICU admission is associated with subsequent ARDS development and mortality.
Keywords: sepsis, bilirubin, ARDS, clinical factors, UGT1A1
Sepsis is the second most common cause of death in non-coronary intensive care unit (ICU) and is among the top 10 causes of death for all hospitalized patients.1 Patients with sepsis are at the highest risk of developing acute respiratory distress syndrome (ARDS).2, 3 However, little is known about biomarkers predictive of ARDS development and mortality in patients with sepsis.
Bilirubin, the end product of heme catabolism in mammals, is generally considered a lipid-soluble waste product that needs to be excreted. However, growing evidences have suggested that bilirubin at high concentrations can induce inflammation, apoptosis, and oxidative stress.4–8 Hyperbilirubinemia, or jaundice, is a well-known complication of sepsis or non-bacterial infection.9 Sepsis and bacterial infection account for 20% of jaundice cases in patients of all ages in community hospital settings.10 But there are no data from large prospective studies on the exact incidence and prognostic relevance of hyperbilirubinemia in adults with sepsis.11 Since most physicians view hyperbilirubinemia as a late event in critical illness, low-grade hyperbilirubinemia is often overlooked in patients not presenting with clinically evident jaundice.12–13 Although hyperbilirubinemia has been associated with overall poor outcomes in critical illness,12–14 the associations of bilirubin with ARDS risk, and factors influencing bilirubin variations in sepsis remain largely unknown.
In humans, bilirubin is mainly metabolized by uridine diphosphase glucuronosyltransferase 1A1 (UGT1A1) that contributes to bilirubin glucuronidation and thus enhances bilirubin elimination.15 The gene encoding for UGT1A1 is located in chromosome 2 (2q37) and spans approximately 160 kb. Individual genetic variations in the UGT1A1 gene, such as the -53 ~-42(TA)6–7 (UGT1A1*28, rs8175347), -3279T>G (UGT1A1*60, rs4124874 ), 211G>A (UGT1A1*6, rs4148323), and -3156G>A (rs10929302) have been reported to affect UGT1A1 gene expression, enzyme activity, and serum bilirubin levels.16–18 However, conflicting associations with these polymorphisms have also been reported.19–21 The apparent discrepancy suggests that single polymorphism may not be sufficient enough to define the contribution of UGT1A1 variants to serum bilirubin levels. No systematic studies have addressed the association of overall genetic variation of the UGT1A1 gene with circulating bilirubin levels in critically ill patients.
The aims of the present study were to evaluate, firstly, whether serum bilirubin levels on ICU admission were associated with sepsis-related ARDS risk and mortality; secondly, whether clinical factors and UGT1A1 genetic variants contribute to inter-individual serum bilirubin variations in sepsis; and finally, whether UGT1A1 polymorphisms were associated with ARDS risk and mortality that were consistent with their effects on bilirubin levels.
METHODS
Study subjects
Study patients were drawn from a prospectively enrolled cohort assembled for the Molecular Epidemiology of ARDS Study.22 Consecutive admissions to the ICUs at the Massachusetts General Hospital (MGH, Boston, MA) were screened for sepsis from September 1999 to November 2006. Sepsis was diagnosed according to the criteria of the American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference.23 Exclusion criteria included age <18 years, diffuse alveolar hemorrhage, chronic lung diseases, directive to withhold intubation, immunosuppression except if secondary to corticosteroid, and treatment with granulocyte colony-stimulating factor (G-CSF). Alcohol abuse was defined as the history of active alcohol abuse, or diagnosis for alcohol detoxification and alcoholism in the past year. ARDS was defined according to American-European Consensus Conference (AECC) criteria.24 Organ dysfunction was measured by the ARDS Network criteria.25 Patients were followed daily for all-cause of 28- and 60-day mortalities. Baseline clinical and laboratory information were collected in the first 24 hrs of ICU admission. The MGH Human Subjects Committee approved the study and informed written consent was obtained from all subjects or surrogates.
Laboratory analysis
Serum total bilirubin and other biomarkers were measured on ICU admission, using the Roche Hitachi 917 analyzer with reagents from Roche Diagnostics (Indianapolis, IN). Serial serum bilirubin levels were measured from day 1 of ARDS diagnosis until ICU discharge or death, for a period of 28 days.
Selection criteria for tagging SNPs (tSNPs) of UGT1A1 were r2 ≥0.8 and minor allele frequency >5.0% across the entire UGT1A1 gene based on the database of the International HapMap Project (http://www.hapmap.org). DNA was extracted from whole blood using PureGene kits (Gentra Systems, Inc. Minneapolis, MN). Genotyping was determined using the Taqman assay with a ABI 7900HT sequence detector system (Applied Biosystems, Foster City, CA). The primer and probe sequences for each SNP are available on request. A total of 10% of samples were genotyped in duplicate for quality control and showed 100% concordance.
Statistical analysis
We compared baseline variables using χ2 test, Fisher’s exact test, Student’s t-test, or Wilcoxon test, as appropriate. Univariate and stepwise multivariable linear regression models were used to test the associations of clinical factors and genetic variants with bilirubin levels. Since serum bilirubin levels had a skewed distribution, bilirubin levels were naturally log transformed and log bilirubin values were used in linear regression models as dependent variables. Multivariable logistic regression was used to evaluate associations of serum bilirubin levels and genetic variants of UGT1A1 with ARDS development and mortality. Association with ARDS development was adjusted for covariates including age, gender, pneumonia, aspiration, multiple transfusion, diabetes, chronic liver diseases, alcohol abuse, history of steroid use, septic shock, modified APACHE III scores (excluding bilirubin component).2 While association with ARDS mortality was adjusted for age, gender, sepsis shock, liver cirrhosis, history of alcohol use, history of steroid use, diabetes, modified APACHE III scores, organ dysfunctions (respiratory, cardiovascular, renal, and hematological), and positive end-expiratory pressure (PEEP, defined as treatment with PEEP >5 cm H2O on ICU admission).26 Survival probability was estimated using Kaplan and Meier log-rank test. Serial measurements were analyzed by generalized Estimating Equation (GEE) model.
