Abstract
The objective of this study was to evaluate whether indicator microbes and physical-chemical parameters were correlated with pathogens within a tidally influenced estuary. Measurements included the analysis of physical-chemical parameters (pH, salinity, temperature, and turbidity), measurements of bacterial indicators (enterococci, fecal coliform, E. coli, and total coliform), viral indicators (somatic and MS2 coliphage), viral pathogens (enterovirus by culture), and protozoan pathogens (Cryptosporidium and Giardia). All pathogen results were negative with the exception of one sample which tested positive for culturable reovirus (8.5 MPN/100 L).. Notable physical-chemical parameters for this sample included low salinity (<1 ppt) and high water temperature (31 °C). Indicator bacteria and indicator virus levels for this sample were within average values typically measured within the study site and were low in comparison with levels observed in other freshwater environments. Overall results suggest that high levels of bacterial and viral indicators were associated with low salinity sites.
1. Introduction
The recreational safety of water bodies is established through the microbiological examination of water samples. The premise for measurements is based upon identifying waters impacted by sanitary sewage. Sanitary sewage, since it carries human waste, can be a source of intestinal pathogens which can cause disease. However, the direct examination of water samples for pathogens is tedious, difficult, and time-consuming. A simpler and accepted method to evaluate potential sewage impacts is to measure levels of “indictor” microorganisms instead. Indicator microbes are much easier to analyze and are expected to be present in high numbers when a water body is impacted by sewage. The indicator microorganisms most frequently used are enterococci, Escherichia coli, and fecal coliforms. Total coliforms have also been historically used to establish the recreational safety of waters. Recommended guideline levels for each of these indicator microbes are summarized in table 1.
Table 1.
Federal and state regulations for indicator microbes in recreational waters.
| Indicator microbe | Regulatory Level | Regulator* |
|---|---|---|
| Fecal coliform | A monthly geometric mean of 200 CFU/100 ml (at the minimum, 10 samples over a 30-day period), and 400 CFU/100 ml in 10% of the samples. 800 CFU/100 ml on a single sample. |
USEPA (1976) FDEP FDOH |
| Total coliform | A monthly geometric mean of 1,000 CFU/100 mL (at the minimum, 10 samples over a 30-day period). 2,400 CFU/100 mL in 20% of the TC single samples. |
USEPA (1976) |
| Enterococci | 35 CFU/100 ml, geometric mean 104 CFU/100 ml, single sample For marine water. |
USEPA (1986) |
| E. coli | 126 CFU/100 ml, geometric mean 235 CFU/100 ml, single sample Only for fresh water. |
USEPA (1986) |
Florida Department of Environmental Protection (FDEP) and Florida Department of Health (FDOH)
Research, however, has shown that non-sewage sources of indicator microbes are also found in the environment, with extensive documentation that soils and beach sands serve as important reservoirs for indicators (Fujioka et al. 1999; Hardina and Fujioka, 1991; Roll and Fujioka 1997; Whitman and Nevers 2003; Shibata et al. 2004; Sherer et al. 1992; Howel et al. 1996). In some cases, the fine sediments and underlying sediments in recreational beach areas have been implicated as a source of enterococci (Indest 2003; Howell et al. 1996; Obiri-Danso and Jones 2000; Sherer et al. 1992). In particular, the re-suspension of these underlying sediments can increase the microbial loads in the water (Graczyk et al. 2007; Indest 2003; Brookes et al. 2004; US Geological Survey 2006a, b). Anderson et al. (1997), who found high densities of enterococci in marine sediments, suggest environmental sources of contamination. Specifically in South Florida, river bank soils and beach sands have been implicated as the source of indicator microbes to the water column (Desmarais et al. 2002; Rogerson et al. 2003; Shibata et al. 2004). The ultimate source of indicators to soils and sands can include regrowth in situ (Desmarais et al. 2002), deposition of animal feces, and possibly microorganisms shed from humans (Papadakis et al. 1997; Elmir et al. 2007).
Interpretation of microbial indicator presence is thus very difficult, since their source is not exclusively from sewage. Given these uncertainties, direct measurements for pathogens may be warranted to establish whether or not a water body is truly impacted by wastes that potentially contain disease causing microorganisms.
The objective of the current study was to evaluate whether indicator microbes were correlated with pathogens in the study waterway and to evaluate physical-chemical parameters that may be associated with pathogen presence. Specifically for the current study, water was analyzed from selected locations along the St. Lucie River Estuary for bacterial indicators (enterococci, fecal coliform, E. coli, and total coliform), viral indicators (somatic and MS2 coliphage), viral pathogens (enterovirus by culture), protozoan pathogens (Cryptosporidium and Giardia) and physical-chemical parameters (pH, salinity, temperature, and turbidity).
2. Materials and Methods
2.1 Site Description
The St. Lucie River Estuary is characterized by intermittent exceedences of fecal indicator bacteria, in particular during the period after the 2004 hurricane season. The Estuary does not receive point sources of sewage and so the apparent increase in exceedences after 2004 were suspected to have been caused by the redistribution of non-point sources through potential changes in the river banks as a result of the four hurricanes which impacted the coastline that year. The site was ideal for evaluating potential relationships between fecal indicator microbes and pathogens in an area impacted by non-point sources of pollution. The St. Lucie River Estuary is located on the eastern coast of Florida about 200 km north of Miami and 250 km southeast of Tampa. The St. Lucie River Estuary is fed by the St. Lucie River, which obtains its water from Lake Okeechobee, located about 55 km to the west. The area drained by the Estuary is approximately 1800 square kilometers. Water from the estuary flows into the Indian River Lagoon to the east, which then discharges into the Atlantic Ocean. The St. Lucie River is characterized by two forks, the North and South Fork. The South Fork receives water from a major freshwater discharge canal from Lake Okeechobee. The North Fork receives water from canals that drain both residential and agricultural areas. The water within the St. Lucie River Estuary is thus brackish, containing a mix of ocean water and freshwater from Lake Okeechobee and local rainwater runoff.
