Abstract
Motivation: Rapidly expanding repositories of highly informative genomic data have generated increasing interest in methods for protein function prediction and inference of biological networks. The successful application of supervised machine learning to these tasks requires a gold standard for protein function: a trusted set of correct examples, which can be used to assess performance through cross-validation or other statistical approaches. Since gene annotation is incomplete for even the best studied model organisms, the biological reliability of such evaluations may be called into question.
Results: We address this concern by constructing and analyzing an experimentally based gold standard through comprehensive validation of protein function predictions for mitochondrion biogenesis in Saccharomyces cerevisiae. Specifically, we determine that (i) current machine learning approaches are able to generalize and predict novel biology from an incomplete gold standard and (ii) incomplete functional annotations adversely affect the evaluation of machine learning performance. While computational approaches performed better than predicted in the face of incomplete data, relative comparison of competing approaches—even those employing the same training data—is problematic with a sparse gold standard. Incomplete knowledge causes individual methods' performances to be differentially underestimated, resulting in misleading performance evaluations. We provide a benchmark gold standard for yeast mitochondria to complement current databases and an analysis of our experimental results in the hopes of mitigating these effects in future comparative evaluations.
Availability: The mitochondrial benchmark gold standard, as well as experimental results and additional data, is available at http://function.princeton.edu/mitochondria
Contact: ogt@cs.princeton.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Methods for protein function prediction and inference of biological networks have recently been of interest due to the growing availability of highly informative genomic data. Many different learning models have been applied to the problem, including kernel methods (Barutcuoglu et al., 2006; Lanckriet et al., 2004), Bayesian networks (Jansen et al., 2003; Sachs et al., 2005; Troyanskaya et al., 2003) and graph-based approaches (Karaoz et al., 2004; Lee et al., 2004; Nabieva et al., 2005). In these methods (and in this article), the task of protein and/or gene function prediction refers to associating proteins with specific biological processes at the cellular level. Many of the methods used for this problem are similar to those used in predicting the biochemical function(s) of a gene (such as kinase activity), but here we focus specifically on the problem of predicting involvement in biological processes (such as DNA damage repair) rather than molecular functions.
All of these approaches fall into the broad category of supervised machine learning classifiers. As such, each method requires trusted sets of examples from the classes it is learning about (e.g. a set of known DNA repair genes for learning about the response to DNA damage). These gold standard sets of genes are typically derived from repositories of gene annotations such as the Gene Ontology (GO; Ashburner et al., 2000), KEGG (Kanehisa et al., 2008) or MIPS (Ruepp et al., 2004) databases. Given such a gold standard and a collection of training data, classifiers can be learned from the data using an algorithm of interest. The same gold standard is typically used for both learning (training) and assessing classifier performance (testing), usually through techniques such as hold-out testing or cross-validation (Russell and Norvig, 2003). New and improved function prediction algorithms are often then justified based on their performance relative to existing methods in such evaluations.
Clearly, these gene annotation databases play a central role in the successful application of machine learning techniques to gene function prediction. In fact, this is one of the major reasons why many published methods have been developed and applied in well-characterized model organisms. Gene annotations are generally considered to be more complete for such organisms, largely because the biological systems themselves are better understood and because of annotation efforts in model organism communities [e.g. SGD for yeast (Hong et al., 2008)]. However, even in Saccharomyces cerevisiae, one of the most extensively curated organisms, ∼20% (∼1100 of ∼5800) of genes have no annotations below the root of the biological process branch of the GO. Furthermore, the majority of annotated genes (∼60%) have only a single GO term annotation, which often indicates incomplete annotation, since many genes are expected to serve in multiple cellular roles. While these examples refer to the GO, other functional catalogs such as KEGG and MIPS are similarly sparse—not because curation efforts are lacking, but due to the large amount of novel biology remaining to be discovered even in simple model organisms. The situation in higher eukaryotes such as mouse and human reflects an even greater degree of incompleteness.