Hardy-Weinberg equilibrium was determined using χ2 test. Haplotypes were calculated using SAS macro HAPPY programs.22 Genetic covariates were analyzed by additive (wildtype, heterozygotes, and homozygotes were coded as 0, 1, and 2, respectively) and haplotype models. False-discovery rate (FDR) was assessed to account for multiple comparison.27 Colinearity test was performed by SAS PROC PRINCOMP procedure.
All analyses were conducted using SAS version 9.1 (SAS Institute, Cary, NC). A p value less than 0.05 was considered statistically significant.
RESULTS
Characteristics of septic patients
Although individuals of all races were screened for this study, we restricted our analysis to Caucasians since 92% of ICU admissions during the study period at MGH were Caucasians. A total of 26580 consecutive ICU admissions were screened. Among them, 1931 patients meeting the criteria for sepsis and without exclusion criteria were recruited. Consent was obtained from 1224 septic patients (consent rate = 63.4%). 217 (18%) patients without bilirubin records on ICU admission and 1 patient with genotyping failure were excluded, leaving 1006 patients for analyses. Based on the sample size of 1006, the power of this study to detect a minimum OR = 1.50 (two-sided alpha = 0.05) was >80%. Of the 1006 patients with sepsis, 326 (32.4%) developed ARDS, and 144 (44.2%) of them died within 60-day follow-up. No differences were noted between patients with and without ARDS with respect to gender, prevalence of Gram-positive and Gram-negative bacteria infection, mean creatinine levels, or steroid use (table 1). Patients developed ARDS were younger, had higher APACHE III scores and lower platelet counts and lower prevalence of diabetes. Pneumonia, chronic liver disorder, and septic shock were more frequent in patients with ARDS. Serum bilirubin levels as well as hyperbilirubinemia rate were significantly higher in septic patients with ARDS than those without ARDS (table 1).
Table 1.
Characteristics of 1006 patients admitted to ICU with sepsis
| Characteristics | All patients (n = 1006) | Patients developed ARDS(n = 326) | Patients did not develop ARDS (n = 680) | p value |
|---|---|---|---|---|
| Age, yrs | 62.3 ± 17.1 | 59.8 ± 17.7 | 63.5 ± 16.7 | 0.001 |
| Female | 402 (40.0%) | 134 (41.1%) | 268 (39.4%) | 0.631 |
| APACHE III score | 74.8 ± 23.8 | 82.6 ± 23.2 | 71.0 ± 23.3 | <0.001 |
| History of alcohol abuse | 127 (12.6%) | 51 (15.6%) | 76 (11.2%) | 0.054 |
| History of steroid use | 97 (9.7%) | 33 (10.1%) | 64 (9.4%) | 0.733 |
| Serum bilirubin (mg/dL) | 0.7 (0.4–1.3) | 0.9 (0.5–1.8) | 0.6 (0.4–1.2) | <0.0001a |
| Bilirubin ≥2.0 mg/dl | 169 (16.8%) | 73 (22.4%) | 96 (14.1%) | 0.002 |
| Serum creatinine (mg/dL) | 1.4 (0.9–2.4) | 1.4 (0.9–2.4) | 1.3 (0.9–2.4) | 0.525a |
| Platelets (×1000/μL) | 215.8 ± 133.5 | 205.0 ± 145.2 | 220.9 ± 127.3 | 0.005 |
| Sepsis shock | 612 (60.8%) | 234 (71.8%) | 378 (55.6%) | <0.001 |
| Pneumonia | 593 (59.0%) | 249 (76.4%) | 344 (50.6%) | <0.001 |
| Multiple transfusion | 35 (3.5%) | 11 (3.4%) | 24 (3.5%) | 1.00 |
| End-stage renal disease | 64 (6.4%) | 23 (7.1%) | 41 (6.0%) | 0.540 |
| Trauma | 9 (0.9%) | 5 (1.6%) | 4 (0.6%) | 0.159 |
| Diabetes history | 254 (25.4%) | 63 (19.3%) | 191 (28.2%) | 0.003 |
| Liver cirrhosis/failure | 58 (5.8%) | 26 (8.0%) | 32 (4.7%) | 0.043 |
| PEEP >5 cm H2O | 313 (31.1%) | 176 (54.0%) | 137 (20.5%) | <0.0001 |
| Microorganisms | ||||
| Gram-positive | 432 (42.9%) | 130 (39.9%) | 302 (44.4%) | 0.174 |
| Gram-negative | 317 (31.5%) | 110 (33.7%) | 207 (30.4%) | 0.292 |