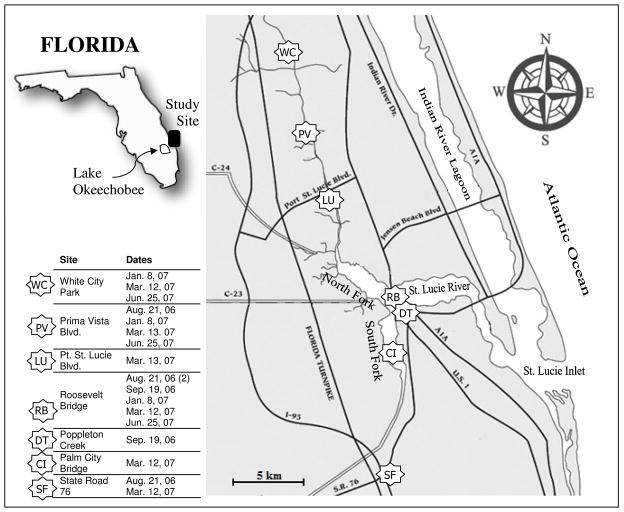
Samples were collected from a total of seven sites (Figure 1). These sites were uniformly distributed through the northern and southern areas of the St. Lucie River Estuary and were chosen to coincide with sampling locations used by regulatory agencies, which showed elevated levels of indicators in the past. The Roosevelt Bridge (RB) site, the site closest to the ocean, was sampled during each sampling trip to provide a baseline for this particular site which is closest to the beaches that run along the coast. This site also has the largest and most direct ocean access with a bridge length of approximately 1.6 km. The close proximity to the ocean allows for strong tidal influences making this sampling site characterized by the highest levels of salinity recorded. The waterway then proceeds from the Roosevelt Bridge to the west before splitting both to the north and south. At this junction the river is hereafter referred to either the North or South Fork of the St. Lucie River. Sampling sites were chosen in order to establish a spatial representation of the water characteristics in the estuary, with three of the sites (WC, PV, and LU) evenly distributed along the North Fork, and the remainder 3 sites (DT, CI, and SF) evenly distributed along the South Fork. The distance between the northern most site and southern site was approximately 28 km. These locations were situated 7 to 11 km from the eastern Florida coastline.
Fig. 1.
Sampling locations for the St. Lucie River Estuary and dates of each sample collection event.
2.2 Sample Collection and Measurement of Physical-chemical Parameters
A total of 18 sampling events were conducted over the course of 5 sampling trips. The total number of samples was limited due to significant time and cost commitments related to pathogen analysis. Additionally, logistical considerations including distance between sampling sites and accessibility at different times influenced the sampling location and times. RB, the central site, was sampled during each of the trips. Water from this site was also used for pathogen matrix spike analysis. At each site, filters were collected for protozoan analysis (Giardia and Cryptosporidium) and for virus analysis (enterovirus, somatic coliphage, and MS2 coliphage). In addition, 200 ml water samples were collected for bacteria analysis (enterococci, E. coli, fecal coliform, and total coliform) and for turbidity measurements using a nephelometer (Hach Model 66120-200, Newark, DE). Other physical-chemical parameters (water temperature, pH, and salinity) were documented using a YSI probe (YSI Model 650-01m Environmental Monitoring Systems, Yellow Springs, OH).
All results were compiled and analyzed. In order to compare physical-chemical parameters and indicators, the different values measured were utilized for calculation of Pearson correlation coefficients (R). “R” values greater than and absolute value of 0.5 were considered to represent strong correlations. Statistical difference analysis was based upon the t-Test: Paired Two Sample for Means with alpha set at 0.05 (corresponds to 95% confidence limits).
All field and laboratory equipment for microbial sample collection and analysis was pre-sterilized (as purchased or through autoclaving) or disinfected (with 0.5% sodium hypochlorite solution—10% household bleach solution—followed by neutralization of the chlorine with 2% sodium thiosulfate solution) (U.S. EPA 2001). Disinfection was used for large-scale equipment and field supplies which could not be autoclaved.
2.2.1 Collection and Analysis Procedures for Bacteria
Samples for bacteria analysis were collected in pre-sterilized bottles. These bottles were then sealed in Ziplock bags, placed in a cooler on freezer packs, transported to a local NELAP-certified laboratory (Harbor Branch Environmental Laboratories Inc., Ft. Pierce, FL), and processed within 6 hours of sample collection. Bacteria samples were analyzed for enterococci by membrane filtration using U.S. EPA Method 1600 (U.S. EPA 2002). Total coliform, fecal coliform, and E. coli were analyzed using a multiple tube fermentation technique (Method 9221 B, APHA 2005).
