Abstract
Localized prostate cancer (CaP) can be cured using several strategies. However, the need to identify active substances in advanced tumor stages is tremendous, as the outcome in such cases is still disappointing. One approach is to deliver human tumor antigen-targeted therapy, which is recognized by T cells or antibodies. We used data mining of the Cancer Immunome Database (CID), which comprises potential immunologic targets identified by serological screening of expression libraries. Candidate antigens were screened by DNA microarrays. Genes were then validated at the protein level by tissue microarrays, representing various stages of CaP disease. Of 43 targets identified by CID, 10 showed an overexpression on the complementary DNA array in CaP metastases. The RHAMM (CD168) gene, earlier identified by our group as an immunogenic antigen in acute and chronic leukemia, also showed highly significant overexpression in CaP metastases compared with localized disease and benign prostatic hyperplasia. At the protein level, RHAMM was highest in metastatic tissue samples and significantly higher in neoplastic localized disease compared with benign tissue. High RHAMM expression was associated with clinical parameters known to be linked to better clinical outcome. Patients with high RHAMM expression in the primaries had a significantly lower risk of biochemical failure. The number of viable cells in cell cultures was reduced in blocking experiments using hormone-sensitive and hormone-insensitive metastatic CaP cell lines. Acknowledging the proven immunogenic effects of RHAMM in leukemia, this antigen is intriguing as a therapeutic target in far-advanced CaP.
Introduction
In western countries, the prevalence of prostate cancer (CaP) is high and has been seen to increase with age. One in six men will have their conditions diagnosed with CaP during their lifetime, and thus, CaP is the leading cause of male cancer-related death, second only to lung cancer [1–3]. Prostate cancer is curable when the tumor is localized to the prostate gland and when treated at an early stage. Unsuspected extracapsular or metastatic disease may significantly increase the risk of primary treatment failure. This risk is especially high if the patient has one or more of the following risk factors: a serum prostate-specific antigen (PSA) level greater than 20 ng/ml, a Gleason score higher than 7, locally advanced disease (clinical stage T3/T4), and extensive disease on prostate biopsy [4].
New therapeutic approaches for localized CaPs and for locally advanced or metastatic diseases have been proposed for research. Nowadays, the primary concept in metastatic CaP is control of tumor growth based on hormonal withdrawal. Luteinizing hormone-releasing hormone agonists act the same way as orchidectomy; other drugs are peripheral inhibitors of androgens. The role of androgen deprivation by orchidectomy is eclipsed by the medicamentous suppression of androgen production. If resistance occurs, tumor cells show an androgen-independent progression. Current systemic therapies with proven efficacy are limited to docetaxel-containing chemotherapy. In combination with low-dose prednisone, docetaxel is recommended for patients with metastatic hormone-refractory CaP (HRPC). This regimen showed a limited survival benefit and is often limited by toxicity in the elderly [5]. The median survival of patients with HRPC is still between 18 and 20 months [6]. Despite new promising results of platin-based chemotherapy (satraplatin) [7–11], it is crucial to intensify research in targeted therapy. At present, advanced CaP is an incurable disease and treatment objective focuses on palliation.
Consequently, the development of new treatment modalities especially for metastatic or locally advanced CaP is important. The development of low-toxicity therapeutic options in HRPC is an unmet medical need.
Novel targets and therapeutic approaches in advanced CaP have changed during the last decade. Some naturally occurring substances, such as retinoids or butyrates, show effects on CaP [12], which can induce cell cycle arrest, differentiation, and apoptosis in many tumor cell types, while having a favorable safety profile in humans [13]. New therapeutic options such as nucleotid-based targeted therapies, small-molecule inhibitors [14–17], antiangiogenic agents such as sunitinib [18–21], and novel cytotoxic therapeutics such as satraplatin [8,9,11] were developed that show promise in the management of patients with advanced CaP and are in clinical trials or approved. Likewise, new-generation expression profile tools give better insight into CaP signatures and unclose potential new targets [22].
Another approach illuminates the immunologic site of cancer [23–26]. In other malignancies, such as breast cancer and melanoma, the immunologic approach has shown encouraging results. Immunotherapeutical approaches are of interest in advanced stages of CaP. They are also used in adjuvant setting after curative therapy, when some tumor load is still present, and hormonal manipulation is used to reduce the risk of recurrent disease.
Database analysis using Cancer Immunome Database (CID)1 yields numerous tumor-associated antigens (TAAs) in different kinds of cancer. Similar to other targeted therapies, an immunomodulatory approach could deploy CaP-specific characteristics. Recent studies have shown autoantibodies against peptides derived from CaP tissue that could be used as basis for a screening test [27,28]. New immunologic approaches could also deliver promising therapeutic targets in the future. A potential target in CaP called the receptor of hyaluronan-mediated motility (RHAMM, also known as CD168 or HMMR) has been identified. This had been described earlier as an immunogenic antigen in acute and chronic leukemia [29] and breast cancer [30] and was shortly reported in a pathway with androgen receptor in CaP [31,32]. The potential use of RHAMM as target for an immunologic therapy is presented in phase 1 studies including hematological diseases such as acute myeloid leukemia, myelodysplastic syndrome, and multiple myeloma. The study performed by Schmitt et al. [33] shows RHAMM as a promising target by observed immunologic and clinical response using RHAMM R3 vaccination.
Every new systemic approach should mandate that new targets are selectively overexpressed in tumor cells to minimize the adverse effects of knockdown, such as CML 28 [34] or RHAMM [31,32] for an immunologic approach. A number of recently developed approaches to antitumoral immunotherapy are being investigated as potential treatments for advanced CaP [35].
On the basis of published experience of our coworkers, we set out in this study to investigate the potential immunogenic role of RHAMM in patients with CaP disease in the clinical setting.