| Anaerobic | 47 (4.7%) | 9 (2.8%) | 38 (5.6%) | 0.047 |
| Fungus | 53 (5.3%) | 25 (7.7%) | 28 (4.1%) | 0.018 |
| Virus | 12 (1.2%) | 2 (0.6%) | 10 (1.5%) | 0.241 |
| Unknown | 336 (33.4%) | 122 (37.4%) | 214 (31.5%) | 0.061 |
Data are presented as n (%), mean ± SD, or median (lower quartile-upper quartile);
Wilcoxon test
Associations of serum total bilirubin levels with ARDS development and mortality
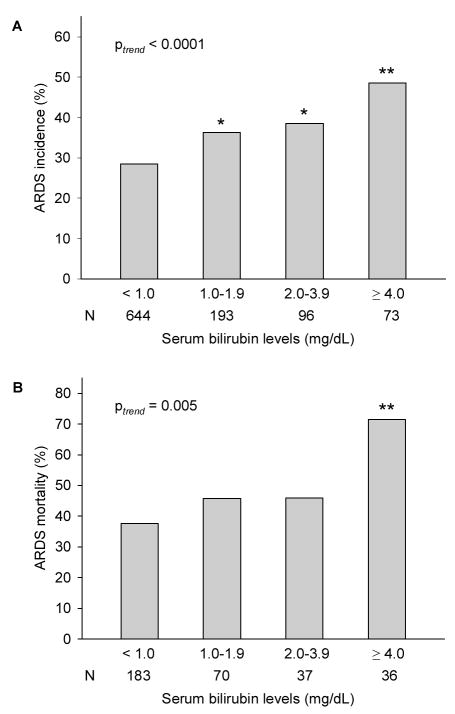
Among 1006 septic patients, 169 (16.8%) had admission bilirubin ≥2.0 mg/dL (hyperbilirubinemia), and the remaining 837 patients had bilirubin <2.0 mg/dL. Higher serum bilirubin levels on ICU admission were significantly associated with increased ARDS incidence (ptrend <0.0001) and ARDS 60-day mortality (ptrend = 0.0002). Interestingly, the incidence of ARDS in patients with modest bilirubin levels (≥1.0 and <2.0 mg/dL) was significantly higher than those with bilirubin <1.0 mg/dL (p = 0.037) (fig 1). For each 1.0 mg/dL increase in admission bilirubin, ARDS risk increased by 7% (OR = 1.07; 95% CI, 1.03–1.13; p = 0.003), ARDS 28-day mortality increased by 20% (OR = 1.20; 95% CI 1.07–1.35; p = 0.002) and ARDS 60-day mortality increased by 18% (OR = 1.18; 95% CI 1.05–1.31; p = 0.004). Similarly, log bilirubin values were significantly associated with ARDS development and mortalities (table 2). Bilirubin levels on ARDS diagnosis were also associated with increased mortality of sepsis-related ARDS. In sensitivity analyses by restricting the analysis to subjects without history of chronic liver disorder (n = 948) and patients who did not have ARDS on ICU admission (n = 867), the associations of admission bilirubin levels with sepsis-related ARDS risk and mortality remained unchanged (data not presented).
Figure 1.
Distribution of ARDS incidence (A) and ARDS 60-day mortality (B) by strata of serum bilirubin levels on ICU admission. * p<0.05 as compared with patients with bilirubin <1.0 mg/dL; ** p<0.01 as compared with patients with bilirubin <1.0 mg/dL. N (number of subjects).
Table 2.
Associations of serum total bilirubin levels with sepsis-related ARDS risk and mortality: Multivariate logistic regression analysis
| ARDS development |
ARDS 28-day mortality |
ARDS 60-day mortality |
||||
|---|---|---|---|---|---|---|
| Serum bilirubin levels | ORa (95% CI) | p value | ORb (95% CI) | p value | ORb (95% CI) | p value |
| On ICU admission | ||||||
| Bilirubin | 1.07 (1.03–1.13) | 0.003 | 1.20 (1.07–1.35) | 0.002 | 1.18 (1.05–1.29) | 0.004 |
| Log-bilirubin | 2.17 (1.46–3.25) | 0.0001 | 2.86 (1.36–6.01) | 0.005 | 2.70 (1.30–5.61) | 0.008 |
| Bilirubin ≥2.0 mg/dL | 2.12 (1.37–3.27) | 0.0007 | 2.24 (1.14–4.38) | 0.019 | 2.09 (1.12–3.90) | 0.020 |
| On ARDS diagnosis | ||||||
| Bilirubin | - | - | 1.19 (1.06–1.32) | 0.002 | 1.16 (1.05–1.28) | 0.003 |
| Log-bilirubin | - | - | 3.32 (1.57–7.06) | 0.002 | 3.29 (1.57–6.89) | 0.002 |
| Bilirubin ≥2.0 mg/dL | - | - | 2.50 (1.23–5.08) | 0.012 | 2.54 (1.33–4.87) | 0.005 |
Abbreviations: OR, odds ratio; CI, confidence interval.
Adjustment for age, gender, clinical risks for ARDS (trauma, pneumonia, aspiration, multiple transfusions), modified APACHE III scores, sepsis shock, chronic liver diseases, history of alcohol use, history of steroid use, and diabetes.
Adjustment for age, gender, sepsis shock, chronic liver diseases, history of alcohol use, history of steroid use, diabetes, modified APACHE III scores, PEEP, and organ dysfunctions (respiratory, cardiovascular, renal, and hematological).
