Introduction
Cysteine peptidases (CP) are ubiquitous enzymes that play fundamental roles in many cellular metabolic pathways.1 In mammalian cells they are highly regulated in apoptotic pathways related to cancer and other severe disorders.2,3 In bacterial cell division, cell-growth and lysis peptidases are employed in the cleavage of peptidoglycans (autolysin).4,5 As secreted antigens and toxins, CPs are key to virulence in Gram-positive pathogens,6 and used to attack competing species in bacterial warfare.7 Viral genomes encode forms of the enzyme for the purpose of capsid trimming during maturation.8 CPs have been classified in clans based on their evolutionary relationships,9 in particular, the CHAP (cysteine, histidine-dependent amidohydrolases/peptidases) domain (pfam: PF05257, MEROPS ID: C51) is classified as an endopeptidase and part of the CA clan of peptidases (CL0125). This 27-member superfamily includes synthetase/amidases, peptidases, viral proteinases, the NPLC/P60 families and other papain-related families.10 Pfam 22.0 lists 602 members for the CHAP domain family, including 475 from the bacterial kingdom (396 in the firmicutes phylum without any known 3D structure), 39 from eukaryota, and 81 from viruses and phage (predominantly involving Gram-positive phage). CHAP domains are often associated with other peptidases, bifunctional glutathionyl spermidine (GSP) amidases (type 2 and 3), choline binding, and with SH3 and/or Von Willebrand (VWA) domains to form multidomain systems that act cooperatively as versatile machineries for murein septum processing while anchored to the cell surface. Deletion of the CHAP-containing cse gene in Streptococcus thermophilus results in impaired separation of cells during mitosis, demonstrating its involvement in cell division.11 The staphylococcal phage ϕ11 hydrolase, a CHAP domain-containing enzyme, exhibits D-alanyl-glycyl endopeptidase and N-acetylmuramyl-L-alanyl amidase activity.12
In this note we present the solution NMR structure of CHAP domain encoded by gene SSP0609 of Staphylococcus saprophyticus [SWISS-PROT ID: Q49ZM2_STAS1; NESG target ID: SyR11]. S. saprophyticus, one of the three major human Gram-positive pathogens, possesses anchoring fimbriae that help colonize the urinary tract resulting in infection.13 Bacterial antigens are known virulence agents7, and SSP0609 is a secreted antigen with potential roles in the onset of S. saprophyticus infection. The protein comprises a type-I signal peptide in the N-terminal region (res. 1 - 47) and a globular CHAP domain in the C-terminal region (res. 48 - 155). The SSP0609 CHAP domain was found to have a highly-conserved Cys – His – Glu – Asn proteolytic relay active site.
Materials and Methods
Uniformly 13C,15N- and 5%-13C, U-15N-enriched Staphylococcus saprophyticus SSP0609 samples were produced following standard protocols of the NESG consortium;14 A complete description of the molecular biology, protein purification, sample preparation, NMR data acquisition, analysis and structure calculation and validation methods used in this work is given in Supplementary Material. The 16.98 kDa protein construct studied here includes the full-length protein sequence with a C-terminal affinity tag (LEHHHHHH), starting at position 156. Protein samples for NMR spectroscopy were concentrated to 0.7 to 0.9 mM in 95% H2O/5% D2O solution containing 20 mM MES, 100 mM NaCl, 10 mM DTT, 0.02% NaN3, and 5 mM CaCl2 at pH 6.5. NMR measurement of the T1 and T2 relaxation rates (τc = 7.1 ± 0.1 at 25 °C) and gel-filtration chromatography with mass detection by static light scattering confirmed the monomeric state of SSP0609 (Supplementary Figs. S1 and S2). Triple resonance NMR data were collected at 25 °C on Bruker AVANCE 600 and 800 MHz NMR spectrometers. Data analysis, including largely automated backbone assignment, manual sidechain assignments, and manual NOESY peak-picking from 15N and 13C-edited NOESY in H2O and 2H2O, provided peak list files necessary to run CYANA 2.1 and AutoStructure 2.1.1 (Supplementary Fig. S3).15-17 Following iterative cycles of noise/artifact peak removal and minor assignment modifications, a complete NOESY peak list set was obtained. This set was used in a final CYANA 2.1 run in which the resulting 20 lowest target function structures were further refined with constrained molecular dynamics in explicit water.18 The resulting final structures underwent the extensive NESG pre-submission validation protocols.19,20 Structural and biochemical characterization of the active site was carried out by NMR. The tautomeric state and the pKa of His109 were determined by 1H-15N HMQC (Fig. 2A)21 and by pH titration of the U-13C,15N SSP0609 while monitoring the changes in chemical shift by 2D NMR (Fig 2B and further information in Supplementary Materials), respectively. Details of electrostatic calculations are outlined in the Supplementary Materials. Structural bioinformatic analysis of the resulting structure was facilitated using the Mark-Us server.22
Figure 2.
