Abstract
Leukemia associated Rho guanine nucleotide exchange factor (LARG) activates RhoA in response to signals received by specific classes of cell surface receptors. The catalytic core of LARG is a Dbl homology (DH) domain whose activity is modulated by an adjacent pleckstrin homology (PH) domain. In this study, we used a transcriptional assay and confocal microscopy to examine the roles of several novel structural features of the LARG DH/PH domains, including a conserved and exposed hydrophobic patch on the PH domain that mediates protein-protein interactions in crystal structures of LARG and its close homolog PDZ-RhoGEF. Mutation of the hydrophobic patch has no effect on nucleotide exchange activity in vitro, but abolished the ability of LARG to activate RhoA and to induce stress fiber formation in cultured cells. The activity of these mutants could be rescued by fusion with exogenous membrane targeting domains. However, because membrane recruitment by activated Gα13 subunits was not sufficient to rescue activity of a hydrophobic patch mutant, the LARG PH domain cannot solely contribute to membrane targeting. Instead, it seems likely the domain is involved in regulatory interactions with other proteins near the membrane surface. We also show that the hydrophobic patch of the PH domain is likely important for the activity of all Lbc family RhoGEFs.
Keywords: Gα13, RhoGEF, Lbc, Pleckstrin homology domain, Actin stress fibers, Membrane recruitment
1. Introduction
Rho family small molecular weight GTPases belong to the Ras superfamily and control a wide variety of cellular processes including cytoskeleton reorganization, cell polarity, microtubule dynamics, cell cycle progression, membrane trafficking, and gene expression [1]. Like all GTPases, Rho family members oscillate between inactive, GDP-bound and active, GTP-bound states. In the cell, at least three distinct families of regulatory proteins control this transition. Rho guanine nucleotide exchange factors (RhoGEFs) stimulate nucleotide exchange on Rho GTPases to generate the activated, GTP-bound form, which is then capable of recognizing downstream effector proteins. GTPase activating proteins accelerate the intrinsic GTPase activity of Rho family members, and guanine nucleotide dissociation inhibitors interact with the prenylated, GDP-bound form of Rho GTPases to control cycling between the membrane and cytosol.
RhoGEFs are typically large and multi-domain proteins characterized by a Dbl homology (DH) domain that is almost always followed in the primary sequence by a pleckstrin homology (PH) domain [2]. The DH domain forms the majority of the interface with the bound GTPase and is responsible for exchange activity. The role of the PH domain is more complex and varies among RhoGEFs. The domain can play a role in targeting RhoGEFs to membranes or to other sub-cellular structures, or in regulating nucleotide exchange efficiency [2]. Such PH-mediated regulation can either be positive [3–5], or negative, thereby inhibiting the intrinsic GEF activity until the appropriate signaling state is achieved [6–8].
Leukemia associated RhoGEF (LARG) was discovered as a fusion partner with the mixed lineage leukemia protein in a patient with acute myeloid leukemia [9] and belongs to a subfamily of three RhoA-selective RhoGEFs that also includes p115RhoGEF and PDZ-RhoGEF [10]. The most distinctive feature of these enzymes is a regulator of G protein signaling (RGS) homology (RH) domain situated N-terminal to the DH/PH domains that binds with high affinity to activated heterotrimeric G protein α12/13 (Gα12/13) subunits [11, 12]. Hence, we refer to them as the RH-RhoGEFs. Although p115RhoGEF is the only member of the family whose activity has been convincingly shown to be regulated by Gα12/13 in vitro [13], the RH domain-mediated interaction clearly contributes to membrane targeting in cells which would presumedly bring the enzyme into close proximity with RhoA [14–17]. Both LARG and PDZ-RhoGEF have been shown to be activated by Gα12/13-coupled receptors, presumably through their interactions with Gα12/13 subunits, either directly by siRNA knockdown of the RhoGEFs [18], or indirectly through use of dominant negative forms of the enzymes [19–22].
In addition to the RH domain, LARG contains an N-terminal postsynaptic density protein, Drosophila disc large tumor suppressor, and zonula occludens-1 protein (PDZ) domain that has been shown to localize LARG to the membrane via interactions with the insulin-like growth factor [23] and Plexin B1 [24, 25] receptors. RhoA-dependent signaling appears to be induced through these interactions. The PH domain of RH-RhoGEFs also appears important for membrane localization, although it does not appear to strongly localize proteins to membranes on its own [17, 26]. Thus, RH-RhoGEFs contain multiple domains that can mediate membrane targeting. Because p115RhoGEF activity is enhanced when fused to the CAAX box from K-Ras [14], membrane localization represents one mechanism by which RH-RhoGEFs can be activated in response to extracellular signals. Interaction via one or more membrane targeting motifs (mediated by PDZ, RH, or PH domains) may be necessary for full activation in cells [26].
The RH-RhoGEF PH domain appears to play a positive role in the regulation of GEF activity [3, 17, 27, 28]. The crystal structures of LARG [28] and PDZ-RhoGEF [27] in complex with RhoA suggest at least one mechanism by which this might occur. In these models, residues in the β1 strand and C-terminal helix of the PH domain form direct contacts with RhoA. LARG thus forms a bidentate interaction with RhoA through its DH and PH domains, which as a result constrains the conformation of residues in the hinge that connects the two domains. These hinge residues also directly interact withRhoA. Site-directed mutagenesis of the residues in the PH domain and in the hinge between the DH and PH domains that contact RhoA reduced GEF activity of DH/PH fragment to the level of the DH domain alone in vitro [28].
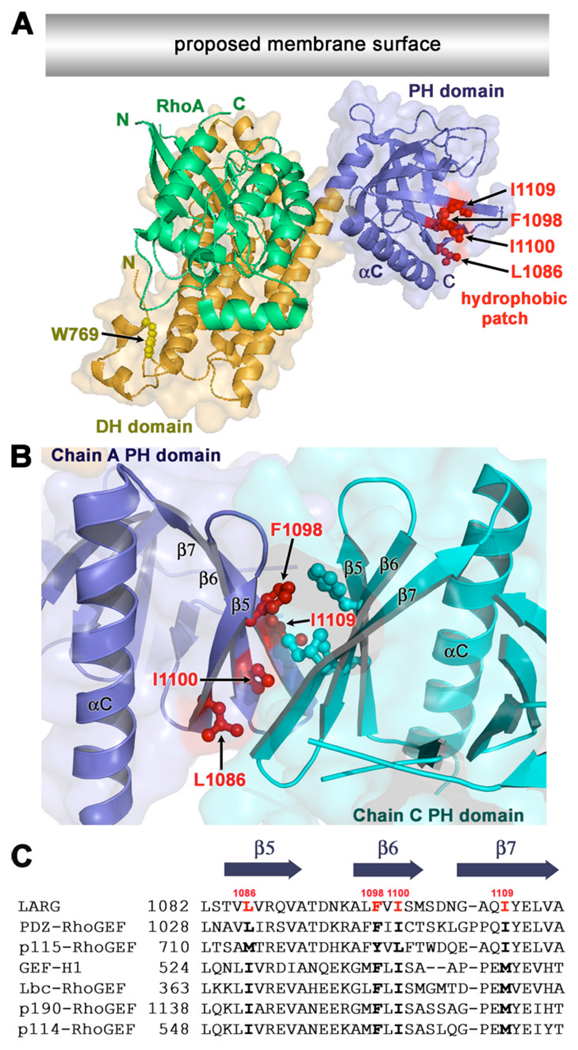
Interestingly, in the crystal structures of the LARG DH/PH and DH/PH-RhoA [28] and PDZ-RhoGEF DH/PH-RhoA complexes, every PH domain crystallized such that it forms a quasi two-fold interface with another PH domain. Each of these interfaces buries a total of ~800 Å2 of surface area. In each case, this interface consists of a hydrophobic patch on the β5-β7 sheet of the PH domain that includes the side chains of Leu-1086, Phe-1098, Ile-1100, and Ile-1109 in LARG (L1032, F1044, I1046, and I1056 in PDZ-RhoGEF) (Fig. 1 A–B). All four PH domains in the asymmetric unit of the LARG DH/PH-RhoA structure and both PH domains in the PDZ-RhoGEF DH/PH-RhoA structure dimerize such that the β5-β7 strands of each PH domain are roughly parallel in orientation (Fig. 1 B). In the LARG DH/PH structure, the same hydrophobic patch forms the interface except that the β5-β7 strands of each PH domain that forms the interface are roughly perpendicular. The residues that comprise this patch on the PH domain are highly conserved not only within the RH-RhoGEF family, but also within a larger subfamily of RhoA-selective RhoGEFs that includes Lbc, GEF-H1 (Lfc), p114-RhoGEF, and p190-RhoGEF (referred to herein as the Lbc subfamily). Unlike the RH-RhoGEFs, these latter RhoGEFs lack obvious RH domains. Based on the LARG and PDZ-RhoGEF crystal structures, we hypothesized that the hydrophobic patch could act as a docking site for other proteins or lipids [28]. Indeed, models of RhoGEF-RhoA complexes at the plasma membrane [2, 28] suggest that the hydrophobic patch could mediate lateral interactions with other proteins at the cell surface (Fig. 1 A).
Figure 1. An exposed hydrophobic patch on the PH domain is conserved within Lbc Family of RhoGEFs.