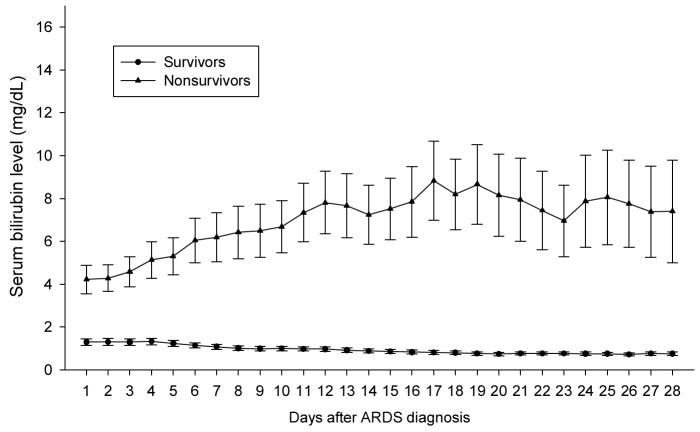
Among patients with sepsis-related ARDS, serial daily serum bilirubin levels in non-survivors were persistently higher than that in survivors from day 1 of ARDS diagnosis to the end of the entire observation period (fig 2. GEE test, p<0.0001). Kaplan-Meier survival analysis showed that patients with hyperbilirubinemia had lower survival rates than patients with admission bilirubin ≤ 2.0 mg/dL (p = 0.002) (fig 3).
Figure 2.
Serial mean serum bilirubin levels after ARDS diagnosis over the observation period between survivors (lower) and non-survivors (upper) in patients with sepsis-related ARDS (n = 326) (p<0.0001, GEE analysis). Day 1 represents the day of ARDS diagnosis. Error bars are SE.
Figure 3.
Estimated survival probability in patients with sepsis-related ARDS by admission bilirubin levels.
Clinical correlates of serum total bilirubin levels on ICU admission in patients with sepsis
In univariate linear regression analysis, age, gender, diabetes, chronic liver disorder, pneumonia, multiple transfusion, alcohol abuse, and respiratory/renal/hematological dysfunctions were significantly associated with serum bilirubin levels in patients with sepsis (table 3). In stepwise multivariable linear regression analysis, chronic liver diseases, male gender, respiratory/renal/hematological dysfunctions were positively associated with ICU admission bilirubin levels, while diabetes and pneumonia were inversely related to ICU admission bilirubin levels. Clinical correlates explained 29.5% of the inter-individual variation (R2 = 0.295) in serum bilirubin levels. Colinearity test suggested that colinearity among clinical factors was weak (condition number = 7.48) When analysis was restricted to patients without history of chronic liver disorders, the direction and magnitude of associations were virtually identical to results reported above (data not presented).
Table 3.
Clinical correlates of serum total bilirubin levels in 1006 patients with sepsis: Linear regression analysis
| Univariate analysis |
Stepwise multivariate analysis |
||||
|---|---|---|---|---|---|
| Clinical variables | Estimates (SE) | p value | Estimates (SE) | p value | |
| Age | −0.0023 (0.0008) | 0.004 | - | NS | |
| Sex, female vs. male | −0.1314 (0.0278) | <0.0001 | −0.0920 (0.0244) | <0.0001 | |
| Septic shock | 0.0539 (0.0281) | 0.055 | - | NS | |
| Diabetes | −0.1033 (0.0315) | 0.001 | −0.1118 (0.0275) | 0.0002 | |
| Chronic liver disorder | 0.7569 (0.0538) | <0.0001 | 0.5780 (0.0543) | <0.0001 | |
| Pneumonia | −0.1362 (0.0276) | <0.0001 | −0.1241 (0.0248) | <0.0001 | |
| Aspiration | −0.0704 (0.0491) | 0.152 | - | NS | |
| Multiple transfusions | 0.3756 (0.0741) | <0.0001 | - | NS | |
| Trauma | −0.0512 (0.1460) | 0.700 | - | NS | |
| History of alcohol abuse | 0.2591 (0.0405) | <0.0001 | - | NS | |
| History of steroid use | −0.0824 (0.0465) | 0.077 | - | NS | |
| PEEP >5 cm H2O | 0.0579 (0.0296) | 0.051 | - | NS | |
| Organ dysfunction | NS | ||||
| Respiratory | 0.1000 (0.0294) | 0.0007 | 0.0928 (0.0257) | <0.0001 | |
| Cardiovascular | 0.0638 (0.0313) | 0.042 | - | NS | |
| Renal | 0.1117 (0.0289) | 0.0001 | 0.0619 (0.0256) | 0.022 | |
| Hematological | 0.4932(0.0398) | <0.0001 | 0.3018 (0.0400) | <0.0001 | |
Abbreviations: SE, standard error; NS, not significant.
Associations of UGT1A1 variants with serum total bilirubin levels in septic patients
The genotyping success rates for the 10 tSNPs ranged from 99.0% to 99.5%. All tSNPs in this study population were consistent with Hardy-Weinberg equilibrium (p>0.05, χ2 goodness-of-fit). The LD pattern across the UGT1A1 locus is shown in fig S1. Three regions of strong LD were identified: block 1 (~8 kbp), block 2 (~kbp), and block 3 (~2 kbp). Compared with the major alleles, minor alleles of rs3755319, rs887829, and rs6742078 in LD block 1 significantly associated with higher serum bilirubin levels (FDR p = 0.005, 0.0005 and 0.0005, respectively; table 4). Minor alleles at rs17864705, and rs1018124 in LD block 1 also showed a trend of association with higher levels of bilirubin, but these associations did not reach statistically significance. The tSNPs explained 7.5 % of the serum bilirubin variation (R2 = 0.075) in models that included all clinical covariates. No associations between serum bilirubin and tSNPs in LD block 2 and block 3 were observed.
Table 4.