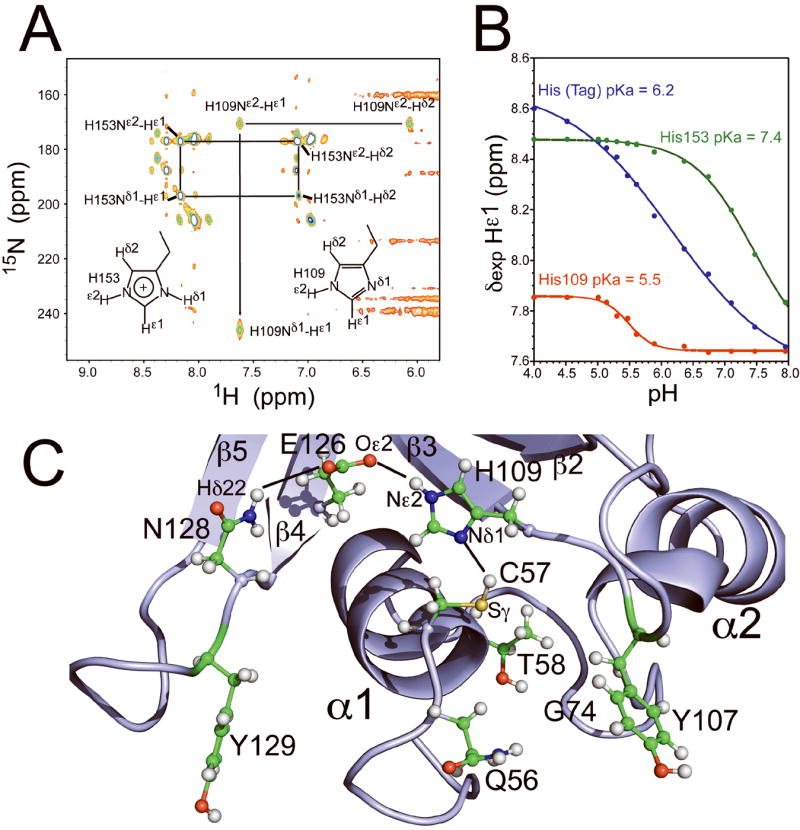
(A) Characterization of His109 and His153 tautomer state by 2D 1H-15N HMQC spectrum of [U-15N, 5%13C] SSP0609 at pH 6.5 and 25 °C, obtained on an 800 MHz NMR spectrometer. Acquisition parameters: 15N carrier at 205 ppm, 2J(15N-1H) = 22Hz, 2 sec relaxation delay, acquisition dimensions 2048 × 200 complex points, sweep widths 20 ppm (1H) × 200 ppm (15N), and 128 transients per t1 increment. (B) Plots of histidine Hε1 chemical shift vs. pH for U-13C,15N S. saprophyticus SSP0609 at 298 K, obtained by 2D 1H-13C HSQC NMR. Red, His109; green, His153 (surface exposed); blue, C-terminal His tag (control); pKa values for each are indicated in the plot. C) Detailed view of SSP0609 active site showing the cysteine peptidase catalytic triad formed by Cys57, His109, Glu126 and the additional participating group Asn128. Showing here are the strongly conserved Gln56, Thr58 and Gly74 in the active site. Key hydrogen bond distances: Cys57 Sγ – His109 Nδ1 = 3.3 Å, His109 Nε2 – Glu126 Oε2 = 3.2 Å, Glu126 Oε1 – Asn128 Nδ2 = 4.1 Å.