The incomplete state of current gene annotations immediately raises at least two questions: how does this affect our ability to develop effective machine learning approaches, and how can we accurately estimate their performance when much of the ground truth is yet to be established? We address these issues by constructing and analyzing an experimentally based gold standard through a comprehensive validation of gene function predictions related to mitochondrion organization and biogenesis (MOB) in S.cerevisiae; the biological and computational methodologies employed in these experiments are detailed in the study by Hess et al. (2009) and Hibbs et al. (2009), respectively. Briefly, we used three published approaches for predicting gene function from large collections of microarray (Hibbs et al., 2007; Huttenhower et al., 2006) and other genomic data (Myers and Troyanskaya, 2007). The most confident predictions from all the three methods were tested, along with a collection of positive controls (genes known to play a role in mitochondrial function). In all, we tested 241 unique genes for association with mitochondrial function, and this experimentally confirmed set serves as the basis for answering fundamental questions about classifier performance; they have also been contributed to SGD for incorporation into the S.cerevisiae GO annotations. In this article, we focus on the affect of existing gold standards on the field of function prediction, and we provide an analysis of these results as a benchmark for the experimentally validated evaluation of function prediction methods.
In the analysis performed here, we find that machine learning approaches can learn effectively even from limited functional annotations; our classification accuracy as confirmed through laboratory experiments is much higher than estimated for all the three methods (an average of 68% higher precision at 10% recall). However, we also observe substantial discrepancies in the estimated and actual relative performance of different prediction methods, even those based on exactly the same training data. These discrepancies have serious implications in comparative prediction evaluation, which we discuss below.
The organization of this article is as follows: we first summarize the details of our experimental validation, including a brief description of prediction methods and the experimental assays used to test mitochondrial function. We then focus on a comparison of estimated classifier performance (based on cross-validation) with actual classification accuracy (based on experimental results) and discuss striking discrepancies between the two evaluations. Finally, we conclude with a discussion of these results, their implications for the general task of predicting gene function and a benchmark gold standard assembled from this data for use in future evaluations.
2 METHODS
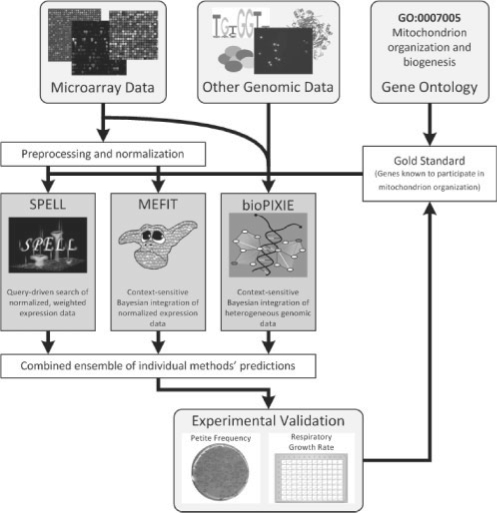
To successfully combine computational gene function prediction with medium-throughput experimental validation, we employed a pipeline detailed in the study by Hess et al. (2009) and Hibbs et al. (2009) and summarized in Figure 1. The system was initialized by generating predictions from three computational methods (detailed below) that use information from the GO (Ashburner et al., 2000) as a portion of their input. These three methods generated ranked lists of genes to be assigned to the MOB term, which were then combined into a master list of testable predictions. The first evaluation was performed on these predictions using only information currently in GO.
Fig. 1.
Overview of the system employed for computational function prediction and medium-throughput experimental validation. We used three computational data integration systems to predict S.cerevisiae genes functioning in the area of mitochondrion biogenesis. An initial gold standard was generated from the GO and used to train two of the machine learning systems: MEFIT, which integrates microarray data, and bioPIXIE, which integrates other diverse genomic data. SPELL was queried using mitochondrial genes from the same gold standard. Genes predicted to function in mitochondrion biogenesis after training or as the result of queries were combined and used to select candidate genes for experimental validation. Genes that significantly perturbed mitochondrion biogenesis when deleted were added to the gold standard, the three prediction methods were retrained, and a second round of experimental validation was performed. In addition to discovering many genes not previously known to participate in mitochondrial biogenesis, this process revealed striking discrepancies between the computational methods' abilities to predict experimental results and their apparent performance based on standard machine learning cross-validation.
Genes predicted to function in mitochondrial biogenesis were then further validated using medium-throughput laboratory experiments: assays covering several hundred genes over the course of several person-months with the accuracy of low-throughput techniques. In the case of MOB, this consisted of a petite frequency assay (detailed below) supplemented with semi-automated liquid growth rate measurements, both yielding statistically rigorous results. Genes verified in this manner to function in the mitochondrion were added to the GO-derived positive standard, augmenting the information available to the computational methods and allowing more accurate predictions to be generated. This allowed a second evaluation of our predictions to be performed incorporating the results of our laboratory experiments.