Associations of UGT1A1 tagging SNPs with serum total bilirubin levels in septic patients: Linear regression analysis
| LD block | Tagging SNPs | Bilirubin levels by genotypesa |
Univariate analysis |
Stepwise multivariate analysis |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | Estimates (SE) | p value | FDR p | Estimates (SE) | p value | FDR p | ||
| rs3755319 | 282 | 513 | 204 | |||||||
| 1.64 ± 0.26 | 1.62 ± 0.16 | 1.86 ± 0.24 | 0.0534 (0.0199) | 0.0073 | 0.024 | 0.0493 (0.0171) | 0.0043 | 0.005 | ||
| rs887829 | 438 | 455 | 108 | |||||||
| 1.60 ± 0.19 | 1.60 ± 0.18 | 2.26 ± 0.37 | 0.0694 (0.0208) | 0.0009 | 0.0045 | 0.0609 (0.0178) | 0.0001 | 0.0005 | ||
| rs6742078 | 436 | 456 | 109 | |||||||
| 1.58 ± 0.18 | 1.60 ± 0.18 | 2.18 ± 0.37 | 0.0709 (0.0207) | 0.0006 | 0.0045 | 0.0696 (0.0178) | 0.0001 | 0.0005 | ||
| rs3771342 | 757 | 227 | 21 | - | ||||||
| 1.75 ± 0.15 | 1.36 ± 0.13 | 2.02 ± 1.29 | −0.0220 (0.0282) | 0.436 | 0.110 | - | NS | - | ||
| rs17864705 | 870 | 122 | 8 | - | ||||||
| 1.64 ± 0.13 | 1.72 ± 0.30 | 4.61 ± 3.34 | 0.0608 (0.0375) | 0.105 | 0.210 | - | NS | - | ||
| rs1018124 | 870 | 114 | 7 | - | ||||||
| Block 1 | 1.68 ± 0.13 | 1.43 ± 0.19 | 4.50 ± 3.85 | 0.0172 (0.0390) | 0.660 | 0.943 | - | NS | - | |
| Block 2 | rs11888492 | 773 | 218 | 13 | - | |||||
| 1.67 ± 0.14 | 1.58 ± 0.27 | 3.06 ± 1.70 | −0.0002 (0.0300) | 0.884 | 0.982 | - | NS | - | ||
| rs8330 | 601 | 328 | 73 | - | ||||||
| 1.75 ± 0.17 | 1.41 ± 0.15 | 2.02 ± 0.58 | − 0.0103 (0.0218) | 0.637 | 0.943 | - | NS | - | ||
| rs1500482 | 620 | 335 | 49 | - | ||||||
| 1.71 ± 0.17 | 1.45 ± 0.15 | 2.55 ± 0.84 | 0.0003 (0.0230) | 0.989 | 0.989 | - | NS | - | ||
| rs4663972 | 610 | 341 | 55 | - | ||||||
| Block 3 | 1.76 ± 0.17 | 1.35 ± 0.14 | 2.62 ± 0.77 | −0.0070 (0.0230) | 0.761 | 0.951 | - | NS | - | |
Abbreviations: LD, linkage disequilibrium; SE, standard error; FDR, false discovery rate.
Data are n or mean ± SD; genotype 0, 1, 2 represent the wildtype, heterozygotes, and homozygotes, respectively. Missing genotyping is not included.
In LD block 1, 4 common haplotypes with frequencies ≥5% were reconstructed (table 5). Using the most common haplotype ATTTTG (52.8%) as the reference, haplotype CCGTTG (32.8%) that harbored the minor alleles of rs3755319, rs887829, and rs6742078 was significantly associated with higher bilirubin levels (adjusted p = 0.0002; global test p<0.0001). No haplotype in LD block 3 was associated with bilirubin levels.
Table 5.
Associations of UGT1A1 haplotypes with serum total bilirubin levels: Linear regression analysis
| LD block | Haplotype | Haplotype frequency | Univariate analysis |
Multivariate analysisc |
||
|---|---|---|---|---|---|---|
| Estimates (SE) | p value | Estimates (SE) | p value | |||
| ATTTTG | 52.8% | 1.0 | - | 1.0 | - | |
| CCGTTG | 32.8% | 0.0698 (0.0215) | 0.0012 | 0.0712 (0.0189) | 0.0002 | |
| Block 1a | CTTGTG | 5.7% | −0.0169 (0.0422) | 0.689 | −0.0089 (0.0368) | 0.807 |
| CTTGGA | 5.6% | 0.0737 (0.0410) | 0.073 | 0.0321 (0.0356) | 0.380 | |
| Others | 3.1% | - | - | - | - | |
| GTT | 73.6% | 1.0 | - | 1.0 | - | |
| CCC | 20.4% | −0.0153 (0.0235) | 0.593 | −0.0240 (0.0206 ) | 0.244 | |
| Block 3b | ||||||
| CTT | 2.53% | −0.0313 (0.0585) | 0.581 | − 0.0179 (0.0513) | 0.727 | |
| Others | 3.47% | - | - | - | - | |
Abbreviations: LD, linkage disequilibrium; SE, standard error.
Polymorphisms are in the order of: rs3755319-rs887829-rs6742078-rs3771342-rs17864705-rs1018124.
Polymorphisms are in the order of: rs8330-rs1500482-rs4663972.
Adjustment for age, gender, sepsis shock, chronic liver diseases, history of alcohol use, history of steroid use, diabetes, modified APACHE III scores, and organ dysfunctions (respiratory, cardiovascular, renal, and hematological).