Results and Discussion
Structural statistics for the solution NMR structure of S. saprophyticus SSP0609 are listed in Supplementary Table S1; the structure shows excellent structure quality scores. In agreement with disorder prediction analysis,23 NMR shows the N-terminal portion of the sequence up to residue 49 to be unstructured in solution. A stereoview of the lowest energy structure representative from the final SSP0609 ensemble (residues 50 - 155) is shown in Fig. 1A. The SSP0609 CHAP domain features the typical ααβββββαβ papain topology. Two short alpha helices (α1, Cys57 - Lys64; α2, Trp76 - Ala86) packed against a six-strand beta sheet saddle (β1, Thr89 - Asn91; β2, Ser98 - Ser101; β3, Val110 - Val116; β4, Val122 - Glu127; β5, Ser136 - Ile141; β6, Asn150 - His153). Linking β5 to β6 is a short helical segment (α3, Ala143 - Ala146) confirmed by 13C chemical shift and NOESY data. The β-sheet core is formed by four highly conserved hydrophobic residues: Pro94, Val110, Val113, and Val116. The fold, characteristic of the cysteine peptidase superfamily clan CA, as well as the location of the active site in the CHAP domain (described below), were accurately predicted.24
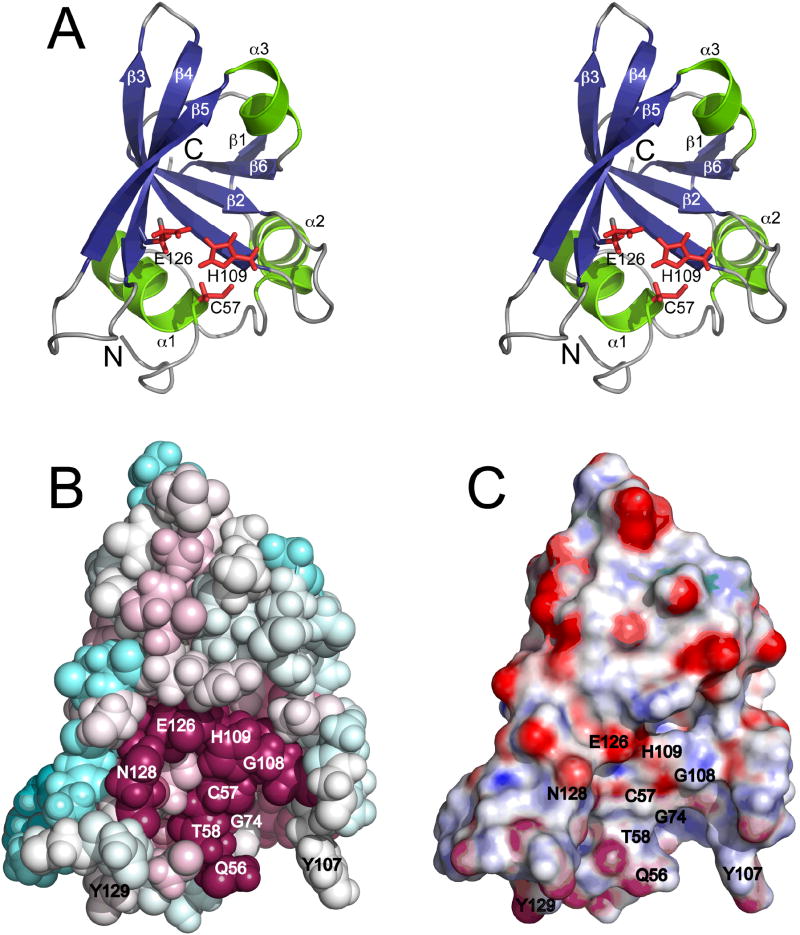
Figure 1.