(A) Model of the LARG DH/PH domains in complex with RhoA at the cell membrane [28]. The side chains of Trp769 (yellow) in the hydrophobic core of the αN1/αN2 N-terminal extension, and Leu1086, Phe1098, Ile1100, and Ile1109 (red) in the hydrophobic patch of the PH domain are shown as ball-and-stick models. The latter residues form a hydrophobic patch oriented parallel to the predicted membrane surface (grey rectangle). The accessible surface of the DH/PH domains is shown semi-transparently. (B) A dimer interface mediated by the hydrophobic patch of the PH domain in the LARG DH/PH-RhoA crystals. The side chains of hydrophobic patch residues are shown as ball-and-stick models colored according to their subunit. In this interface, the β5, β6, and β7 strands of each PH domain are oriented in roughly parallel orientation. (C) Alignment of PH domain sequences spanning the hydrophobic patch residues from different members of the Lbc RhoGEF family. GenBank identifiers are as follows: LARG (34395525), PDZ-RhoGEF (34395516), p115-RhoGEF (34395524), GEF-H1 (6919894), Lbc-RhoGEF (6016482), p190-RhoGEF (172046113), and p114-RhoGEF (190358159). The positions of Leu1086, Phe1098, Ile1100, and Ile1109 are highlighted in red in the LARG sequence, and conserved residues in other Lbc family members are shown with bold text.
In this study, we explore the roles of Lbc-subfamily specific features of the LARG catalytic DH/PH domains, in particular the hydrophobic patch. We show that although mutations in the hydrophobic patch do not have a significant effect on in vitro activity, they greatly reduce LARG and Lbc activity in cells. Because these mutants can be rescued by fusion of LARG with two different membrane-targeting motifs, the PH domain of RH-RhoGEF and Lbc subfamily members appears to be involved in membrane targeting via a novel protein-protein interaction mechanism that does not appear to involve classical PH domain-phospholipid interactions. As has been observed for p115RhoGEF, membrane targeting of LARG leads to a significant enhancement of RhoA activity, supporting the hypothesis that membrane targeting can play a significant role in the regulation of RH-RhoGEF activity.
2. Materials and methods
2.1. Plasmids
A pCGN-hygro vector bearing FL WT human LARG was a gift from Dr Channing Der (University of N. Carolina), and was altered to fix a frame shift run-on mutation at the C-terminus. To generate GFP fusions, LARG-FL and its mutants were subcloned into the pEGFP-C1 vector (Clontech laboratories, Inc.) using flanking BamHI restriction sites. LARG-DH/PH (residues 666–1138), -PH (residues 982–1138), -ΔC (residues 1–1138), -ΔPH/C (residues1–989), and -DH (residues 666–989) were made as GFP fusions in pEGFP-C1 using the XhoI and BamHI sites. To generate LARG fusions with the PLC-δ1 tandem PH (residues 11–140) domains, we first sub-cloned the fragment containing tandem (2x) PH domains from the pPP1851 vector (a gift from Dr. P.M. Pryciak, University of Massachusetts, Worcester, MA) into the pEGFP-C1 vector using XmaI to generate pEGFP-2xPLCδ1 PH, and then cloned PCR fragments containing different LARG truncations into pEGFP-2xPLCδ1 PH using the XhoI and KpnI sites. The pRGS2-YFP-Ras vector was a gift from Dr. S. Heximer (University of Toronto, Ontario CA) and used as a PCR template to generate a fragment containing the Ras polybasic and CAAX prenylation sequence (residues 172–188) with flanking KpnI and BamHI sites. LARG-CAAX fusions were then created by replacing the 2xPLCδ1 PH fragment in the pEGFP-LARG-2xPLCδ1 PH constructs with the generated Ras PCR fragment. SRE.L firefly luciferase reporter, pRL-thymidine kinase renilla luciferase reporter, and pcDNA-human Gα13-WT and -Q226L plasmids were described previously [29]. E. coli expression constructs for LARG RH/DH/PH (residues 341–1138) and DH/PH (residues 765–1138) were described previously [28, 30]. Human Lbc-RhoGEF in the pSRαNeo plasmid was a gift from Dr D. Toksoz (Tufts-New England Medical Center, Boston, MA) and was used as PCR template to generate Lbc-DH/PH and sub-clone it into pEGFP-C1. Human RhoA-G17A for pull downs was created from the WT pMAL-RhoA vector [28]. All site-directed mutants were created using QuikChange (Qiagen) and confirmed by DNA sequencing.
2.2. Cell culture and transfection
HEK293T, COS-7, and NIH3T3 cells were maintained in high glucose Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% heat-inactivated new born calf serum (NCS) in the presence of 100 units/mL penicillin and 100 µg/mL streptomycin and were incubated at 37 °C in a humidified atmosphere with 5% CO2. Cells were transfected using Lipofectamine 2000 (Invitrogen) or FuGENE 6 (Roche Applied Science) according to the manufacturer’s instructions.
2.3. SRE.L Luciferase Reporter Assay
HEK293T cells were seeded into 96 well plate 24 h before transfection. In antibiotic free medium, cells were co-transfected with the indicated plasmids, pSRE.L, pRL-TK, and pcDNA3.1 (to adjust total DNA to 100 ng). One day after transfection, cells were starved in DMEM containing 0.5% NCS, and incubated for an additional 24 h. Cells were then washed with PBS, lysed, and then assayed for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol. Luminescence was read on a Victor plate reader with dual injectors (Perkin-Helmer). Firefly activities were measured in triplicate for each transfection mix, and then normalized to renilla activities and reported as fold activation over the basal level corresponding to the activity of the empty (pCGN or pEGFP) vector. Data were analyzed for significance in Prism using ANOVA with a Bonferroni post-test. For comparison of the expression of LARG constructs in HEK293T, we used an anti-HA monoclonal antibody (Covance).
2.4. Confocal microscopy and stress fiber staining
HEK293T, COS-7 or NIH3T3 cells were plated on poly-D lysine (Sigma) coated cover slips in 12 well plates. After attachment, cells were transfected with the indicated plasmids. After serum starvation in DMEM plus 0.5% NCS, cells were washed with PBS, fixed with 4% paraformaldehyde, and quenched with 50 mM glycine in PBS. For localization studies in HEK293T or COS-7 cells, coverslips were directly mounted on glass slides using Vectashield Mounting Medium (Vector Laboratories, Burlingame, CA). For actin stress fiber staining, fixed NIH3T3 cells were permeabilized with 0.1% Triton X-100 in PBS, blocked with 1% BSA plus 1% goat serum in PBS, and stained with Alexa Fluor 594 Phalloidin (Invitrogen) before mounting. Images were captured using an Olympus FluoView 500 laser scanning confocal microscope (Olympus America, Inc., Center Valley, PA) with a 60x oil immersion objective (Nikon, Japan), and processed with Adobe Photoshop (Adobe system, Inc. Mountain view, CA).
2.5. Cycloheximide treatment
Transfected HEK293T cells in 6 well plates were treated with 50 µM cycloheximide (Sigma) for the indicated period of time and washed with PBS before adding ice-cold lysis buffer supplemented with EDTA free protease inhibitor cocktail (Roche Applied Science). Extracts were then resolved on SDS-PAGE and blotted with anti-GFP (Santa Cruz) and anti-actin (Cytoskeleton) antibodies.
2.6. Protein preparation
LARG RH/DH/PH and DH/PH were expressed in E. coli and purified as described previously [28]. Recombinant RhoA-G17A was purified using Ni-NTA beads and size exclusion chromatography before biotinylation with biotin N-hydroxy succinimide ester (Sigma) for pull down assays. N-terminally hexahistidine tagged Gαi/13 was expressed in High Five insect cells and purified as described [31] before labeling for FCPIA.
2.7. RhoA-G17A pull-down assay and immuno-precipitation
Plasmids encoding GFP or GFP-LARG DH/PH (WT or F1098D) were transfected in HEK293T cells seeded in 60 mm dishes. One day after transfection, cells were lysed in a buffer containing 50 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% (w/v) NP-40, and fresh protease inhibitor cocktail (Roche Applied Science). For the RhoA G17A pull-down, HEK293T extracts were incubated with streptavidin-coated agarose beads (Invitrogen) that had been incubated with biotinylated RhoA-G17A. For tubulin or actin immuno-precipitations, HEK293T extracts were incubated with anti-GFP antibody (Santa Cruz) coupled to protein A-agarose beads (Upstate, NY). After washing with lysis buffer, the beads were suspended in SDS loading buffer. Input, retained protein, and control samples were resolved on SDS-PAGE and analyzed by Western blot. Anti-GFP antibody (Santa Cruz) was used to blot RhoA-G17A pull-down samples, and anti-tubulin (Abcam) or anti-actin antibodies were used to blot anti-GFP immuno-precipitates.