Association of UGT1A1 variants with the development and mortality of sepsis-related ARDS
Because UGT1A1 variants were strongly associated with higher serum bilirubin levels, and higher bilirubin levels were associated with increased ARDS development and mortality, we evaluated whether UGT1A1 tSNPs or haplotypes are associated with ARDS development or mortality. The distributions of UGT1A1 tSNP genotypes were not significantly different between patients with and without ARDS (all p values >0.05). In logistic regression using the major allele homozygotes as referent genotypes, the rs17864705 and rs1018124 (in LD block 1) were marginally associated with increased risk of ARDS (ORadj = 1.52; p = 0.037 for rs17864705. ORadj = 1.48; p = 0.058 for rs1018124). But these associations were no longer significant in FDR analysis (table S3). Similarly, two haplotypes were marginally associated with increased ARDS risk in overall analysis (table S3). No significant associations were detected between any UGT1A1 tSNPs or haplotypes and ARDS mortalities.
DISCUSSION
This study shows that higher bilirubin on ICU admission is associated with subsequent sepsis-related ARDS development and mortality. Furthermore, our study shows that serum bilirubin levels in sepsis are mainly influenced by clinical factors and partly by genetic variants of UGT1A1.
Although sepsis is a major risk for ARDS,28 no prior study has described predictive biomarkers for ARDS development in septic patients. Our results showed that a slight increase in bilirubin on ICU admission was associated with marked increase of ARDS risk and mortality in septic patients, suggesting that serum bilirubin is an early and sensitive biomarker of sepsis-related ARDS. The fact that adjustments for multiple covariates did not change these associations further suggested that bilirubin is an independent predictor of sepsis-related ARDS. Previous studies on prognostic value of bilirubin in sepsis have focused mainly on survival as the major outcome measure.29, 30 Although the ultimate outcome measure for any patient is survival, death is not the only outcome measure of ICU treatment. In critically ill patients, identification of risk biomarkers for ARDS development may help clinicians in both diagnostic evaluation and management. For instance, hyperbilirubinemia has been reported to predispose surgical ICU patients to infection, indicating a need for higher level of vigilance for infection in ICU patients with hyperbilirubinemia.12, 31
Our results from a large study population confirmed the findings in previous small studies that hyperbilirubinemia is associated with worse survival in ARDS patients.29, 30 Unlike previous reports, ARDS diagnosis in our study was determined prospectively using the widely accepted AECC definition, and therefore, phenotype misclassification was minimized. The use of individual phenotype rather than critical illness from mixture causes reduces confounding from any possible associations between the various causes and outcomes. Moreover, the outcomes of ARDS were measured for 60-day mortality, therefore bias in survival calculation due to incomplete observation was minimized.
Although physiological levels of bilirubin are considered a potent antioxidant,32 increasing evidences have shown that bilirubin at high concentrations may be an active participant in the disease process. Bilirubin in the blood can induce cell lysis of erythrocytes.33 Elevated Bilirubin can stimulate oxidative stress and decrease cell survival 34–35. Bilirubin promotes apoptosis in cultured cells.5 In addition, bilirubin can also induce inflammatory response, which is further increased when cells are simultaneously exposed to lipopolysaccharide.
Consistent with a previous report in critically ill patients,13 we observed that several clinical factors including pneumonia, pre-admission steroid use, diabetes history, and organ dysfunctions (respiratory, renal and hematological) were associated with serum bilirubin levels. In contrast to previous study using univariate analysis, we defined the contributions of clinical factors to bilirubin variations by multivariate regression models and demonstrated that clinical factors explained 29.5 % of the overall variation of serum bilirubin levels in septic patients. It should be pointed out that although chronic liver disease is strongly associated with increased serum bilirubin levels, hepatic dysfunction is not the only organ dysfunction associated with hyperbilirubinemia in sepsis. Cholestasis, circulating endotoxin and hemolysis may also play important roles in the etiology of hyperbilirubinemia in sepsis.11 In subjects without chronic liver disorders, we found that other clinical factors were also significantly associated with serum bilirubin levels. It has been reported that elevated bilirubin was often seen in sepsis and septic shock patients in the absence of primary liver or biliary disease.31
Despite the evidence that genetic variants of UGT1A1 were significantly associated with serum bilirubin levels, the overall contribution of these variants was small compared with clinical factors. Similar to the moderate impact of genetic variants of UGT1A1 on serum bilirubin levels, the strengths of overall associations between UGT1A1 polymorphisms with sepsis-related ARDS risk were also modest. Considering that less than 8% of the variance in bilirubin levels is explained by UGT1A1 polymorphisms, it is not surprising that we did not detect a strong association of these polymorphisms with ARDS risk or mortality, although the association of serum bilirubin level with ARDS risk and mortality is relatively strong.
The large sample size, the adjustment for multiple clinical covariates, comprehensive evaluation of common variants surrounding the UGT1A1 gene, and serial measurements of bilirubin levels are the strengths of this study. Despite these assets, our study has some limitations. First, our study design did not allow us to define whether bilirubin is an effecter molecule in the pathogenesis of ARDS or merely a marker of systemic injury. Future research is needed to elucidate the potential pathogenic role of bilirubin in ARDS. Second, this study could not exclude the possibility of medication influence on bilirubin levels. However, a recent study including 17 drugs potentially inducing hepatotoxicity has shown that medication administrations during ICU stay did not significantly contribute to serum bilirubin levels.13 Third, we were unable to exclude the possibility that our cohort might include subjects with existing Gilbert’s syndrome because all subjects in this study were critically ill patients. Whether Gilbert’s syndrome might be associated with sepsis-related ARDS requires further investigation. Fourth, 217 septic patients were excluded from analysis due to no ICU admission bilirubin records. Since most of these patients had bilirubin measurements <2.0 mg/dL analyzed before ICU admission, they were considered as patients with normal bilirubin levels and were not tested for bilirubin by ICU clinicians. Comparison analysis showed that patients without ICU admission bilirubin values had lower severity of illness and less organ failure than those with admission bilirubin and were more similar to those with bilirubin <1.0 mg/dL than those patients with bilirubin >1.0 mg/dL (table S2). Therefore, we assumed that patients without ICU admission bilirubin values were probably among subjects with bilirubin <1.0 mg/dL and exclusion of these patients from our analyses did not change the results, and probably diluted the strength of associations detected in this study.