(A) Stereoview of the ribbon representation of the lowest energy conformer (lowest CNS energy) from the ensemble of deposited solution NMR structures of full length SSP0609 (PDB_ID: 2K3A). Only residues 50 - 155 are shown. The secondary structural elements are labeled and the triad sidechain residues (Cys57, His109, Glu126) are highlighted as red sticks. (B) ConSurf images of SSP0609 for identical size and orientation as (A). ConSurf analysis was conducted for the entire CHAP protein domain family, standard ConSurf residue coloring reflecting the degree of residue conservation over the entire family were used (color scheme: magenta = highly conserved, cyan = variable). (C) Electrostatic potential surface diagrams for SSP0609 shown in the same orientation as (A) and (B).
Analysis of the molecular surface of SSP0609 by ConSurf25 [Fig. 1B] shows seven highly conserved residues at or near the surface, Gln56, Cys57, Thr58, Gly74, Gly108, His109, Glu126, and Asn128, forming a very broad and shallow cavity. Residues Cys57, His109, and Glu126, highlighted in Fig. 1A, are arranged in the typical clan CA sequence order and topology. This residue triplet was shown to be a viable proteolytic triad found in pyroglutamyl peptidases, a family (C15) of otherwise unrelated cysteine peptidases (clan CF).26 In SSP0609 the extended charge transfer relay includes Asn128 as an additional stabilizing group. Cys57 is positioned at the beginning of helix α1, followed by His109 on strand β3, Glu126 at the end of strand β4 and Asn128 toward on the BB4 loop (Fig. S4). Electrostatic surface potential27 [Fig. 1C and Suppl. Fig. S5] images of SSP0609 show the active site cleft with several polar residues consistent with peptidase active sites. Overall, the molecule exhibits weak and diffused negative charge.
More evidence for the peptidase motif emerges from the structural and biochemical characterization of the SSP0609 active site (Fig. 2A,B). The 1H-15N HMQC pattern clearly shows His109 in the neutral state. The structure quality is significantly improved upon by the introduction of the correct His109 tautomer in NMR structure-determination protocol. In the NMR conditions (pH 6.5), the resulting NOE-based models indicate the Cys57 Sγ and His109 Nδ1 to be within hydrogen bond distance (3.3 Å), evidently sharing the Cys57 Hγ proton (Fig. 2C). The pKa of His109 was found to be 5.5 (Fig. 2B), which is indicative of a buried histidine in a highly cooperative hydrogen bond network and typical of such residue in proteolytic triads. By contrast, the surface exposed His153 sidechain (pKa = 7.4) and histidines in the purification tag (pKa = 6.2) are more basic and protonated (on Nδ1 and Nε2) at NMR conditions. The slightly elevated pKa of His153 is explained by the formation of a salt bridge between the positively charged imidazole Nδ1Hδ1 and the negatively charged Glu95 sidechain Oε (His153 Nδ1- Glu95 Oε = 2.9 Å).