2.8. Flow Cytometry Protein Interaction Assay (FCPIA)
E. coli purified LARG RH/DH/PH was biotinylated with biotin N-hydroxysuccinimide ester (Sigma) and coupled to xMap LumAvidin microspheres (Luminex). Gαi/13 was amine labeled with Alexa Fluor 532 carboxylic acid succinimide ester (Invitrogen) at a 3:1 fluorophore/protein ratio in 20 mM HEPES pH 8.0, 100 mM NaCl, 5 mM MgCl2, 0.1% (v/v) lubrol, 2 mM DTT, 50 µM GDP, 20 µM AlCl3, and 10 mM NaF). Excess fluorophore was removed by filtration through a Zeba Desalt spin column (Pierce). AlCl3 and NaF were omitted from samples to measure non-specific binding. For FCPIA direct binding experiments, increasing amounts of fluorescently labeled Gαi/13 were added to biotinylated RH/DH/PH-WT on beads, incubated for 30 min, and the median fluorescence intensity (MFI) of bead-associated fluorescence was measured in a Luminex 96-well flow cytometer. The data was processed using GraphPad Prism. For FCPIA competition binding experiments, we used increasing amounts of unlabeled LARG proteins (RH/DH/PH-WT, -F1098D or -E354K) to compete with 20–40 nM fluorescently-labeled Gαi/13 bound to biotinylated RH/DH/PH-WT on beads. To detect LARG binding to Gβγ or tubulin, LARG RH/DH/PH was either fluorescently labeled (in solution) or biotinylated (on beads). Pure bovine tubulin (Cytoskeleton) was fluorescently labeled in a buffer containing 80 mM PIPES pH 6.9, 2 mM MgCl2 and 0.5 mM EGTA. Mixtures were incubated 30 min before measuring MFI.
2.9. FP RhoA guanine nucleotide exchange assay
The change in FP of BODIPY FL GTPγS (excitation/emission maxima ~503/512 nm; Invitrogen) was measured as it binds RhoA [6]. Briefly, 1 µM of BODIPY FL GTPγS was added to a reaction mixture containing 2 µM RhoA-GDP in 20 mM HEPES pH 8.0, 150 mM NaCl, 10% glycerol, 10 mM MgCl2, and 1 mM DTT. Samples in 384-well plates were excited with plane polarized light, and the increase in millipolarization was monitored over time using a BMG Labtech PHERAstar. Data was processed using GraphPad Prism.
2.10. Protein lipid overlay assay
Overlay assays were performed by using PIP Strip membranes (Echelon biosciences, Salt lake City, UT) according to the manufacturer’s protocol. After 1 hr blocking with 1% nonfat-dry milk in PBS at room temperature, strip membranes were incubated overnight at 4 °C with purified LARG MBP-DH/PH or DH/PH at 1 µg/ml in blocking buffer. Strips were then analyzed by Western blot using penta-His HRP (Qiagen).
3. Results
3.1. The role of Lbc family-specific structure features for LARG mediated RhoA activation in cells
We previously reported two crystal structures of LARG DH/PH domains alone and in complex with nucleotide-free RhoA [28]. These structures revealed several notable differences from other DH/PH domains. The first was an N-terminal extension of the DH domain composed of two additional α helices (αN1 and αN2). The second was the presence of a conserved, exposed hydrophobic patch on the PH domain. These features were also observed in the structure of the PDZ-RhoGEF DH/PH-RhoA complex [27], and, by sequence homology, are expected to exist in all Lbc subfamily members. Using site directed mutagenesis and an in vitro FRET-based nucleotide exchange assay, we demonstrated that disruption of the N-terminal helices (e.g. via W769A/D mutations or by N-terminal truncation) lead to reduced catalytic efficiency (~7-fold for the W769D mutant). The importance of the hydrophobic patch was not tested.
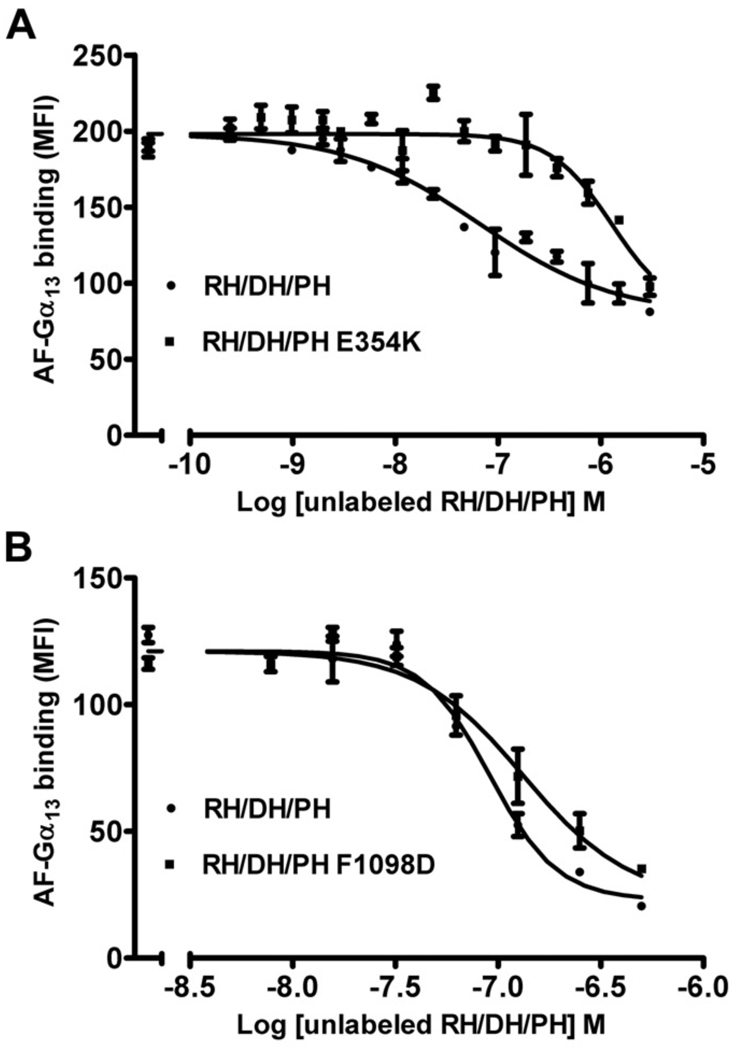
To test the role of the novel N-terminal extension and the PH hydrophobic patch in living cells, we generated the W769D, F1098D, I110E, and I1109D mutants of either HA- or GFP-tagged full-length (FL) LARG. Because a major route of activation for LARG is expected to be through Gα12/13 subunits, we also wanted to be able to assess the contribution of Gα subunits to activity measured in cells. We therefore created the LARG-E354K mutant, which alters a key residue immediately N-terminal to the RH domain that, like the analogous p115RhoGEF-E27K mutant [32], is expected to be required for high affinity binding to Gα13. An in vitro flow cytometry protein interaction assay (FCPIA) indicated that this substitution in LARG increases the Kd of LARG for Gα13 to 540 nM, or ~23-fold (Fig. 2 A). Thus, the SRE.L response using this variant would likely be inhibited if signal transduction involved Gα12/13.
Figure 2. FCPIA competition experiments to measure the affinity of LARG proteins for Gα13 subunits.
Competition of biotinylated RH/DH/PH binding to fluorescently labeled Gα13 with increasing amounts of unlabeled LARG RH/DH/PH (A) (WT or E354K) or (B) unlabeled LARG RH/DH/PH (WT or F1098D). Shown is one representative experiment. The error bars represent the SD of each data point measured in duplicate. The calculated Ki’s for RH/DH/PH-WT, -F1098D, and -E354K are 20 ± 4 nM (n=4), 20 ± 9 nM (n=3) and 540 ± 20 nM (n=2), respectively.
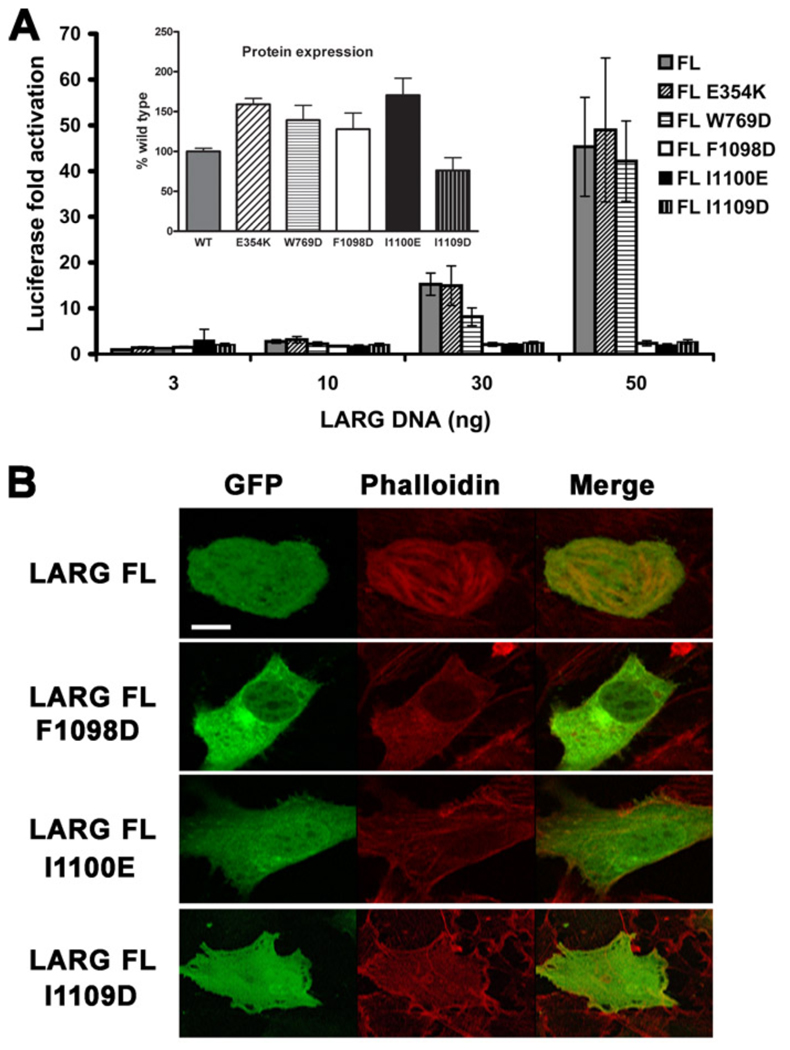
The ability of wild-type (WT) and mutant LARG constructs to activate gene transcription in serum-starved HEK293T cells was then measured using a Rho-specific serum response element (SRE.L) luciferase reporter assay. Surprisingly, alteration of any of the residues from the hydrophobic patch almost completely abrogated SRE.L activity in the context of either the HA- (Fig. 3 A) or GFP- (Fig. S1 A) tagged proteins, whereas the HA-tagged E354K and W769D mutants retained essentially full activity. The WT activity exhibited by the E354K mutant is consistent with a lack of G protein regulation under these assay conditions. To confirm the results with the hydrophobic patch mutants, we also tested them in the background of the LARG DH/PH fragment fused to GFP.
