Abstract
The global regulatory protein CsrA binds to the 5′-untranslated leader of target transcripts and alters their translation and/or stability. CsrA is a symmetrical homodimer containing two identical RNA-binding surfaces. Gel shift assays with model RNA substrates now show that CsrA can bind simultaneously at two target sites within a transcript (bridging or dual site binding). An intersite distance of ∼18 nucleotides (nt) was optimal, although bridging occurred with an intersite distance of 10 to ≥ 63 nt. The close 10-nt spacing reduced the stability of dual site binding, as competition for one site by a second CsrA dimer readily occurred. Both RNA-binding surfaces of a single CsrA protein were essential for efficient in vitro repression of a glgC′-‘lacZ translational fusion that contains four CsrA target sites within the untranslated leader. Heterodimeric CsrA (HD-CsrA) containing a single R44A replacement, which was defective for binding at its mutant surface but bound RNA normally at its wild-type (WT) surface, was ∼14-fold less effective at repression than homodimeric WT-CsrA. Furthermore, deletion of a CsrA target site of glgC that lies upstream from the Shine-Dalgarno sequence did not affect regulation by HD-CsrA but decreased regulation by WT-CsrA, confirming a regulatory role of dual site binding. Finally, we propose a mechanism whereby a globular ribonucleoprotein complex is formed between CsrA and its noncoding RNA antagonist, CsrB. Because many target sites of CsrB are located closer together than is optimal for bridging, binding to non-adjacent sites should be energetically favored, causing multiple CsrA dimers to tether CsrB into the observed globular form rather than an extended CsrA-CsrB complex.
Introduction
RNA-binding proteins participate in a myriad of cellular processes and are necessary for all known forms of life to exist.1; 2 The abundance of these proteins mirrors their diverse functions, yet only a relatively small number of known RNA-binding motifs are distributed throughout this large family.2; 3; 4 The disparity is due to modularity in protein design and multimerization of RNA binding motifs. Multiple binding sites within the same protein can increase target specificity and RNA-binding avidity.5 Both bacterial and eukaryotic RNA-binding proteins with multiple RNA binding motifs are known.6; 7; 8; 9 For example, the eukaryotic proteins Dicer and U2AF35 contain 3 and 4 different RNA-binding motifs, respectively, while Vigilin has 14 copies of the same motif;4; 8 the bacterial transcriptional terminator NusA contains an S1 and dual K-Homology domains (KH)10; 11 while the circular, homo-undecameric trp RNA-binding attenuation protein (TRAP) is composed of 11 identical surfaces capable of binding (G/U)AG triplet repeats.12
The molecular interactions between RNA and RNA-binding proteins are guided by the nature of the protein motifs that are present; however, the spacing of RNA-binding motifs and their target sequences is also essential for tight protein-RNA association and proper gene regulation. For example, TRAP was unable to bind RNA when the distance between trinucleotide repeats was changed from the optimum of 2 nt to either 1, 3 or 4 nt.13 Conversely, deletion of the linker between RRM2 and RRM3 in the mammalian CUG-binding protein (CUG-BP) severely compromised rUrG interaction, presumably by inhibiting cooperativity between two regions of the protein.14 The present study addresses questions about the spacing of RNA target sites and the biological necessity of multiple RNA-binding surfaces in the E. coli RNA-binding protein CsrA.
CsrA (and its orthologs RsmA, RsmE) is a global regulatory protein that controls carbon metabolism,15; 16 biofilm formation,17; 18 motility,19; 20 peptide uptake,21 quorum-sensing22 and virulence functions23; 24; 25; 26 of many eubacteria. Regulation is accomplished by specific binding of CsrA to conserved sequences in the 5′-untranslated leader of target mRNAs, leading to effects on their translation and/or stability.17; 19 In addition to its mRNA targets, CsrA interacts with small noncoding RNAs, CsrB27 and CsrC28 in E. coli, that sequester and antagonize this protein. These sRNAs are predicted to form multiple stem-loops containing conserved sequences resembling CsrA target sites in mRNA.
Studies of CsrA have culminated in the recent determination of its 3D-structure. Solution NMR studies29 and crystallography30; 31 demonstrated that CsrA represents a new class of RNA-binding protein containing a unique RNA-binding motif. It is assembled via the interdigitation of 5 β-strands of two identical polypeptides, forming a bilaterally symmetrical protein that is stabilized by a hydrophobic core and extensive hydrogen bonding. Two identical RNA-binding surfaces are located on opposite sides of the protein, each of which is composed primarily of the N-terminal β-strand (β1) of one polypeptide and the parallel β5 strand of the second polypeptide. These surfaces contain several amino acid residues that are needed for optimal RNA binding and regulation, the most important of which was R44.32 NMR studies of a CsrA ortholog (RsmE) complexed with target RNAs confirmed the protein-RNA contacts that were implied by alanine substitution studies.33 Thus, as in TRAP12; 34; 35 and MBNL1,36 an RNA binding surface is formed by the interface of two interacting polypeptide subunits.
Electrophoretic mobility shift assays (EMSA), RNA footprinting and other studies have shown that multiple CsrA proteins can bind to a single mRNA or sRNA transcript that contains multiple binding sites.17; 37 In conjunction with the evidence that CsrA contains two RNA-binding surfaces, we hypothesized that a single CsrA dimer might bridge two target sites on the same RNA.32 The mirrored orientation of the dual CsrA binding surfaces combined with the flexibility of single-stranded RNA should dictate the positioning of the second RNA target site after initial contact is made. Furthermore, the local concentration of a secondary target should be greatly increased upon CsrA binding to a primary site38, thus increasing the probability of a secondary intramolecular interaction, which should increase binding avidity.
Here, we demonstrate that a CsrA dimer can indeed bind two target sites on a single RNA simultaneously, resulting in a compact “bridge complex”, whose stability depended on the distance separating the RNA target sites. Our findings further revealed that both RNA-binding surfaces of CsrA must function simultaneously for efficient post-transcriptional regulation, and in glgC repression, they facilitate bridging from a high-affinity stem-loop in the untranslated leader to a downstream inhibitory site that overlaps the SD sequence. A model for CsrA binding to RNAs with multiple target sites is presented and its implications for regulation of mRNA translation and CsrA sequestration by regulatory sRNAs are discussed.
Results
WT-CsrA dimer binds two RNA oligonucleotides simultaneously
We previously demonstrated that a CsrA dimer contains two identical surfaces that are critical for RNA binding and regulation of gene expression.32 Subsequently, 3D-NMR analysis revealed that a CsrA ortholog can bind two RNA oligonucleotides in solution.33 To further examine CsrA binding properties, the WT-CsrA protein was tested for binding to a high affinity, SELEX-derived oligonucleotide,39 referred to as RNA-1A (Table 1). In this EMSA experiment, both a major and a minor complex were formed (Fig.1a). Because these reactions were conducted at a substantial excess of protein vs. RNA, and yet both complexes were relatively abundant, the binding may have been cooperative, as has been reported. 37 The addition of increasing amounts of specific unlabeled RNA competitor to the binding reaction with WT-CsrA resulted in the formation of the faster mobility minor complex at the expense of the slower moving major complex (Fig. 1b, lanes 2-6). Because this exchange was due to the addition of the RNA oligonucleotide to the complex, we concluded that the faster moving species was composed of one CsrA protein bound to two independent RNAs and that the slower moving species was composed of one CsrA dimer bound to one RNA. The increased mobility of the larger complex (1 CsrA:2 RNA) must have been due to the greater influence of the negative charge conferred by the binding of a second RNA relative to its effect on the Stokes radius.
Table 1.
Synthetic RNA oligonucleotides used in this study.a
| Name | Intersite Distance | Linker size (N) | Sequence |
|---|---|---|---|
| RNA-1A | |||
| RNA-1B | |||
| RNA-2A | |||
| RNA-2B | |||
| RNA10-2BS | 10 | 1 | |
| RNA10-1BS | 10 | 1 | |
| RNA14-2BS | 14 | 1 | |
| RNA14-1BS | 14 | 1 | |
| RNA18-2BS | 18 | 5 | |
| RNA18-1BS | 18 | 5 | |
| RNA28-2BS | 28 | 15 | |
| RNA28-1BS | 28 | 15 | |
| RNA43-2BS | 43 | 30 | |
| RNA43-1BS | 43 | 30 | |
| RNA63-2BS | 63 | 50 | |
| RNA63-1BS | 63 | 50 |
Unless otherwise noted, the linker (N) was composed of a poly (A) tract. The intersite distance is measured in ribonucleotides starting on either side of the invariant “GGA” motif. Arrows under each sequence indicate annealing strands of a stem in a likely stem-loop structure as modeled by MFOLD.47; 74 The conserved GGA nucleotides of high affinity binding targets are highlighted in gray. Oligoribonucleotide nomenclature reflects both the intersite distance and the number of functional target sites (ie., intersite distance of 10 nt with 2 binding sites = RNA10-2BS). RNAs were supplied by Integrated DNA Technologies (Coralville, IA) or synthesized using the Megashortscript Kit (Ambion, Austin, TX). During the course of experiments, GGA replacements were changed from CCC to AAA when it was revealed that a CCC trinucleotide could potentially facilitate unwanted RNA oligonucleotide secondary structures as predicted by MFOLD47; 74.