Presence of the substrate would simultaneously trigger the deprotonation of Cys57 Sγ by His109 Nδ1 and Cys57 thiolate attack the C′ peptidyl leading to the tetrahedral anionic intermediate.28 The resulting charged species is sequestered by a pair of hydrogen bonds within the active site cavity (oxyanion hole). A variety of suitable hydrogen-bond donors in peptidases are known,29 In the structure of SSP0609, the Gln56 sidechain, and/or the backbone NH groups of Gly74 and Gly108 are positioned to fulfill this role. These residues are highly conserved and likely essential to enzymatic activity. Acquisition of Cys57 SγH by His109 establishes a positive charge on the indole ring, which is distributed to the carboxyl sidechain (Oε) of Glu126 via interaction with the His109 Hε2-Nε2 hydrogen-bond donor (His109 Nε2 – Glu126 Oε2 = 3.2 Å) and further distributed to the Asn128 Nδ2 moiety (Glu126 Oε1 – Asn128 Nδ2 = 4.1 Å).30 Although still well over 90% conserved, the Glu126 residue is the least conserved member of the Cys57-His109-Glu126-Asn128 quartet across the CHAP domain family. In firmicutes, the most common variant for Glu at position 126 is the chemically-similar Asp. Interestingly Glu126Gly substitutions are found in eight pathogenic species, six of which are S. aureus strains. Based on the detailed knowledge of the active site gathered in this work we postulate a substantial rearrangement of the active site in these limited number of firmicute variants.
In S. saprophyticus, the immediate vicinity of the active site lacks a set of large, cavity-defining hydrophobic residues that would confer specificity to the binding site. However, two exposed aromatic groups, Tyr107 and Tyr129, are positioned to possibly function as “rails” at either side of the active site, and are likely to have substrate anchoring function. Specifically, Tyr107 is located on the β2 - β3 (BB2) loop and in close proximity to Cys57 while Tyr129 is located on the β4 - β5 (BB4) loop. Tyr107 is > 90% conserved in Gram-positive firmicutes, and Tyr129 is highly conserved as an aromatic type (~70/30 Tyr, Trp/Val) across the entire family.
A search for structurally similar proteins in the Protein Data Bank using the DALI31 server produces several hits of sizeable Z-score, but minimal aligned sequence identity (see full DALI report in Suppl. Table S1). These include several structures of proteins from the same peptidase superfamily including E. coli bifunctional glutathionyl spermidine (GSP) CHAP domains (PDB_IDs 2IOB, 2IO9; sequence_identity 17% and 18%, respectively) 32, A. variabilis NLP/P60 (PDB_ID 2HBW; sequence_identity 7%) and N. punctiforme COG0791 (PDB_ID 2EVR; sequence_identity 8%) cell wall-associated hydrolases, and E coli lipoprotein Spr 33 (PDB_ID 2K1G; sequence_identity 9%). The novel modeling leverage for SSP0609 (PDB_ID: 2K3A) is 86 models (UniProt 12.8);34 i.e. some 86 protein sequences can be homology modeled using the 3D structure of SSP0609 that could not be modeled using the structures available in the PDB on the date of its deposition.
Structure-based sequence alignment [Suppl. Fig. S4A] and overlay [Suppl. Fig. S4B] of SSP0609 with the E. coli GSP CHAP domain structure shows a much more secluded cysteine active site in the GSP CHAP domain. While the overall papain fold is identical, the sequence homology is low (18%) and several difference in the structure are present. Most notably the longer BB2, BB4 and BB5 (substitutes α3 in 2IOB) loops surround the active site with non-polar residues and produce a well defined, deep binding pocket. Also, in the E. coli GSP CHAP domain the Asn128 equivalent lacks structural alignment with the SSP0609 protein discounting its active site involvement. These observations indicate a different substrate binding and scope for SSP0609. Although SSP0609’s role as proteolytic enzyme is clearly indicated by the 3D structure of its active site, characterization substrate and exact scope of this enzyme in the life cycle of S. saprophyticus awaits further structural and biochemical studies.
Supplementary Material
Acknowledgments
We thank Gaohua Liu and Alexander Eletski for helpful discussions. Resonance assignments, raw NMR data, and the 3D NOESY peak lists have been deposited in the BioMagResDB (BMRB ID: 15335), and atomic coordinates have been deposited into the Protein Data Bank (PDB_ID, 2K3A). This work was supported by a grant from the Protein Structure Initiative of the National Institutes of Health (U54-GM074958).