2.2.2 Collection and Analysis Procedures for Protozoa
Procedures for sample collection and analysis for Cryptosporidium and Giardia followed U.S. EPA Method 1623 (U.S. EPA, 2001) with the exception that an overflow valve was installed upstream of the filter to provide for more control on the flow rate and filter pressure. Prior to installation of the filter, the system was purged with 19 L of sample water. The filter (Filtamax, IDEXX Inc., Westbrook, Maine) was then placed inside the filter holder and filtration of the source water was continued until either filter clogging or until 200L of sample water passed through the filter. Two different types of Filtamax filters were used due to a change in the elution equipment. During the collection of the first 12 samples the filter (FMC 10603) for a manual elution station was used and during the last 6 samples the filter (FMC 6001) for an automatic wash station was used. As a result of this change, consistent with filter manufacturer instructions, the maximum sample collection flow rate used during sample collection was 4 L/min for the first 12 samples and 2 L/min for the last 6 samples. The maximum operating pressure in both cases was 75 psi. Upon completion of the filtration process, the inlet end of the filter holder was disconnected while maintaining the level of the inlet fitting above the level of the outlet fitting to prevent backwashing and the loss of oocysts and cysts from the filter. The filter and water retained in the filter holder was poured into a pre-sterilized Ziplock bag. Shipping methods for protozoan filters were based on standard protocols (U.S. EPA 2001). Filter cartridges were triple-bagged in pre-sterilized Ziplock bags and placed on 6–8 small ice packs (pre-frozen at −20°C). The samples were shipped overnight to the Florida Department of Health Laboratory, Tampa, (FDOH) and analyzed within 72 h from the commencement of sample collection. Upon receipt at the FDOH laboratory, samples were eluted and analyzed as per standard protocols (Method 1623, U.S. EPA 2001).
2.2.3 Collection and Analysis Procedures for Viruses
Virus collection methods were based on a standardized method (U.S. EPA 2001 – Manual of Methods for Virology, U.S. EPA 1996 – ICR Microbial Laboratory Manual) with two exceptions. First, as for protozoan sample collection, an overflow valve was installed upstream of the filter to provide more control on flow rate and filter pressure. Second, a different filter type was used as per Method 9510C (APHA 2005) to allow for the sorption of the virus from brackish water. In brief, the method involved pH and salt adjustment of the source water prior to filtration. An equilibration chamber was used for the source water adjustment. This chamber was first rinsed with source water and then filled to 220 L. A separate 1 L sample of the source water was titrated to a pH of 4 using a 1 M HCl solution. The amount of acid used for the 1 L sample was then scaled to estimate the amount of acid needed to adjust the pH of the source water in the equilibration chamber. The calculated amount of acid was added cautiously to the chamber with stirring while measuring the pH of the water (SympHony™, SP70P, West Chester, PA) until a pH of 4.0 was reached. Two kilograms of MgCl2 were added to each chamber to increase the salt content contained within the chamber by an additional 0.1 N MgCl2. Once the water in the chamber was pH and salt adjusted, 19 L of this pH/salt-adjusted water was used to rinse the filtration system prior to the installation of the filter. After rinsing, a negatively charged filter (Filterite, P/N DFNT 0.45–10, US System Components Corp., Ocala, FL) was aseptically inserted in the filter holder. The water in the chamber was pumped through the system at a rate of 3–7 L/min and at a pressure of less than 30 psi. At the end of the filtration, air was pumped through the system to expel excess water; the filter was removed aseptically and placed in a pre-sterilized Ziplock bag with ~50 ml of source water (not pH or salt adjusted) to avoid filter drying and to help buffer the pH of the water during sample shipment. Shipping methods for virus filters were also based on standard protocols (U.S. EPA 2001). Filter cartridges were triple-bagged in pre-sterilized Ziplock bags and placed on 6–8 small ice packs (pre-frozen at −20°C). The samples were then shipped overnight to the Florida Department of Health Laboratory, Tampa, (FDOH) and analyzed within 72 h from the commencement of the sample collection. For virus analysis, the samples were held at 4°C until processing. Filters were eluted with beef extract, virus concentrated by organic flocculation, and the concentrate assayed for total culturable enteroviruses using BGM cells as the host culture (U.S. EPA 2001). Cultures were examined periodically for 14 days post inoculation, frozen at −70°C, thawed rapidly at 37°C and 10% of the media from each culture flask was passaged onto a new (< 5 day) culture. These cultures were examined for 14 days before the specimen was considered “no virus isolated”. A total of 40 flasks (25 cm2) were used for primary inoculation from each water filter sample.
In addition to the total culturable enterovirus analysis, the eluate sample was also analyzed for coliphage (somatic and MS2). Coliphage analysis was based on U.S. EPA method 1602 (U.S. EPA, 2001).
The sample that tested positive for culturable enterovirus was further evaluated microscopically and via PCR assay to confirm that the virus was a reovirus. Microscopic analysis was based upon a unique cytopathology in BGM cells, which shows highly granulated sometimes spindle shaped BGM cells; one end may float freely while the other remains attached to the plastic of the flask. The PCR assay included RNA purification (Qiagen One-step RTPCR kit, cat # 210212) and amplification using primers from a conserved region (Spinner and Giovanni, 2001).
In addition to the regular sampling there was one matrix spike sample for enterovirus and two matrix spike samples for protozoa. This was done by spiking a portion of the sample water with Cryptosporidium oocysts or Giardia cysts and a portion of the pH/salt modified water with enterovirus. After the first 12 samples when the filter for the protozoan analysis was changed (from that used for a manual wash station to an automated wash station) another matrix spike sample was evaluated for Cryptosporidium and Giardia. Results from the matrix spike samples were satisfactory showing recovery percentages between 25 and 41% for Cryptosporidium and Giardia and 23% for enterovirus.
3. Results
3.1 Physical-Chemical Parameters and Indicators
Results for physical chemical parameters and indicators at each of the seven sites were variable (Table 2). Physical-chemical parameters were within expected ranges for a tidally influenced estuary. The overall average pH value measured at near neutral, 7.6 (6.1 minimum and 8.7 maximum). Water temperature (26.3 °C mean) decreased an average 9 °C from summer to winter. Salinity (12 ppt mean) was dependent upon location, with the sites located furthest from the junction of the North and South Forks (e.g. WC and PV) characterized by relatively low salinities (< 3 ppt), and those located closest (e.g. RB) characterized by salinities in the range of 10 to 34 ppt. Water turbidity was also generally low, characterized by a mean of 7.3 NTU (3.4 NTU minimum and 15 NTU maximum).