Fig. 1.
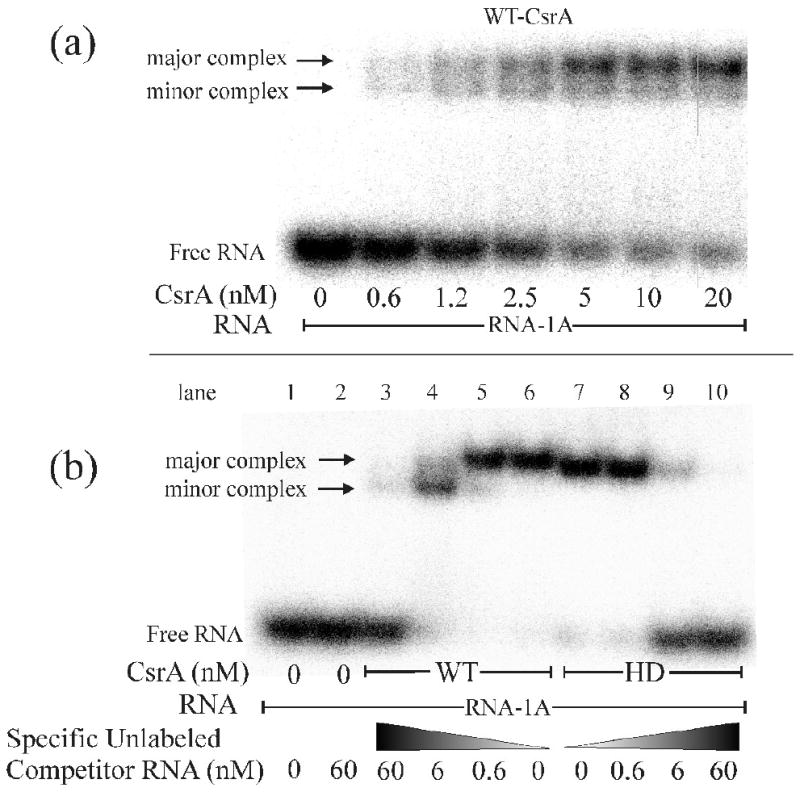
Electrophoretic Mobility Shift Assay (EMSA) demonstrating that WT-CsrA contains 2 independent RNA binding surfaces while the HD-CsrA contains only one. a) EMSA with WT-CsrA protein showing the presence of a “major” and “minor” shifted complex (arrows), as well as “free RNA”, suggesting that CsrA can bind 2 independent RNAs. b) Competition experiment with increasing concentrations of specific unlabeled competitor RNA. Note that addition of competitor RNA caused a redistribution of the WT-CsrA complex to the lower “minor” position, while HD-CsrA failed to exhibit a secondary complex with additional competitor,
RNA binding by a CsrA heterodimer (HD-CsrA) containing mutant and wild type RNA-binding surfaces
To better understand how CsrA interacts with RNAs containing multiple target sites we prepared a heterodimeric CsrA protein that is composed of one wild-type polypeptide and one his-tagged polypeptide containing an alanine substitution (R44A). This substitution reduces the affinity of the homodimer mutant protein for a high-affinity RNA target by ∼150-fold.32
To test whether both RNA-binding surfaces of HD-CsrA were functional, increasing amounts of unlabeled RNA-1A were added to a binding reaction containing HD-CsrA. Unlike the reaction with WT-CsrA, the HD-CsrA protein never exhibited a second shift (Fig. 1b, lanes 7-10), confirming that it was unable to bind two RNAs simultaneously under our reactions conditions. The mobility of the HD-CsrA/RNA complex was slightly faster than the WT-CsrA complex on native polyacrylamide, presumably due to the R44A substitution, which results in a net increase in negative charge. HD-CsrA bound to RNA-1A with approximately one third of the binding affinity (Kd = 3.4 nM ± 0.4) of WT-CsrA protein (Kd = 1.25 nM ± 0.3) (Fig. 2).
Fig. 2.
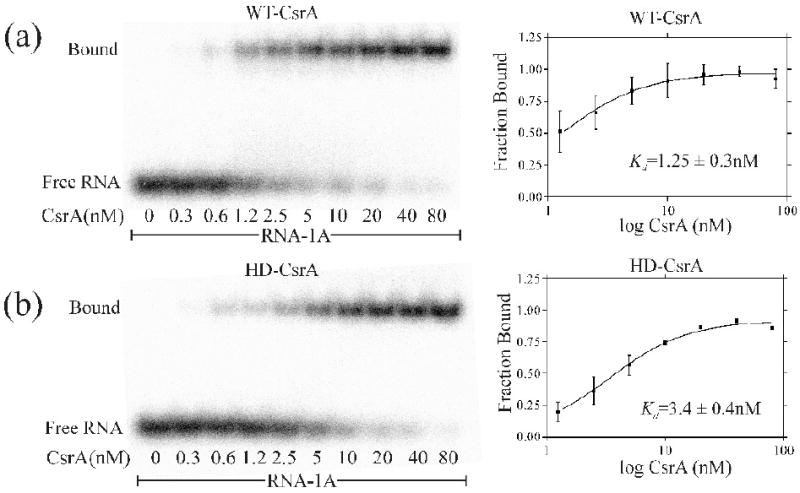
EMSA demonstration that HD-CsrA binds to a single-site RNA with approximately one third the avidity of WT-CsrA. a,b) EMSA of WT-CsrA and HD-CsrA proteins and their corresponding binding curves. The avidity of WT-CsrA for RNA, as reflected in the apparent equilibrium binding constant (Kd = 1.3 nM), is greater than that of the HD-CsrA (Kd = 3.4 nM) for the same labeled RNA oligonucleotide. Note that the shorter development time and contrast adjustment in this image vs. Fig. 1 deemphasized the “minor complexes”. Graphpad Prism version 3 was used for graphical representations and calculations
Binding of the CsrA dimer to a series of RNAs containing dual target sites separated by varying distances
Most RNAs bound by CsrA (RsmA) contain multiple target sites that are related to the SELEX-derived consensus sequence “RUACARGGAUGU” where the underlined nucleotides were 100% conserved and were present in the loop of a hairpin structure.17; 21; 27; 28; 37; 39; 40; 41; 42; 43; 44 In native RNA molecules, the GGA motifs of sequential CsrA target sites may be as close as 6 or 7 nt, as predicted in CsrC28 and CsrB,27 respectively, or they may be more than 50 nt apart, as determined in the 5′-leader of the pgaABCD message.17 Nevertheless, the way in which the respective RNA binding sites of a CsrA dimer interact with paired target sites has not been determined for any of these RNAs. Furthermore, we have proposed that the symmetrical structure of CsrA, with two identical binding surfaces on opposite sides of the dimer, appears to account for the observation that CsrB, which contains ∼18-22 target sequences, forms a globular complex with ∼9-10 CsrA dimers.32 The implication is that all or most of the CsrA dimers of this complex should form noncovalent bridges between two target sites on the same RNA.
To test the hypothesis that a CsrA dimer bridges two binding sites in target RNAs, and to probe the minimum and maximum intersite distances at which this occurs, we designed a series of RNA oligonucleotides containing dual high-affinity target sites set at varying distances from each other (Table 1: RNA10-2BS, RNA14-2BS, RNA18-2BS, RNA28-2BS, RNA43-2BS and RNA63-2BS). These RNAs were examined by EMSA for the binding of WT-CsrA or HD-CsrA (Fig. 3a-f). A second series of RNAs were prepared, identical to the first series, except that the 3′-high-affinity binding sites were altered (i.e., the central GGA was changed to AAA or CCC; see Table 1 for an explanation of the use of CCC or AAA) to prevent CsrA binding (Table 1: RNA10-1BS, RNA14-1BS, RNA18-1BS, RNA28-1BS, RNA43-1BS and RNA63-1BS). The single-site RNAs were expected to form only a 1:1 complex with HD-CsrA, in which the WT surface of the protein is bound to the high affinity GGA site, providing a series of standard shifts to which the other complexes can be compared (Fig. 3a-f, complex 6). Complexes formed by two CsrA dimers bound to a single RNA should migrate considerably slower than the standards. Complexes that travel faster than the 1:1 standards should be more compact, consistent with dual site bridging by CsrA. The design of all oligonucleotides except RNA10-2BS and RNA10-1BS included 16 nt RNA hairpins with stems of 6 base pairs, which was previously established to be a high affinity target for CsrA.32; 45 To decrease the distance between CsrA target sites, we constructed RNA10-2BS and RNA10-1BS with 12 nucleotide hairpins (4 base pair stems). The in silico analysis46; 47 of smaller RNA stems did not predict the formation of stable hairpin structures at a reaction temperature of 37 °C.