Figure 3. Mutation of the hydrophobic patch abrogates LARG-mediated transcription and stress fiber formation.
(A) Normalized luciferase activities of increasing amounts of HA-tagged LARG FL (WT and mutant) plasmids as measured in a luciferase reporter assay. Firefly luciferase activities were normalized to renilla activities, and reported as fold activation over the basal level. Data shown is reported as the mean ± SD and representative of three independent experiments, each measured in triplicate. Inset: the expression level of each protein was obtained by Western analysis and reported as percentage of WT level. Error bars are the SDs from three independent experiments, each measured in duplicate. (B) GFP fusions of LARG FL (WT, F1098D, I1100E, and I1109D) were transfected into NIH3T3 cells. Serum starved cells were then fixed and stained with Alexa fluor 594-Phalloidin to detect stress fibers. Only WT LARG was able to induce stress fiber formation. Scale bar: 10 µm.
These mutants were also deficient in RhoA-mediated transcriptional activation (Fig. S1 B). Because it was possible that the substitution of charged residues in the hydrophobic patch could destabilize the fold of the PH domain, we also created the more conservative F1098A, I1100A, and L1109A mutants in FL LARG, but these also failed to induce RhoA-dependent gene transcription in cells (Fig. S1 C). Thus, the hydrophobic patch appears critical for basal, Gα12/13-indepdendent signaling by LARG.
The polymerization of actin into stress fibers is an intermediate step in the RhoA pathway leading to gene transcription [33] and can also be used to assess RhoA activation in cells. We tested the ability of GFP fusions of LARG and the three PH hydrophobic patch mutants to induce actin stress fiber formation, as imaged by confocal microscopy of phalloidin-stained NIH3T3 cells (Fig. 3 B). Only WT LARG was able to induce stress fibers, and thus the deficiency in signaling caused by the PH domain mutants occurs upstream of actin polymerization. The hydrophobic patch mutants in the context of the LARG DH/PH fragment fused to GFP were also defective in actin stress fiber formation (Fig. S1 D).
3.2. The PH domain hydrophobic patch mutants do not affect GEF activity in vitro
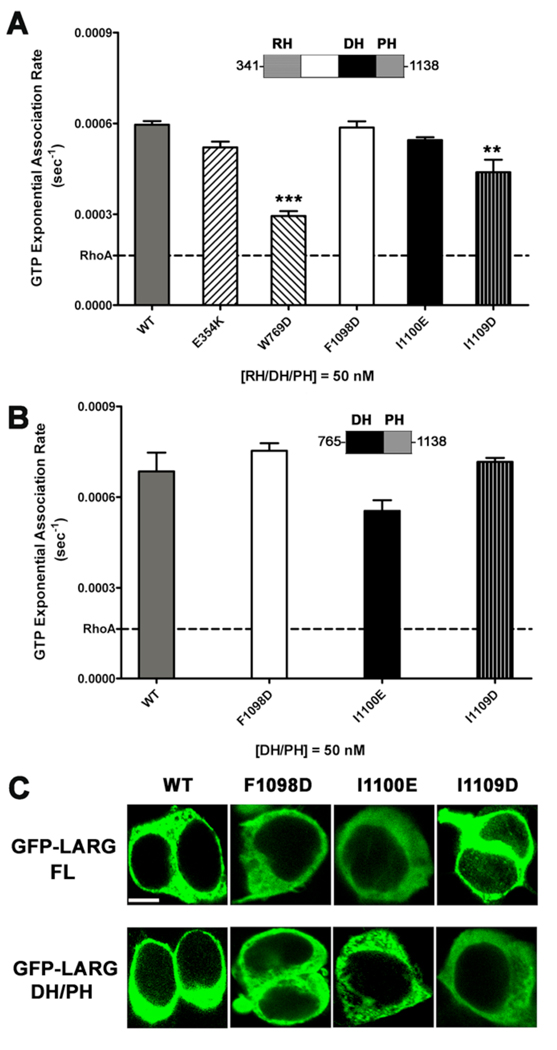
Although the hydrophobic patch is remote from the RhoA binding site (Fig. 1 A), it remained possible that mutations in the patch could influence the intrinsic GEF activity of LARG. We therefore measured the in vitro nucleotide exchange activity of the hydrophobic patch mutants. For this purpose, we generated the E354K, W769D, F1098D, I1100E, and I1109D mutants in the context of an RH/DH/PH fragment expressed in E. coli, and compared their nucleotide exchange activity using a fluorescence polarization (FP) assay that monitors the binding of BODIPY-FL GTPγS to RhoA [6, 34]. The RH/DH/PH-F1098D and -I1100E mutants were as active as LARG RH/DH/PH-WT (Fig. 4 A). The RH/DH/PH-W769D mutant had significantly reduced activity, consistent with prior fluorescence resonance energy transfer (FRET)-based nucleotide exchange assays of the DH/PH-W769D mutant [28]. Of the hydrophobic patch mutants, only I1109D mutant had reduced exchange activity. DH/PH-E354K showed no defect in exchange activity.
Figure 4. Hydrophobic patch mutations of LARG do not affect activity in vitro nor localization in cells.
(A) GEF activities of RH/DH/PH fragments of LARG. The activity of 50 nM LARG and its mutants were measured as the increase in millipolarization (mP) of BODIPY FL GTPγS upon binding to RhoA in an FP assay. The data from three independent experiments, each measured in triplicate, were fitted with exponential association rates (sec−1), with the error bars giving the SD. W769D had significantly lower activity (***, p<0.001) than WT. The lower activity of I1109D (**, p<0.01) is likely because of its instability (see panel B). (B) GEF activities of LARG DH/PH fragments. The activity of 50 nM WT or hydrophobic patch mutants was measured as in panel (A), and data represents the mean ± SD of two (I1100E and I1109D) or three (WT and F1098D) independent experiments, each measured in triplicate. All three hydrophobic mutants, including DH/PH-I1109D, were not significantly different from WT. (C) Confocal microscopy images of GFP fusions of LARG FL and DH/PH in HEK293T cells. Both FL and DH/PH LARG show diffuse cytoplasmic distribution with no obvious membrane localization. No differences in distribution were observed for the hydrophobic patch mutants. Scale bar: 10 µm.
Some of the RH/DH/PH proteins contained degradation products that could not be separated, and these fragments could potentially influence the measured exchange rates [28]. We therefore tested the in vitro activity of the hydrophobic patch mutants in the context of the more stable DH/PH fragment of LARG. In this case, all of the mutants were as active as DH/PH-WT (Fig. 4 B). Thus, LARG proteins with mutations in the hydrophobic patch appear fully competent to catalyze nucleotide exchange on RhoA.
3.3 The hydrophobic patch mutants are structurally intact in cells
Although Western blots indicated that all the PH domain mutants expressed similarly to WT in mammalian cells (Fig. 3 A inset), these proteins could be unstable or have folding defects that abrogate function in cells. We therefore tested the ability of LARG DH/PH-WT and DH/PH-F1098D expressed in HEK293T cells to interact with purified RhoA-G17A, a nucleotide-binding deficient mutant of RhoA [35]. Both WT and F1098D proteins could bind RhoA-G17A equally well (Fig. S2 A), indicating that the F1098D mutant expressed in mammalian cells was still capable of functional interactions with RhoA. To verify the stability of hydrophobic patch mutants in HEK293T cells, we used cycloheximide to block protein synthesis, and compared the loss of DH/PH-WT and DH/PH-F1098D proteins over time. However, there was no apparent difference between the lifetimes of these proteins (Fig. S2 B).
3.4. Cellular distribution of LARG with mutations in the hydrophobic patch
Because the hydrophobic patch mutants were defective in cells despite being expressed equally well, retaining GEF activity in vitro, and being able to bind RhoA, we hypothesized that the mutations led to a defect in cellular localization, such that LARG is unable to access the pools of RhoA responsible for gene transcription. To test this hypothesis, GFP fusions of LARG-FL and -DH/PH were transiently transfected into HEK293T cells, and their distribution analyzed by confocal microscopy. However, there was no apparent difference among the proteins, which exhibited a homogeneous cytoplasmic distribution (Fig. 4 C) similar to that previously reported for WT LARG, p115-RhoGEF, and PDZ-RhoGEF [16, 36, 37]. Also consistent with a prior report [37], truncation of the LARG C-terminal tail (residues 1139–1544) (LARG-ΔC), redistributes both WT and hydrophobic patch mutants to the nucleus (Fig. S3 A). Despite this redistribution, WT LARG-ΔC retained its ability to induce RhoA dependent gene transcription in HEK293T cells, and the F1098D, I1100E, and I1109D mutations in this context still had greatly reduced activity (Fig. S3 B).