Table 2.
Arithmetic means of physical-chemical parameters and bacterial and viral indicators by site.
| Site1 | Physical-Chemical Parameters | Bacterial Indicators (CFU or MPN/100 ml)2 |
Viral Indicators (PFU/100 ml) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| pH | Temp (°C) |
Salinity (ppt) |
Turbidity (NTU) |
Total coliform |
Fecal coliform |
E. coli | Enterococci | Somatic coliphage |
MS coliphage |
|
| CI (n=1) | 8.05 | 22.8 | 22.4 | 8.5 | 23 | 14 | 23 | 17 | <0.3 | 0.3 |
| DT (n=1) | 7.33 | 29.4 | 5.5 | 4.3 | 1700 | 500 | 26 | 77 | 68.4 | 2.5 |
| LU (n=1) | 7.02 | 23.1 | 27.1 | 6.6 | 30 | 23 | 23 | 13 | <0.3 | <0.3 |
| PV (n=4) | 8.11 | 26.7 | 4.4 | 7.3 | 700 (650) | 53 (41) | 53 (41) | 28 (25) | 6.15 | 13 |
| RB (n=6) | 7.50 | 27.3 | 21.8 | 7.2 | 217 (57) | 19 (8) | 17 (5) | 15 (5) | 3.5 | 1.8 |
| SF (n=2) | 7.40 | 25.9 | 3.7 | 7.5 | 725 (620) | 105 (104) | 95 (94) | 153 (142) | <0.5 | 34.1 |
| WC (n=3) | 7.62 | 25.2 | 1.4 | 8.3 | 800 (794) | 177 (157) | 160 (125) | 84 (65) | 7.0 | 8.9 |
|
| ||||||||||
| Regulatory Guideline (Mean) 3 | 200 | 126 | 35 | |||||||
| Regulatory Guideline (Single Sample)3 | 800 | 235 | 104 | |||||||
Number in parenthesis by the site identifier corresponds to the number of samples.
Number in parenthesis corresponds to the geometric mean. Cells bold correspond to data which exceeds the regulatory guidelines. Numbers outside parenthesis correspond to the arithmetic means for sites with n>1.
EPA Regulatory Guideline for Designated Beach Area (E. coli and enterococci) or the FDEP Regulatory Level for a Class III Recreational Water.
For all 18 measurements combined, the geometric mean for each indicator bacteria group (total coliforms – 205 MPN/100 mL, fecal coliforms – 35 MPN/100 mL, E. coli – 25 MPN/100 mL, and enterococci – 20 CFU/100 mL) was below the regulatory guidelines for recreational water. Indicator virus levels were characterized by geometric means of 20 PFU/100 mL for somatic coliphage and 4 PFU/100 mL for MS coliphage for all sites, collectively.
However, when considering sites individually, the geometric means for individual sites did exceed regulatory bacteria guideline levels in some cases. These sites included WC, SF, and DT. WC (n = 3) was characterized by a geometric mean level of 125 MPN/100 ml for E. coli and 65 CFU/100 ml for enterococci. Both of these levels exceeded the federal mean guideline for recreational marine waters. The geometric-mean fecal coliform level at this site (157 MPN/100 ml), however, was below the state regulatory guideline (200 MPN/100 ml) for a Class III recreational water. Of note is that the somatic and MS coliphage level was elevated for this site (>7 PFU/100 ml) in comparison to other sites. SF (n = 2) was characterized by the highest geometric mean enterococci level (142 CFU/100 ml) which clearly exceeded the federal recommended level. MS coliphage was also elevated (34 PFU/100 ml) at this site. DT (n = 1) characterized by the highest fecal coliform level (500 MPN/100 ml) and by an enterococci level of 77 CFU/100 ml, both of which exceeded the regulatory mean guideline levels. Somatic coliphage (68 PFU/100 ml) was also elevated at this site in comparison to the other sites.
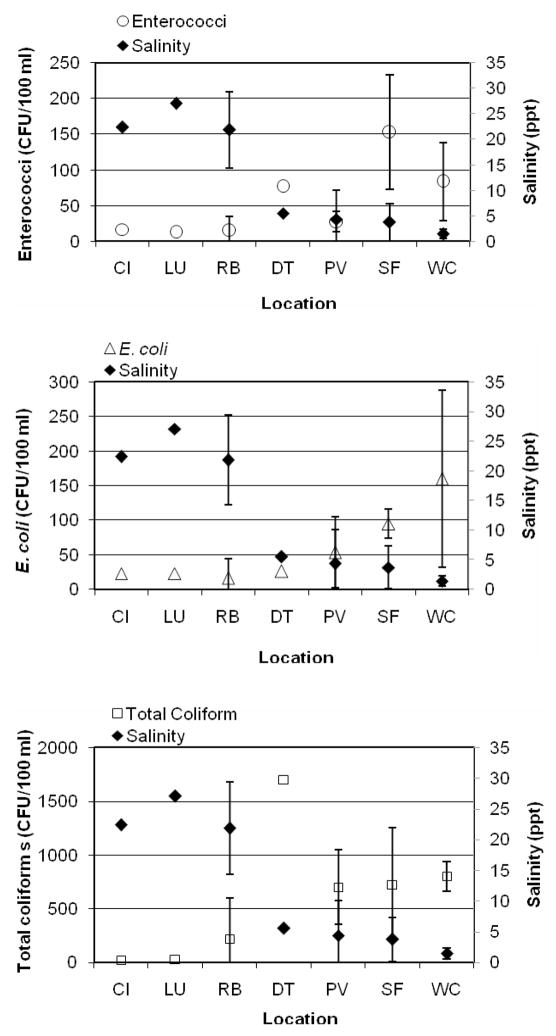
A common characteristic of sites WC, SF, and DT is that they are located at a larger distance from the junction of the North and South Forks; therefore they have reduced access to ocean water mixing and consequently, relatively lower salinities. None of the more saline sites located closer to this junction of the forks (CL, LU, and RB) exceeded regulatory limits, and viral indicator levels were also relatively low (< 2 PFU/100 mL on average). Overall the concentration of microbes was generally higher in those inland sites where salinity was low (Figure 2).