Fig. 3.
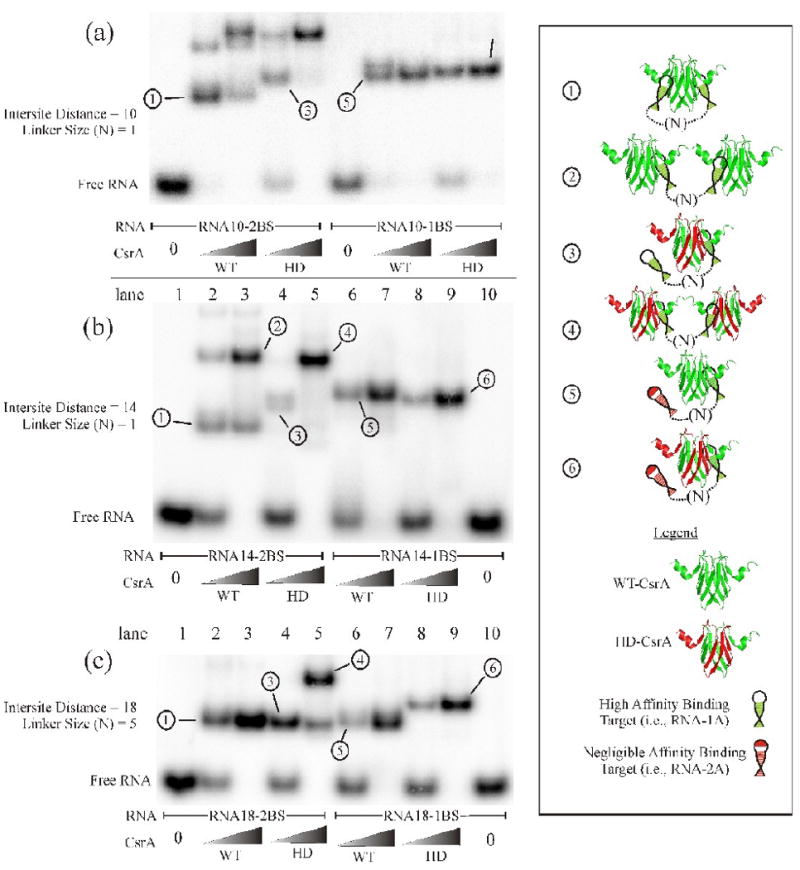
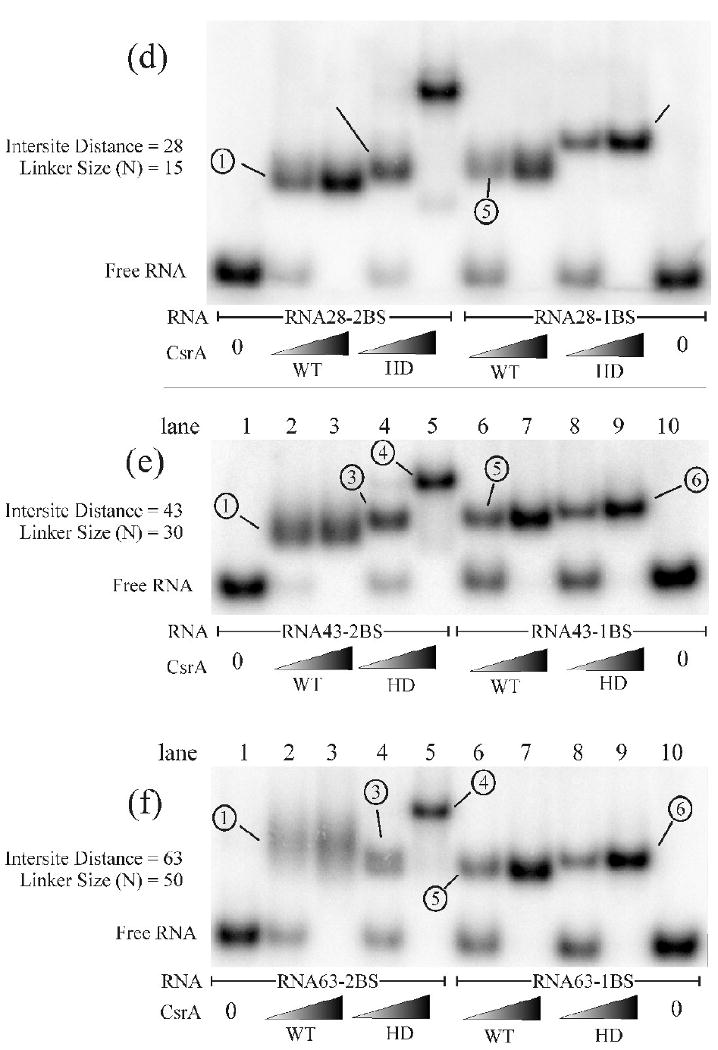
EMSA analysis of the optimal CsrA binding site spacing using a series of synthetic RNA oligonucleotides. EMSAs were performed with the RNAs listed in Table 1. Two concentrations of WT-CsrA or HD-CsrA protein were employed for each series of binding reactions, the first reaction in each binding set (i.e., gel “a” lane 2) contains a low concentration (2.5 nM) of the protein listed, while the reaction to the right of it (i.e., gel “a” lane 3) contains a high concentration (62.5 nM) of CsrA protein. RNAs containing either one or two high affinity binding sites are also shown. Important shifted species are numbered, described in the results, and correspond to the structural interpretations in the boxed area to the right. The intersite distance and linker size (N) for each RNA oligo used is measured in nucleotides. Experiments were performed at least twice to confirm banding patterns and migration distances.
To characterize the target sites of the model RNAs, we initially confirmed that WT-CsrA bound with equal affinity to the target sites at the 5′ and 3′-termini of dual target RNAs by synthesizing each stem loop independently (Table 1: RNA-1A and RNA-1B) and testing them for binding (Fig. 1 and data not shown). We also established that target sites that were designed to have negligible affinity for CsrA (Table 1: RNA-2A and RNA-2B) showed no binding up to 160 nM CsrA by EMSA (data not shown).
All reactions containing HD-CsrA with a single-target RNA revealed a single complex, which was concluded to be composed of the WT RNA-binding surface of HD-CsrA bound to the high affinity stem-loop of the RNA (a 1:1 standard complex, labeled “6” in Fig. 3a, lanes 9 and 10; 3b-f, lanes 8 and 9). Strikingly, most of the dual site RNAs with intersite distances from 10 to 43 nt exhibited a WT-CsrA:RNA band (complex 1) of greater mobility (more compact) than the 1:1 standard complexes formed by HD-CsrA and the single-site RNAs (complex 6). This finding suggested that WT-CsrA can bridge closely adjoining RNA target sites in addition to distantly separated ones. At low HD-CsrA concentrations, every binding reaction with a dual high affinity target RNA (Fig. 3a-f, lane 4) exhibited a fast moving shift (complex 3) similar to the 1:1 complex 6. As the concentration of HD-CsrA was increased, a complex with considerably slower mobility (Fig. 3a-f, lane 5, complex 4) was formed at the expense of complex 3, implying that it is composed of 2:1 CsrA:RNA. This complex was expected, because the affinity of the R44A substitution, which is present at mutant binding surface the HD-CsrA, for a high-affinity target site is very low (Kd of the R44A homodimer was ∼180 nM) 32 and should not compete effectively with the WT surface of a second HD-CsrA molecule for RNA-binding. The concentration of free HD-CsrA required to fully compete for the second high-affinity site after the initial binding event was between 2.5 nM and 62.5 nM for every RNA (complex 4) except for RNA18-2BS (Fig. 3). Thus, the second high affinity target site on RNA18-2BS appears to have a tighter association with the initially bound HD-CsrA dimer compared to the other RNAs, resulting in a complex that apparently hindered competition by the second HD-CsrA dimer (Fig. 3c, lane 5, complex 4). Notably, the mobility of complex 3 (HD-CsrA bound to a dual site RNA) was not consistently equal to that of complex 6 (HD-CsrA bound to single-site RNA) for every pair of synthetic oligonucleotides, as might have been predicted (compare Fig. 3a, lane 4 with 3d, lane 4). It is known that while the R44A mutation severely reduces the binding affinity of a CsrA homodimer double mutant for RNA (∼180 nM), it does not eliminate it.32 Therefore, this variation may be due to the residual affinity between the mutant RNA binding surface of CsrA and the second high affinity binding site of the RNA (e.g., Fig. 3d lane 4).