Materials and Methods
Sample Collection
Clinical samples were provided from the radical prostatectomy series and Rapid Autopsy Program at the University of Michigan. The specimens were processed within 20 minutes after surgical resection. Alternate sections of the prostate gland were submitted for histologic review. The remaining sections were snap-frozen and stored. All samples used for complementary DNA (cDNA) expression array analysis and Western Blot analysis were evaluated by the study pathologists. Clinical and pathology data for all patients were acquired with approval from the institutional review board at the University of Michigan and maintained on a secure relational database [36]. The Rapid Autopsy Program, which allowed men with advanced CaP to consent to an autopsy immediately after death, was developed. This procedure has previously been described in detail [37].
Primary CaP samples, as well as lymph node metastases, were contributed from the University of Ulm, Germany, for the construction of tissue microarrays (TMAs) [38].
Identification of Potential Immunologic Targets
Data mining in CID-related databases was performed to identify potential immunologic target structures. Consequently, candidate antigens were screened by their own cDNA microarrays [39–41] and cross-checked using expression profile databases of several research groups that are available for the research community (www.oncomine.org) [42,43].
cDNA Array Expression Profile
RNA was isolated from 71 prostate tissue samples representing benign prostatic hyperplasia, localized CaP, and metastatic CaP. The construction of the cDNA microarrays has been previously explained [39,44]. In brief, plasmid templates for a maximum of 20K transcripts were isolated from bacterial clones and inserts amplified by polymerase chain reaction. Purified polymerase chain reaction fragments were printed onto glass slides and cross-linked with the DNA targets. cDNA generated from localized CaP samples and metastatic tissue samples were labeled with a distinguishable fluorescent dye and hybridized to the cDNA microarray. The cDNA microarray was analyzed using a scanner, and fluorescence ratios were determined for each gene. Color intensities were converted into ratios of gene expression. These ratios were imported into a database for further analysis.
Tissue Microarray
Formalin-fixed, paraffin-embedded tissue blocks with tumor samples from CaP and lymph node metastases were selected. Each case was identified using patient reports as part of this institutional review board-approved project (University of Ulm and University of Michigan). One pathologist (M.A.R.) reviewed the hematoxylin and eosin-stained slides and was responsible for circling areas of CaP, metastases or benign prostate, which were then used as template for the TMA.
To take tumor heterogeneity into account, a minimum of three TMA cores (0.6 mm in diameter) were sampled from each donor block using a validated sampling method [45].
TMAs were constructed using a manual tissue arrayer (Beecher Instruments, Silver Spring, MD) as previously described [46] with a total of approximately 900 tissue cores from 15 benign prostate samples, 220 primary CaP, and 47 lymph node metastases. For tissue processing, standard biotin-avidin complex immunohistochemistry was performed using a RHAMM antibody (E-19; sc-16170; Santa Cruz Biotechnology, Santa Cruz, CA). Staining intensity was determined by the semiautomated quantitative image analysis system (ACIS II; Clarient Chromavision Medical Systems, San Juan Capistrano, CA) [47]. The output data of RHAMM expression are represented by a continuous scale of 3,3′-diaminobenzidine (DAB) brown area and average DAB staining intensity for each core, ranging from 0 to 255. The RHAMM expression intensity of each array was normalized by calculating the z-score for each core. The z-score normalization functions to equalize the immunohistochemical experiments on different TMAs because of the variability of staining. The converted z-scores were then aggregated into one large data set. Tissue cores of some patients were represented on more than one TMA, and after combining all z-values for each patient, the mean z-score intensity of DAB staining for each patient was determined.
Cell Lines and Cell Cultures
In cell culture experiments, hormone-sensitive (LNCaP) and hormone-independent metastatic (PC3 and DU145) cancer cell lines were used. The cell lines were cultured in RPMI 1640 (PromoCell, Heidelberg, Germany) including 5% FBS.
Growth Inhibition and Cell Viability Assay
LNCaP (5 x 103), PC3 (2 x 103), and DU145 (4 x 103) cells were grown in microtiter plates and treated with increasing concentrations (0, 5, and 10 µg/ml) of RHAMM-specific antibody E-19 (sc-16170; Santa Cruz Biotechnology) on day 1 for 72 hours. According to the manufacturer's information, on day 4, growth inhibition was correlated to absorbance determined using the XTT assay Cell Proliferation Kit (Roche Applied Science, Mannheim, Germany). The control was set to 100%. The percentage of viable cells as mean values and SE is given for repeated experiments.
Statistical Analysis
The RHAMM protein expression was evaluated using the mean score result for each prostate tissue type per patient. To test for significant differences, analysis of variance (ANOVA) was performed. To determine association with clinical parameters, Mann-Whitney U tests were performed. Statistical analyses were performed using SPSS (SPSS, Inc, Chicago, IL). P values ≤ .01 were considered statistically significant.
Kaplan-Meier analysis was performed for RHAMM protein expression in the primary localized CaP and its association with biochemical recurrence-free survival.
Results
CID and cDNA Analysis
CID data mining and own expression cloning experiments identified 43 genes with potential humoral response to CaP. Of 43 the CID identified targets, 10 showed an overexpression on the Michigan cDNA 20K chip in CaP metastases in compared with localized disease and benign prostatic hyperplasia (Table 1). One of these targets was RHAMM.
Table 1.
Overexpression on the 20K cDNA Array of Targets and Corresponding P Values in CaP Metastases Compared with Localized Disease and Benign Prostatic Hyperplasia.
| Target | ANOVA | Post hoc (Scheffé) | |
| Benign Prostate Tissue vs CaP Metastases | Localized CaP vs CaP Metastases | ||
| BAP1 | .094 | NS | NS |
| BRCA1 | .156 | NS | NS |
| PSG2 | .002 | <.01 | <.05 |
| MPV17 | .002 | <.01 | <.05 |
| Stau2 | .001 | .01 | <.01 |
| Mark3 | .025 | NS | <.05 |
| MCM3 | .019 | NS | <.05 |
| PMSCL2 | .010 | NS | .01 |
| MAPK3 | .109 | NS | NS |
| RHAMM | .004 | <.05 | .01 |
For statistical analysis, a multifactorial ANOVA was performed.