Fig. 2.
Salinity and enterococci (top), E. coli (middle), total coliforms (bottom) sorted by site. Error bars correspond to standard deviations for sites with more than one data point.
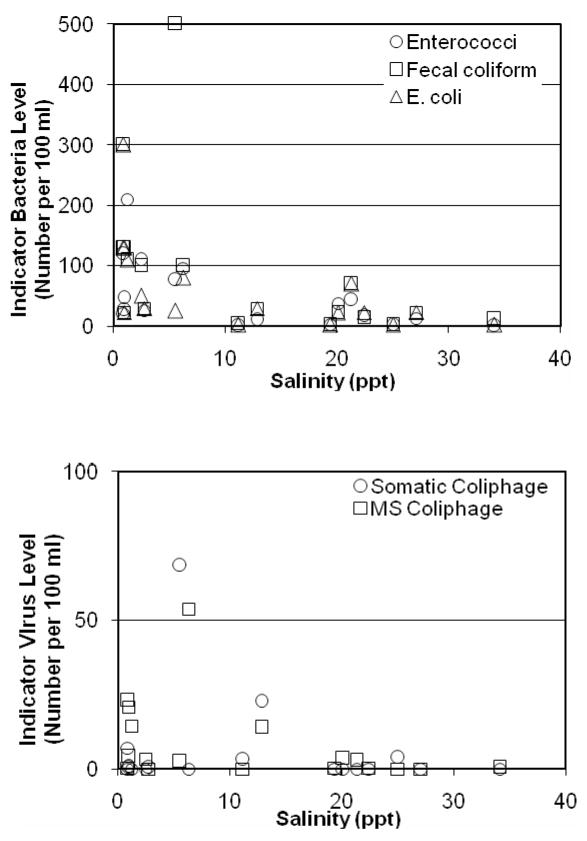
When comparing indicators, the strongest correlations were observed between fecal coliform and total coliform (R = 0.76), and fecal coliform and somatic coliphage (R = 0.75) (Table 3). Other notable correlations between indicators include total coliform and somatic coliphage (R = 0.59), total coliform and enterococci (R = 0.56), enterococci and E. coli (R = 0.53). Of the physical-chemical parameters measured in this study only one, salinity, showed significant negative correlations with indicator microbes (Figure 3). Specifically, correlations were observed between salinity and total coliform (R = −0.65), E. coli (R = −0.54), and enterococci (R = −0.56).
Table 3.
Correlation (R) values between parameters measured.
| pH | Temp | Salinity | Turbidity | Total coliform |
Fecal coliform |
E. coli | Enterococci | Somatic coliphage |
MS coliphage |
|
|---|---|---|---|---|---|---|---|---|---|---|
| pH | 1 | |||||||||
| Temp | −0.19 | 1 | ||||||||
| Salinity | −0.11 | −0.20 | 1 | |||||||
| Turbidity | −0.21 | −0.03 | −0.03 | 1 | ||||||
| Total coliform | −0.09 | 0.38 | −0.65 | 0.04 | 1 | |||||
| Fecal coliform | −0.10 | 0.12 | −0.46 | −0.07 | 0.76 | 1 | ||||
| E. coli | 0.01 | −0.07 | −0.54 | 0.14 | 0.37 | 0.48 | 1 | |||
| Enterococci | −0.28 | −0.03 | −0.56 | 0.31 | 0.56 | 0.45 | 0.53 | 1 | ||
| Somatic coliphage | −0.13 | 0.19 | −0.15 | −0.20 | 0.59 | 0.75 | −0.13 | 0.04 | 1 | |
| MS coliphage | 0.37 | −0.42 | −0.37 | −0.07 | 0.01 | 0.15 | 0.37 | 0.47 | −0.09 | 1 |
Fig. 3.
Indicator bacteria (top) and indicator virus (bottom) versus salinity.
3.2 Statistical Analysis of Temperature and Salinity
The 18 individual measurements were split into two groups, one set of 9 measurements characterized by warmer water temperatures (>28.7 °C) and a second set of 9 measurements characterized by cooler temperatures (<23.6 °C). No statistical differences were observed in the indicator microbe levels between each of these sets of data (p value of 0.46 for enterococci, 0.49 for E. coli, 0.27 for fecal coliform, 0.44 for somatic coliphage, and 0.16 for MS coliphage).
Similarly, the 18 individual measurements were split in terms of salinity, with one set of 9 characterized by salinity values less than 6.3 ppt and a second set of 9 characterized by salinity values greater than 11.3 ppt. In this case the indicator bacteria levels were significantly different between the groups (p value of < 0.01 for enterococci, p = 0.01 for E. coli, and p = 0.01 for fecal coliform). The indicator viruses however, were not statistically different between the two groups at 95% confidence limits (p value of 0.06 for MS2 coliphage and p = 0.27 for somatic coliphage).
3.3 Pathogens
Viral and protozoan pathogens measured in the study were below detection limits (<0.5 MPN/100L and <1.6 to <9.6 cysts and oocysts/100 L respectively), with the exception of one sample. This sample (WC collected on June 25, 2007) was positive (8.5 MPN/100L) for culturable virus. Confirmatory tests indicated that the culturable virus was reovirus.