Unlike the HD-CsrA, addition of the higher concentration of WT-CsrA to RNAs with two high affinity binding sites only led to putative 2:1 CsrA:RNA complexes with the two shortest RNAs, RNA10-2BS and RNA14-2BS (Fig. 3a, b, lanes 2 and 3, complex 2). At an intersite distance of 10 nt, WT-CsrA appears to be only weakly associated to one of the two target sites to which it is bound, allowing a second WT-CsrA to successfully compete for binding at one site. Because dual site RNAs with intersite distances ≥ 18 nt (RNA18-2BS, RNA28-2BS, RNA43-2BS and RNA63-2BS) did not show any evidence of forming 2:1 complexes, they apparently associate rapidly and tightly with a single WT-CsrA protein at both GGA targets, preventing competition by a second CsrA protein for binding to either target site. We conclude that RNAs with target sites separated by only 10 nt do not form stable bridge complexes with both RNA binding surfaces of CsrA, presumably due to steric restrictions. Curiously, the smallest synthetic RNA tested (RNA10-2BS) exhibited a distinct shifted species (Fig. 3a, lane 2, top band) that is not consistent with complexes 1 through 6. This may represent an alternate protein-RNA conformation or more likely, a complex of higher stoichiometry (2:2 CsrA:RNA). Recall from Fig. 1 that addition of a small RNA to a CsrA-RNA complex can cause increased gel mobility.
At target separation distances ≥ 18 nt there was apparently no steric restriction imposed by the protein or RNA for dual-site binding. Therefore, 18 nt may be near the optimum spacing distance between RNA targets. As seen in Figure 3c, the WT-CsrA/RNA18-2BS shift (lanes 2 and 3, complex 1) exhibited the same apparent mobility as the HD-CsrA/RNA18-2BS (lanes 4 and 5, complex 3) and WT-CsrA/RNA18-1BS (lanes 6 and 7, complex 5). These three complexes exhibited greater mobility than the 1:1 complex formed by HD-CsrA and RNA18-1BS (lanes 8 and 9, complex 6), which contained a single high-affinity site. The implication of this observation is that the high affinity target sites of RNA18-2BS, as well as the high and low affinity targets of RNA18-1BS, are optimally spaced and situated for dual binding. Thus, near-optimal spacing may counteract the effect of the R44A mutation in the HD-CsrA/RNA18-2BS complex, as well as the effect of the low affinity target site on RNA18-1BS as it binds WT-CsrA. Since the mobility of these shifts resembles that of the WT-dual site reaction (Fig. 3c, lanes 2 and 3, complex 1), these complexes probably represent bridge complexes in which relatively weak protein-RNA interactions are facilitated by tethering (see Discussion). Nevertheless, the combined effects of an R44A protein substitution and a low affinity RNA target site in the HD-CsrA/RNA18-1BS combination (Fig. 3c, lanes 8 and 9, complex 6) apparently prevented the formation of a bridge complex despite near optimal spacing.
At intersite distances greater than 28 nt the WT-CsrA:dual site RNA complexes began to smear in a native polyacrylamide gel and the exact composition of the species was unclear (Fig. 3e and f, lanes 2 and 3, complex 1). Smearing may be due to the extensive linker size of RNA43-2BS and RNA63-2BS (30 and 50 nt respectively), which allows multiple degrees of freedom in the single-stranded intersite region when both target sites are bound. However, we cannot eliminate the possibility that smearing is caused by loss of a compact structure due to transient dissociation of one interaction.
Theoretically, when WT-CsrA is combined with a single-target RNA (e.g., RNA14-1BS), two simple complexes may form, a 1:1 protein-RNA and a 1:2 protein-RNA combination. The presence of only one species (Fig. 3b-f, lanes 6 and 7, complex 5) in all but one of these reactions suggests that although WT-CsrA has the capacity to bind two oligonucleotides, binding of one RNA may sterically inhibit the binding of a second RNA at the free surface (i.e. there is bridging to the low affinity site). An exception to this observation was RNA10-1BS (Fig. 3a, lane 7), which formed a second shift at low protein concentrations that migrated slightly slower than the apparent 1:1 HD-CsrA:RNA complex (Fig. 3a, lanes 9 and 10). We have not determined the basis of the latter observation. However, we suspect that the small size of RNA10-1BS decreases steric (bridging) constraints and allows binding of two RNAs to the same protein. In this case, the RNA oligonucleotide appears to be of sufficient size such that the relative influence of the Stokes radius of the putative 1:2 CsrA:RNA outweighs that of the increased negative charge, resulting in a slower migrating shift.
Stoichiometry of CsrA:RNA complexes
The experiments described above suggested that fast moving complexes formed between WT-CsrA and RNAs with two high-affinity binding sites differed in conformation but not in stoichiometry from the 1:1 complexes formed between HD-CsrA and single-site RNAs. To test this hypothesis, the protein:RNA stoichiometries of four representative CsrA-RNA complexes were examined by EMSA. As described in the Materials and Methods, [γ-32P]-labeled RNAs in gels were detected by phosphorimager analysis, followed by transfer of CsrA protein to a PVDF membrane and subsequent immunoblotting using a polyclonal antibody for CsrA.48 The stoichiometries of WT-CsrA and HD-CsrA complexes with RNA28-2BS and RNA28-1BS were chosen for this analysis because they yielded unambiguous shifts. Under the chosen conditions, the HD-CsrA:RNA28-1BS reaction yielded only a single shift (Fig. 4, lane 1), which was concluded to represent the 1:1 complex of HD-CsrA dimer bound to RNA, and its RNA:protein ratio was assigned a value of 1.0. The stoichiometries of the other shifted complexes were determined by comparison to this one (Fig. 4). The stoichiometry of the slow-moving complex formed by HD-CsrA:RNA28-2BS (Fig. 4, lane 3) was determined to be 1.9 ± 0.1, indicating that it contains two HD-CsrA dimers, each of which is bound to a high-affinity site of this RNA. The fast-moving WT-CsrA:RNA28-1BS complex (Fig. 4, lane 2) was present at a protein-RNA ratio of 1.1 ± 0.2. This species traveled significantly faster than HD-CsrA:RNA28-1BS (lane 1). This difference was likely due to an additional weak association between the low affinity RNA target site from RNA28-1BS and the free RNA binding surface of the WT-CsrA protein to which it was tethered. The fastest migrating complex, WT-CsrA:RNA28-2BS (Fig. 4, lane 4), had a protein-RNA ratio of 0.96 ± 0.1, consistent with one WT-CsrA protein bound to a single RNA. This experiment demonstrated conclusively that although WT-CsrA/RNA28-2BS exhibits greater mobility, it nevertheless contains the same ratio of protein to RNA as the apparent 1:1 HD-CsrA/RNA28-1BS complex. These results support the interpretations from gel shifts (Fig. 3) that WT-CsrA bridges both high affinity target sites of RNA28-2BS forming a compact structure that migrates faster than HD-CsrA/RNA28-1BS. By extrapolation, the fast-moving complexes formed by the other dual site RNAs and WT-CsrA are also inferred to represent bridge complexes (Fig. 3 complex 1).
Fig. 4.
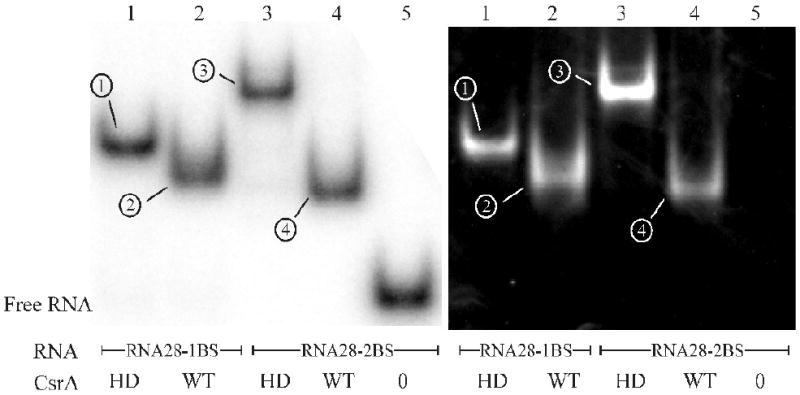
Stoichiometric measurements of CsrA:RNA ratios within shifted complexes. EMSA experiments were conducted similar to Figure 3 with WT-CsrA or HD-CsrA in combination with either RNA28-2BS or RNA28-1BS. After detection of labeled RNA by PhosporImager analysis (left panel, “RNA-EMSA”), protein was transferred to a PVDF membrane by electroblotting and probed for CsrA protein (right panel, “Western Analysis”) using a CsrA-specific polyclonal antibody48; the exact protocol is outlined in the Materials and Methods. RNA and protein bands were quantified using Quantity One software (Biorad, Hercules, CA) and the HD-CsrA:RNA28-1BS (complex 1) ratio was set to 1.0 (explained in materials and methods and results). All other protein:RNA complexes were compared to HD-CsrA:RNA28-1BS for determination of their relative CsrA:RNA ratios as follows: WT-CsrA:RNA28-1BS (complex 2) = 1.1 ± 0.2; HD-CsrA:RNA28-2BS (complex 3) = 1.9 ± 0.1; WT-CsrA:RNA28-2BS (complex 4) = 0.96 ± 0.1. Stoichiometric analyses were performed in triplicate and representative EMSA and western blots are shown.
