Abstract
Genetic polymorphisms of encoding antigen B2 gene (AgB2) in Echinococcus granulosus were studied using PCR-RFLP and DNA sequencing among 20 Egyptian isolates. Five isolates from different host origins (humans, camels, pigs, and sheep) were collected and used. All examined isolates of each host group gave very similar patterns of PCR-RFLP after restriction enzyme digestion with AluI, with the gene size of approximately 140 bp and 240 bp for sheep and human isolates, and approximately 150 bp and 250 bp for pig and camel isolates. No digestion pattern was obtained after incubation of all studied isolates with EcoRI. These results reveal high intra-group homogeneity. DNA sequence analysis highlighted that human infecting strain showed 100% identity with respect to sheep infecting isolate, 96% and 99% with pig and camel infecting isolates, respectively.
Keywords: Echinococcus granulosus, AgB polymorphism, genetic diversity, PCR-RFLP, riboprinting pattern, gene sequence
INTRODUCTION
The larval stage of the tapeworm Echinococcus granulosus is the causative agent of cystic echinococcosis (CE), a major health and economic problem in many countries around the world [1,2]. It affects humans and a wide range of livestock species and is characterized by the presence of steadily growing unilocular cysts filled with a hydatid cyst fluid (HCF) in host internal organs, mostly the liver and lungs. E. granulosus larvae synthesize a lipoprotein known as antigen B (AgB) in the tegumental cells of the protoscolices and, to a lesser extent, in the laminated and germinal layer of the brood capsules before being secreted in the HCF [3]. AgB is a thermostable lipoprotein that dissociates in SDS-PAGE into 3 bands of 8-12, 16, and 24 kDa [4]. It is one of the most abundant parasite antigens in HCF and commonly used in immunodiagnosis of CE [5]. It is involved in the evasion of the immune response of the host due to its ability to inhibit elastase activity and neutrophils recruitment and to elicit an immunopathology-associated T-helper 2 (Th2) cell response, predominantly in patients with progressive CE [6].
The nature and quality of AgB are variable among different host species and endemic areas. Rosenzvit et al. [7] suggested that precisely the contact with the host molecules and cells makes AgB proteins prone to antigenic variation which can affect their practical application. Therefore, serologic evaluation of AgB prepared from different E. granulosus hosts in endemic regions and countries according to local facilities must always be considered [6]. AgB is encoded by a gene family which is constituted by 5 gene loci, AgB1 [8], AgB2 [9], AgB3 [10], AgB4, and AgB5 [11], each encoding a related, but distinct AgB subunit. The comparative analysis of the diagnostic potential of antigens encoded by some of these genes showed that the recombinant antigen AgB2 had the best diagnostic performance [5]. Numerous studies have provided evidence that E. granulosus is characterized by extensive genetic variation comprising a number of strains that differ in biological features, such as intermediate host specificity, developmental rate, and infectivity to humans [12].
E. granulosus strains (G1-G10) were named according to their most commonly identified intermediate host. Only 5 strains were known to infect humans; sheep (G1), Tasmanian sheep (G2), cattle (G5), camel (G6), and pig (G7) strains [13]. PCR-based restriction fragment length polymorphism (PCR-RFLP) technique and DNA sequence analysis have now been used extensively to characterize strain groupings within E. granulosus and to detect DNA polymorphism. Unlike systems that depend upon chance recognition of target DNA by random primers, the region of the genome that is analyzed has been purposefully chosen in PCR-RFLP to detect nucleotide variation at enzyme specific sites in the amplified fragments [14]. An alternative and more detailed method involves sequencing. To date, no study has been performed for the analysis of the genetic variation of AgB coding genes in Egyptian E. granulosus isolates.
In the present study, the genetic variability among different Egyptian E. granulosus isolates collected from humans, camels, pigs, and sheep was detected at the AgB2 level using PCR-RFLP and DNA sequencing. The knowledge of this variation is necessary for rational design, application, and standardization of diagnostic tests utilizing AgB in Egypt.