Taking advantage of these experimental results allowed the generation of new, more complete lists of genes predicted to function in mitochondrial biogenesis. These lists were recombined, and genes newly predicted to have mitochondrial function were again experimentally validated. We found that the accuracy of both the individual prediction methods and of the combined predictions was greatly underestimated by the initial GO-derived standard. This implies that while GO provides enough knowledge to enable predictive machine learning, functional annotations alone are insufficient (at least for the MOB term) to fully describe a biological process or to allow comparative evaluation of different methods. The final list of genes participating in mitochondrial biogenesis—from the GO, underannotations (described below) and our experimental validations—were assembled into the benchmark gold standard provided here.
2.1 Computational predictions
The three systems employed to generate computational function predictions were bioPIXIE (Myers and Troyanskaya, 2007; Myers et al., 2005), MEFIT (Huttenhower et al., 2006) and SPELL (Hibbs et al., 2007). The systems' implementation details are provided in their respective publications; in brief, bioPIXIE predicts pairwise functional relationships using a Bayesian framework consuming diverse genomic experimental data. This framework includes one Bayesian classifier per biological context of interest, where in this case, each context was an individual GO term. A positive standard generated from GO was used to learn conditional probability tables specific to MOB. Predicted annotations to this term were derived from the resulting weighted interaction network by finding the significance of each gene's connectivity to known mitochondrial genes:
| (1) |
| (2) |
where ci is gene i's confidence of mitochondrial function, M is the set of 106 genes annotated to MOB, G is the genome, w(i, j) is the predicted probability of functional relationship between genes i and j, HG(w, x, y, z) denotes the hypergeometric probability distribution and {x} indicates that x is rounded to the nearest integer.
MEFIT also predicts pairwise functional relationships using a collection of GO-trained naïve Bayesian classifiers. It consumes gene expression data drawn from ∼2500 microarray conditions drawn mainly from GEO (Barrett et al., 2007), SMD (Demeter et al., 2007) and ArrayExpress (Parkinson et al., 2007). A ranked list of mitochondrial function predictions was derived from the MOB-specific network by calculating each gene's ratio of connectivity to known mitochondrial genes:
| (3) |
where ci, M, G and w(i, j) are as above.
SPELL is a query-driven system that also consumes these ∼2500 microarray conditions. When provided with a set of query genes, SPELL preprocesses each microarray dataset using singular value decomposition and weights them based on the correlations among the query genes in that data. Using these weights, the remainder of the genome is ranked by weighted average correlation with the query genes. To generate a set of predicted mitochondrial genes, the 106 genes annotated to MOB were used as a query. In all the cases, these systems were initially trained and evaluated on the GO structure and annotations from April 15, 2007.
These three prediction methods were also used to produce an ensemble prediction set using estimated precision. To compute estimated precision, a unified standard was formed by considering the 106 genes annotated to MOB to be positive examples, withholding the 80 genes of the mitochondrial ribosome (due to inordinately strong coexpression; see Myers et al., 2006), and considering the remaining 4824 annotated genes in the genome to be negative examples. For each method's predictions, we computed the precision/recall scores using each gene's rank as a positive/negative threshold; this allowed the assignment of standard precision/recall scores to each gene in each method's predictions (each gene thus providing one point on the lines in Fig. 2). These precisions, which map method-specific prediction confidences to the same underlying gold standard, were thus comparable across methods, and the final combined prediction list was generated by ranking each gene by its average precision across the three methods.
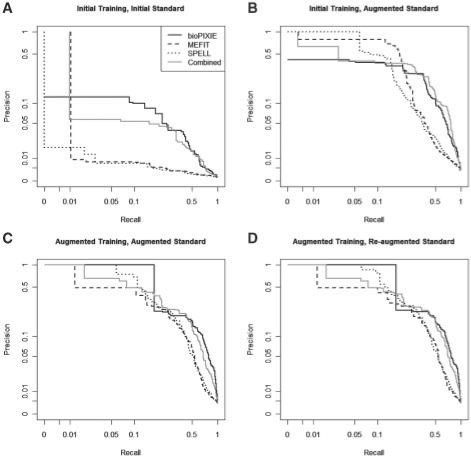
Fig. 2.
Prediction accuracy as estimated by prior knowledge, one round of laboratory validation, and a second iteration of experimental validation. (A) Performance of three function prediction methods and their ensemble as evaluated by a GO-based gold standard. (B) Accuracy of the same predictions as evaluated by a standard augmented with the results of one round of experimental validations (189 tests). (C) New predictions (generated by the same three methods) evaluated using the augmented standard of (B). (D) Accuracy of these predictions evaluated using additional information from a second round of laboratory experiments (52 additional tests). Actual predictive accuracy as evaluated by experimental results is very different than would be expected from a GO-based evaluation.