In conclusion, we show that serum bilirubin level on ICU admission is an independent early predictor of ARDS development and mortality in septic patients. Several clinical characteristics and genetic variants of UGT1A1 independently affect serum bilirubin levels. However, the relationship between UGT1A1 variants and bilirubin levels did not translate largely to an associated change in ARDS outcomes.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge valuable contributions to this study from the following: Marcia Chertok, Janna Frelich, Julia Shin, Andrea Shafer, Lia Shimada, Weiling Zhang, Kelly McCoy, and Thomas McCabe.
Funding: This work was supported by grants from the National Institute of Health (HL60710 and ES00002) and the Flight Attendant Medical Research Institute (FAMRI, 062459_YCSA).
Footnotes
Competing interests: None.
The Corresponding Author has the right to grant on behalf of all authors and does granton behalf of all authors, an exclusive license (or non exclusive for government employees)on a worldwide basis to the BMJ Publishing Group Ltd and its licensees, to permit thisarticle (if accepted) to be published in Thorax and any other BMJ Group products and toexploit all subsidiary rights, as set out in our license.
References
- 1.Martin GS, Mannino DM, Eaton S, et al. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med. 2003;348:546–54. doi: 10.1056/NEJMoa022139. [DOI] [PubMed] [Google Scholar]
- 2.Piantadosi CA, Schwartz DA. The acute respiratory distress syndrome. Ann Intern Med. 2004;141:460–70. doi: 10.7326/0003-4819-141-6-200409210-00012. [DOI] [PubMed] [Google Scholar]
- 3.Hudson LD, Milberg JA, Anardi D, et al. Clinical risks for development of the acute respiratory distress syndrome. Am J Respir Crit Care Med. 1995;151:293–301. doi: 10.1164/ajrccm.151.2.7842182. [DOI] [PubMed] [Google Scholar]
- 4.Noriega GO, Tomaro ML, del Batlle AM. Bilirubin is highly effective in preventing in vivo delta-aminolevulinic acid-induced oxidative cell damage. Biochim Biophys Acta. 2003;1638:173–8. doi: 10.1016/s0925-4439(03)00081-4. [DOI] [PubMed] [Google Scholar]
- 5.Fernandes A, Falcao AS, Silva RF, et al. Inflammatory signalling pathways involved in astroglial activation by unconjugated bilirubin. J Neurochem. 2006;96:1667–79. doi: 10.1111/j.1471-4159.2006.03680.x. [DOI] [PubMed] [Google Scholar]
- 6.Rodrigues CM, Sola S, Brito MA, et al. Bilirubin directly disrupts membrane lipid polarity and fluidity, protein order, and redox status in rat mitochondria. J Hepatol. 2002;36:335–41. doi: 10.1016/s0168-8278(01)00279-3. [DOI] [PubMed] [Google Scholar]
- 7.Alexandra Brito M, Silva RF, Brites D. Bilirubin toxicity to human erythrocytes: a review. Clin Chim Acta. 2006;374:46–56. doi: 10.1016/j.cca.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 8.Chopra M, Reuben JS, Sharma AC. Acute Lung Injury: Apoptosis and Signaling Mechanisms. Exp Biol Med (Maywood) 2009;234:361–71. doi: 10.3181/0811-MR-318. [DOI] [PubMed] [Google Scholar]
- 9.Fuchs M, Sanyal AJ. Sepsis and cholestasis. Clin Liver Dis. 2008;12:151–72. ix. doi: 10.1016/j.cld.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Whitehead MW, Hainsworth I, Kingham JG. The causes of obvious jaundice in South West Wales: perceptions versus reality. Gut. 2001;48:409–13. doi: 10.1136/gut.48.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chand N, Sanyal AJ. Sepsis-induced cholestasis. Hepatology. 2007;45:230–41. doi: 10.1002/hep.21480. [DOI] [PubMed] [Google Scholar]
- 12.Field E, Horst HM, Rubinfeld IS, et al. Hyperbilirubinemia: a risk factor for infection in the surgical intensive care unit[ Am J Surg 2008. 1953046discussion 306–7. [DOI] [PubMed] [Google Scholar]
- 13.Brienza N, Dalfino L, Cinnella G, et al. Jaundice in critical illness: promoting factors of a concealed reality. Intensive Care Med. 2006;32:267–74. doi: 10.1007/s00134-005-0023-3. [DOI] [PubMed] [Google Scholar]
- 14.Harbrecht BG, Zenati MS, Doyle HR, et al. Hepatic dysfunction increases length of stay and risk of death after injury. J Trauma. 2002;53:517–23. doi: 10.1097/00005373-200209000-00020. [DOI] [PubMed] [Google Scholar]
- 15.Tukey RH, Strassburg CP. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol. 2000;40:581–616. doi: 10.1146/annurev.pharmtox.40.1.581. [DOI] [PubMed] [Google Scholar]
- 16.Lampe JW, Bigler J, Horner NK, et al. UDP-glucuronosyltransferase (UGT1A1*28 and UGT1A6*2) polymorphisms in Caucasians and Asians: relationships to serum bilirubin concentrations. Pharmacogenetics. 1999;9:341–9. doi: 10.1097/00008571-199906000-00009. [DOI] [PubMed] [Google Scholar]