MATERIALS AND METHODS
Animal cysts of E. granulosus were collected from Cairo abattoir, while human HCF was collected by percutanuous aspiration injection reaspiration (PAIR) technique under sterile conditions from patients in the Abdominal Ultrasonographic Unit of Tropical Medicine Department, Kasr El-Aini Hospital. One fertile cyst was considered as an isolate. Fertile cyst of E. granulosus was identified on the basis of the presence of protoscolices. A total of 20 isolates were collected; 5 isolates from camels (2 pulmonary and 3 hepatic cysts), 5 isolates from pigs (3 pulmonary and 2 hepatic cysts), and 5 isolates from sheep out of 30 sheep examined (2 pulmonary and 3 hepatic cysts). In addition, 5 isolates from 5 patients were used as representatives of human group. Hydatid fluid from each cyst separately was centrifuged at 500 rpm for 3-5 min. The deposited protoscolices were rinsed in PBS, fixed in 95% (v/v) ethanol and preserved at -70℃ until required [15]. The protoscolices were rinsed several times with distilled water to remove the ethanol, and then total DNA was extracted using Easy Quick Blood DNA extraction kit (Genomix, Cairo, Egypt). Spectrophotometric determination of DNA concentration at A260 was done according to Karcher [16].
PCR was performed in a reaction volume of 25 µl containing 50 ng DNA sample, 10 pmol of each of 2 primers described by Fernandez et al. [9]); F5'GGATCCTTCGTGGCCGTCGTTCAAGC3' and R5'TCGACAAATCATGTGTCCCGACGCA3'(Jena Bioscience, Jena, Germany) and 12.5 µl of 2x SuperHot PCR Master Mix (Bioron, Ludwigshafen, Germany) . PCR was carried out by initial denaturation at 95℃ for 3 min, followed by 35 cycles each at 95℃ for 1 min, annealing temperature at 55 for 1 min, extension temperature at 72℃ for 1.5 min, and final extension at 72℃ for 10 min. The amplified DNA fragments were separated on 1.5% agarose gel (Bioshop Canada, Burlington, Ontario, Canada), stained with ethidium bromide (Bioshop Canada), visualized on a UV Transilluminator, photographed by Gel Doc. BIORAD 2000 and analyzed with software data analysis for BioRad Model 620 USA. The specific band approximately 400 bp was cut from the gel and purified by gel purification kit (Jena Bioscience) according to the manufacturer's instructions.
The PCR products were digested with 2 separate restriction endonucleases; used individually (ALUI and EcoRI) as described previously [17] using the buffers recommended by the manufacturer (Bioron). Generally, the incubation period at the enzyme's optimal temperature (37℃) varied from 4 to 12 hr to ensure the complete digestion. The digestion reaction was inactivated by heating to 65℃ for 20 min. Fragments were separated on 2% agarose gel (Bioshop Canada), stained with ethidium bromide (0.5 µg/ml) (Bioshop Canada), and photographed under UV light with a gel documentation system.
The PCR-products of one isolate from each group were purified from excess primers and nucleotides by the use of AxyPrep PCR Clean-up kit (AXYGEN Biosciences, Union City, California, USA) and directly sequenced as described previously [18] using the same primers as described for the amplification process. The products were sequenced using the Big Dye Terminator Cycle Sequencing Ready Reaction Kit (ABI Applied Biosystems, Foster City, California, USA) on a 3130XL Genetic Analyzer (Applied Biosystems).
PCR-RFLP bands were defined by their molecular weights estimated from the size standards and using the Gel Analyzer 3 Egy-Gene Program (Egypt). Polymorphisms were scored for presence (1) or absence (0) of the bands. The data were transferred to a statistical software program, Statistical Package for Social Science, version 10.00 (SPSS Inc, Chicago, Illinois, USA) to obtain analytical statistics in the form of Jaccard's similarity coefficient (S) showing the genetic similarity among different isolate groups of E. granulosus based on pair-wise comparison. The dendrogram was constructed using the Average Linkage between groups statistical system. The laboratory data were recorded on an investigative report form and then transferred to Finch program which can translate the results of sequencing to a chromatogram. The sequences of the isolates were then transferred to a blasting program which aids alignment of the nucleotides to translate each sequence to the specific genetic origin. Then comparison between the different isolates was done. Data can be reviewed with DNA sequencing analysis software, SeqScape®software, or GeneMapper® software.