Both CsrA RNA-binding surfaces are necessary for efficient regulation
Having established parameters by which WT-CsrA protein can bind simultaneously to two target sites on a single RNA, we next explored the biological relevance of this arrangement. The presence of two or more RNA target sites within a CsrA-regulated transcript could cause tighter binding of a single CsrA protein, the formation of an RNA secondary structure such as a loop, and/or the recognition of a low affinity target by tethering of CsrA at a nearby high affinity site. Each of these consequences may result in increased translational inhibition with respect to a CsrA protein with a single binding surface. To test whether both CsrA binding surfaces of a dimer were necessary for efficient gene regulation, we performed S30-directed in vitro coupled transcription-translation49 experiments using either WT-CsrA or HD-CsrA protein (Fig. 5). The 5′-untranslated leader of glgCAP contains two previously characterized CsrA target sites; one that overlaps the SD sequence and one that is situated 28 nt upstream.37 Supercoiled plasmids containing a wild-type glgC′-‘lacZ translational fusion (pCZ3-3) or the same construct with a 3 nt deletion of the upstream CsrA target site (glgCΔGGA′-‘lacZ, pCSB25),37 were used as DNA templates for the reactions. An analysis of the expression data at 50% relative GlgC-LacZ polypeptide synthesis revealed that WT-CsrA was ∼14 fold more efficient at repressing expression from the wild-type glgC template (IC50 ≅ 22 nM) compared to the HD-CsrA (IC50 ≅ 323 nM) (Fig. 5a). When this value is corrected for the number of RNA-binding surfaces per dimer, repression by the WT-CsrA was ∼7-fold greater than HD-CsrA. When the upstream CsrA binding site of the glgC′-lacZ fusion was deleted from the template, repression by HD-CsrA was unaffected (IC50 ≅ 310 nM). In contrast, translational repression by the WT-CsrA protein decreased to ∼7-fold (IC50 ≅ 44 nM) compared to the HD-CsrA, or ∼3.5-fold when adjusted for the number of binding surfaces per dimer (Fig. 5b). These results indicated that both RNA-binding surfaces of CsrA and both CsrA binding sites of the glgCAP 5′-leader are required for maximal regulation of this operon.
Fig. 5.
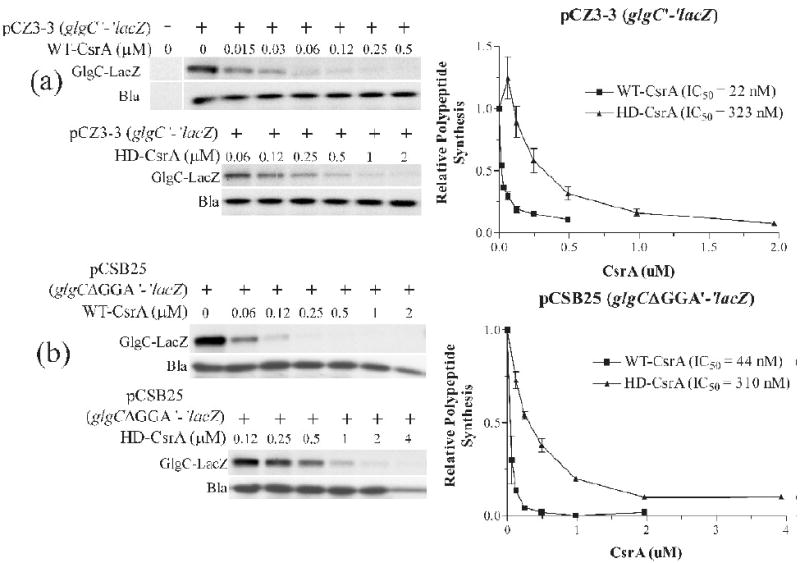
Effects of WT-CsrA or HD-CsrA on the expression of glgC′-‘lacZ or glgCΔGGA′-‘lacZ translational fusions in S-30 coupled transcription-translation reactions. a) Reactions contained 2 μg of supercoiled plasmid pCZ3-3 (glgC′-‘lacZ) and either WT-CsrA (top panel) or HD-CsrA (bottom panel) at the concentrations indicated; right panel graphically displays quantified results. b) Similar reactions were conducted with 2 μg of supercoiled plasmid pCSB25 (glgCΔGGA′-‘lacZ). The concentration of CsrA that leads to a 50% inhibition of gene expression (IC50) was calculated based on a simple point-to-point curve and a Hill-Slope (4PL) model, both of which gave similar values. Simultaneous expression from the bla gene of these plasmids (Bla) served as an internal control for nonspecific inhibition. Graphpad Prism version 3 was used for graphical representations and IC50 calculations. In vitro transcription-translation assays were conducted at least twice for each set, with similar results.
Additional, uncharacterized CsrA target sites in the glgCAP 5′-leader RNA
HD-CsrA was expected to repress protein expression at similar levels from the WT and mutant glgC′-‘lacZ templates because binding to the upstream CsrA target site should not block translation, and both templates contained the same CsrA binding site overlapping the SD sequence. However, a greater difference in IC50 might have been anticipated for WT-CsrA with these templates if the upstream binding site of glgC mRNA was primarily responsible for differential repression by the two proteins (because of the ability of WT-CsrA to bridge). A possible explanation was that the WT glgCAP leader contained additional CsrA binding sites, which could mediate bridging of WT-CsrA to the SD sequence in the mutant transcript. Therefore, we searched for additional CsrA target sites in the glgC leader. While previous footprinting results identified only two CsrA binding sites in the glgCAP leader transcript (Fig. 6b, BS2 and BS4), a position-weighted matrix (pwm) search tool identified a total of four potential CsrA binding sites, including the two identified by footprinting.37; 45 As protection at BS1 and BS3 (Fig. 6b) was not observed by footprinting, a 3′-boundary analysis was performed to search for evidence of these additional CsrA binding sites. The information derived from a boundary experiment differs from the information obtained from a footprint analysis. Whereas a footprint reveals the nucleotides that are protected from enzymatic or chemical probes, 3′-boundary experiments reveal the 3′-nucleotides that are required for binding. Thus, the two techniques are complementary. After RNAs were 5′-end-labeled, base hydrolyzed and incubated in the presence of 1 μM CsrA, samples were fractionated through a native polyacrylamide gel followed by autoradiography. Four diffuse bands were observed corresponding to CsrA-glg leader RNA complexes and a single band containing unbound RNA. The RNA from each of these bands was gel purified and subsequently fractionated side by side under denaturing conditions. The cutoffs for the 3′-boundary analysis were relatively sharp and revealed the 3′-boundaries of CsrA binding sites at positions C86, U97, G113 and G128 (Fig. 6a and b). Each of these binding sites (BS1-BS4) corresponds to those identified by RNA footprinting and/or the pwm. Thus, boundary analysis confirmed that two CsrA target sequences (BS1 and BS3) lie on either side of the originally characterized upstream site within a stem loop (BS2), for a total of four target sites. The two new sites also are reasonable matches to the SELEX-derived consensus of 5′-RUACARGGAUGU-3′39 and an RNA secondary structure algorithm predicted that BS1 may form a stem loop structure (Fig. 6c).50; 51 The presence of two additional sites strongly suggests a redundant binding mechanism, wherein the deletion of a high affinity CsrA target site, which was identified by footprinting, was partially compensated by one or both of the nearby sites that remained in the mutant glgC transcript, reducing the overall impact of the GGA deletion on CsrA-mediated translational repression.
Fig. 6.
