Abstract
Summary: PIQA is a quality analysis pipeline designed to examine genomic reads produced by Next Generation Sequencing technology (Illumina G1 Genome Analyzer). A short statistical summary, as well as tile-by-tile and cycle-by-cycle graphical representation of clusters density, quality scores and nucleotide frequencies allow easy identification of various technical problems including defective tiles, mistakes in sample/library preparations and abnormalities in the frequencies of appearance of sequenced genomic reads. PIQA is written in the R statistical programming language and is compatible with bustard, fastq and scarf Illumina G1 Genome Analyzer data formats.
Availability: The PIQA pipeline, installation instructions and examples are available at the supplementary web site (http://bioinfo.uh.edu/PIQA).
Contact: yfofanov@bioinfo.uh.edu
1 INTRODUCTION
Next Generation Sequencing machines, e.g. Illumina Genome Analyzer (Illumina Inc., San Diego, CA, USA) and SOLiD (Applied Biosystems, Foster City, CA, USA), are capable of producing millions of relatively short (20–55 bases) genomic subsequences (reads) in one (2–3 days) run (Illumina, 2008a, b, 2009a). Efficient and less expensive than traditional Sanger sequencing, the Illumina Genome Analyzer (G1) has drawn many authors in sequencing (and analyzing) hundreds of genomic samples (Illumina, 2009a; Kathryn et al., 2008; Srivatsan et al., 2008). Due to the large amount of data produced, the final user of the data may find difficult accomplishing important tasks such as estimating how much useful data were produced, detecting defective tiles/lanes present on the flow cell, or identifying the maximum length of reads which can be used without compromising base calls quality. The study of quality issues of the data produced is an active area of research for next generation sequencing with efforts elsewhere (Dolan and Denver, 2008) that could be complementary to the work presented here.
Early detection of various problems that can appear during the sample preparation and sequencing process significantly reduce time and effort required for more sophisticated steps of analysis, such as de novo sequence assembly or mapping reads to reference sequences. Herein, we present a simple quality analysis pipeline (PIQA), developed to be used on regular desktop PCs or as an ‘extension’ of the standard Illumina Genome Analyzer software (Illumina pipeline). PIQA reads data in the bustard, fastq and scarf formats and eases the visual identification of technical problems, including mistakes in sample/library preparations, defective tiles/lanes and abnormalities in the frequencies of appearance of genomic reads.
2 FEATURES
2.1 Structure of the data
Each sequencing run of an Illumina Genome Analyzer G1 uses a single glass flow cell consisting of eight independent lanes (each lane may contain a different DNA sample). Each lane is populated with randomly fragmented genomic DNA previously capped at both ends with two types of DNA subsequences (adapters). One type of adapters allows the sequence to be attached to the surface of the flow cell and it is also used during the amplification phase to form clusters of the same type of sequence on the surface. A second type of adapters attach to the opposite end of the sequences acting as primers from which the sequencing-by-synthesis starts (Bentley, 2006; Church, 2006; Illumina, 2008a, 2009b). The sequencing phase may consist of 20–50 cycles (Illumina, 2008b), limited by the probability (or the corresponding quality score) with which each nucleotide can be identified. The quality score decreases as the number of cycles increases. The standard Illumina quality score is represented by an integer value that ranges from −40 to +40; an average score above +10 is considered acceptable. The total number of sequencing cycles corresponds to the length of sequences produced (reads). During each cycle, every lane is imaged four times using a different wavelength for each nucleotide. These images are collected as the camera sweeps up and down each lane three times, covering 100–110 (depending on the Illumina software release used) non-overlapping tiles on each sweep. Once the sequencing process is finished, the image files go into the image analysis stage of the Illumina pipeline (called FIRECREST). The pipeline continues to a base-calling stage (called BUSTARD) that assigns quality scores and determines the cluster's sequence. The final stage of the pipeline (GERALD) filters out low-quality reads and trims the sequences by excluding low-quality prefixes and/or suffixes of all the reads, while still keeping the length of all the reads equal.
In a successful run, the average number of reads produced for a single lane varies from 2 to 10 million for unfiltered data (1–6 million for filtered data). The size of the text files containing 5 M reads of 36 nt. varies from ≈245 Mb (bustard format) if only sequencing reads are included, to ≈725 Mb in fastq format, if the quality of each nucleotide is included.
Various problems which may occur in each step of the sample preparation and sequencing can be detected by an analysis of the variation of clusters density; base-call proportions; and base quality across cycles of the run, and across tiles and lanes of the flow cell.
PIQA processes data of one flow cell lane at a time and outputs three HTML documents. The main page of the report (PIQA_report.html) consists of a sequencing summary showing general information about the run, a set of graphs and links to two complementary HTML pages. The graphs displayed in PIQA_report.html assess the clusters density per tile, the base-calls proportions per tile and per cycle, and finally the base-calls quality per tile and per cycle. The complementary HTML documents show the proportion of base-calls per tile for each cycle and the average quality of base-calls per tile for each cycle.