Group factors: expression in benign prostate tissue samples, localized CaP, and CaP metastases. Significance was approved with post hoc tests (Scheffé). On cDNA level, no significant differences between localized CaP and benign prostate tissue were found.
NS indicates not significant.
On the basis of previous group experience and the reported value of RHAMM in acute and chronic leukemia [29], as well as in breast cancer [30], we focused our study on RHAMM. Own cDNA analysis showed highly significant overexpression of RHAMM in CaP metastases compared with localized disease and benign prostate tissue (Figure 1). Data mining in Oncomine (www.oncomine.org) yielded a similar expression profile with an overexpression of RHAMM in other platforms. Yu et al. [48] found an overexpression of RHAMM in metastatic CaP compared with normal prostate tissue or prostate carcinomas (P < .001). An overexpression of RHAMM on messenger RNA level in CaP compared with normal prostate tissue was also shown by La Tulippe et al. (P < .001) [49].
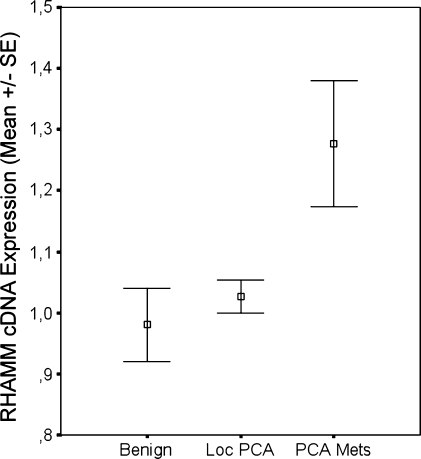
Figure 1.
cDNA microarray expression analysis of RHAMM in benign prostatic tissue (Benign), localized (Loc PCA), and metastatic CaP specimen (PCA Mets). Significant overexpression of RHAMM in metastases compared with localized prostatic cancer (P = .007) or benign prostate tissue (P < .001).
TMA for RHAMM (CD168)
For verification of RHAMM overexpression at the protein level, TMAs were constructed with approximately 900 individual tissue cores of localized CaP, CaP metastases, and benign prostatic hyperplasia. Immunohistochemistry using anti-RHAMM antibody (E-19) showed highest levels in metastatic tissue samples and significantly higher expression in neoplastic localized disease compared with benign tissue (P = .003). Staining intensity was determined with ACIS II as a marker for protein expression of RHAMM. Staining resulted in a deep brown color for high RHAMM protein levels as shown in Figure 2.
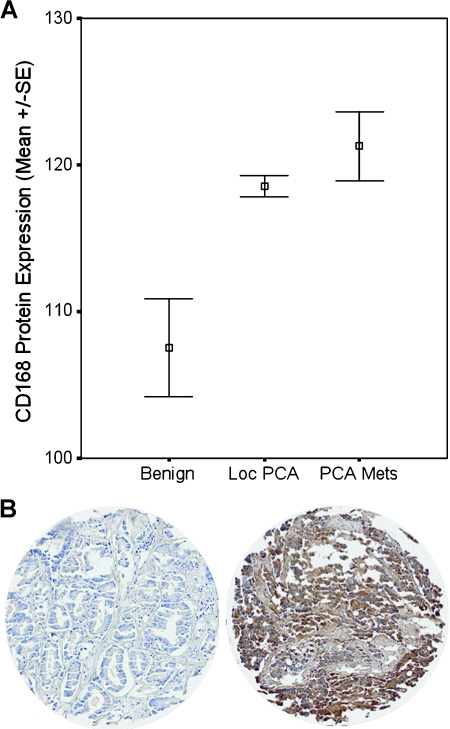
Figure 2.
(A) Mean protein expression of RHAMM in benign tissue, localized CaP, and metastatic tissue is shown. Significant higher protein levels of RHAMM were detected in localized CaP (P = .003) and metastases (P < .001) compared with benign prostatic hyperplasia. RHAMM was expressed higher in CaP metastases compared with protein levels in localized CaP, although this did not reach statistical significance. (B) Representative cores of low (left) and high RHAMM expression (right) in on the TMA.
Association of RHAMM Expression and Clinical Parameters
Within the significantly RHAMM-overexpressing primary tumor samples on the TMA, patients with higher RHAMM protein expression levels in primaries showed significantly lower risk of biochemical failure (Figure 3). In addition, a higher expression of RHAMM was associated with negative lymph node status, lower T stadium, and lower serum PSA at the time of diagnosis (Figure 4). These clinical parameters are also known to be associated with better outcome.
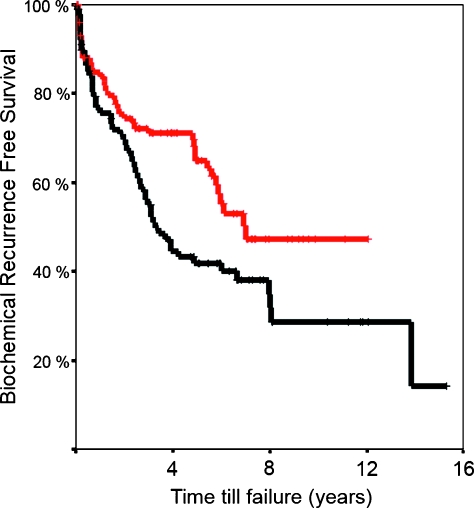
Figure 3.
Kaplan-Meier analysis showed significant lower risk of biochemical failure (log rank, P = .023; relative risk = 0.621) in patients with high RHAMM expression in primary localized CaP (red = high expression/black = low expression).