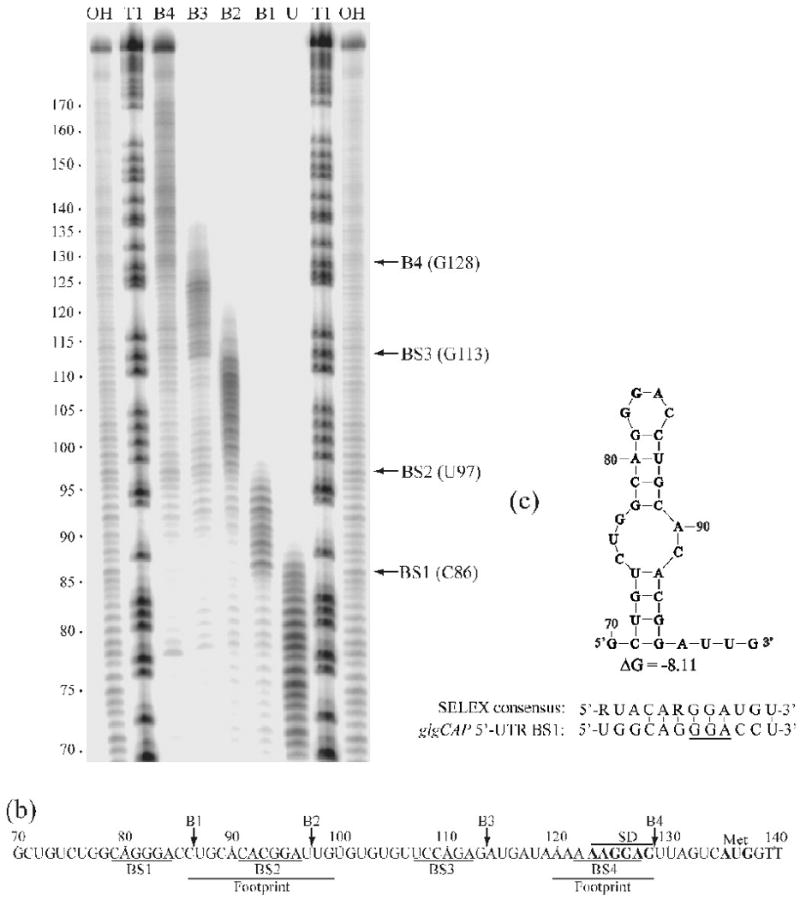
Boundary Analysis revealing additional uncharacterized CsrA target sites in the glgCAP 5′-leader. A) Limited alkaline hydrolysis ladders of glg RNA incubated with 1 μM CsrA. CsrA-RNA complexes were separated from unbound RNA on a native gel and subsequently fractionated through a 10% denaturing gel (shown). Lanes corresponding to distinct bound complexes (B1, B2, B3 or B4) and unbound (U) RNA are shown. Lanes corresponding to limited base hydrolysis (OH) and RNase T1 digestion (T1) ladders are indicated. Positions of the boundaries are marked with arrows on the right. Numbering on the left is from the start of glgCAP transcription. b) The glgCAP leader transcript contains four CsrA binding sites (BS1-BS4). Positions of the 3′ boundaries (B1-B4), as well as the previously identified CsrA footprints are shown.37 Positions of the glgC SD sequence and translation initiation codon (Met) are also shown. c) UNAfold model (an algorithm based on Mfold68) of RNA structure that includes BS1 within the single-stranded region of a hairpin loop. Note that hairpins containing BS1 and BS2 will not form simultaneously because they share common stem sequences. ΔG of this structure and a comparison of BS1 with the SELEX-derived CsrA consensus sequence is shown.39
Discussion
CsrA represents a novel class of RNA-binding global regulatory proteins that possess two binding surfaces located on opposite sides of a symmetrical dimer.29-33 The amino acids that mediate RNA binding have been thoroughly defined, the most important being R44.30; 32 The RNA target to which CsrA binds has also been well characterized, consisting of a semi-conserved sequence surrounding a nearly invariant “GGA” motif, which is often centered in the single-stranded loop of a hairpin.37; 39; 42 The present study examined the higher order binding properties of the CsrA dimer. The intervening spacing between RNA target-sites was found to be important for the stability of the resulting ribonucleoprotein complexes, and the presence of both RNA-binding surfaces of CsrA was determined to be important for its function as a translational repressor in a well-defined in vitro model system. Thus, our findings further define the complex manner in which CsrA interacts with RNA targets and ultimately regulates gene expression.
By comparing recombinant CsrA proteins containing one (HD-CsrA) or two (WT-CsrA) functional RNA-binding surfaces, we demonstrated that a WT-CsrA dimer can bind to either one or two RNA molecules. Furthermore, CsrA can form a bridge complex wherein one protein is bound to two target sites of an RNA when they are located as close together as 10 nt or as distant as 63 nt. An intersite distance of 18 nt may provide near optimal spacing between target sites because this distance appeared to compensate for defects in either a secondary RNA target site or a CsrA binding surface, but not both. A spacing of < 18 nt was detrimental for tight bridging, as binding to one of the target sites was easily displaced by the addition of excess CsrA to form a tripartite complex containing two CsrA dimers and one RNA molecule. Conversely, RNAs with intersite distances of ≥ 18 nt formed stable bridge complexes with WT-CsrA, and neither of the protein-bound target sites could be displaced by free CsrA. We note that the RNAs used in these studies were designed to contain CsrA binding sites within single-stranded loops of hairpins, the preferred recognition site.39 It is possible that spacing constraints may differ in unstructured or alternatively structured RNAs. Presumably, the sequence recognition and spacing constraints of CsrA have evolutionary implications, wherein gene expression may be fine-tuned by altering the sequence of a CsrA target or by increasing or decreasing the distance between binding sites in the 5′-leader of a transcript.
Tethering of CsrA at one RNA target should lead to an increase in the local concentration of secondary sites in the RNA, favoring formation of a bridge complex.38 It follows that such binding may create an RNA repression loop or other secondary structure. Research by Rippe (2001) supporting such a model calculated that the optimum interaction distance between two proteins bound to the same RNA is 15-20 nt, and the local protein concentration increases to approximately 1 mM in this case.38 As this distance deviates from the optimum, the apparent local protein concentration decreases. While this cited work was based on protein-protein association on single-stranded RNA, our studies examined RNAs containing some secondary structure and dual target sites tethered by a single protein. Nevertheless, the conclusions can be generally applied to the present study. Thus, we estimate that given an RNA containing two optimally spaced target sites, initial binding of CsrA at one site should produce a local concentration on the order of 1 mM for the remaining target site, facilitating low affinity binding interactions. This principle was illustrated by the apparent formation of bridge complexes between HD-CsrA and a dual site RNA with 18 nt spacing (Fig. 3c, d, lane 4) or between WT-CsrA and RNA with a single high-affinity binding site (Fig. 3c, d, lanes 6 and 7).
Based on previous studies,48 we calculated the CsrA binding surface concentration in the cytoplasm to be ∼20 μM (data not shown), which is nearly two orders of magnitude below the theoretical local concentration of CsrA when it is bound to a dual-site transcript. Consequently, CsrA binding at one site almost certainly leads to a cooperative interaction at an adjacent site, as opposed to contact by a second free CsrA dimer, under physiological conditions. Indeed, cooperative CsrA-RNA interaction has been observed previously.17; 21; 28 Note that our calculation for CsrA surfaces does not distinguish between surfaces bound to RNA (CsrB/C and mRNA) or RNA-free surfaces. The actual free concentration is likely much lower (sub-micromolar), because most target transcripts identified to date are bound with Kd (apparent equilibrium binding constant) values in the range of 1-40 nM, making bridging interactions even more certain.17; 21; 28; 37
The observed spacing requirement for optimal binding may explain a long-standing observation that the CsrA-CsrB complex is globular in shape.27 E. coli CsrB contains as many as 22 possible CsrA target sites with an average spacing of 12.25 nt, and all but 2 pairs of sites are separated by fewer than 18 nt (Table 3). This short average spacing distance suggests that a single CsrA protein would not tightly associate with most adjacent target sites. Instead, it is more likely that CsrA usually bridges non-adjacent sites (i.e., sites separated by ≥ 18 nt), giving rise to an energetically stable globular complex. A review of other small regulatory RNAs in the CsrA/RsmA circuitry of related bacteria revealed a similar pattern of short intersite distances (Table 3). Consequently, V. cholerae CsrB, P. fluorescens CHA0 RsmX and RsmZ, and P. aeruginosa PrrB would be expected to adopt a globular form when bound by CsrA/RsmA. We previously found that turnover of E. coli CsrB and CsrC RNAs requires a non-nucleolytic specificity factor, CsrD, in conjunction with the endonuclease RNase E.52 Whether the formation of a globular complex with CsrA affects turnover of CsrB through the CsrD-RNaseE pathway or other biological properties of this complex remains an open question. In contrast, CsrA/RsmA-regulated mRNAs contain a higher proportion of target sites separated by ≥ 18 nt compared to the noncoding regulatory RNAs (Table 3), strongly suggesting that CsrA often binds to adjoining target sites in CsrA-regulated mRNAs.
Table 3.
RNA targets of CsrA (or its orthologs) and predicted or experimentally determined spacinga.
| RNA targeta | Sourceb | Target sites predicted or determined | Average distance between adjacent sites (nt) | Pairs of sites separated by <18 nt | Pairs of sites separated by ≥18 nt | Target overlap with SD? | Reference |
|---|---|---|---|---|---|---|---|
| CsrB | E. coli | ∼21 | 12.25 | 18 | 2 | N/A | 27 |
| CsrC | E. coli | ∼9 | 11.4 | 7 | 1 | N/A | 28 |
| CsrB | V. cholerae | ∼25 | 11.6 | 21 | 3 | N/A | 22 |
| RsmX | P. fluorescens | ∼5 | 10 | 4 | 0 | N/A | 77 |
| RsmZ | P. fluorescens | ∼7 | 9.8 | 6 | 0 | N/A | 78 |
| RsmY | P. fluorescens | ∼6 | 12.8 | 3 | 2 | N/A | 42 |
| PrrB | P. aeruginosa | ∼6 | 11 | 5 | 0 | N/A | 43 |
| hag | B. subtilis | 2 | 26 | 0 | 1 | Yes | 40 |
| ycdT | E. coli | 2 | 29 | 0 | 1 | Noc | 53 |
| ydeH | E. coli | 2 | 11 | 0 | 1 | Yesc | 53 |
| hcnA | P. fluorescens | 5 | 15.5 | 2 | 2 | Yes | 44 |
| glgC | E. coli | 4 | 11.3 | 3 | 0 | Yes | 37 |
| cstA | E. coli | 4 | 14 | 2 | 1 | Yes | 21 |
| pgaA | E.coli | 6 | 29.4 | 3 | 2 | Yes | 17 |
sRNAs that bind to CsrA are capitalized (CsrB – PrrB); mRNA leaders are italicized.
