Abstract
Summary: Haplotype assembly is becoming a very important tool in genome sequencing of human and other organisms. Although haplotypes were previously inferred from genome assemblies, there has never been a comparative haplotype browser that depicts a global picture of whole-genome alignments among haplotypes of different organisms. We introduce a whole-genome HAPLotype brOWSER (HAPLOWSER), providing evolutionary perspectives from multiple aligned haplotypes and functional annotations. Haplowser enables the comparison of haplotypes from metagenomes, and associates conserved regions or the bases at the conserved regions with functional annotations and custom tracks. The associations are quantified for further analysis and presented as pie charts. Functional annotations and custom tracks that are projected onto haplotypes are saved as multiple files in FASTA format. Haplowser provides a user-friendly interface, and can display alignments of haplotypes with functional annotations at any resolution.
Availability: Haplowser, written in Java, supports multiple platforms including Windows and Linux. Haplowser is publicly available at http://embio.yonsei.ac.kr/haplowser
Contact: sanghyun@cs.yonsei.ac.kr; lilei@usc.edu
Supplementary information: Supplementary data are available at http://embio.yonsei.ac.kr/haplowser
1 INTRODUCTION
Comparative genomics and comparative genome browsers have focused on comparing haploid sequences of multiple organisms and visualizing haploid sequences. Although haplotypes were inferred in the highly polymorphic genomes of Ciona intestinalis and Ciona savignyi by using computational methods (Kim et al., 2007; Vinson et al., 2005), an effective way of comparing haplotypes and its visualization has yet to be explored. While haplotypes inferred from the genome assembly of an individual human showed promising information (Levy et al., 2007; Wang et al., 2008), haplotype comparisons of personal genomes will play significant roles in the forthcoming era of ‘Personal Genomics’ (Church, 2005; Wheeler et al., 2008). In metagenomics, whole-genome shotgun sequencing of environmental samples and application of genomic techniques to those samples are routinely conducted to unravel population structures and evolutionary features. If haplotypes are inferred from metagenome assemblies of multiple environmental samples, the resulting haplotype information can be compared to reveal the distinct characteristics of microbial communities.
Although web servers or stand-alone programs are widely used to visualize comparative data (Engles et al., 2006; Frazer et al., 2004; Karolchik et al., 2008), none of them display haplotypes. Haplower is a comparative browser for comparing and visualizing haplotypes from individual genomes and metagenomes. In addition to aligned haplotypes, Haplowser is capable of displaying functional annotations and user-defined custom tracks that are cross-linked to alignments.
2 IMPLEMENTATION
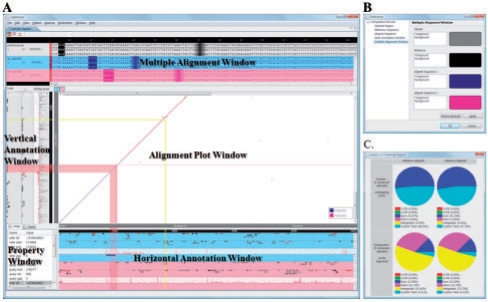
The main view of Haplowser consists of five windows [Alignment Plot (AP) Window, Multiple Alignment (MA) Window, Vertical Annotation (VA) Window, Horizontal Annotation (HA) Window and Property Window]; see Figure 1A. Haplowser imports alignment data in MUMmer (Kurtz et al., 2004) or AVID format (Bray et al., 2003), displaying them in the AP Window (Figure 1A). Alignments run as colored diagonal lines in the AP Window, where the vertical axis indicates the genomic coordinates of the reference sequence and the horizontal axis indicates the genomic coordinates of the aligned sequence. After multiple haploid sequences are aligned to a single haploid reference sequence, haplotypes can be easily aligned to the haploid reference sequence and haploid aligned sequences because haplotypes are of the same length as their haploid sequence (Kim et al., 2007). Whole-genome alignments of a haploid reference sequence and haploid aligned sequences are first generated by using either MUMmer or AVID. As alignments of a haploid reference sequence and haploid aligned sequences are imported, haplotypes are imported and aligned to the haploid reference sequence and haploid aligned sequences, both of which are imported as well. The AP Window can be zoomed in or out by using the mouse operation (scroll wheel) or the scale controller. The resolution of the AP Window varies from the entire haplotype level up to the base level.
Fig. 1.