RESULTS
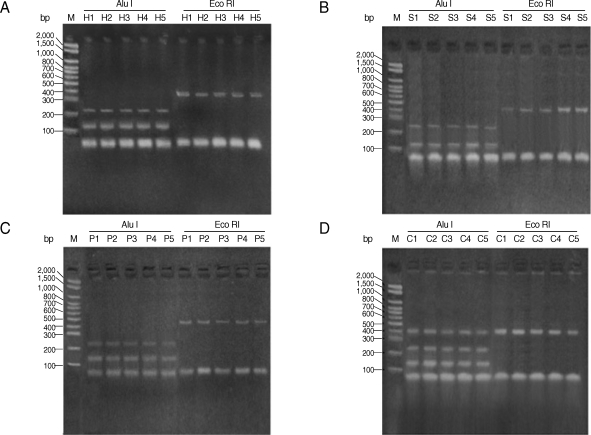
All studied isolates gave a PCR product representing a fragment of the AgB2 gene (approximately 400 bp) (Fig. 1). The 400 bp of PCR amplified fragment of the AgB2 gene was digested separately with (AluI & Eco RI) restriction enzymes, respectively (Fig. 2) for all isolates. Nearly all examined isolates within each group (human, pig, camel, and sheep) (lanes H, P, C, and S, respectively) gave very similar patterns of PCR-RFLP after enzyme digestion with AluI; approximately 140 bp and 240 bp for sheep and human isolates (Fig. 2A, B) and approximately 150 bp and 250 bp for pig and camel isolates (Fig. 2C, D). No digestion pattern was obtained after incubation with EcoRI with all studied isolates.
Fig. 1.
PCR-amplified Ag B2 gene fragments from representative isolates of E. granulosus: (A) human (H) and sheep (S) isolates, (B) pig (P) and camel (C) isolates. M = marker, bp = base pair. Arrow points to specific bands.
Fig. 2.
PCR-RFLP patterns of AgB2 genes of E. granulosus Egyptian isolates using restriction endonuclease enzymes AluI and EcoRI. (A) human (H); (B) sheep (S); (C) pig (P); (D) camel (C). M = marker, bp = base pair.
The results of comparative analysis of band patterns obtained by PCR-RFLP with enzyme AluI using Jaccard's similarity coefficient showed that the highest genetic similarity was observed between human and sheep isolates (100%) followed by human and camel isolates, and camel and pig isolates (66.7%), while the lowest was observed between human and pig isolates (42.9%). The dendrogram showed that the sheep strain is the most relevant strain related to humans; indeed they are genetically typical (Fig. 3). The sequence analysis highlighted that human strain showed identity of 100% to sheep strain and 96% and 99% with pig and camel isolates, respectively (Fig. 4).
Fig. 3.
Dendrogram using average linkage (between groups) generated by Jaccard's similarity coefficient based on RFLP with enzymes AluI and EcoRI showing the relationship among different E. granulosus isolate groups on the basis of AgB2. var = variable, var 1 = human, var 2 = pig, var 3 = camel, var 4 = sheep.
Fig. 4.
Sequences producing significant alignments comparative between human and sheep (A), human and pig (B), and human and camel (C) isolates.
DISCUSSION
Kamenetzky et al. [19] mentioned that no polymorphism in AgB2 genes was found in each strain but there was a substantial level of inter-strain variation in AgB2 related genes and that cysts from the same strain shared more genomic DNA than cysts from different strains. The high genetic similarity between the human and sheep isolates were confirmed by Jaccard's similarity coefficient and the dendrogram. This is consistent with the results obtained by Zhang et al. [20] who found close similarities between human and sheep isolates with distinctive PCR-RFLP pattern between them and camel isolate. In order to validate the PCR-RFLP technique, the 4 isolates (one from each group) were analyzed by DNA sequencing. AgB sequencing data denotes and further confirms PCR-RFLP that the human and sheep isolates of E. granulosus are the closest relatives amongst the examined isolates. Comparison of results from the current study and previous reports are limited due to dissimilarities in methodology. Using PCR and RT-PCR, followed by cloning and sequencing, Arend et al. [3] characterized a high degree of AgB2 sequence polymorphism between strains. Kamenetzky et al. [19] using PCR-SSCP (single strand conformation polymorphism) followed by DNA sequencing to evaluate sequence variation and transcription profile of AgB2-coding genes in several human infecting strains of E. granulosus found a substantial level of inter-strain variation in AgB2-related genes between G1/G2 strain on one hand and G6/G7 and G5 on the other hand. Their finding was supported by a previous study using mitochondrial or nuclear markers which also grouped G1/G2 strains and G6/G7 strains as 2 separate groups leaving G5 strain outside but closer to the G6/G7 group [21].