Notable physical-chemical parameters for the sample that measured positive for reovirus included low salinity (< 1ppt) and high water temperature (31 °C). Turbidity was relatively low (3.5 NTU) and ambient water pH was within normal range (7.5 units). Indicator bacteria (total coliform – 700 MPN/100 mL, fecal coliform – 130 MPN/100 mL, E. coli – 130 MPN/100 mL, and enterococci – 21 CFU/100 mL) and indicator virus levels (somatic coliphage – 7 PFU/100 mL, MS coliphage – 0.3 PFU/100 mL) were within average values as typically measured for the entire system and were relatively low for the low salinity sites measured in this study. There were no extraordinary observations associated with WC except that it measured positive for culturable virus.
4. Discussion
Bacterial indicator levels (101 to 102 CFU or MPN/100 ml), viral indicators (< 102 PFU/100 ml), and pathogens (no detects for protozoa and one detect for virus) measured were within ranges reported for ocean waters (≤103/100 ml, ≤102/100 ml, and <102/100 L respectively) (Griffin et al. 1999, Muscillo et al. 1997, Katayama et al. 2004, Sobsey et al. 2003, Lipp et al. 2001, Wetz et al. 2004, Fan et al. 2001) and generally lower than those reported for river water (≤104/100 ml, ≤104/100 ml, ≤104/100 L respectively) (Obi et al. 2003, Lodder and Husman 2005, Westrell et al. 2006, Tani et al. 1992, Hachich et al. 2004). Although the literature may be skewed towards measurements in impacted areas (as generally monitoring efforts focus on such sites), the results nevertheless suggest that the St. Lucie River Estuary is characterized by normal indicator levels for waters within urban areas. The positive detect for culturable reovirus, however, merits more discussion and investigation. This virus is found in sewage and in animal fecal waste and can cause illness in humans—more specifically, it affects the gastrointestinal system and respiratory tract— (McConnell et al. 1984, Norman and Lee 1987).
Overall, results suggest that salinity relates to microbial contamination. Sites characterized by lower salinities were, in general, also characterized by higher levels of bacterial indicators. This association was supported by correlation analysis, which showed that, among all of the physical-chemical parameters evaluated, salinity was most strongly correlated with bacterial indicators. Similar observations associating lower salinity with higher levels of indicator bacteria have been documented in the literature (He et al. 2007, Goyal et al. 1977). However, laboratory based studies focusing on the impacts of salinity on microbe survival have shown that increased salinity does not impact fecal indicator bacteria or virus survival rates (Borst and Selvakumar 2004, Gantzer et al. 1998), but it does increase the inactivation of Giardia cysts (Johnson et al. 1997). The implications of salinity for fecal indicator microbes observed in this study may be due to a greater contribution of sources within upstream/fresher waters. In other words, sites may simply coincide with the primary sources of fecal indicator microbes, with the impacts of salinity on survival rates playing a secondary role. Further sampling within this and other tropical and subtropical estuaries is recommended in order to establish a better understanding of the significance of correlations between microbial contamination and salinity levels.
Of particular significance is that for the only sample that measured positive for culturable virus, the indicator bacterial levels (enterococci 21 CFU/100 ml, E. coli 130 MPN/100 ml, fecal coliform 130 MPN/100 ml) were within regulatory limits for single sample analysis, adding to the extensive literature that reveals a lack of correlation between fecal indicator bacteria and pathogens within waterways affected exclusively by non point sources. The indicator virus levels within the site (7 PFU/100ml somatic coliphage, 0.3 PFU/100ml MS2 coliphage) were also relatively low, especially in comparison to measurements from other low salinity samples. If the reovirus contamination was caused by recent community-scale sewage contamination, indicators should have also been elevated. In this case, indicators may have died-off more rapidly, with the virus persisting in the environment. Another possible explanation could be that the source of the reovirus was limited to a single or a few individuals whose fecal bacteria contributions were outnumbered by the reovirus released (e.g. in the case of localized individual fecal releases). In any case, there was a lack of correlation between pathogen presence and fecal indicator levels. The only common factor between sites characterized by elevated fecal indicators levels and pathogen presence was low salinity at upstream locations potentially due to reduced mixing of the freshwater with the ocean water. The elevated indicator levels do not necessarily coincide in time with the presence of pathogens.
Further work is recommended for this river estuary to identify the source of reovirus and indicator bacteria/viruses. A sanitary survey should be conducted including an evaluation of local and upstream sources, as some evidence suggests the source may be removed from the actual sampling site (due to the relatively low levels of indicators associated with the sample that was positive for reovirus). The fact that one sample in 18 tested positive for reovirus suggests that the source of culturable virus is intermittent, adding to the complexity of source identification and characterization. The recurrence of this intermittent source should be further evaluated with a focus on measurements at WC and other more inland, less saline, sites.
A risk assessment is also recommended in order to better establish the human health significance of the positive reovirus measurement. This risk assessment would likely require establishing the range and recurrence, if any, of enteric virus through additional measurements. The risk assessment, if pursued, should be put within the context of the intended use of the site. The WC site is used predominantly for boating activities. Such use will likely allow for higher acceptable levels of virus and bacteria in comparison to a site used predominantly for swimming.