2.2 Density of clusters
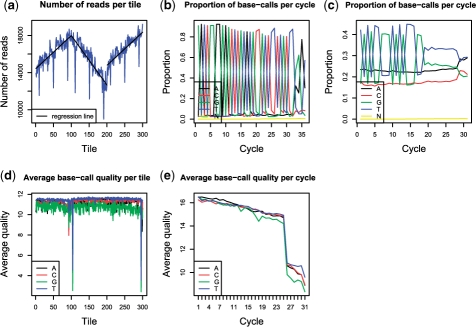
The total number of clusters (reads) across the lane can serve as an indicator of the overall success of a sequencing run (Fig. 1a). Various mechanical, optical and sample preparation issues can, however, significantly disturb the expected pattern. Frequently observed abnormalities include: poor sample quality and/or poor accuracy in DNA quantitation, leading to cluster densities either too high or too low across the entire lane; problems with the optical system, causing decreased cluster density (usually for all the lanes on the flow cell); and finally, mechanical defects such as oil drops and cracks causing decrease in the density of clusters across neighboring tiles.
Fig. 1.
Example output of the PIQA program.
2.3 Base calls
Ideally, since each lane is populated by randomly fragmented genomic DNA, the proportion of nucleotides (denoted by A, T, C, G and N—for unknown base calls) observed in each tile, lane and cycle is expected to be identical. Deviation from this pattern could be a signature of major technical and/or sample preparation problems, for instance, optical failures or sequence bias introduced during the sample preparation. In Figure 1b, too many adapter sequences were introduced during sample preparation, causing the sequencing of adapters instead of the sample. In Figure 1c, the first 20 nt were affected by the primer used during whole genome amplification.
PIQA produces a simple but useful quality analysis of the data. For example, considering quality scores below + 10 as unacceptable, tile-by-tile (Fig. 1d) and cycle-by-cycle (Fig. 1e) plots of the average quality score for each base allows one to make an educated decision about which tiles need to be excluded from consideration and if the trimming of reads (exclusion of prefix and/or suffix parts of reads) is required.
3 IMPLEMENTATION
The PIQA package is implemented in R (R Development Core Team, 2007) and C++. Versions for Windows-32; LINUX-32 and 64; and Mac OS X-32 and 64, are available for download on the Supplementary web site. PIQA makes use of the R2HTML (Lecoutre, 2008) library, and it is implemented as a pipeline in two stages: the first part generates a text summary file (also available to the user for further analyses), the second part uses this summary to produce an HTML report.
ACKNOWLEDGEMENTS
PIQA was developed in collaboration between the Bioinformatics Laboratory of the University of Houston and Eureka Genomics Corp.
Funding: Department of Homeland Security Science and Technology Directorate (award NBCHC070063 to Y.F.); Texas Learning and Computation Center (to Y.F.); training fellowship from the Keck Center Biomedical Discovery Training Program of the Gulf Coast Consortia (NIH Grant No. 1 T90 DA022885-02 to C.F.); CONACYT, Mexico for scholarship support (to E.B.).
Conflict of Interest: none declared.
REFERENCES
- Bentley DR. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Church GM. Genomes for ALL. Sci. Am. 2006;294:46–54. doi: 10.1038/scientificamerican0106-46. [DOI] [PubMed] [Google Scholar]
- Dolan P, Denver D. TileQC: a system for tile-based quality control of Solexa data. BMC Bioinformatics. 2008;9:250. doi: 10.1186/1471-2105-9-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illumina Inc. Illumina Genome Analyzer Brochure. 2008a Available at http://www.illumina.com/downloads/GenomeAnalyzer_Brochure.pdf (last accessed date July 28, 2009) [Google Scholar]
- Illumina Inc. Specification sheet: Illumina sequencing. 2008b Available at http://www.illumina.com/downloads/GenomeAnalyzer_SpecSheet.pdf (last accessed date July 28, 2009) [Google Scholar]
- Illumina Inc. Illumina News Releases. 2009a Available at http://investor.illumina.com/phoenix.zhtml?c=121127&p=irol-news&nyo=0 (last accessed date July 28, 2009) [Google Scholar]
- Illumina Inc. Illumina Sequencing Technology. 2009b Available at http://www.illumina.com/downloads/SS_DNAsequencing.pdf (last accessed date July 28, 2009) [Google Scholar]
- Kathryn EH, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat. Genet. 2008;40:987–993. doi: 10.1038/ng.195. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecoutre E. R2HTML: HTML exportation for R objects. R package version 1.58. 2008 Available at http://www.feferraz.net/en/R2HTML.html http://www.r-project.org http://www.stat.ucl.ac.be/R2HTML/ (last accessed date July 28, 2009) [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria: 2007. ISBN 3-900051-07-0, Available at http://www.R-project.org. (last accessed date July 28, 2009) [Google Scholar]
- Srivatsan A, et al. High-Precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 2008;4:e1000139. doi: 10.1371/journal.pgen.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]