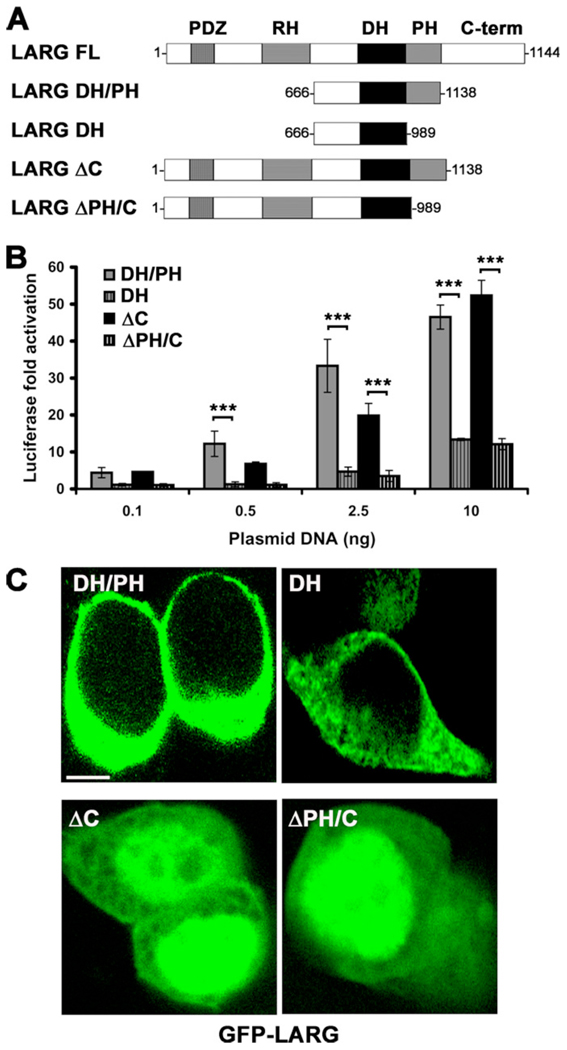
Because the hydrophobic patch mutants generated non-functional LARG proteins in cells, we next decided to re-examine the activity and cellular distribution of LARG constructs that lack the PH domain (Fig. 5 A). Consistent with a positive signaling role for the PH domain and the hydrophobic patch mutants, the DH domain alone and a fragment of LARG C-terminally truncated after the DH domain (ΔPH/C) showed significantly less activity than the corresponding DH/PH and ΔC fragments (Fig. 5 B). There was, however, no obvious difference in localization among LARG-DH and -DH/PH proteins, and no difference between the LARG-ΔC and -ΔPH/C truncations, which both exhibited nuclear localization (Fig. 5 C).
Figure 5. Deletion of the PH domain decreases LARG activity in cells.
(A) Schematic representation of LARG constructs used in cell-based assays. (B) Normalized SRE.L luciferase activities of the indicated constructs. Data represents the mean ± SD of three independent experiments, each measured in triplicate (***, p<0.001). (C) Distribution of LARG constructs in HEK293T cells. GFP-ΔC and GFP-ΔPH/C show pronounced nuclear distribution. Scale bar: 10 µm.
3.5. Membrane targeting conditionally rescues the activity of hydrophobic patch mutants in cells
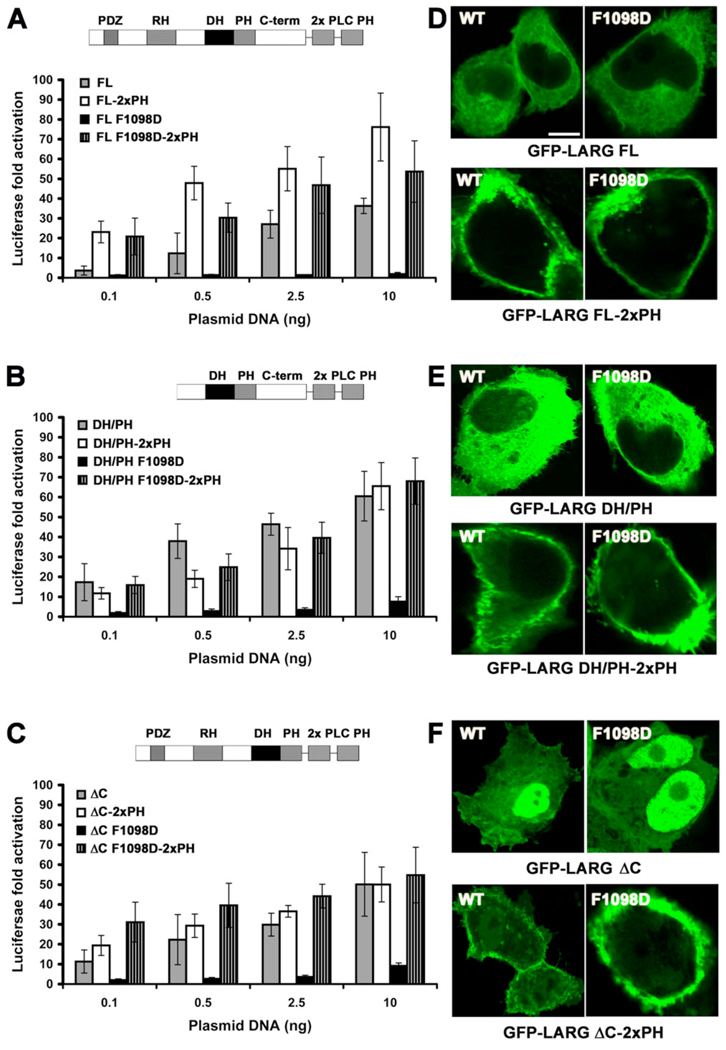
Because we observed no clear membrane association of LARG over-expressed in resting cells (Fig. 4 Cs), we decided to test whether artificially targeting LARG and its PH domain mutants to the plasma membrane would have an effect on SRE.L activity. We therefore created LARG-WT and -F1098D fusions with either two tandem PH domains from PLCδ1, which binds with high affinity to phosphatidyl inositol-4,5-bisphosphate head groups [38], or with the C-terminal polybasic region and CAAX box of K-Ras [39]. In each case, the membrane targeted proteins exhibited WT or higher SRE.L activities (Fig. 6 A–C, Fig. S4 A – C), similar to that reported for membrane-targeted p115RhoGEF [14]. Strikingly, the activity of membrane-targeted F1098D mutant was fully rescued, not only in the context of LARG-FL, but also in the context of LARG-DH/PH and LARG-ΔC (Fig. 6 A–C, Fig. S4 A – C). Confocal microscopy confirmed that all forms of LARG-2xPH or -CAAX tested (both WT and the F1098D mutant) were predominately localized at the cell membrane, including the ΔC truncation that exhibits nuclear localization in the absence of the targeting fusion domain (Fig. 6 D–F, Fig. S4 D – F).
Figure 6. Fusion with two tandem PLCδ1 PH domains rescues the activity of the F1098D mutant.
(A–C) Normalized SRE.L luciferase activities of the LARG (A) FL, (B) DH/PH, and (C) ΔC proteins (WT and F1098D) in the presence or absence of two PH domains from PLCδ1 fused to the C-terminus. Data represents the mean ± SD of 2 independent experiments, each measured in triplicate. (D–F) Confocal microscopy images of COS-7 cells transfected with GFP-LARG (D) FL, (E) DH/PH, and (F) ΔC proteins. Scale bar: 10 µm.
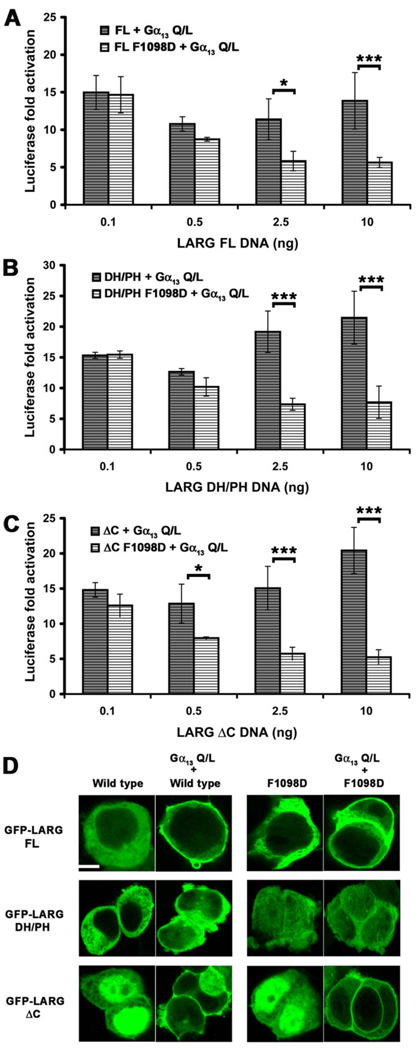
LARG can also be targeted to cell membranes via their RH domains by activated Gα12/13 subunits, and thus we next examined whether this interaction would likewise rescue the hydrophobic patch mutants of LARG. We first attempted to examine how the hydrophobic patch mutants affect synergy between Gα13 and LARG in the SRE.L assay. Previous studies have either reported no augmentation of LARG activity in cells by active forms of Gα12/13 subunits [3], or at most very mild synergy [29, 40]. We were unable to observe augmentation of SRE.L activity when LARG was co-expressed with sub-saturating amounts of constitutively active Gα13-Q226L. Instead, we observed less than additive responses from WT LARG constructs. At higher plasmid concentrations, the contribution of the LARG F1098D mutant inhibited Gα12/13 activity, even in the context of the DH/PH fragment, which lacks an RH domain (Fig. 7 A–C). Similar decreases in SRE.L activity mediated by Gα13-Q226L were observed in the presence of increasing amounts of plasmid expressing GFP alone (Fig. S5 A – C) and in the presence of LARG lacking an RH domain (DH/PH) (Fig. S5 B), suggesting that this inhibition is non-specific and not a “dominant negative” phenotype mediated by the hydrophobic patch mutants. However, the SRE.L response of LARG WT was significantly higher than for the mutant proteins, which followed a similar trend to the GFP alone control, indicating that the activity of the hydrophobic patch mutants cannot be rescued by activated Gα13.