Abbreviations: E., Escherichia; V., Vibrio; P., Pseudomonas.
Putative mRNA target site, not tested experimentally. Note that ycdT lacks a predicted target site at SD, but contains one overlapping the putative initiation codon, which should also inhibit translation.
Our studies provide the first experimental demonstration of the function of dual RNA-binding sites of CsrA in regulation. The glgCAP 5′-leader contains two previously characterized CsrA target sites, one overlapping the SD sequence and the other 28 nt upstream in an RNA hairpin. 37 Two additional target sites were identified in the present study, which are located on either side of the original upstream hairpin. We found that while the HD-CsrA protein displayed approximately one third the affinity for a single-site oligoribonucleotide target compared to the WT-CsrA (Fig. 2), it was ∼14-fold less efficient (7-fold based on molar equivalence of RNA binding surfaces) in repressing a glgC′-‘lacZ reporter fusion (Fig. 5). When the GGA sequence of the upsteam RNA target site (Fig. 6, BS2) was deleted, regulation by HD-CsrA was unaffected, while the relative difference in regulation between the WT-CsrA and HD-CsrA proteins decreased to 7-fold (∼3.5-fold based on equal numbers of binding surfaces) (Fig. 5). This finding supports a model of repression wherein two RNA target sites mediate translational repression by interacting simultaneously with one CsrA dimer (Fig 7). Apparently, binding of HD-CsrA only inhibits translation if it blocks the SD sequence, but not if it binds to an upstream stem-loop. For that reason, deletion of one upstream site negatively impacted WT-CsrA activity but did not affect regulation by HD-CsrA. WT-CsrA exhibited a modest, yet still significant, two-fold decrease in activity after deletion of the original stem-loop target (BS2) in the glgCAP leader. Deletion of the conserved GGA at BS2 was previously found to decrease, but did not abolish, CsrA binding to this leader (Kd was increased from 39 to 150 nM).37 The unexpectedly modest effect of this deletion on regulation by WT-CsrA might be partially explained by residual affinity for the deleted upstream site. More likely, the presence of two additional upstream targets (BS1 and BS3), including one that could form an RNA hairpin (Fig. 6), compensates for the loss of the apparent high-affinity binding site. Given the distances between adjacent binding sites in the glgCAP 5′-leader (Table 3), bridging by CsrA is likely restricted to BS1 and BS3, BS1 and BS4, or BS2 and BS4.
Fig. 7.
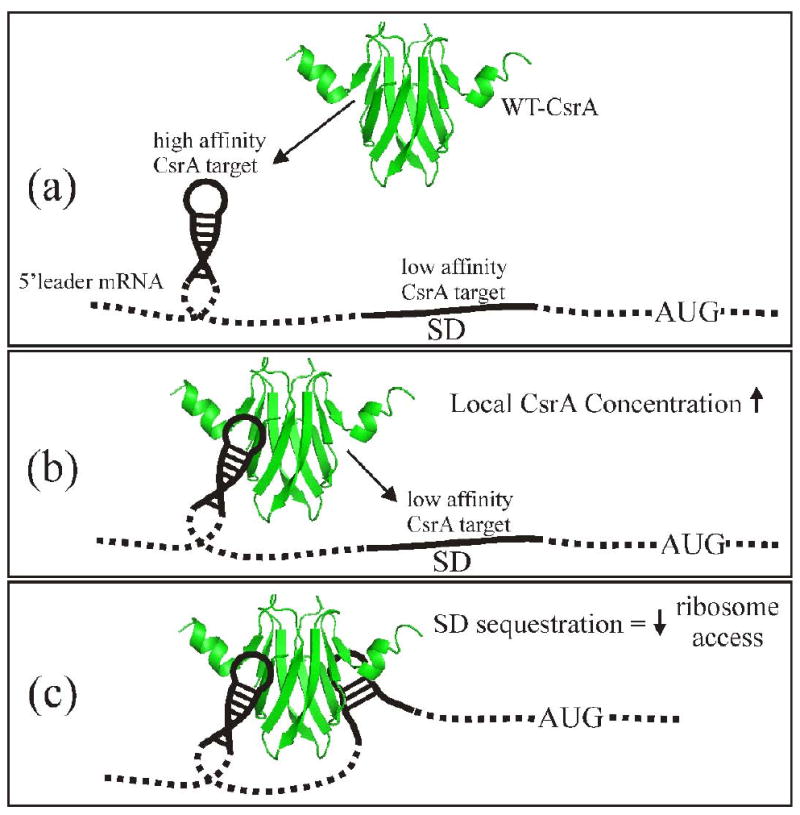
A model for CsrA regulation by binding to the 5′ leader of repressed transcripts, based on results from RNA gel shifts and glgC S30 transcription-translation. a) CsrA initially binds at a high affinity to a target site, commonly located within a hairpin structure that lies proximal to the SD. b) After initial binding, the increased local concentration of CsrA allows the free RNA-binding surface to interact with the downstream low affinity target site overlapping the SD sequence. c) Binding of the low affinity target site sequesters the SD sequence, thereby blocking ribosome loading and decreasing translational initiation. SD = Shine Dalgarno; AUG = methionine start codon.
While binding of CsrA at two sites was needed for complete repression in the glgC studies, our data suggest that it was not necessarily the occlusion of both target sites by one protein that is essential. Rather, efficient interaction with a low affinity target that directly overlaps the SD may require the tethering of CsrA to a nearby high affinity target within a stem-loop to increase the local concentration of this protein. Note that while primary sequence is critical for RNA recognition by CsrA, localization of a target sequence within the loop of a hairpin, as observed in the glgC leader, substantially improves binding to RNA.39 Thus, the apparent way in which CsrA regulates glgC expression may reflect a general repression mechanism that involves initial binding to a high affinity primary RNA target site (Fig. 7a), in the case of glgC regulation, this is the upstream hairpin (BS2) that was observed in previous footprinting studies or the alternate upstream hairpin (BS1) identified by boundary analysis.37 A tethered CsrA protein would then recognize and bind to the possibly lower affinity target that overlaps the SD sequence (Fig. 7b, c). Considering that CsrA regulates expression of horizontally acquired virulence factors of many species, an important evolutionary implication of this mechanism is that the SD sequence need not be altered extensively to accommodate weak CsrA binding, which might otherwise affect intrinsic translational capacity of the translation initiation region.
Most negatively-regulated transcripts studied to date contain at least one CsrA target site in the 5′-leader mRNA in addition to a site that overlaps the SD (Table 3).17; 21; 40; 53 While our model in Figure 7 describes the mechanism for an mRNA with two CsrA binding sites, several mRNA targets contain four or more binding sites, including glgCAP (Table 3). The presence of four CsrA binding sites in the glgCAP leader suggests that two CsrA dimers could be bound to any given transcript. While BS3 does not overlap the SD sequence, it is part of the ribosome binding site. Thus, the presence of one CsrA dimer bound to BS1 and BS3 and a second dimer bound to BS2 and BS4 should further reduce ribosome binding, and hence GlgC synthesis.
Because tethering increases the probability of molecular interaction and thus proper regulation, it is not surprising that examples can be found in both eukaryotes and prokaryotes. For instance, translational initiation in eukaryotes involves a large, complex structure which includes the poly(A)-binding protein (PABP), that is tethered to the mRNA 5′-leader and interacts with the poly(A) tail and other regulatory proteins found at the 3′-untranslated region (UTR), such as the GAIT complex.2 In addition, the bacterial endoribonuclease RNaseE may specifically recognize monophosphates and tether itself to the 5′-termini of transcripts (and possibly to the 3′-poly(A) tail via bacterial PABP) while it cleaves internal targets.54; 55 Additional examples of tethering include the classic prokaryotic transcriptional regulators CI (λ phage repressor), GalR (galactose repressor), LacI (lactose repressor) and AraC (arabinose activator/repressor) which bind operator sequences near DNA promoters. Tethering promotes protein self-association, bringing non-contiguous stretches of double stranded DNA together, creating repression loops in the case of gal56; 57 and lac,58 activation loops for ara59; 60 and a combination lytic repressing/lysogenic activating loop for λ.61; 62 Similar to these transcriptional regulators, CsrA may alter the structure of the 5′-leader of a message by tethering distant sites, which by itself, could affect ribosome access and translational capacity. However, we suspect the primary regulatory capability of CsrA is based on RNA-binding and direct occlusion of the translation initiation region of repressed genes. In support of this interpretation, we recently demonstrated that CsrA binds and modestly regulates hfq mRNA translation using only a single CsrA target site that overlaps the hfq SD sequence.45 A role of higher order RNA structural alterations in CsrA-mediated activation processes, such as stabilization of flhDC mRNA,19 is also possible, but at the present time, no molecular model exists for activation.