2.2 Laboratory experiments
The primary assay used to validate our mitochondrial predictions was a measurement of petite colony frequency, supplemented with a measurement of growth rate in liquid medium. Detailed methods for these assays can be found elsewhere (Hess et al., 2009); in summary, we performed all the assays on haploid deletion mutants drawn from the S.cerevisiae heterozygous deletion collection (Tong and Boone, 2006). Knockout strains corresponding to genes with predicted mitochondrial function were drawn from the collection, sporulated, selected for haploids and assayed as follows.
‘Petite’ yeast colonies form from yeast lacking functioning mitochondria (specifically, mitochondrial DNA). Yeast mitochondrial DNA is naturally somewhat unstable, and wild-type S.cerevisiae forms petites in our assay with a base frequency of ∼23%. To compare this base rate with that of each deletion mutant, we sporulated the heterozygous deletion collection, isolated six independent deletion mutants for each gene tested and grew these strains in media requiring aerobic respiration. The resulting plates were stained with tetrazolium, turning respiring colonies red and leaving petite colonies white (Ogur et al., 1957). This allowed colony types to be counted manually; these counts were converted into percentages, which were then compared against wild-type for significance using the Mann–Whitney U test.
Growth curves in liquid media (a measurement of optical colony density over time) were determined using a Tecan GENios plate reader and incubator to record colony densities in 96-well plates every 15 min over 42 h. Each plate contained 12 mutants with six replicates each plus 24 wild-type replicates. Growth rates were derived from these curves by using Matlab (MathWorks Natick, MA, USA) to fit an exponential model:
| (4) |
This model was fit over each whole curve, the first two-thirds, or the first half, whichever yielded the best fit (to avoid plateau effects and to model only exponential growth). Wells with an adjusted R2 <0.9 were marked as non-growing and growth rates for the remaining wells were determined by subtracting the row, column and plate means for each well from the exponential parameter c. This yielded a rate c′ for each well, and each knockout's c′s were tested for significance against the wild-type population using a Mann–Whitney U test.
To detect colonies growing exponentially but with significant differences in fitness, smoothed maximum densities d were calculated for all wells deemed exponential. Wells in which the maximum density was less than twice the minimum were marked as non-growing. From the remainder, plate, row and column averages were subtracted from each well, generating adjusted maxima d′. Each mutant's d′s were again compared with the wild-type values using a Mann–Whitney U test. In both exponential growth and maximum saturation measurements, mutants with more than one outlier were deemed inconclusive and excluded from the results.
All defects specific to respiratory growth (i.e. significant in glycerol but not glucose) were considered. Mutants that failed to grow by both exponential growth and maximum density measurements were assigned a severe phenotype; mutants that failed to grow by one measurement or were significantly defective in both were assigned a moderate phenotype. Mutants with a significant defect by only one measurement were assigned a weak phenotype, and all other mutants received no phenotype.
2.3 Validation methods and criteria
Each stage of our experimental validation relied on a combination of controls and replicates to ensure statistical rigor. Several categories of mutants were tested, beginning with independently isolated wild-type control colonies. We chose positive controls for the various experimental assays from among the 106 genes annotated to MOB. Finally, three types of predictions were tested: underannotated genes with literature support for mitochondrial function (but not annotated to MOB; these were treated as positive controls), known genes with some GO annotation outside of MOB (and no current literature support for mitochondrial function) and unknown genes with no current GO annotation. See Hess et al. (2009) for a complete list of the 48 positive controls (six from MOB, 42 underannotated), 75 knowns and 118 unknowns tested in our assays.
The results of experimental assays were deemed significant enough to validate a gene's involvement in MOB only after passing stringent statistical requirements. In the case of the petite frequency assay, any mutant differing from the wild-type controls with effect size >20% and P <0.05 was deemed to be verified to MOB. These genes were added to the augmented standard used for retraining and in Figure 2. The growth rate assay was used to explore more specific subprocesses of the general MOB term, e.g. respiratory growth as discussed below.
3 RESULTS
We provide a benchmark gold standard of 341 S.cerevisiae genes participating in the process of mitochondrial biogenesis, collected from 106 existing annotations in the GO, 135 underannotated genes and 100 genes confirmed by our experimental results. We show that, in the absence of such an experimentally based gold standard, the sparsity of current functional catalogs can lead to misleading machine learning evaluations. Specifically, both absolute estimates of predictive performance (e.g. using cross-validation) and comparative evaluations (among different function prediction methods) can produce spurious results when based on a gold standard with substantial missing information. However, machine learning techniques can still predict gene function accurately even when trained on a sparse gold standard.