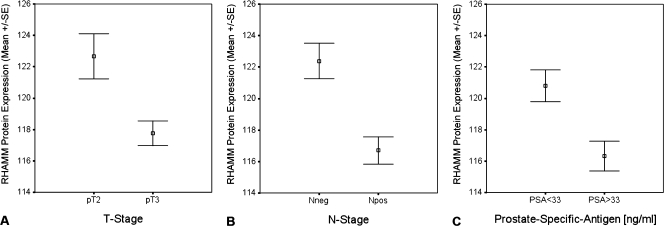
Figure 4.
Protein expression of RHAMM on the TMAs was analyzed for the association with clinical parameters such as tumor stage (T stage; A), lymph node status (N stage; B), and PSA (C) categorized using the mean PSA level as cutoff value. P values were calculated based on the Mann-Whitney U test. (A) There is a strong association between RHAMM protein expression and tumor stage defined as T2 and T3. Pathologic T3 tumors known to have worse outcome have a significant lower RHAMM expression (P = .01). (B) Lymph node-positive patients have a significant lower protein expression in the primaries compared with patients without lymph node metastases at the time of radical prostatectomy (P < .0001). (C) Patients with high PSA levels, known as an independent predictor for worse outcome, have a significantly lower protein expression of RHAMM (P < .001).
Growth Inhibition and Cell Viability
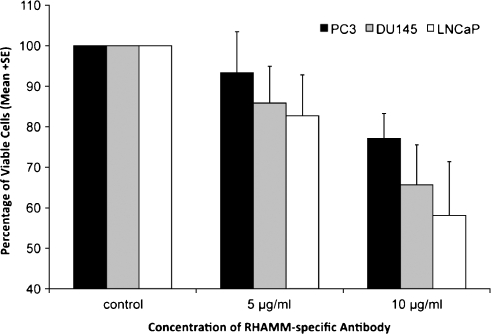
In cell culture experiments, the number of viable cells determined by XTT assay was reduced in blocking experiments using hormone-sensitive (LNCaP) and hormone-independent metastatic (PC3 and DU145) cancer cell lines. This was done by applying increasing concentrations of RHAMM-specific antibody (E-19). Higher antibody concentrations resulted in a reduced number of viable cells in all cell lines (Figure 5). The highest growth inhibition was found in LNCaP cells with 82.7% viable cells at a concentration of 5 µg/ml and 58.1% at 10 µg/ml of RHAMM-specific antibody.
Figure 5.
Decreasing numbers of viable cells determined by the XTT assay by exposure to increasing concentrations (5 and 10 µg/ml) of a RHAMM-specific antibody (E-19) in LNCaP, PC3, and DU145 cell lines. Reduced viable cells occurred with increasing antibody concentration. Relative cell amounts are shown. The control was set to 100% of viable cells.
Discussion
Initially, in this study, we used a reverse immunology approach to identify immunogenic structures on CaP. This was possible courtesy of the Ludwig Institute for Cancer Research, which provided open World Wide Web access to SEREX (serological identification of antigens by recombinant expression cloning)-defined TAAs. The proposal that humoral responses can occur only with T-cell help has been proven already in the first description of the SEREX method. This method also picked the T-cell-activating antigens tyrosinase and MAGE-1 by this serological screening approach [50]. Expression of the genes was rechecked with potential humoral responses for messenger RNA expression based on a 20K cDNA chip provided by our collaborators at the University of Michigan. The outcome resulted in 10 intersecting genes, which fulfilled both criteria. However, of these immunoreactive molecules, only a very few are suited as targets for immunotherapy.
There are several proteins within this list (Table 1), which play an important role in the physiological cell metabolism or contribute to the correct cell division such as MPV17 or MARK3, respectively [51,52]. This indicates that the presence of autoantibodies may result from unintended cell damage and suggests that this is rather a bystander effect rather than a true augmentable immune response against malignant cells. Thus, this observation gives rise to the question if this reverse immunology approach, which has been used, is valid for the identification of viable target antigens.
Conversely, autoantibody signatures might be useful in enhancing the specificity and sensitivity of serological screening tests. This was described by researchers from the University of Michigan, who found autoantibodies against α-methylacyl-CoA racemase in CaP patients and showed that the specificity and sensitivity of α-methylacyl-CoA racemase was superior to that of PSA, especially in those patients with intermediate PSA levels [53]. In a more recent attempt to use CaP-specific auto-antibodies as biomarkers in CaP patients, the same group developed a 22-phage peptide detector, which significantly added to the diagnostic power of PSA alone, and this might be important in patients exhibiting low PSA levels [27]. As a clinical impact, the number of unnecessary biopsies might be reduced.
RHAMM has been added to the list as a potentially immunogenic antigen in CaP for three reasons. First, it has been described as a TAA in the year 2002, picked also by a SEREX-derived method, and it displayed humoral responses in a wide range of tumors [29]. Second, it showed a favorable expression profile with the placenta, thymus, and testis, being the only normal tissues expressing this antigen. Finally, after the identification of immunogenic peptides, RHAMM R3 has made its way into a clinical phase 1 to 2 study in patients with hematological malignancies and led to immunologic effects in most patients and to some clinical benefit [33,54].
There are several aspects of RHAMM, which are of particular interest. RHAMM is a glycosaminoglycan and extracellular matrix molecule that plays an essential role in cell growth, differentiation, and motility. Overexpression of RHAMM is essential for ras-mediated transformation, and it is associated with the development of metastases [55]. Moreover, signaling through RHAMM and its downstream signal molecules, including ROK1, Gab-1, PI3K*p110a, and eIF4E3, particularly enhanced the progression of androgen-independent CaP cell lines in several aspects [32].
Therefore, immune therapies targeting RHAMM might target not only an immunogenic antigen but also a gene critically involved in cell cycle, differentiation, and proliferation [56].