- 17.Sai K, Saeki M, Saito Y, et al. UGT1A1 haplotypes associated with reduced glucuronidation and increased serum bilirubin in irinotecan-administered Japanese patients with cancer. Clin Pharmacol Ther. 2004;75:501–15. doi: 10.1016/j.clpt.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 18.Saeki M, Saito Y, Sai K, et al. A combinatorial haplotype of the UDP-glucuronosyltransferase 1A1 gene increases total bilirubin concentrations in Japanese volunteers. Clin Chem. 2007;53:356–8. doi: 10.1373/clinchem.2006.077990. [DOI] [PubMed] [Google Scholar]
- 19.Miners JO, McKinnon RA, Mackenzie PI. Genetic polymorphisms of UDP-glucuronosyltransferases and their functional significance . Toxicology. 2002;181–182:453–6. doi: 10.1016/s0300-483x(02)00449-3. [DOI] [PubMed] [Google Scholar]
- 20.Toffoli G, Cecchin E, Corona G, et al. The role of UGT1A1*28 polymorphism in the pharmacodynamics and pharmacokinetics of irinotecan in patients with metastatic colorectal cancer. J Clin Oncol. 2006;24:3061–8. doi: 10.1200/JCO.2005.05.5400. [DOI] [PubMed] [Google Scholar]
- 21.Peterkin VC, Bauman JN, Goosen TC, et al. Limited influence of UGT1A1*28 and no effect of UGT2B7*2 polymorphisms on UGT1A1 or UGT2B7 activities and protein expression in human liver microsomes. Br J Clin Pharmacol. 2007;64:458–68. doi: 10.1111/j.1365-2125.2007.02923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhai R, Gong MN, Zhou W, et al. Genotypes and haplotypes of the VEGF gene are associated with higher mortality and lower VEGF plasma levels in patients with ARDS. Thorax. 2007;62:718–22. doi: 10.1136/thx.2006.069393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bone RC, Balk RA, Cerra FB, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101:1644–55. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 24.Bernard GR, Artigas A, Brigham KL, et al. Report of the American-European consensus conference on ARDS: definitions, mechanisms, relevant outcomes and clinical trial coordination. The Consensus Committee. Intensive Care Med. 1994;20:225–32. doi: 10.1007/BF01704707. [DOI] [PubMed] [Google Scholar]
- 25.The Acute Respiratory Distress Syndrome Network. Ventilation with lower tidal volumes as compared with traditional tidal volumes for acute lung injury and the acute respiratory distress syndrome. N Engl J Med. 2000;342:1301–8. doi: 10.1056/NEJM200005043421801. [DOI] [PubMed] [Google Scholar]
- 26.Rubenfeld GD, Herridge MS. Epidemiology and outcomes of acute lung injury. Chest. 2007;131:554–62. doi: 10.1378/chest.06-1976. [DOI] [PubMed] [Google Scholar]
- 27.Benjamini Y, Yekutieli D. Quantitative trait Loci analysis using the false discovery rate. Genetics. 2005;171:783–90. doi: 10.1534/genetics.104.036699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fein AM, Calalang-Colucci MG. Acute lung injury and acute respiratory distress syndrome in sepsis and septic shock. Crit Care Clin. 2000;16:289–317. doi: 10.1016/s0749-0704(05)70111-1. [DOI] [PubMed] [Google Scholar]
- 29.Schwartz DB, Bone RC, Balk RA, et al. Hepatic dysfunction in the adult respiratory distress syndrome. Chest. 1989;95:871–5. doi: 10.1378/chest.95.4.871. [DOI] [PubMed] [Google Scholar]
- 30.Hebert PC, Drummond AJ, Singer J, et al. A simple multiple system organ failure scoring system predicts mortality of patients who have sepsis syndrome. Chest. 1993;104:230–5. doi: 10.1378/chest.104.1.230. [DOI] [PubMed] [Google Scholar]
- 31.Franson TR, LaBrecque DR, Buggy BP, et al. Serial bilirubin determinations as a prognostic marker in clinical infections. Am J Med Sci. 1989;297:149–52. doi: 10.1097/00000441-198903000-00003. [DOI] [PubMed] [Google Scholar]
- 32.Minetti M, Mallozzi C, Di Stasi AM, et al. Bilirubin is an effective antioxidant of peroxynitrite-mediated protein oxidation in human blood plasma. Arch Biochem Biophys. 1998;352:165–74. doi: 10.1006/abbi.1998.0584. [DOI] [PubMed] [Google Scholar]
- 33.Brito MA, Silva R, Tiribelli C, Brites D. Assessment of bilirubin toxicity to erythrocytes. Implication in neonatal jaundice management. Eur J Clin Invest. 2000;30:239–47. doi: 10.1046/j.1365-2362.2000.00612.x. [DOI] [PubMed] [Google Scholar]
- 34.Cesaratto L, Calligaris SD, Vascotto C, et al. Bilirubin-induced cell toxicity involves PTEN activation through an APE1/Ref-1-dependent pathway. J Mol Med. 2007;85:1099–112. doi: 10.1007/s00109-007-0204-3. [DOI] [PubMed] [Google Scholar]
- 35.Ostrow JD, Pascolo L, Brites D, Tiribelli C. Molecular basis of bilirubin-induced neurotoxicity. Trends Mol Med. 2004;10:65–70. doi: 10.1016/j.molmed.2003.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.