Figure 7. Constitutively active Gα13 targets LARG to the membrane but does not rescue the F1098D mutant.
Normalized SRE.L luciferase activities of LARG (A) FL, (B) DH/PH and (C) ΔC (WT and F1098D) co-expressed with Gα13-Q226L in HEK293T cells. LARG and Gα13 do not exhibit synergy under these conditions, and the F1098D mutant in each context appears to diminish RhoA activation. Shown are the results from a representative experiment (from 2–3 total experiments each) with the error bars representing the SD of each data point measured in triplicate. (D) Confocal images of HEK293T cells co-transfected with expression plasmids for GFP-LARG (WT or F1098D) and Gα13-Q226L. For LARG FL and ΔC, 80% and 60% of the cells containing a GFP signal clearly had membrane-localized LARG, respectively. As expected from the lack of an RH domain, none of the LARG DH/PH transfected HEK293T cells appeared to have membrane-localized LARG. Scale bar: 10 µm. The confocal image of GFP-LARG ΔC is the same as in Fig. 4C and is shown here for comparison.
In contrast, confocal microscopy of transfected HEK293T cells showed that LARG-FL and -ΔC were recruited to the plasma membrane when co-expressed with Gα13-Q226L (Fig. 7 D). LARG-DH/PH was not recruited, consistent with the RH domain mediating the primary interaction with Gα13. The F1098D mutant localized similarly to WT in each construct tested, indicating that the hydrophobic patch is not required for Gα13-mediated membrane recruitment. Thus, although binding to activated Gα13 can localize LARG to the membrane, it is not sufficient to rescue the activity of the F1098D mutant, and that membrane recruitment is not the sole requirement for LARG activation by Gα13.
3.6. Potential interaction partners with the LARG PH domain
Although our results using membrane-targeting fusions of LARG imply that the hydrophobic patch of the PH domain is involved in membrane recruitment, the fact that membrane-recruitment mediated by Gα13 appears insufficient to rescue SRE.L activity of hydrophobic patch mutants suggests that the PH domain interacts with other molecules at the cell membrane to mediate Gα13 signaling.
One potential target, of course, is activated Gα12/13, which have been reported to bind not only to the RH domain of RH-RhoGEFs, but also to the DH/PH region of p115RhoGEF, although not to the PH domain alone [41]. There is also some evidence that some other members of the Lbc subfamily, such as Lbc, can be directly regulated by Gα12/13 subunits [42–45]. Thus, the PH domain could form at least part of a binding site for activated Gα subunits. We used FPCIA to test whether we could observe a loss in equilibrium binding to the RH/DH/PH fragment of LARG if we introduced a mutation into the hydrophobic patch of the PH domain. However, AlF4 − -activated Gα13 binds the biotinylated LARG RH/DH/PH-WT with as high an affinity as RH/DH/PH-F1098D (Ki = 20 nM) (Fig. 2 B). No binding of the LARG DH/PH fragment to Gα13 was detected in this assay (data not shown). Thus, competition binding experiments do not support the hypothesis that the hydrophobic patch contributes significantly to interactions with Gα13.
We also tested for interactions between the LARG PH domain and other reported interaction partners for Lbc subfamily RhoGEFs, including actin [46], tubulin [47], and Gβγ subunits [48]. We however could not detect specific binding between GFP-LARG DH/PH and tubulin or actin using immuno-precipitation of HEK293T cell lysates, nor between LARG RH/DH/PH and tubulin or Gβγ using FCPIA (data not shown). Lastly, we examined whether the hydrophobic patch mutants perturbed the ability of the LARG PH domain to bind phospholipids. Using an overlay lipid-binding assay, we found that there was no detectable affinity binding to any of the lipids tested (data not shown), consistent with the structures of the LARG and PDZ-RhoGEF PH domains, which lack conventional phospholipid binding determinants [49].
If the PH domain binds to a specific protein target in cells, then over-expression of the domain might be expected to have a dominant negative phenotype. Indeed, a dominant negative phenotype mediated by a RhoGEF PH domain has previously been reported for Dbl RhoGEF [50], which could reflect competition for binding with ezrin [51]. Therefore, we transfected increasing amounts of the LARG PH domain in HEK293T cells and tested activity using the luciferase reporter transcription assay. Although the WT PH domain did appear to inhibit activity in this assay, the F1098D mutant did not abrogate this ability (Fig. S5 D – F). This data is consistent with the effects of the F1098D mutant when co-expressed with constitutively active Gα13 (Fig. 7 A–C, Fig. S5 A – C), wherein a non-functional LARG protein can suppress the SRE.L response stimulated by Gα13. Taken together, these experiments show that increasing amounts of co-transfected plasmids can have a non-specific inhibitory effect on transcription activity in this assay format.
3.7. Implications for other Lbc subfamily members
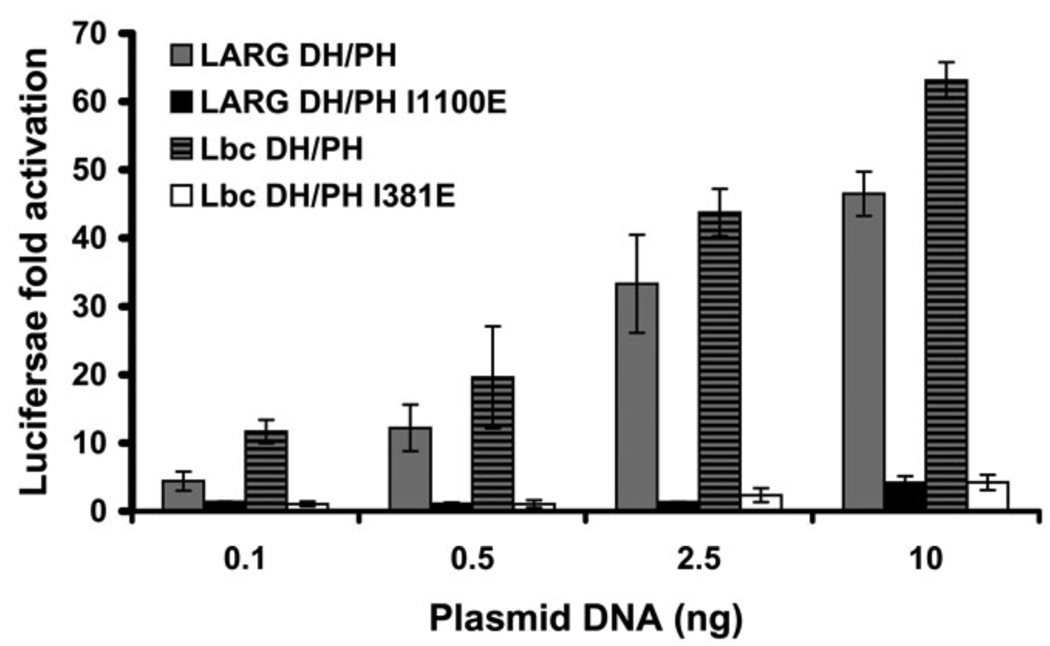
To test whether the hydrophobic patch is important for non-RH-RhoGEF members of the Lbc subfamily (Fig. 1 C), we created the I348E mutant in the DH/PH fragment of Lbc (corresponding to I1100E in LARG) and compared its ability to activate transcription with that of WT Lbc-DH/PH. Similar to LARG DH/PH-I1100E, the Lbc DH/PH-I381E mutant induced RhoA activation at levels far below those of the WT protein (Fig. 8). By extension, the hydrophobic patch is expected to play a similarly important role in all Lbc subfamily members.
Figure 8. Disruption of the Lbc hydrophobic patch also abolishes RhoA activation in cells.
Normalized SRE.L luciferase activities induced by DH/PH fragments of LARG (WT or I1100E) and Lbc-RhoGEF (WT or I381D) reveal that mutation of the hydrophobic patch leads to a similar defect in both LARG and Lbc. Data represents the mean ± SD of three independent experiments, each measured in triplicate.