3.1 Experimentally validated accuracy is higher than predicted
It is striking that even in S.cerevisiae, one of the organisms most thoroughly annotated in current functional catalogs, publicly available high-throughput experimental data provide a wealth of gene function information not yet captured by the GO MOB term. Such information can be extracted by classifiers trained on a curated gold standard (such as GO) to identify additional genes with potential roles in this function. Figure 2 contrasts the estimated accuracy of our three function prediction systems (and of the combined consensus predictions) before and after multiple rounds of experimental validation. Our initial predictions were generated using only pre-existing experimental data and GO annotations; scoring these against GO (without held-out test data) yields Figure 2A. Figure 2B evaluates the same predictions using an answer set augmented with the results of our first round of experimental validation and determination of underannotations. Figure 2C and D show the equivalent difference after the prediction methods are retrained on this augmented standard and after the standard is augmented again by a second round of experimental validation.
Of particular note is the difference in performance between Figure 2A's GO-based standard and Figure 2B's experimentally verified standard, also captured in the expected versus actual phenotype counts of Figure 3. The precision/recall curves in Figure 2A and B are generated using the same set of computational predictions made using only existing high-throughput data and the GO—but evaluation using GO alone vastly underestimates their accuracy. While there is some bias in this evaluation due to the focus on testing novel predictions (which, when verified, will boost the precision of the predictor), it accounts for only a small fraction of the difference in evaluation (e.g. see Hibbs et al., 2009) for an analysis of randomly selected genes). One may conclude from this that, at least in certain functional areas, functional catalogs such as GO currently possess sufficient depth to direct accurate machine learning in large datasets, but they may not have sufficient breadth to fully characterize novel predictions generated in this way from experimental data. Supplementary Figure 1 provides a similar comparison using the MIPS database, confirming that this finding is not specific to the GO; KEGG contains insufficient information on mitochondrial biogenesis to perform a comparison. We stress that this is not a fault of any functional catalog or of biological database curators in general; it is simply a product of the large amount of biology left to discover even in time-honored model organisms.
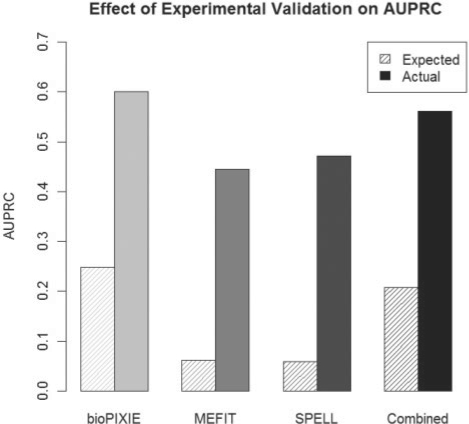
Fig. 3.
Comparison of phenotype frequencies expected from a computational gold standard versus experimentally validated frequency. Expected mitochondrial phenotype area under the precision/recall curves (AUPRCs) were calculated from Figure 2A using only the GO; experimentally validated AUPRCs use the augmented standard of Figure 2D. Phenotype frequencies and accuracy of computational predictions are much higher in all cases than anticipated by pre-existing functional catalogs.
3.2 Comparative evaluation of predictions can be misleading
This variation in coverage within a gold standard, when combined with similar variations in prediction methods, can substantially misrepresent function prediction accuracy (Figs 2 and 3). As indicated by the experimentally validated standard of Figure 2D and the experimental phenotypes in Figure 3, our three prediction systems perform with roughly equivalent overall accuracies, particularly in low-recall/high precision areas of biological interest. However, there is sufficient diversity in the three prediction sets that they overlap quite differentially with the existing 106 GO MOB annotations. Prior to experimental validation, for example, bioPIXIE ranks actin-related proteins such as ARP2 and ARP3 very highly; these are present in the original MOB term and thus raise bioPIXIE's precision in Figure 2A. However, genes such as YMR157C and YMR098C were ranked highly by MEFIT and SPELL, but not initially annotated to MOB. Our experimental assays found that these genes do indeed function in the mitochondria, revealing in Figure 2B that all the three prediction methods were performing quite well despite their initially low-apparent precision.