The key point in the present study is the analysis of RHAMM expression on cDNA and protein levels in a large amount of samples. In our cDNA expression profiles, we found a highly significant overexpression of RHAMM in CaP metastases compared with localized CaP or benign prostate tissue. This finding was consistent with data revealed by data mining from several other microarray platforms accessible through the Oncomine database (www.oncomine.org) [42,43]. To confirm the Genomics data with Proteomics, we subjected an even larger cohort of 282 tissue samples from CaP patients to TMAs and again found a significant overexpression of RHAMM primary tumor samples and even higher in lymph node metastases, although it did not reach statistical significance. These data suggest that RHAMM is upregulated in the malignant phenotype and in disease progression, and it might additionally be involved in the development of metastases. These results are consistent to TMA data from Lin et al. [31,32], who also found an overexpression of RHAMM in samples of CaP compared with benign prostatic tissue. The same group impressively demonstrated the role of RHAMM downstream through the ROK-PI3K-eIF4E signaling cascade in clinical staging, cell proliferation, cell invasion, and metastasis of advanced CaP [32].
Overexpression of RHAMM seems not to be specific for CaP only because different studies have shown that RHAMM might be an interesting target in different kinds of solid tumors such as head and neck squamous cell carcinomas [57], breast cancer [30], or even colorectal cancer [58].
When comparing RHAMM protein expression with clinical data, we observed contradictory results at a first glance because further analysis in primary tumors revealed that higher expression levels are associated with a longer time window of biochemical recurrence-free survival. However, this observation could be explained by the concepts of immunosurveillance and immunoediting. The oncogenesis of an individual tumor is the result of a continuous interaction between potentially dividing malignant cells and host lymphocytes, which could display several states of responsiveness. In an immunoediting process, three phases of interaction have been proposed: tumor cell elimination, equilibrium, and tumor cell escape [59]. After removal of the primary tumor, potentially widespread single cells that express a significant amount of RHAMM peptides in the context of a surface major histocompatibility complex molecule and sufficient costimulatory molecules might be eliminated by the host defense, that is, by RHAMM-specific cytotoxic T lymphocytes. This could have led to sustained tumor cell elimination and subsequently to the prolonged biochemical failure-free survival.
Similarly, in patients experiencing acute myeloid leukemia, the overexpression of at least 1 TAA (RHAMM, CA9/G250, or PRAME) improved even overall survival, and this might also reflect sustained immunologic control of minimal residual disease [60].
The fact that CaPs with higher T and N stages displayed a significantly lower amount of RHAMM protein in primaries on the TMAs might be also explained with the long-lasting interaction of RHAMM and host cytotoxic T lymphocyte, finally leading to tumor escape with an outgrowth of RHAMM-negative tumor cells. The well-known heterogeneity within CaPs with, that is, different Gleason patterns, may facilitate such events of immunoselection. Because we did not provide proof of a strong T-cell pressure against RHAMM in CaP patients by the means of enzyme-linked ImmunoSpot assay or tetramer staining, in the present study, the proposed mechanism of T-cell-based immunoselection remains a model so far.
However, if RHAMM expression truly leads to a prolonged time to progression after the resection of the primary due to immunologic control, an augmentation of the T-cell response against these antigens by a vaccination protocol might be of further benefit for the patients. It is most likely that the patients' greatest benefit would be seen in the adjuvant setting with a low tumor burden being present, that is, small metastases in lymph nodes or a positive margin. Another potential therapeutic consequence of RHAMM overexpression is founded on the observation that incubation of CaP cell lines with a RHAMM-specific antibody reduced the viability of both hormone-sensitive and hormone-insensitive CaP cells cell lines. Consistently, it was demonstrated by Lin et al. [31,32] that downstream blockade of the RHAMM pathway by the ROCK inhibitor Y-27632 or anti-ROCK1 micro RNA was able to abolish cancer cell proliferation, invasion, and metastasis in the same cell lines used in our study.
Recently, the same group showed that androgen stimulation leads in overexpression of RHAMM and hyaluronan-stimulated activation of the RHAMM-ROCK1 cascade in LNCaP cells, suggesting that RHAMM plays a role in androgen-dependent as well as in castration-resistant stage of CaP [31].
Summarizing, the overexpression of RHAMM at the protein level was found in both localized CaP and metastatic sites. Whereas higher protein levels in primaries are associated with better clinical outcome, highest levels were found in lymph node metastases, an advanced stage of disease. This illustrates that RHAMM might also be a target suitable for monoclonal antibody therapy in advanced CaP.
Conclusions
We identified 10 differentially overexpressed potential immunogenic targets in CaP by data mining and own cDNA array analysis. RHAMM (CD168) was overexpressed at the cDNA level as well as at the protein level in the metastatic aspect compared with localized CaP or benign tissue. The protein overexpression of RHAMM was associated with a lower risk of biochemical failure. Expression of RHAMM also had an association with clinical parameters known to be associated with better clinical outcome such as favorable T stage, no lymph node metastases, and lower PSA serum level.
In cell culture, numbers of viable cells were reduced in blocking experiments with the RHAMM-selective antibody (E-19) using hormone-sensitive (LNCaP) and hormone-insensitive metastatic (PC3 and DU145) CaP cell lines.
The pathways involved in the intracellular signaling cascade mediated by RHAMM in CaP are currently under analysis.
Acknowledging
the proven immunogenic effects of RHAMM in clinical trials for hematological diseases, this antigen might be a promising therapeutic target in the adjuvant setting or even in advanced CaP.
Abbreviations
- CaP
prostate cancer
- cDNA
complementary DNA
- CID
Cancer Immunome Database
- DU145
hormone-independent CaP cell line
- LNCaP
hormone-sensitive CaP cell line
- PC3
hormone-independent CaP cell line
- PSA
prostate-specific antigen
- RHAMM/CD168
receptor of hyaluronan-mediated motility (HMMR)
- SEREX
serological identification of antigens by recombinant expression cloning
- TMA
tissue microarray
Footnotes
All authors of the article have declared that there are no commercial affiliations, stock interests, or patent licenses of potential conflict of interest with the presented work. There are no financial interests of a company whose product has been used for studies in the work presented in the article.