4. Discussion
Both in vitro and cell-based studies have shown that the PH domains of Lbc subfamily RhoGEFs make a substantial contribution to their activity [3, 17, 27, 28, 52]. Additionally, our studies show that mutations in the hydrophobic patch of the PH domains of LARG and Lbc dramatically reduce activity in cells, and even more than when the PH domain is entirely deleted. The hydrophobic patch may therefore be the dominant feature of the PH domain that positively contributes to GEF activity in living cells. Our in vitro data show that these mutations do not cause a loss of RhoA nucleotide exchange activity, and the fact that these mutant proteins can bind to Rho-G17A indicates that they are capable of binding RhoA when over-expressed in mammalian cells.
Because the activity of LARG with mutations in the hydrophobic patch can be rescued by membrane targeting via the tandem PH domains from PLCδ1 or the polybasic C-terminal region of K-Ras, one might conclude that the hydrophobic patch is a low-affinity membrane-targeting motif, such that one cannot easily observe a defect in membrane targeting of over-expressed proteins by confocal microscopy. However, the fact that membrane targeting by Gα13 could not rescue activity indicates that the defect caused by mutation of the hydrophobic patch is more complex. Our data suggests that upon binding to Gα13, the hydrophobic patch of the PH domain of LARG is somehow required to efficiently mediate signal transduction. This requirement can perhaps be bypassed when LARG is nonspecifically targeted to the membrane, which would give LARG access to pools of RhoA that it would not normally encounter. Although a direct interaction between the hydrophobic patch of the PH domain and Gα12/13 subunits is an attractive model, we have not been able to document any significant differences between the affinity of Gα13 for the WT or F1098D mutants of the LARG RH, RH/DH/PH and RH/DH/PH fragments by equilibrium binding, nor could we detect Gα13 binding by fragments of LARG that lack the RH domain. A recent paper using surface plasmon resonance suggested that Gα13 could weakly interact with the DH/PH and C-terminal regions of LARG [53]. The differences between their report and ours may be because of differences in the assay format, or in the specific protein fragments used.
Based on the crystal structures of LARG and PDZ-RhoGEF and models such as shown in Fig. 1 A, the hydrophobic patch most likely exerts its effects via other proteins found at the cell membrane. To identify a direct cellular target of the LARG PH domain, we tested potential leads from the literature (i.e. Gα13, Gβγ, actin, tubulin and phospholipids), but we have thus far failed to demonstrate any convincing high affinity interactions. The relatively small size of the hydrophobic patch on the PH domain may only allow for only low affinity interactions, and that an interaction can only be easily observed under conditions where there is a high local concentration of the target (e.g. actin filament, or PIP2 in various lipid micro-domains). Alternatively, it could be that the interactions of the LARG PH domain require simultaneous interactions with both a membrane surface and the target protein, akin to the behavior of the G protein-coupled receptor kinase 2 PH domain, which binds to both anionic phospholipids and Gβγ subunits [54]. The fact that the hydrophobic patch forms a quasi-two fold interface in crystal lattices might suggest a role in RhoGEF dimerization. However, this crystalline interface is not identical among the three crystal structures, and there is as of yet no evidence that the PH domain can mediate dimerization in vitro.
If the PH domain mediates interactions with a discrete cellular target, then over-expression of the PH domain should lead to a dominant negative effect on LARG signal transduction. Indeed, the LARG PH domain exhibited such an effect in SRE.L response assays (Fig. 7 A–C, Fig. S5 D–F). However, both WT and the F1098D mutant inhibited transcription to the same level, and our data suggests that this mild inhibition could be an artifact. If so, then our inability to observe a dominant negative phenotype is consistent with the idea that the LARG PH domain mediates only a low affinity interaction, and that signaling mediated by LARG depends on multiple interactions at the cell membrane.
In the context of an SRE.L activity assay, a functional interaction between Gα13 and LARG has been difficult to observe. At least one other lab has reported no synergy between co-transfected Gα13 and LARG [3], and other groups have reported at best a mild synergy (~1.5–3 fold) [29, 40]. Our F1098D constructs appeared to inhibit activity induced by constitutively active Gα13 (Fig. 7 A–C) in a manner consistent with the dominant negative effects we observed for the LARG PH domain in the absence of the Gα subunit (Fig. S5 D – F). Therefore, the reason we did not observe synergy between Gα13 and LARG in our SRE.L assays is not due to the fact cells were not being transfected with both plasmids at the same time. Although we cannot verify that the amount of expressed Gα13 was constant with increasing co-transfected plasmid, a significant difference is obtained when one co-expresses a WT versus a hydrophobic patch mutant of LARG. This result supports the conclusion that Gα13 can recruit LARG to the membrane so long as it has an RH domain, but it still fails to activate LARG when it has a hydrophobic patch mutation.
The conservation of the residues in the hydrophobic patch within the Lbc subfamily of RhoGEFs suggests that their PH domains operate by a common mechanism. Indeed, disruption of the hydrophobic patch in Lbc PH domain by mutation of Ile381, which is analogous to LARG-Ile1100, also greatly diminishes the ability of Lbc to induce RhoA activation in cells (Fig. 8).
5. Conclusion
In the present work, we have identified a novel functional motif of an RH-RhoGEF PH domain that is critical for function in cells. This motif consists of a conserved hydrophobic patch at the surface formed by the β5-β7 strands. Our analysis suggests that the PH hydrophobic patch is involved in interactions with other proteins at the membrane surface to help facilitate nucleotide exchange on RhoA. This report describes also similar role of this hydrophobic motif in related, non RH-containing RhoGEFs such as Lbc.
Supplementary Material
Acknowledgements
We thank Dr. Gary Reuther (University of South Florida) for his assistance in early stages of the project, and Mark Nance, Marc Ridilla, and Cassandra Coco for technical assistance. Support was provided by American Cancer Society Research Scholar grant 04-185-01 and National Institute of Health grants HL071818 and HL086865 (to JJGT), and National Institutes of Health grant 5R01GM039561 (to RRN).
Abbreviations
- DH
Dbl homology
- FCPIA
flow cytometry protein interaction assay
- FL
full-length
- FP
fluorescence polarization
- FRET
fluorescence resonance energy transfer
- GEF
guanine nucleotide exchange factor
- LARG
leukemia associated RhoGEF
- MFI
median fluorescence intensity
- NCS
new born calf serum
- PDZ
N-terminal post synaptic density protein, Drosophila disc large tumor suppressor, and zonula occludens-1 protein
- PH
pleckstrin homology
- RGS
regulator of G protein signaling
- RH
RGS homology
- RhoGEF
Rho guanine nucleotide exchange factor
- SRE
serum response element
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Etienne-Manneville S, Hall A. Nature. 2002;420(6916):629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 2.Rossman KL, Der CJ, Sondek J. Nat Rev Mol Cell Biol. 2005;6(2):167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 3.Reuther GW, Lambert QT, Booden MA, Wennerberg K, Becknell B, Marcucci G, Sondek J, Caligiuri MA, Der CJ. J Biol Chem. 2001;276(29):27145–27151. doi: 10.1074/jbc.M103565200. [DOI] [PubMed] [Google Scholar]
- 4.Liu X, Wang H, Eberstadt M, Schnuchel A, Olejniczak ET, Meadows RP, Schkeryantz JM, Janowick DA, Harlan JE, Harris EA, Staunton DE, Fesik SW. Cell. 1998;95(2):269–277. doi: 10.1016/s0092-8674(00)81757-2. [DOI] [PubMed] [Google Scholar]