This differential masking of performance by incomplete standards has clear implications in comparative evaluation of biological function predictions. Due to the highly complex nature of systems biology and the amount of knowledge still missing from even the best curated functional catalogs, it becomes possible—even likely—to learn ‘real biology’ that is not reflected in a gold standard and thus degrades, rather than improves, apparent performance. Conversely, it is equally possible to overfit a standard, improve performance on computational evaluations and produce fewer experimentally verifiable predictions. It is thus essential that, until a greater understanding of the breadth of systems biology allows the construction of more complete functional catalogs, computational predictions are validated using appropriately designed and scaled laboratory experiments or through experimentally based benchmarks.
3.3 Genes with multiple cellular roles complicate the selection of gold standard negatives
Of the 144 mutants validated to MOB by our petite frequency assay, 100 were novel predictions with no prior literature support of mitochondrial function. Thirty-nine of these novel predictions, almost 40%, already possessed annotations to non-mitochondrial functions within the GO. Proteins with known, unrelated biological roles are often used as gold standard negatives for bioinformatic machine learning tasks; when such proteins also have multiple unannotated functions in related processes, this can contaminate the gold standard and artificially decrease apparent performance. Many genes participate in multiple biological processes (Blencowe, 2006), and while GO and other functional catalogs are specifically designed to encode such characteristics, their significance and commonality has perhaps not been fully appreciated. Since undiscovered secondary functions behave as incorrect negatives when present in a gold standard, a partial solution is to focus evaluations on known positive examples and thus downweight the influence of gold standard negatives [e.g. precision at low recall, discussed more extensively in Myers et al. (2006)]. This underrepresentation of functional plurality in current standards can thus, like the lack of coverage discussed above, obscure or bias comparative evaluation of gene function predictions.
3.4 Medium-throughput experiments validate predictions
The fact that these experimental validations were done in medium throughput is a key to achieving both coverage and reliability in our augmented standards. In addition to the global evaluative power of our petite frequency assay demonstrated in Figures 2 and 3, directed medium-throughput experiments such as the growth rate assay can indicate specific sub-functions for particular genes. For example, the yeast myosins MYO3 and MYO5 are often considered to be redundant, and few high-throughput assays can differentiate their biological roles (Moseley and Goode, 2006). More specific, medium-throughput assays can reveal subtle differences in gene sub-functionalization, however. In our results, MYO3 and the functionally uncharacterized AIM8 both show much higher than expected petite frequencies (150%, P < 10−3 and 136%, P < 10−3). However, a myo3Δ mutant shows no significant growth defect in liquid media, while aim8Δ is significantly impaired (achieving neither exponential growth nor a single doubling of culture density). While the petite frequency assay alone could not differentiate these genes' activities within MOB, the more targeted growth rate assay suggests that AIM8 may function specifically within respiratory growth. Additionally, this myo3Δ phenotype is in interesting contrast with MYO5, which leaves petite frequency essentially unchanged when deleted (108%, P > 0.2); to our knowledge, these two myosins have not previously been shown to act differentially in mitochondrial inheritance, and these results may indicate a specific role for MYO3 in mitochondrial motility.
4 DISCUSSION
We have examined the impact of an initially incomplete standard on machine learning behavior and evaluation using a large-scale experimental validation of gene function predictions; we also provide the results of this validation as a standalone benchmark for gene function prediction. While we used mitochondrial organization in yeast as a model system, there are important global lessons we can derive from the results. We have demonstrated that, while a variety of machine learning methods can discover novel biology based on incomplete gold standards, a lack of functional coverage in these standards can seriously bias subsequent evaluations of these learning methods. This difficulty in evaluation emphasizes the importance of validating computational predictions not only through curated gold standards, but also using quantitative experimental results. This underscores the need to develop such experimental benchmarks for a variety of diverse biological processes and model organisms.
The benchmark resulting from this analysis is available at http://function.princeton.edu/mitochondria. This includes four tiers of experimentally validated proteins participating in S.cerevisiae mitochondrial biogenesis: 106 initially available from the GO, 135 generated by guided literature curation, 83 confirmed by our first set of experimental results and 17 by the second set. Approximately 4500 high-confidence negatives are also included, i.e. proteins known to function in other biological processes and with no evidence for mitochondrial involvement; however, this benchmark is by no means perfect or complete, since even tested negatives may include genes with a redundant role in mitochondrial biogenesis. In tandem with existing curated standards, these results provide experimentally driven training and test sets for development of future machine learning, data integration and function prediction methodologies.