References
- 1.Barrett AJ, Rawlings ND. Evolutionary lines of cysteine peptidases. Biol Chem. 2001;382(5):727–733. doi: 10.1515/BC.2001.088. [DOI] [PubMed] [Google Scholar]
- 2.Dickinson DP. Cysteine peptidases of mammals: their biological roles and potential effects in the oral cavity and other tissues in health and disease. Crit Rev Oral Biol Med. 2002;13(3):238–275. doi: 10.1177/154411130201300304. [DOI] [PubMed] [Google Scholar]
- 3.Vermeulen K, Van Bockstaele DR, Berneman ZN. Apoptosis: mechanisms and relevance in cancer. Ann Hematol. 2005;84(10):627–639. doi: 10.1007/s00277-005-1065-x. [DOI] [PubMed] [Google Scholar]
- 4.Smith TJ, Blackman SA, Foster SJ. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology. 2000;146(Pt 2):249–262. doi: 10.1099/00221287-146-2-249. [DOI] [PubMed] [Google Scholar]
- 5.Rigden DJ, Jedrzejas MJ, Galperin MY. Amidase domains from bacterial and phage autolysins define a family of gamma-D,L-glutamate-specific amidohydrolases. Trends Biochem Sci. 2003;28(5):230–234. doi: 10.1016/s0968-0004(03)00062-8. [DOI] [PubMed] [Google Scholar]
- 6.Kubica M, Guzik K, Koziel J, Zarebski M, Richter W, Gajkowska B, Golda A, Maciag-Gudowska A, Brix K, Shaw L, Foster T, Potempa J. A potential new pathway for Staphylococcus aureus dissemination: the silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS ONE. 2008;3(1):e1409. doi: 10.1371/journal.pone.0001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee VT, Schneewind O. Protein secretion and the pathogenesis of bacterial infections. Genes Dev. 2001;15(14):1725–1752. doi: 10.1101/gad.896801. [DOI] [PubMed] [Google Scholar]
- 8.Weber JM. Synthesis and assay of recombinant adenovirus protease. Methods Mol Med. 2007;131:251–255. doi: 10.1007/978-1-59745-277-9_18. [DOI] [PubMed] [Google Scholar]
- 9.Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2008;36(Database issue):D320–325. doi: 10.1093/nar/gkm954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bateman A, Rawlings ND. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem Sci. 2003;28(5):234–237. doi: 10.1016/S0968-0004(03)00061-6. [DOI] [PubMed] [Google Scholar]
- 11.Layec S, Decaris B, Leblond-Bourget N. Characterization of proteins belonging to the CHAP-related superfamily within the Firmicutes. J Mol Microbiol Biotechnol. 2008;14(1-3):31–40. doi: 10.1159/000106080. [DOI] [PubMed] [Google Scholar]
- 12.Navarre WW, Ton-That H, Faull KF, Schneewind O. Multiple enzymatic activities of the murein hydrolase from staphylococcal phage phi11. Identification of a D-alanyl-glycine endopeptidase activity. J Biol Chem. 1999;274(22):15847–15856. doi: 10.1074/jbc.274.22.15847. [DOI] [PubMed] [Google Scholar]
- 13.Kuroda M, Yamashita A, Hirakawa H, Kumano M, Morikawa K, Higashide M, Maruyama A, Inose Y, Matoba K, Toh H, Kuhara S, Hattori M, Ohta T. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc Natl Acad Sci U S A. 2005;102(37):13272–13277. doi: 10.1073/pnas.0502950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Acton TB, Gunsalus KC, Xiao R, Ma LC, Aramini J, Baran MC, Chiang YW, Climent T, Cooper B, Denissova NG, Douglas SM, Everett JK, Ho CK, Macapagal D, Rajan PK, Shastry R, Shih LY, Swapna GV, Wilson M, Wu M, Gerstein M, Inouye M, Hunt JF, Montelione GT. Robotic cloning and Protein Production Platform of the Northeast Structural Genomics Consortium. Methods Enzymol. 2005;394:210–243. doi: 10.1016/S0076-6879(05)94008-1. [DOI] [PubMed] [Google Scholar]
- 15.Moseley HN, Monleon D, Montelione GT. Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Methods Enzymol. 2001;339:91–108. doi: 10.1016/s0076-6879(01)39311-4. [DOI] [PubMed] [Google Scholar]