In concordance with the previous data, Muzulin et al. [22] suggested that AgB2-related sequences may be subjected to selective pressure to present as a functional gene exclusively in G1/G2 strains and as non-functional genes (can not be translated into a functional protein) in G5 and the G6/G7 cluster suggesting inter-strain variation. Furthermore, analysis of E. granulosus isolates from different geographical areas, including Egypt by Frosch et al. [23] revealed AgB1 nucleotide sequence differences at conserved positions that resulted in altered amino acid sequences. They reported that the observed sequence heterogeneity of the AgB genes in different geographic origin could reflect the occurrence of distinct clonal populations of which differed in the use of their final and intermediate hosts and morphological, biochemical, and genetic characteristics. Their finding is supported by Mamuti et al. [6] who concluded that the genetic variability in different host and/or different level of host immune responses against AgB may be relevant factors in the variability that is observed in the immunogenecity of AgB among different CE patients. In addition, Rosenzvit et al. [7] reported that analyzing the variation of AgB-coding genes may provide a specific correlation of strain genetic diversity with AgB functional and practical significance. The knowledge of variation in AgB genes would help the understanding of the fine mechanisms of adaptation of helminth parasites to their hosts and is necessary for the rational design and application of diagnostic tests, anthelmintic drugs, and immunoprophylaxis reagents.
The combined use of PCR-RFLP and DNA sequencing detects the polymorphism of AgB2 related genes among human, camel, pig, and sheep E. granulosus Egyptian isolates. Human and sheep isolates are the most AgB2 related pair. Whether the polymorphism of AgB-genes among different Egyptian isolates affects its diagnostic performance also remains to be determined with further experimentations.
ACKNOWLEDGEMENTS
We are grateful to the center of Genetic Engineering and Biotechnology Ain Shams University, Cairo, Egypt, for granting free facilities of laboratory and equipment throughout this work. Herewith we declare that the experiments comply with the current laws and regulations of the Egyptian Ministry of Higher Education and Scientific Research.
References
- 1.Rausch RL, Thompson J. Life cycle patterns and geographic distribution of Echinococcus species. In: Thompson RCA, Lymbery AJ, editors. Echinococcus and Hydatid Disease. Wallingford, Oxon: CAB International; 1995. pp. 89–134. [Google Scholar]
- 2.Dinkel A, Njoroge EM, Zimmermann A, Wälz M, Zeyhle E, Elmahdi IE, Mackenstedt U, Romig T. A PCR system for detection of species and genotypes of the Echinococcus granulosus-complex, with reference to the epidemiological situation in Eastern Africa. Int J Parasitol. 2004;34:645–653. doi: 10.1016/j.ijpara.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 3.Arend AC, Zaha A, Ayala FJ, Haag KL. The Echinococcus granulosus antigen B shows a high degree of genetic variability. Exp Parasitol. 2004;108:76–80. doi: 10.1016/j.exppara.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 4.Maddison SE, Slemenda SB, Schantz PM, Fried JA, Wilson M, Tsang VCW. A specific diagnostic antigen of Echinococcus granulosus with an apparent molecular weight of 8 kDa. Am J Trop Med Hyg. 1989;40:377–383. doi: 10.4269/ajtmh.1989.40.377. [DOI] [PubMed] [Google Scholar]
- 5.Virginio VG, Hernández A, Rott MB, Monteiro KM, Zandonai AF, Nieto A, Zaha A, Ferreira HB. A set of recombinant antigens from Echinococcus granulosus with potential for use in the immunodiagnosis of human cystic hydatid disease. Clin Exp Immunol. 2003;132:309–315. doi: 10.1046/j.1365-2249.2003.02123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mamuti W, Sako Y, Xiao N, Nakaya K, Nakao M, Yamasaki H, Lightowlers MW, Craig PS, Ito A. Echinococcus multilocularis: developmental stage-specific expression of antigen B 8-kDa-subunits. Exp Parasitol. 2006;113:75–82. doi: 10.1016/j.exppara.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Rosenzvit MC, Camicia F, Kamenetzky L, Muzulin PM, Gutierrez AM. Identification and intra-specific variability analysis of secreted and membrane-bound proteins from Echinococcus granulosus. Parasitol Int. 2006;55:S63–S67. doi: 10.1016/j.parint.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 8.Shepherd JC, Aitken A, McManus DP. A protein secreted in vivo by Echinococcus granulosus inhibits elastase activity and neutrophil chemotaxis. Mol Biochem Parasitol. 1991;44:81–90. doi: 10.1016/0166-6851(91)90223-s. [DOI] [PubMed] [Google Scholar]