A key observation from our study is that gene function prediction methods are much more reliable in this context than anticipated from a purely computational evaluation. For instance, using only the GO, we estimated that 5–25% of the genes we tested would be true positives (the range of average precisions of the three individual methods). However, we confirmed mitochondrial phenotypes for 52%, an increase of 2- to 10-fold over expected. These confirmed phenotypes include both genes of previously unknown function as well as genes with known involvement in other processes (but no known MOB annotation). Moreover, these confirmations are not just peripherally related to mitochondrial function, but some appear to play crucial roles in core mitochondrial activities [e.g. respiration or mitochondrial inheritance (Hess et al., 2009)]. These results indicate that computational methods were able to correctly find novel biological function for 100 proteins and to assign underannotated functions to 135 additional proteins, even when trained using a relatively sparse initial gold standard.
A second striking observation resulting from this validation process is the amount of novel biology remaining to be discovered, even in well-annotated areas and organisms such as yeast mitochondrial biology. Our work began in April 2007, at which point, there were 106 S.cerevisiae genes associated with the GO-term MOB. Manual examination of each method's top predictions revealed another 135 genes that had ample evidence in the literature for mitochondrial function but had no existing annotation to the MOB GO term. Of the 193 additional novel predictions we tested experimentally, we confirmed mitochondrial impairment phenotypes for 100 proteins (52%), bringing the total number of MOB annotations to 341. This has effectively increased the number of annotations to this GO term by 220% with only a few months of computationally directed experiments and literature review.
A less optimistic conclusion of this study is the difficulty of relative performance comparisons between function prediction methods using the currently available incomplete gold standards. Our evaluation considered three different methods, an initial curated gold standard, and our final benchmark gold standard augmented with our experimental validations. We found that the methods' apparent relative performance across these two evaluations was strikingly different, qualitatively as well as quantitatively; for example, using the initial gold standard, bioPIXIE's AUPRC was far higher than MEFIT's, while their performances were more comparable using the final, augmented gold standard [see Hibbs et al. (2009) for additional details]. At the opposite extreme, a hypothetical machine learning method that overfit its sparse training data might have looked excellent in an initial evaluation but far worse after laboratory validation. Thus, at least in the case of mitochondrial biogenesis, a comparative evaluation based solely on existing annotations was misleading due to incomplete knowledge.
While functional annotation repositories represent a valuable source for comparative evaluations of prediction methods outside of resource-intensive experimental validation, it is critical to supplement such annotations with laboratory results or experimentally based benchmarks, whenever possible. Experimental collaboration is, of course, non-trivial; experimental studies based on computational predictions require special attention from computational groups and, obviously, substantial commitment from experimental labs. However, our findings draw into question the field's ability to accurately resolve performance differences between competing approaches using only incomplete gold standards, particularly when such differences are small. This observation suggests that the application of existing gene function prediction methods in a laboratory setting can produce more tangible biological results than can the incremental refinement and development of new computational approaches. Experimental results produced by such validations can further be made publicly available as discrete benchmarks and submitted to functional annotation catalogs such as the GO to improve future efforts across the community.
There are, of course, a variety of limitations to the generalizability of these results. We have evaluated a single GO biological process term in a specific organism, and while there is every reason to believe that qualitatively similar issues will arise for other organisms and processes, their quantitative degree remains unknown. Within yeast itself, we have found comparable preliminary results for other GO terms (Hess et al., 2009). As mentioned above, GO, KEGG, MIPS and other functional catalogs all provide excellent coverage of a variety of model organisms when analyzed appropriately (Myers et al., 2006), and computational techniques represent a complementary method for exploring unannotated biology; mitochondrial processes are highly conserved across eukaryotes, and yeast mitochondria have been heavily used as a model system. Thus, annotated knowledge in this area should be representative of general biological processes. However, extending these results to other systems does require the availability of and resources for appropriate laboratory assays, an obviously non-trivial requirement. Since not every computational results can be tested experimentally, it remains a challenge for the bioinformatician to consider the generalizability and biological validity of comparative machine learning evaluations, in part by techniques such as cross-validation using this and other laboratory-based standards.
Here, we provide a benchmark for computational gene function prediction derived from experimental validation of mitochondrial biogenesis genes in yeast. During the validation of this benchmark, we have demonstrated that the current incompleteness of gene annotation repositories does not necessarily impair computational function prediction, but it does hamper comparative performance evaluation of different techniques. We anticipate that this combination of computational effort with rapid laboratory validation can be applied to a variety of other biological processes (e.g. DNA repair) to generate more complete, area-specific functional catalogs. These would in turn provide more accurate bases for the comparative evaluation of computational techniques, although this is still not a substitute for the depth, precision and scientific potential of collaborative computational and laboratory investigation. While such validations are unlikely to be performed for every method or every biological process, our hope is that the combination of several experimental benchmarks with curated standards will increase the accuracy of comparative evaluations and enable the continued improvement of computational techniques.