In 2002, the Academy of Cancer Immunology and the Ludwig Institute for Cancer Research became the cosponsors of a new database, the Cancer Immunome Database (CID), which replaced the SEREX database (http://ludwig-sun5.unil.ch/CancerImmunomeDB/).
References
- 1.Horwich A, Parker C. Prostate cancer: ESMO clinical recommendations for diagnosis, treatment and follow-up. Ann Oncol. 2007;18(Suppl 2):ii36–ii37. doi: 10.1093/annonc/mdm028. [DOI] [PubMed] [Google Scholar]
- 2.Weir HK, Thun MJ, Hankey BF, Ries LA, Howe HL, Wingo PA, Jemal A, Ward E, Anderson RN, Edwards BK. Annual report to the nation on the status of cancer, 1975–2000, featuring the uses of surveillance data for cancer prevention and control. J Natl Cancer Inst. 2003;95:1276–1299. doi: 10.1093/jnci/djg040. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Tiwari RC, Murray T, Ghafoor A, Samuels A, Ward E, Feuer EJ, Thun MJ. Cancer statistics, 2004. CA Cancer J Clin. 2004;54:8–29. doi: 10.3322/canjclin.54.1.8. [DOI] [PubMed] [Google Scholar]
- 4.Namiki K, Rosser CJ. Neoadjuvant therapy and prostate cancer: what a urologist should know. Curr Opin Urol. 2007;17:188–193. doi: 10.1097/MOU.0b013e3280e08802. [DOI] [PubMed] [Google Scholar]
- 5.Graham J. Chemotherapy for metastatic disease: current status. Clin Oncol (R Coll Radiol) 2005;17:572–578. doi: 10.1016/j.clon.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 6.Hadaschik BA, Sowery RD, Gleave ME. Novel targets and approaches in advanced prostate cancer. Curr Opin Urol. 2007;17:182–187. doi: 10.1097/MOU.0b013e3280dd8a4f. [DOI] [PubMed] [Google Scholar]
- 7.Armstrong AJ, George DJ. Satraplatin in the treatment of hormone-refractory metastatic prostate cancer. Ther Clin Risk Manag. 2007;3:877–883. [PMC free article] [PubMed] [Google Scholar]
- 8.Kerr C. Satraplatin for hormone-refractory prostate cancer. Lancet Oncol. 2007;8:290. doi: 10.1016/s1470-2045(07)70096-7. [DOI] [PubMed] [Google Scholar]
- 9.McKeage MJ. Satraplatin in hormone-refractory prostate cancer and other tumour types: pharmacological properties and clinical evaluation. Drugs. 2007;67:859–869. doi: 10.2165/00003495-200767060-00003. [DOI] [PubMed] [Google Scholar]
- 10.Sternberg CN. Satraplatin in the treatment of hormone-refractory prostate cancer. BJU Int. 2005;96:990–994. doi: 10.1111/j.1464-410X.2005.05799.x. [DOI] [PubMed] [Google Scholar]
- 11.Sternberg CN, Whelan P, Hetherington J, Paluchowska B, Slee PH, Vekemans K, Van Erps P, Theodore C, Koriakine O, Oliver T, et al. Phase III trial of satraplatin, an oral platinum plus prednisone vs. prednisone alone in patients with hormone-refractory prostate cancer. Oncology. 2005;68:2–9. doi: 10.1159/000084201. [DOI] [PubMed] [Google Scholar]
- 12.Kuefer R, Genze F, Zugmaier W, Hautmann RE, Rinnab L, Gschwend JE, Angelmeier M, Estrada A, Buechele B. Antagonistic effects of sodium butyrate and N-(4-hydroxyphenyl)-retinamide on prostate cancer. Neoplasia. 2007;9:246–253. doi: 10.1593/neo.06766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen JS, Faller DV, Spanjaard RA. Short-chain fatty acid inhibitors of histone deacetylases: promising anticancer therapeutics? Curr Cancer Drug Targets. 2003;3:219–236. doi: 10.2174/1568009033481994. [DOI] [PubMed] [Google Scholar]
- 14.Holder S, Zemskova M, Zhang C, Tabrizizad M, Bremer R, Neidigh JW, Lilly MB. Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase. Mol Cancer Ther. 2007;6:163–172. doi: 10.1158/1535-7163.MCT-06-0397. [DOI] [PubMed] [Google Scholar]
- 15.Sherk AB, Frigo DE, Schnackenberg CG, Bray JD, Laping NJ, Trizna W, Hammond M, Patterson JR, Thompson SK, Kazmin D, et al. Development of a small-molecule serum- and glucocorticoid-regulated kinase-1 antagonist and its evaluation as a prostate cancer therapeutic. Cancer Res. 2008;68:7475–7483. doi: 10.1158/0008-5472.CAN-08-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tolcher AW, Mita A, Lewis LD, Garrett CR, Till E, Daud AI, Patnaik A, Papadopoulos K, Takimoto C, Bartels P, et al. Phase I and pharmacokinetic study of YM155, a small-molecule inhibitor of survivin. J Clin Oncol. 2008;26:5198–5203. doi: 10.1200/JCO.2008.17.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M, Tan W, Zhou J, Leow J, Go M, Lee HS, Casey PJ. A small molecule inhibitor of isoprenylcysteine carboxymethyltransferase induces autophagic cell death in PC3 prostate cancer cells. J Biol Chem. 2008;283:18678–18684. doi: 10.1074/jbc.M801855200. [DOI] [PubMed] [Google Scholar]
- 18.Guerin O, Formento P, Lo Nigro C, Hofman P, Fischel JL, Etienne-Grimaldi MC, Merlano M, Ferrero JM, Milano G. Supra-additive antitumor effect of sunitinib malate (SU11248, Sutent) combined with docetaxel. A new therapeutic perspective in hormone refractory prostate cancer. J Cancer Res Clin Oncol. 2008;134:51–57. doi: 10.1007/s00432-007-0247-4. [DOI] [PubMed] [Google Scholar]