- 16.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 17.Huang YJ, Tejero R, Powers R, Montelione GT. A topology-constrained distance network algorithm for protein structure determination from NOESY data. Proteins. 2006;62(3):587–603. doi: 10.1002/prot.20820. [DOI] [PubMed] [Google Scholar]
- 18.Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50(3):496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 19.Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66(4):778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 20.Huang YJ, Powers R, Montelione GT. Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. J Am Chem Soc. 2005;127(6):1665–1674. doi: 10.1021/ja047109h. [DOI] [PubMed] [Google Scholar]
- 21.Pelton JG, Torchia DA, Meadow ND, Roseman S. Tautomeric states of the active-site histidines of phosphorylated and unphosphorylated IIIGlc, a signal-transducing protein from Escherichia coli, using two-dimensional heteronuclear NMR techniques. Protein Sci. 1993;2(4):543–558. doi: 10.1002/pro.5560020406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer M, Honig B. Mark-Us. Columbia University; 2006. ( http://luna.bioc.columbia.edu/honiglab/mark-us/) [Google Scholar]
- 23.Huang YJ, Montelione GT. DisMeta. 2008 ( http://www-nmr.cabm.rutgers.edu/bioinformatics/disorder/)
- 24.Anantharaman V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 2003;4(2):R11. doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33(Web Server issue):W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sokabe M, Kawamura T, Sakai N, Yao M, Watanabe N, Tanaka I. The X-ray crystal structure of pyrrolidone-carboxylate peptidase from hyperthermophilic archaea Pyrococcus horikoshii. J Struct Funct Genomics. 2002;2(3):145–154. doi: 10.1023/a:1021257701676. [DOI] [PubMed] [Google Scholar]
- 27.Rocchia W, Sridharan S, Nicholls A, Alexov E, Chiabrera A, Honig B. Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies: applications to the molecular systems and geometric objects. J Comput Chem. 2002;23(1):128–137. doi: 10.1002/jcc.1161. [DOI] [PubMed] [Google Scholar]
- 28.Lau EY, Bruice TC. Consequences of breaking the Asp-His hydrogen bond of the catalytic triad: effects on the structure and dynamics of the serine esterase cutinase. Biophys J. 1999;77(1):85–98. doi: 10.1016/S0006-3495(99)76874-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szeltner Z, Renner V, Polgar L. Substrate- and pH-dependent contribution of oxyanion binding site to the catalysis of prolyl oligopeptidase, a paradigm of the serine oligopeptidase family. Protein Sci. 2000;9(2):353–360. doi: 10.1110/ps.9.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korza HJ, Bochtler M. Pseudomonas aeruginosa LD-carboxypeptidase, a serine peptidase with a Ser-His-Glu triad and a nucleophilic elbow. J Biol Chem. 2005;280(49):40802–40812. doi: 10.1074/jbc.M506328200. [DOI] [PubMed] [Google Scholar]
- 31.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233(1):123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 32.Pai CH, Chiang BY, Ko TP, Chou CC, Chong CM, Yen FJ, Chen S, Coward JK, Wang AH, Lin CH. Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase. EMBO J. 2006;25(24):5970–5982. doi: 10.1038/sj.emboj.7601440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aramini JM, Rossi P, Huang YJ, Zhao L, Jiang M, Malagqui M, Xiao R, Locke J, Nair R, Rost B, Acton TB, Inouye M, Montelione GT. Solution NMR structure of the NlpC/P60 domain of Spr lipoprotein from Escherichia coli: Structural evidence for a novel cysteine peptidase catalytic triad. Biochemistry. 2008 doi: 10.1021/bi8010779. in press. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, Montelione GT, Rost B. Novel leverage of structural genomics. Nat Biotechnol. 2007;25(8):849–851. doi: 10.1038/nbt0807-849. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.