Acknowledgments
We would like to thank the Florida Department of Environmental Protection for financial, logistical, and administrative support for this project. In particular we would like to thank Darrel Graziani and Timothy Gray for their administrative and field sampling efforts. We also would like to thank St. Lucie and Martin County Departments of Health for logistical support with respect to delivering samples for bacteria analysis. This project was also supported financially by the University of Miami NSF-NIEHS OHH Center (NSF OCE0432368 and NIEHS P50 ES12736-01), and the NSF-REU Program (OCE 0432368) for additional financial contributions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- American Public Health Association. Standard methods for the examination of water and wastewater. 21. American Public Health Association; Washington, D.C: 2005. [Google Scholar]
- Anderson SA, Turner SJ, Lewis GD. Enterococci in the New Zealand environment: Implications for water quality monitoring. Water Science and Technology. 1997;35:325–331. [Google Scholar]
- Borst M, Selvakumar A. Microorganism Die-Off Rates Under Various Conditions. USEPA Science Forum; Washington, DC: 2004. Retrieved 8/13/2008 from http://www.epa.gov/ORD/NRMRL/scienceforum/selvakumar_a.pdf. [Google Scholar]
- Brookes JD, Antenucci J, Hipsey M, Burch MD, Ashbolt NJ, Ferguson C. Fate and transport of pathogens in lakes and reservoirs. Environ Int. 2004;30:741–759. doi: 10.1016/j.envint.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Desmarais TR, Solo-Gabriele HM, Palmer CJ. Influence of Soil on Fecal Indicator Organisms in a Tidally Influenced Subtropical Environment. Appl Environ Microbiol. 2002;68:1165–1172. doi: 10.1128/AEM.68.3.1165-1172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmir SM, Wright ME, Solo-Gabriele HM, Abdelzaher A, Fleming LE, Miller G, Rybolowik M, Shih M-TP, Pillai SP, Cooper JA, Quaye EA. Quantitative Evaluation of Bacteria Released by Bathers in a Marine Water. Water Research. 2007;41:3–10. doi: 10.1016/j.watres.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Xiao-Jun, Chen Pei-Tang, Chen Cheng-Zhang, Albinet F. Investigation on the occurrence of giardia and cryptosporidium in water supply system and sea water in Macao region. China Water & Wastewater. 2001;17(11):32. [Google Scholar]
- Fujioka RS, Sian-Denton C, Borja M, Castro J, Morphew K. Soil: the environmental source of Escherichia Coli and Enterococci in Guam’s stream. J Appl Microbiol Sympos Suppl. 1999;85:83S–89S. doi: 10.1111/j.1365-2672.1998.tb05286.x. [DOI] [PubMed] [Google Scholar]
- Gantzer C, Dubois E, Crance J, Billaudel S, Kopecka H, Schwartzbrod L, Pommepuy M, Le Guyader F. Influence of environmental factors on the survival of enteric viruses in seawater. Oceanologica Acta. 1998;21(6):983–993. [Google Scholar]
- Graczyk TK, Sunderland D, Tamang L, Shields TM, Lucy FE, Breysse PN. Quantitative Evaluation of the Impact of Bather Density on Levels of Human-Virulent Microspordian Spores in Recreational Waters. Applied Environ Microbiology. 2007;73:4095–4099. doi: 10.1128/AEM.00365-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin Dale W, Gibson Charles J, III, Lipp Erin K, Riley Kelley, Paul John H, III, Rose Joan B. Detection of viral pathogens by reverse transcriptase PCR and of microbial indicators by standard methods in the canals of the Florida keys. Applied and Environmental Microbiology. 1999 September;65(9):4118–4125. doi: 10.1128/aem.65.9.4118-4125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachich EM, Sato MIZ, Galvani AT, Menegon JRN, Mucci JLN. Giardia and Cryptosporidium in source waters of Sao Paulo State, Brazil. Water Science and Technology. 2004;50(1):239–245. [PubMed] [Google Scholar]
- Hardina CM, Fujioka RS. Soil: the environmental source of E. coli and enterococci in Hawaii’s streams. Env Toxicol Water Quality. 1991;6:185–195. [Google Scholar]
- He L-M, Lu J, Shi W. Variability of fecal indicator bacteria in flowing and ponded waters in southern California: Implications for bacterial TMDL development and implementation. Water Research. 2007;41(14):3132–3140. doi: 10.1016/j.watres.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Howell JM, Coyne MS, Cornelius PL. Effect of Sediment Particle Size and Temperature on Fecal Bacteria Mortality Rates and the Fecal Coliform/Fecal Streptococci Ratio. Journal of Environ Qual. 1996;21:1216–1220. [Google Scholar]
- Indest K. Interim guidance on assessing the risk posed by pathogens associated with dredged material. 2003 (No. ERDC/TN EEDP-01-49) [Google Scholar]
- Johnson D, Enriquez C, Peper I, Davis T, Gerba C, Rose J. Survival of Giardia, Cryptosporidium, poliovirus, and Salmonella in marine waters. Wat Sci Technol. 1997;35(11–12):p 261–268. [Google Scholar]
- Katayama H, Okuma K, Furumai H, Ohgaki S. Series of surveys for enteric viruses and indicator organisms in Tokyo Bay after an event of combined sewer overflow. Water Science and Technology. 2004;50(1):259–262. [PubMed] [Google Scholar]
- Lipp EK, Farrah SA, Rose JB. Assessment and impact of microbial fecal pollution and human enteric pathogens in a coastal community. Marine Pollution Bulletin. 2001;42(4):286–293. doi: 10.1016/s0025-326x(00)00152-1. [DOI] [PubMed] [Google Scholar]
- Lodder WJ, De Roda Husman AM. Presence of noroviruses and other enteric viruses in sewage and surface waters in The Netherlands. Applied and Environmental Microbiology. 2005 March;71(3):1453–1461. doi: 10.1128/AEM.71.3.1453-1461.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell LK, Sims RC, Barnett BB. Reovirus Removal and Inactivation by Slow-rate Sand Filtration. Appl Environ Microbiol. 1984 October;48(4):818–825. doi: 10.1128/aem.48.4.818-825.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscillo Michele, Carducci Annalaura, La Rosa Giuseppina, Cantiani Laura, Marianelli Cinzia. Enteric virus detection in Adriatic seawater by cell culture, polymerase chain reaction and polyacrylamide gel electrophoresis. Water Research. 1997 Aug;31(8):1980–1984. [Google Scholar]
