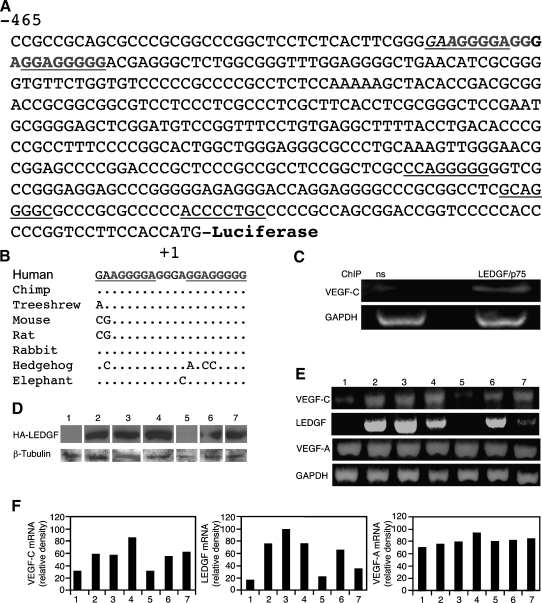
Figure 1.
The VEGF-C gene contains putative LEDGF/p75 binding sites. (A) Nucleotide sequence of the 468-bp 5′-flanking region of human VEGF-C gene. The nucleotide sequences are numbered in relation to the ATG, which is designated “+1” R. Underlining indicates an STRE, the putative LEDGF binding site. (B) Conservation among species of a putative binding unit for LEDGF was identified using the conservation track of the UCSC genome browser, and the positions of mismatches are indicated. The binding site runs from -426 to -407 (human chr4:177,950,885-177,950,866). (C) ChIP assays were performed using specific primers for the 468-bp VEGF-C promoter (A) and nonspecific antibodies (ns) or anti-LEDGF/p75 antibodies to demonstrate specific binding of LEDGF/p75 in H1299 cells. Evaluation of total genomic DNA was carried out with GAPDH primers. (D) H1299 human lung cancer cells stably transfected either with a control pIRES vector or with a construct encoding rat LEDGF tagged with an influenza virus hemagglutinin epitope (pIRES-HA-LEDGF). Puromycin-resistant pools of cells and individual clones were selected from each of the lines. Immunoblot assay of LEDGF expression levels of puromycin-resistant stably transfected pools (lanes 1 and 2) and individual clones (clones 3–7) using anti-HA antibodies. Protein amounts were quantified using anti-β-tubulin antibodies. (E) RT-PCR analysis of the expression of human VEGF-C, VEGF-A, GAPDH, and transfected rat LEDGF genes was carried out on total RNA extracted from the previously mentioned stably transfected pools and individual clones. (F) Relative densities of the scanned bands normalized against GAPDH.