- 9.Fernández V, Ferreira HB, Fernández C, Zaha A, Nieto A. Molecular characterization of a novel 8-kDa subunit of Echinococcus granulosus antigen B. Mol Biochem Parasitol. 1996;77:247–250. doi: 10.1016/0166-6851(96)02602-3. [DOI] [PubMed] [Google Scholar]
- 10.Chemale G, Haag KL, Ferreira HB, Zaha A. Echinococcus granulosus antigen B is encoded by a gene family. Mol Biochem Parasitol. 2001;116:233–237. doi: 10.1016/s0166-6851(01)00316-4. [DOI] [PubMed] [Google Scholar]
- 11.Haag KL, Alves-Junior L, Zaha A, Ayala FJ. Contingent, non-neutral evolution in a multicellular parasite: natural selection and gene conversion in the Echinococcus granulosus antigen B gene family. Gene. 2004;333:157–167. doi: 10.1016/j.gene.2004.02.027. [DOI] [PubMed] [Google Scholar]
- 12.Lavikainen A, Lehtiner MJ, Meri T, Hirvelä-Koski V, Meri S. Molecular genetic characterization of the Fennoscandian cervid strain, a new genotypic group (G10) of Echinococcus granulosus. Parasitology. 2003;127:207–215. doi: 10.1017/s0031182003003780. [DOI] [PubMed] [Google Scholar]
- 13.Bart JM, Bardonnet K, Elfegoun MCB, Dumon H, Dia L, Vuitton DA, Piarroux R. Echinococcus granulosus strain typing in North Africa: comparison of eight nuclear and mitochondrial DNA fragments. Parasitology. 2004;128:229–234. doi: 10.1017/s0031182003004359. [DOI] [PubMed] [Google Scholar]
- 14.Bowles J, Blair D, McManus DP. Genetic variants within the genes Echinococcus identified by mitochondrial DNA sequencing. Mol Biochem Parasitol. 1992;54:165–173. doi: 10.1016/0166-6851(92)90109-w. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Eslami A, Hasseini SH, McManus DP. Indication of the presence of two distinct strains of Echinococcus granulosus in Iran by mitochondrial DNA markers. Am J Trop Med Hyg. 1998;59:171–174. doi: 10.4269/ajtmh.1998.59.171. [DOI] [PubMed] [Google Scholar]
- 16.Karcher SJ. Molecular Biology, A Project Approach. 1st ed. San Diego, New York, Boston: Academic Press; 1995. Polymerase chain reaction; pp. 79–110. [Google Scholar]
- 17.Bowles J, McManus DP. Rapid discrimination of Echinococcus species and strains using a polymerase chain reaction based-RFLP method. Mol Biochem Parasitol. 1993;57:231–239. doi: 10.1016/0166-6851(93)90199-8. [DOI] [PubMed] [Google Scholar]
- 18.Rozas J, Rozas R, Dna SP. DNA sequence polymorphism. 2001. Distributed by the authors, Department of Genetics, University of Barcelona, available at http://www.bio.ub.es/yjulio/DnaSP.html.
- 19.Kamenetzky L, Muzulin PM, Gutierrez AM, Angel SO, Zoha A, Guarnera EA, Rosenzvit MC. High polymorphism in genes encoding antigen B from human infecting strains of Echinococcus granulosus. Parasitology. 2005;131:805–815. doi: 10.1017/S0031182005008474. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Eslami A, Hasseini S, McManus D. Indication of the presence of two distinct strains of Echinococcus granulosus in Iran by mitochondrial DNA markers. Am J Trop Med Hyg. 1998;59:171–174. doi: 10.4269/ajtmh.1998.59.171. [DOI] [PubMed] [Google Scholar]
- 21.Bartholomei-Santos ML, Heinzelmann LS, Oliveira RP, Chemale G, Gutierrez AM, Kamenetzky L, Haag KL, Zaha A. Isolation and characterization of microsatellites from the tapeworm Echinococcus granulosus. Parasitology. 2003;126:599–605. [PubMed] [Google Scholar]
- 22.Muzulin PM, Kamenetzky L, Gutierrez AM, Guarnera EA, Rosenzvit MC. Echinococcus granulosus antigen B gene family: further studies of strain polymorphism at the genomic and transcriptional levels. Exp Parasitol. 2008;118:156–164. doi: 10.1016/j.exppara.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Frosch P, Hartmann M, Mühlschlegel F, Frosch M. Sequence heterogeneity of the echinococcal antigen B. Mol Biochem Parasitol. 1994;64:171–175. doi: 10.1016/0166-6851(94)90145-7. [DOI] [PubMed] [Google Scholar]