- Norman Kara L, Lee Patrick WK. Reovirus as a novel oncolytic agent. Journal of Clinical Investigation. 2000 April;105(8):1035–1038. doi: 10.1172/JCI9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obi C Larry, Potgieter N, Bessong PO, Matsaung G. Scope of potential bacterial agents of diarrhoea and microbial assessment of quality of river water sources in rural Venda communities in South Africa. Water Science and Technology. 2003;47(3):59–64. [PubMed] [Google Scholar]
- Obiri-Danso K, Jones J. Intertidal sediments as reservoirs for hippurate negative camplybacters, salmonellae and faecel indicators in three EU recongnized bathing waters in North West England. Water Research. 2000;34:519–527. [Google Scholar]
- Papadakis JA, Mavridou A, Richardson SC, Lampiri M, Marcelou U. Bather-Related Microbial and Yeast Populations in Sand and Seawater. Water Research. 1997;31:799–804. [Google Scholar]
- Rogerson A, McCorquodale D, Esiobu N. Prevalence and survival of microorganisms in shoreline interstitial waters: a search for indicators of health risks. (EPA # 828830) 2003 Final Report—November 2003. [Google Scholar]
- Roll BM, Fujioka RS. Sources of faecal indicator bacterica in a brackish, tropical stream and their impact on recreational water quality. Water Science and Technology. 1997;35:179–186. [Google Scholar]
- Goyal Sagar M, Gerba Charles P, Melnick Joseph L. Occurrence and distribution of bacterial indicators and pathogens in canal communities along the texas coast. Applied And Environmental Microbiology. 1977 Aug;:139–149. doi: 10.1128/aem.34.2.139-149.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer BM, Miner JR, Moore JA, Buckhouse JC. Indicator bacteria survival in stream sediments. Journal of Environmental Quality. 1992;21:591–596. [Google Scholar]
- Shibata T, Solo-Gabriele HM, Fleming LE, Elmir S. Monitoring marine recreational water quality using multiple microbial indicators in an urban tropical environment. Water Research. 2004;38:3119–3131. doi: 10.1016/j.watres.2004.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobsey MD, Perdue R, Overton M, Fisher J. Factors influencing faecal contamination in coastal marinas. Water Science and Technology. 2003;47(3):199–204. [PubMed] [Google Scholar]
- Spinner ML, Di Giovanni GD. Detection and Identification of Mammalian Reoviruses in Surface Water by Combined Cell Culture and Reverse Transcription-PCR. Applied and Environmental Microbiology. 2001;67(7):3016–3020. doi: 10.1128/AEM.67.7.3016-3020.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani N, Shimamoto K, Ichimura K, Nishii Y, Tomita S, Oda Y. Enteric virus levels in river water. Water Research. 1992 Jan;26(1):45–48. [Google Scholar]
- Westrell Therese, Teunis Peter, van den Berg Harold, Lodder Willemijn, Ketelaars Henk, Stenstrom ThorAxel, de Roda Husman Ana Maria. Short- and long-term variations of norovirus concentrations in the Meuse river during a 2-year study period. Water Research. 40:2613–2620. doi: 10.1016/j.watres.2006.05.019. [DOI] [PubMed] [Google Scholar]
- U.S. Environmental Protection Agency. Quality Criteria for Water. Washington D.C: U.S. Environmental Protection Agency; 1976. EPA-440976023. [Google Scholar]
- U.S. Environmental Protection Agency. Ambient water quality criteria for bacteria. Washington D.C: U.S. Environmental Protection Agency; 1986. EPA A440/5-84-002. [Google Scholar]
- U.S. Environmental Protection Agency. USEPA ICR microbial laboratory manual. U.S. Environmental Protection Agency; Washington, D.C: 1996. EPA/600/R-95/178. [Google Scholar]
- U.S. Environmental Protection Agency. Method 1623: Cryptosporidium and Giardia in Water by Filtration/IMS/FA. U.S. Environmental Protection Agency; Washington, D.C: 2001. EPA-821-R-01-025. [Google Scholar]
- U.S. Environmental Protection Agency. Method 1602: Male-specific (F+) and Somatic Coliphage in Water by Single Agar Layer (SAL) Procedure. U.S. Environmental Protection Agency; Washington, D.C: 2001. EPA 821-R-01-029. [Google Scholar]
- U.S. Environmental Protection Agency. USEPA manual of methods for virology. U.S. Environmental Protection Agency; Washington, D.C: 2001. EPA 600/4-84/013. [Google Scholar]
- U.S. Environmental Protection Agency. Method 1600: Membrane filter test method for enterococci in water. U.S. Environmental Protection Agency; Washington, D.C: 2002. EPA-821-R-02-022. [Google Scholar]
- U.S. Geological Survey. Microbiology and public beach safety: integrated science for the protection of public health. 2006a. FS 2006-3045 ed. [Google Scholar]
- U.S. Geological Survey. Pathogen exposure through recreational water. 2006b Retrieved 10/18/2007 from http://health.usgs.gov/pathogens.
- Wetz Jennifer Jarrell, Lipp Erin K, Griffin Dale W, Lukasik Jerzy, Wait Douglas, Sobsey Mark D, Scott Troy M, Rose Joan B. Presence, infectivity, and stability of enteric viruses in seawater: Relationship to marine water quality in the Florida Keys. Marine Pollution Bulletin. 2004 April;48(7–8):698–704. doi: 10.1016/j.marpolbul.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Whitman RL, Nevers MB. Foreshore Sans as a Source of Escherichia coli in Nearshore Water of a Lake Michigan Beach. Applied and Environmental Microbiology. 2003;69:5555–5562. doi: 10.1128/AEM.69.9.5555-5562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]