- 5.Rossman KL, Worthylake DK, Snyder JT, Siderovski DP, Campbell SL, Sondek J. Embo J. 2002;21(6):1315–1326. doi: 10.1093/emboj/21.6.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lutz S, Shankaranarayanan A, Coco C, Ridilla M, Nance MR, Vettel C, Baltus D, Evelyn CR, Neubig RR, Wieland T, Tesmer JJ. Science. 2007;318(5858):1923–1927. doi: 10.1126/science.1147554. [DOI] [PubMed] [Google Scholar]
- 7.Nimnual AS, Yatsula BA, Bar-Sagi D. Science. 1998;279(5350):560–563. doi: 10.1126/science.279.5350.560. [DOI] [PubMed] [Google Scholar]
- 8.Soisson SM, Nimnual AS, Uy M, Bar-Sagi D, Kuriyan J. Cell. 1998;95(2):259–268. doi: 10.1016/s0092-8674(00)81756-0. [DOI] [PubMed] [Google Scholar]
- 9.Kourlas PJ, Strout MP, Becknell B, Veronese ML, Croce CM, Theil KS, Krahe R, Ruutu T, Knuutila S, Bloomfield CD, Caligiuri MA. Proc Natl Acad Sci U S A. 2000;97(5):2145–2150. doi: 10.1073/pnas.040569197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukuhara S, Chikumi H, Gutkind JS. Oncogene. 2001;20(13):1661–1668. doi: 10.1038/sj.onc.1204182. [DOI] [PubMed] [Google Scholar]
- 11.Mao J, Yuan H, Xie W, Wu D. Proc Natl Acad Sci U S A. 1998;95(22):12973–12976. doi: 10.1073/pnas.95.22.12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozasa T, Jiang X, Hart MJ, Sternweis PM, Singer WD, Gilman AG, Bollag G, Sternweis PC. Science. 1998;280(5372):2109–2111. doi: 10.1126/science.280.5372.2109. [DOI] [PubMed] [Google Scholar]
- 13.Hart MJ, Jiang X, Kozasa T, Roscoe W, Singer WD, Gilman AG, Sternweis PC, Bollag G. Science. 1998;280(5372):2112–2114. doi: 10.1126/science.280.5372.2112. [DOI] [PubMed] [Google Scholar]
- 14.Bhattacharyya R, Banerjee J, Khalili K, Wedegaertner PB. Cell Signal. 2009 doi: 10.1016/j.cellsig.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhattacharyya R, Wedegaertner PB. J Biol Chem. 2000;275(20):14992–14999. doi: 10.1074/jbc.M000415200. [DOI] [PubMed] [Google Scholar]
- 16.Meyer BH, Freuler F, Guerini D, Siehler S. J Cell Biochem. 2008;104(5):1660–1670. doi: 10.1002/jcb.21732. [DOI] [PubMed] [Google Scholar]
- 17.Wells CD, Gutowski S, Bollag G, Sternweis PC. J Biol Chem. 2001;276(31):28897–28905. doi: 10.1074/jbc.M102913200. [DOI] [PubMed] [Google Scholar]
- 18.Wang Q, Liu M, Kozasa T, Rothstein JD, Sternweis PC, Neubig RR. J Biol Chem. 2004 doi: 10.1074/jbc.C400105200. [DOI] [PubMed] [Google Scholar]
- 19.Fukuhara S, Chikumi H, Gutkind JS. FEBS Lett. 2000;485(2–3):183–188. doi: 10.1016/s0014-5793(00)02224-9. [DOI] [PubMed] [Google Scholar]
- 20.Fukuhara S, Murga C, Zohar M, Igishi T, Gutkind JS. J Biol Chem. 1999;274(9):5868–5879. doi: 10.1074/jbc.274.9.5868. [DOI] [PubMed] [Google Scholar]
- 21.Rumenapp U, Asmus M, Schablowski H, Woznicki M, Han L, Jakobs KH, Fahimi-Vahid M, Michalek C, Wieland T, Schmidt M. J Biol Chem. 2001;276(4):2474–2479. doi: 10.1074/jbc.M004957200. [DOI] [PubMed] [Google Scholar]
- 22.Rumenapp U, Blomquist A, Schworer G, Schablowski H, Psoma A, Jakobs KH. FEBS Lett. 1999;459(3):313–318. doi: 10.1016/s0014-5793(99)01270-3. [DOI] [PubMed] [Google Scholar]
- 23.Taya S, Inagaki N, Sengiku H, Makino H, Iwamatsu A, Urakawa I, Nagao K, Kataoka S, Kaibuchi K. J Cell Biol. 2001;155(5):809–820. doi: 10.1083/jcb.200106139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aurandt J, Vikis HG, Gutkind JS, Ahn N, Guan KL. Proc Natl Acad Sci U S A. 2002;99(19):12085–12090. doi: 10.1073/pnas.142433199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swiercz JM, Kuner R, Behrens J, Offermanns S. Neuron. 2002;35(1):51–63. doi: 10.1016/s0896-6273(02)00750-x. [DOI] [PubMed] [Google Scholar]
- 26.Bhattacharyya R, Wedegaertner PB. Biochem J. 2003;371(Pt 3):709–720. doi: 10.1042/BJ20021897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derewenda U, Oleksy A, Stevenson AS, Korczynska J, Dauter Z, Somlyo AP, Otlewski J, Somlyo AV, Derewenda ZS. Structure. 2004;12(11):1955–1965. doi: 10.1016/j.str.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Kristelly R, Gao G, Tesmer JJ. J Biol Chem. 2004;279(45):47352–47362. doi: 10.1074/jbc.M406056200. [DOI] [PubMed] [Google Scholar]
- 29.Evelyn CR, Wade SM, Wang Q, Wu M, Iniguez-Lluhi JA, Merajver SD, Neubig RR. Mol Cancer Ther. 2007;6(8):2249–2260. doi: 10.1158/1535-7163.MCT-06-0782. [DOI] [PubMed] [Google Scholar]
- 30.Kristelly R, Earnest BT, Krishnamoorthy L, Tesmer JJ. Acta Crystallogr D Biol Crystallogr. 2003;59(Pt 10):1859–1862. doi: 10.1107/s0907444903018067. [DOI] [PubMed] [Google Scholar]
- 31.Kreutz B, Yau DM, Nance MR, Tanabe S, Tesmer JJ, Kozasa T. Biochemistry. 2006;45(1):167–174. doi: 10.1021/bi051729t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z, Singer WD, Sternweis PC, Sprang SR. Nat Struct Mol Biol. 2005;12(2):191–197. doi: 10.1038/nsmb888. [DOI] [PubMed] [Google Scholar]
- 33.Ridley AJ, Hall A. EMBO J. 1994;13(11):2600–2610. doi: 10.1002/j.1460-2075.1994.tb06550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evelyn CR, Ferng T, Rojas RJ, Larsen MJ, Sondek J, Neubig RR. J Biomol Screen. 2009;14(2):161–172. doi: 10.1177/1087057108328761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Mata R, Wennerberg K, Arthur WT, Noren NK, Ellerbroek SM, Burridge K. Methods Enzymol. 2006;406:425–437. doi: 10.1016/S0076-6879(06)06031-9. [DOI] [PubMed] [Google Scholar]
- 36.Goulimari P, Knieling H, Engel U, Grosse R. Mol Biol Cell. 2008;19(1):30–40. doi: 10.1091/mbc.E06-11-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grabocka E, Wedegaertner PB. Mol Pharmacol. 2007;72(4):993–1002. doi: 10.1124/mol.107.035162. [DOI] [PubMed] [Google Scholar]
- 38.Baumeister MA, Rossman KL, Sondek J, Lemmon MA. Biochem J. 2006;400(3):563–572. doi: 10.1042/BJ20061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu S, He J, Ho WT, Ramineni S, Thal DM, Natesh R, Tesmer JJ, Hepler JR, Heximer SP. J Biol Chem. 2007;282(45):33064–33075. doi: 10.1074/jbc.M702685200. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki N, Nakamura S, Mano H, Kozasa T. Proc Natl Acad Sci U S A. 2003;100(2):733–738. doi: 10.1073/pnas.0234057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wells CD, Liu MY, Jackson M, Gutowski S, Sternweis PM, Rothstein JD, Kozasa T, Sternweis PC. J Biol Chem. 2002;277(2):1174–1181. doi: 10.1074/jbc.M105274200. [DOI] [PubMed] [Google Scholar]
- 42.Carnegie GK, Soughayer J, Smith FD, Pedroja BS, Zhang F, Diviani D, Bristow MR, Kunkel MT, Newton AC, Langeberg LK, Scott JD. Mol Cell. 2008;32(2):169–179. doi: 10.1016/j.molcel.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diviani D, Soderling J, Scott JD. J Biol Chem. 2001;276(47):44247–44257. doi: 10.1074/jbc.M106629200. [DOI] [PubMed] [Google Scholar]
- 44.Dutt P, Nguyen N, Toksoz D. Cell Signal. 2004;16(2):201–209. doi: 10.1016/s0898-6568(03)00132-3. [DOI] [PubMed] [Google Scholar]
- 45.Majumdar M, Seasholtz TM, Buckmaster C, Toksoz D, Brown JH. J Biol Chem. 1999;274(38):26815–26821. doi: 10.1074/jbc.274.38.26815. [DOI] [PubMed] [Google Scholar]
- 46.Olson MF, Sterpetti P, Nagata K, Toksoz D, Hall A. Oncogene. 1997;15(23):2827–2831. doi: 10.1038/sj.onc.1201594. [DOI] [PubMed] [Google Scholar]
- 47.Ren Y, Li R, Zheng Y, Busch H. J Biol Chem. 1998;273(52):34954–34960. doi: 10.1074/jbc.273.52.34954. [DOI] [PubMed] [Google Scholar]
- 48.Niu J, Profirovic J, Pan H, Vaiskunaite R, Voyno-Yasenetskaya T. Circ Res. 2003;93(9):848–856. doi: 10.1161/01.RES.0000097607.14733.0C. [DOI] [PubMed] [Google Scholar]
- 49.Lemmon MA, Ferguson KM. Biochem Soc Trans. 2001;29(Pt 4):377–384. doi: 10.1042/bst0290377. [DOI] [PubMed] [Google Scholar]
- 50.Zheng Y, Zangrilli D, Cerione RA, Eva A. J Biol Chem. 1996;271(32):19017–19020. doi: 10.1074/jbc.271.32.19017. [DOI] [PubMed] [Google Scholar]
- 51.Vanni C, Parodi A, Mancini P, Visco V, Ottaviano C, Torrisi MR, Eva A. Oncogene. 2004;23(23):4098–4106. doi: 10.1038/sj.onc.1207509. [DOI] [PubMed] [Google Scholar]
- 52.Glaven JA, Whitehead I, Bagrodia S, Kay R, Cerione RA. J Biol Chem. 1999;274(4):2279–2285. doi: 10.1074/jbc.274.4.2279. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki N, Tsumoto K, Hajicek N, Daigo K, Tokita R, Minami S, Kodama T, Hamakubo T, Kozasa T. J Biol Chem. 2009;284(8):5000–5009. doi: 10.1074/jbc.M804073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Touhara K, Koch WJ, Hawes BE, Lefkowitz RJ. J Biol Chem. 1995;270(28):17000–17005. doi: 10.1074/jbc.270.28.17000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.