Main view of Haplowser and analysis window. (A) In the AP Window, the vertical axis corresponds to the reference haplotypes, while the horizontal axis corresponds to the aligned haplotypes. The haplotypes from multiple genomes or metagenomes can be aligned to the single reference haplotypes. (B) The colors of the objects (eg. genes, alignments, custom tracks, etc.) shown in (A) can be configured by a user. (C) The conserved regions or alignments overlapping each category of annotation (e.g. exons, introns, 5′ UTR, 3′ UTR and intergenic regions) are analyzed and displayed in a pop-up window. For details, see text.
The region selected by a mouse click in the AP Window is synchronously displayed in the MA Window, where the multiple alignments of haplotypes are shown. The VA and HA Windows are also synchronized with the AP Window, consisting of 4 panes (Forward-directed Gene Pane, Reverse-directed Gene Pane, SNP Pane and Custom Track Pane); the VA Window represents the annotations of the reference haplotypes and the HA Window represents the annotations of the aligned haplotypes. When a user selects a specific region in the AP Window, the range that corresponds to the selected region is denoted by lines in the VA and HA Windows. Similarly, when a user selects an alignment, the ranges spanned by the alignment are highlighted in the VA and HA Windows.
The VA and HA Windows display genes, SNPs, and custom tracks. Gene annotation can be imported in GFF, UCSC or SGF (Simple Gene Format) format. Genes with different annotation types (e.g. tRNA, rRNA and protein coding genes) are represented in different colors (Figure 1B). When a gene is selected, the gene and the orthologous genes are simultaneously highlighted. The Property Window displays the information related to the selected gene (type, direction, starting point, etc.). Because imported gene sequences are internally mapped to haplotypes, a user can save haplotype-mapped gene sequences in FASTA format. In this case, the number of saved files is equivalent to the number of imported haplotypes.
A user can add custom tracks in a dialog box, or load text files in other gene annotation formats (UCSC or SGF format). Alignment data in BLAST (Altschul et al., 1997) or BLAT (Kent 2002) format can also be displayed in the Custom Track Pane. Similar to gene sequences, a user can save haplotype-mapped custom tracks in FASTA format files because an added custom track is internally mapped to haplotypes as well. For example, a user can save haplotype-mapped enhancer sequences, importing the enhancer sequences as a custom track.
Haplowser enables users to annotate alignments or conserved regions by calculating the fractions of alignments or conserved regions associated with exons, introns, 5′ UTR, 3′ UTR and intergenic regions (Kim et al., 2007). First, the fractions of conserved regions (or alignments) overlapping exons, falling within introns, and falling within intergenic regions are calculated and represented as a pie chart (Figure 1C). Second, the base composition of conserved regions (or alignments) associated with exons, introns, 5′ UTR, 3′ UTR and intergenic regions is calculated and represented as a pie chart (Figure 1C). If a user adds custom tracks (e.g. enhancer sequences), the fractions of conserved regions associated with the custom tracks are also included in the pie charts.
3 CONCLUSION
Haplowser is a comparative browser to compare haplotypes inferred from genome assemblies or metagenome assemblies. It offers a convenient way to navigate haplotype sequences and functional annotations, both of which operate synchronously. Along with zooming, a user can navigate any region of haplotypes and functional annotations at any resolution.
ACKNOWLEDGEMENTS
We thank Daehyun Kim for helpful comments.
Funding: The Korea Science and Engineering Foundation, Ministry of Sciences & Technology (2007-03965 to J.H.K., W.K. and S.P.); The National Institutes of Health, USA (HG002790 to M. S. W. and L. M. L.).
Conflict of Interest: none declared.
REFERENCES
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search program. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N, et al. AVID: a global alignment program. Genome Res. 2003;13:97–102. doi: 10.1101/gr.789803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM. The personal genome project. Mol. Syst. Biol. 2005;1 doi: 10.1038/msb4100040. 2005.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels R, et al. Combo: a whole genome comparative browser. Bioinformatics. 2006;22:1782–1783. doi: 10.1093/bioinformatics/btl193. [DOI] [PubMed] [Google Scholar]
- Frazer KA, et al. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent JW. BLAT-The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, et al. Diploid genome reconstruction of Ciona intestinalis and comparative analysis with Ciona savignyi. Genome Res. 2007;17:1101–1110. doi: 10.1101/gr.5894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson J, et al. Assembly of polymorphic genomes: algorithms and application to Ciona savignyi. Genome Res. 2005;15:1127–1135. doi: 10.1101/gr.3722605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]