- 19.Sonpavde G, Hutson TE, Berry WR, Boehm KA, Asmar L. Phase II trial of sunitinib for the therapy of progressive metastatic castration-refractory prostate cancer after previous docetaxel chemotherapy. Clin Genitourin Cancer. 2008;6:134–137. doi: 10.3816/CGC.2008.n.023. [DOI] [PubMed] [Google Scholar]
- 20.Cumashi A, Tinari N, Rossi C, Lattanzio R, Natoli C, Piantelli M, Iacobelli S. Sunitinib malate (SU-11248) alone or in combination with low-dose docetaxel inhibits the growth of DU-145 prostate cancer xenografts. Cancer Lett. 2008;270:229–233. doi: 10.1016/j.canlet.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 21.Pantuck AJ, Zomorodian N, Belldegrun AS. Phase I, open-label, single-center, multiple-dose, dose-escalation clinical study of SUO11248 (sunitinib) in subjects with high-risk prostate cancer who have elected to undergo radical prostatectomy. Curr Urol Rep. 2007;8:3–4. doi: 10.1007/s11934-007-0014-8. [DOI] [PubMed] [Google Scholar]
- 22.Bibikova M, Chudin E, Arsanjani A, Zhou L, Garcia EW, Modder J, Kostelec M, Barker D, Downs T, Fan JB, et al. Expression signatures that correlated with Gleason score and relapse in prostate cancer. Genomics. 2007;89:666–672. doi: 10.1016/j.ygeno.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Chen T, Tang XD, Wan Y, Chen L, Yu ST, Xiong Z, Fang DC, Liang GP, Yang SM. HLA-A2-restricted cytotoxic T lymphocyte epitopes from human heparanase as novel targets for broad-spectrum tumor immunotherapy. Neoplasia. 2008;10:977–986. doi: 10.1593/neo.08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malyankar UM. Tumor-associated antigens and biomarkers in cancer and immune therapy. Int Rev Immunol. 2007;26:223–247. doi: 10.1080/08830180701402496. [DOI] [PubMed] [Google Scholar]
- 25.Ambs S, Marincola FM, Thurin M. Profiling of immune response to guide cancer diagnosis, prognosis, and prediction of therapy. Cancer Res. 2008;68:4031–4033. doi: 10.1158/0008-5472.CAN-08-0521. [DOI] [PubMed] [Google Scholar]
- 26.Dougan M, Dranoff G. Immune therapy for cancer. Annu Rev Immunol. 2009;27:83–117. doi: 10.1146/annurev.immunol.021908.132544. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Yu J, Sreekumar A, Varambally S, Shen R, Giacherio D, Mehra R, Montie JE, Pienta KJ, Sanda MG, et al. Autoantibody signatures in prostate cancer. N Engl J Med. 2005;353:1224–1235. doi: 10.1056/NEJMoa051931. [DOI] [PubMed] [Google Scholar]
- 28.Bradford TJ, Wang X, Chinnaiyan AM. Cancer immunomics: using autoantibody signatures in the early detection of prostate cancer. Urol Oncol. 2006;24:237–242. doi: 10.1016/j.urolonc.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 29.Greiner J, Ringhoffer M, Taniguchi M, Schmitt A, Kirchner D, Krahn G, Heilmann V, Gschwend J, Bergmann L, Dohner H, et al. Receptor for hyaluronan acid-mediated motility (RHAMM) is a new immunogenic leukemia-associated antigen in acute and chronic myeloid leukemia. Exp Hematol. 2002;30:1029–1035. doi: 10.1016/s0301-472x(02)00874-3. [DOI] [PubMed] [Google Scholar]
- 30.Wang C, Thor AD, Moore DH, II, Zhao Y, Kerschmann R, Stern R, Watson PH, Turley EA. The overexpression of RHAMM, a hyaluronan-binding protein that regulates ras signaling, correlates with overexpression of mitogen-activated protein kinase and is a significant parameter in breast cancer progression. Clin Cancer Res. 1998;4:567–576. [PubMed] [Google Scholar]
- 31.Lin SL, Chang D, Chiang A, Ying SY. Androgen receptor regulates CD168 expression and signaling in prostate cancer. Carcinogenesis. 2008;29:282–290. doi: 10.1093/carcin/bgm259. [DOI] [PubMed] [Google Scholar]
- 32.Lin SL, Chang D, Ying SY. Hyaluronan stimulates transformation of androgen-independent prostate cancer. Carcinogenesis. 2007;28:310–320. doi: 10.1093/carcin/bgl134. [DOI] [PubMed] [Google Scholar]
- 33.Schmitt M, Schmitt A, Rojewski MT, Chen J, Giannopoulos K, Fei F, Yu Y, Gotz M, Heyduk M, Ritter G, et al. RHAMM-R3 peptide vaccination in patients with acute myeloid leukemia, myelodysplastic syndrome, and multiple myeloma elicits immunologic and clinical responses. Blood. 2008;111:1357–1365. doi: 10.1182/blood-2007-07-099366. [DOI] [PubMed] [Google Scholar]
- 34.Yang XF, Wu CJ, Chen L, Alyea EP, Canning C, Kantoff P, Soiffer RJ, Dranoff G, Ritz J. CML28 is a broadly immunogenic antigen, which is overexpressed in tumor cells. Cancer Res. 2002;62:5517–5522. [PMC free article] [PubMed] [Google Scholar]
- 35.Webster WS, Small EJ, Rini BI, Kwon ED. Prostate cancer immunology: biology, therapeutics, and challenges. J Clin Oncol. 2005;23:8262–8269. doi: 10.1200/JCO.2005.03.4595. [DOI] [PubMed] [Google Scholar]
- 36.Manley S, Mucci NR, De Marzo AM, Rubin MA. Relational database structure to manage high-density tissue microarray data and images for pathology studies focusing on clinical outcome: the prostate specialized program of research excellence model. Am J Pathol. 2001;159:837–843. doi: 10.1016/S0002-9440(10)61759-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubin MA, Putzi M, Mucci N, Smith DC, Wojno K, Korenchuk S, Pienta KJ. Rapid (“warm”) autopsy study for procurement of metastatic prostate cancer. Clin Cancer Res. 2000;6:1038–1045. [PubMed] [Google Scholar]