ACKNOWLEDGEMENTS
We would like to thank Kara Dolinski for her helpful comments and conversation about the study. We would also like to acknowledge our collaborators in the initial study who helped to generate the data analyzed here: Alicia Hayes, Jadine Paw, John Clore, Rosa Mendoza, Bryan San Luis, Corey Nislow, Guri Giaever and Michael Costanzo.
Funding: National Institutes of Health (grants R01 GM071966, T32 HG003284), NSF CAREER award (DBI-0546275); National Science Foundation (grant IIS-0513552); a Google Research Award (to O.G.T.); NIGMS Center of Excellence (grant P50 GM071508).
Conflict of Interest: none declared.
REFERENCES
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, et al. NCBI GEO: mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barutcuoglu Z, et al. Hierarchical multi-label prediction of gene function. Bioinformatics. 2006;22:830–836. doi: 10.1093/bioinformatics/btk048. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Demeter J, et al. The Stanford Microarray Database: implementation of new analysis tools and open source release of software. Nucleic Acids Res. 2007;35:D766–D770. doi: 10.1093/nar/gkl1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess DC, et al. Computationally driven, quantitative experiments discover genes required for mitochondrial biogenesis. PLoS Genet. 2009;5:e1000407. doi: 10.1371/journal.pgen.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbs MA, et al. Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics. 2007;23:2692–2699. doi: 10.1093/bioinformatics/btm403. [DOI] [PubMed] [Google Scholar]
- Hibbs MA, et al. Directing experimental biology: a case study in mitochondrial biogenesis. PLoS Comput. Biol. 2009;5:e1000322. doi: 10.1371/journal.pcbi.1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong EL, et al. Gene Ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 2008;36:D577–D581. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, et al. A scalable method for integration and functional analysis of multiple microarray datasets. Bioinformatics. 2006;22:2890–2897. doi: 10.1093/bioinformatics/btl492. [DOI] [PubMed] [Google Scholar]
- Jansen R, et al. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaoz U, et al. Whole-genome annotation by using evidence integration in functional-linkage networks. Proc. Natl Acad. Sci. USA. 2004;101:2888–2893. doi: 10.1073/pnas.0307326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanckriet GR, et al. A statistical framework for genomic data fusion. Bioinformatics. 2004;20:2626–2635. doi: 10.1093/bioinformatics/bth294. [DOI] [PubMed] [Google Scholar]
- Lee I, et al. A probabilistic functional network of yeast genes. Science. 2004;306:1555–1558. doi: 10.1126/science.1099511. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Goode BL. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol. Mol. Biol. Rev. 2006;70:605–645. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers CL, Troyanskaya OG. Context-sensitive data integration and prediction of biological networks. Bioinformatics. 2007;23:2322–2330. doi: 10.1093/bioinformatics/btm332. [DOI] [PubMed] [Google Scholar]
- Myers CL, et al. Finding function: evaluation methods for functional genomic data. BMC Genomics. 2006;7:187. doi: 10.1186/1471-2164-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers CL, et al. Discovery of biological networks from diverse functional genomic data. Genome Biol. 2005;6:R114. doi: 10.1186/gb-2005-6-13-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabieva E, et al. Whole-proteome prediction of protein function via graph-theoretic analysis of interaction maps. Bioinformatics. 2005;21(Suppl. 1):i302–i310. doi: 10.1093/bioinformatics/bti1054. [DOI] [PubMed] [Google Scholar]
- Ogur M, et al. Tetrazolium overlay technique for population studies of respiration deficiency in yeast. Science. 1957;125:928–929. doi: 10.1126/science.125.3254.928. [DOI] [PubMed] [Google Scholar]
- Parkinson H, et al. ArrayExpress—a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35:D747–D750. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A, et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S, Norvig P. Artificial Intelligence: A Modern Approach. Upper Saddle River, New Jersey: Prentice Hall; 2003. [Google Scholar]
- Sachs K, et al. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- Tong AH, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- Troyanskaya OG, et al. A Bayesian framework for combining heterogeneous data sources for gene function prediction (in Saccharomyces cerevisiae) Proc. Natl Acad. Sci. USA. 2003;100:8348–8353. doi: 10.1073/pnas.0832373100. [DOI] [PMC free article] [PubMed] [Google Scholar]