- 38.Hofer MD, Kuefer R, Huang W, Li H, Bismar TA, Perner S, Hautmann RE, Sanda MG, Gschwend JE, Rubin MA. Prognostic factors in lymph node-positive prostate cancer. Urology. 2006;67:1016–1021. doi: 10.1016/j.urology.2005.10.055. [DOI] [PubMed] [Google Scholar]
- 39.Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 40.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Dhanasekaran SM, Dash A, Yu J, Maine IP, Laxman B, Tomlins SA, Creighton CJ, Menon A, Rubin MA, Chinnaiyan AM. Molecular profiling of human prostate tissues: insights into gene expression patterns of prostate development during puberty. FASEB J. 2005;19:243–245. doi: 10.1096/fj.04-2415fje. [DOI] [PubMed] [Google Scholar]
- 42.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 45.Rubin MA, Dunn R, Strawderman M, Pienta KJ. Tissue microarray sampling strategy for prostate cancer biomarker analysis. Am J Surg Pathol. 2002;26:312–319. doi: 10.1097/00000478-200203000-00004. [DOI] [PubMed] [Google Scholar]
- 46.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 47.Rubin MA, Bismar TA, Andren O, Mucci L, Kim R, Shen R, Ghosh D, Wei JT, Chinnaiyan AM, Adami HO, et al. Decreased α-methylacyl CoA racemase expression in localized prostate cancer is associated with an increased rate of biochemical recurrence and cancer-specific death. Cancer Epidemiol Biomarkers Prev. 2005;14:1424–1432. doi: 10.1158/1055-9965.EPI-04-0801. [DOI] [PubMed] [Google Scholar]
- 48.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 49.La Tulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, Gerald WL. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- 50.Sahin U, Tureci O, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M. Human neoplasms elicit multiple specific immune responses in the autologous host. Proc Natl Acad Sci USA. 1995;92:11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goransson O, Deak M, Wullschleger S, Morrice NA, Prescott AR, Alessi DR. Regulation of the polarity kinases PAR-1/MARK by 14-3-3 interaction and phosphorylation. J Cell Sci. 2006;119:4059–4070. doi: 10.1242/jcs.03097. [DOI] [PubMed] [Google Scholar]
- 52.Wong LJ, Brunetti-Pierri N, Zhang Q, Yazigi N, Bove KE, Dahms BB, Puchowicz MA, Gonzalez-Gomez I, Schmitt ES, Truong CK, et al. Mutations in the MPV17 gene are responsible for rapidly progressive liver failure in infancy. Hepatology. 2007;46:1218–1227. doi: 10.1002/hep.21799. [DOI] [PubMed] [Google Scholar]
- 53.Sreekumar A, Laxman B, Rhodes DR, Bhagavathula S, Harwood J, Giacherio D, Ghosh D, Sanda MG, Rubin MA, Chinnaiyan AM. Humoral immune response to α-methylacyl-CoA racemase and prostate cancer. J Natl Cancer Inst. 2004;96:834–843. doi: 10.1093/jnci/djh145. [DOI] [PubMed] [Google Scholar]
- 54.Greiner J, Li L, Ringhoffer M, Barth TF, Giannopoulos K, Guillaume P, Ritter G, Wiesneth M, Dohner H, Schmitt M. Identification and characterization of epitopes of the receptor for hyaluronic acid-mediated motility (RHAMM/CD168) recognized by CD8+ T cells of HLA-A2-positive patients with acute myeloid leukemia. Blood. 2005;106:938–945. doi: 10.1182/blood-2004-12-4787. [DOI] [PubMed] [Google Scholar]
- 55.Hall CL, Turley EA. Hyaluronan: RHAMM mediated cell locomotion and signaling in tumorigenesis. J Neurooncol. 1995;26:221–229. doi: 10.1007/BF01052625. [DOI] [PubMed] [Google Scholar]
- 56.Greiner J, Bullinger L, Guinn BA, Dohner H, Schmitt M. Leukemia-associated antigens are critical for the proliferation of acute myeloid leukemia cells. Clin Cancer Res. 2008;14:7161–7166. doi: 10.1158/1078-0432.CCR-08-1102. [DOI] [PubMed] [Google Scholar]
- 57.Schmitt A, Barth TF, Beyer E, Borchert F, Rojewski M, Chen J, Guillaume P, Gronau S, Greiner J, Moller P, et al. The tumor antigens RHAMM and G250/CAIX are expressed in head and neck squamous cell carcinomas and elicit specific CD8+ T cell responses. Int J Oncol. 2009;34:629–639. doi: 10.3892/ijo_00000188. [DOI] [PubMed] [Google Scholar]
- 58.Zlobec I, Terracciano L, Tornillo L, Gunthert U, Vuong T, Jass JR, Lugli A. Role of RHAMM within the hierarchy of well-established prognostic factors in colorectal cancer. Gut. 2008;57:1413–1419. doi: 10.1136/gut.2007.141192. [DOI] [PubMed] [Google Scholar]
- 59.Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21:137–148. doi: 10.1016/j.immuni.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 60.Greiner J, Schmitt M, Li L, Giannopoulos K, Bosch K, Schmitt A, Dohner K, Schlenk RF, Pollack JR, Dohner H, et al. Expression of tumor-associated antigens in acute myeloid leukemia: Implications for specific immunotherapeutic approaches. Blood. 2006;108:4109–4117. doi: 10.1182/blood-2006-01-023127. [DOI] [PubMed] [Google Scholar]