We should note that while a looping mechanism may be common to both CsrA and transcriptional regulators, the persistence length (measure of relative polymer stiffness, also known as Kuhn length) of double stranded DNA (∼150 bp) dictates that it is approximately 20-fold less flexible than a similar length of single-stranded DNA or RNA.38 Therefore, CsrA can bridge targets that are much closer together than those of a dsDNA-binding protein; minimum of 90 bp for LacI58 and 2.4 kb for CI61 vs. ≤ 10 nt for CsrA.
As mentioned above, CsrA inhibits hfq expression by binding to a single RNA target site that blocks access to the SD sequence.45 Is it possible that another RNA could be bound to the second CsrA binding surface that should be available in this case? Because CsrA contains two independent RNA-binding surfaces, a single CsrA dimer might bind two different messages or small RNAs in a manner reminiscent of Hfq itself. Hfq facilitates RNA-RNA interactions through two independent RNA-binding surfaces located on opposite sides of the hexamer; for instance, Hfq facilitates the interaction of DsrA sRNA with the rpoS message.63; 64 Although CsrA can bind simultaneously to two independent oligoribonucleotides, further studies are required to know whether such intermolecular bridging has any biological relevance.
Materials and Methods
Bacterial Strains and Plasmids
All strains and plasmids used in this study are listed in Table 2. For routine culture of bacteria, strains were grown in Luria-Bertani (LB) medium at 37 °C with shaking.65 Antibiotics were added to media as needed at the following concentrations: ampicillin, 100 μg ml-1; kanamycin, 100 μg ml-1; chloramphenicol, 25 μg ml-1; tetracycline, 10 μg ml-1; spectinomycin, 35 μg ml-1; streptomycin, 35 μg ml-1.
Table 2.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Description or relevant genotype | Source or reference |
|---|---|---|
| E. coli K-12 Strains | ||
| TRBW3414 | csrA::kan, rpoS | 75 |
| TRCF7789B-C- | CF7789 csrA::kan ΔcsrB::cam ΔcsrC::tet | 32 |
| Plasmids | ||
| pJMCN4 | aadA gene from pAH144 cloned into pBATC6; StrR/SpcR | This Study |
| pCZ3-3 | ΦglgC′-‘lacZ in pMLB1034, AmpR | 49 |
| pCSB25 | pCZ3-3 with upstream ΔGGA | 37 |
| pAltC4 | T7-driven glgCAP RNA expression construct (+46 to +179 relative to the start of transcription) | 72 |
| pMLB1034 | ‘laZ translational fusion vector; AmpR | 76 |
| pCSRH6-19 | pKK223-3 expressing C-terminal His6-tag CsrA; AmpR | 27 |
| pAH144 | CRIM plasmid, aadA; StrR/SpcR | 67 |
| pTWIN1 | Chitin Binding Domain (CBD) fusion plasmid | NEB |
| pTC14 | CsrA-CBD fusion in pTWIN1 | K. Suzuki |
| pBAD33 | Arabinose inducible vector; CmR | 66 |
| pBATC6 | CsrA-CBD fragment from pTC14 cloned into pBAD33 | This Study |
| pCSRR44A | csrA R44A C-terminal His6 tag, derived from pCSRH6-19 | 32 |
Recombinant DNA techniques
Plasmid pJMCN4 that expresses a wild type CsrA-Intein/Chitin Binding Domain (CBD) fusion monomer was constructed as follows: DNA encoding the exact csrA coding region, minus the termination codon, was cloned into the CBD-fusion plasmid pTWIN1 (NEB, Ipswich, MA) creating the CsrA-CBD fusion construct pTC14 (K. Suzuki, unpublished). The XbaI to PstI DNA fragment from pTC14 containing the CsrA-CBD fusion was subcloned into the arabinose-inducible expression vector pBAD3366 creating plasmid pBATC6 and the insert was then confirmed by sequencing (SeqWright DNA Technology, Houston, TX). This plasmid was then converted to Str/SpcR by cloning a ClaI to NotI DNA fragment from pAH14467 into the ScaI site of pBATC6, creating plasmid pJMCN4. This plasmid was designed such that the CBD was removed from the CsrA-CBD fusion by intein-mediated cleavage.
Protein purification
WT and HD-CsrA proteins were purified from strain TRCF7789B-C- harboring both plasmids pCSRH6-19 and pJMCN4 or pCSRR44A and pJMCN4 as follows: 20 L of culture was grown overnight at 37°C in LB medium with shaking and arabinose (0.2% final concentration) was added 3 hours prior to cell harvest. The cells were concentrated by centrifugation, resuspended in NEB wash buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0) at 1 g per ml and lysed by sonication or French press. The resulting lysate was cleared by centrifugation and combined with 3 ml of Ni-NTA (NEB, Ipswich, MA) for 1 hr to allow binding of CsrA-His6 to the nickel-NTA beads. The lysate-Ni-NTA mix was applied to a gravity flow column and the beads were washed with 30 ml of wash buffer containing 20 mM imidazole. CsrA protein was eluted with 10 ml wash buffer containing 250 mM imidazole and then dialyzed against at total of 8L of CBD Buffer #2 (20 mM Tris-HCl pH 7.0, 500 mM NaCl, 1 mM EDTA, NEB Impact Kit). The dialyzed protein was then passed by gravity flow over a column containing chitin beads (NEB, S6651S) that were previously equilibrated with Buffer #2. 10 ml of Buffer #3 (20 mM Tris-HCl pH 8.5, 500 mM NaCl, 1 mM EDTA, 40 mM DTT) was passed over the column, flow was stopped and the intein cleavage reaction was allowed to continue overnight at 4°C. Flow was continued and the eluant was collected as one fraction. The column was then washed twice with 5 ml Buffer #2 and this was combined with the initial eluate. The pooled eluate was dialyzed against a total of 8L of NEB wash buffer without imidazole. Imidazole was then added to the dialyzed protein to 10 mM and 0.25ml of Ni-NTA slurry was added, incubated at 4°C for 1 hour with gentle shaking and the beads were then applied to a gravity flow column. The beads were washed with wash buffer containing 10 mM imidazole and the HD-CsrA or WT-CsrA were eluted with 5 ml of wash buffer containing 250 mM imidazole and collected in ∼1 ml fractions. Protein size and purity was assessed by SDS-PAGE (15%) and Coomassie blue staining, which for the WT-CsrA dimer revealed two closely migrating protein bands of equal staining intensity and of sizes consistent with a WT CsrA monomer released by intein cleavage (6,855 Da) and a WT CsrA monomer containing a hexa-histidine tag (7,678 Da). The HD-CsrA dimer revealed a similar pair of bands corresponding to a site-directed alanine-substituted CsrA monomer with a C-terminal hexa-histidine tag (7,593 Da) and a WT CsrA monor released by intein cleavage (data not shown). Protein concentration was measured using the bicinchroninic acid (BCA) assay68 (Pierce Biotech, Rockford, IL).
RNA Electrophoretic Gel Mobility Shift Assay
RNA gel mobility shift assays were carried out according to previously published procedures.17; 32 Protein calculations were based on the mass of the CsrA-His6/CsrA WT dimer (14,516 Da) or the CsrAR44A-His6/CsrA heterodimer (14,448 Da). RNA probes were designed based on the high-affinity consensus CsrA binding sequence determined by SELEX analysis.17; 32 RNA probes were either synthesized by Integrated DNA Technologies (Coralville, IA) or synthesized in vitro with the Megashortscript Kit (Ambion, Austin, TX) and 5′-end-labelled using T4 polynucleotide kinase and [γ-32P]-ATP. RNA radiolabeling, gel shift experiments, detection, quantitation and calculations were conducted according to previously published procedures.32; 69 All EMSAs were performed with [γ-32P]-labeled RNA at 0.06 nM final concentration, unless stated otherwise. EMSAs were conducted at least twice for each set of RNA probes to confirm banding patterns and/or Kd measurements.
Protein-RNA Stoichiometry Measurements
RNA28-2BS and RNA28-1BS (Table 1) were 5′-end-labeled with [γ-32P]-ATP and EMSAs were conducted as previously detailed using WT-CsrA and HD-CsrA.17; 32; 39 After electrophoresis, gels were wrapped in plastic and exposed to a flexible Kodak storage phosphor plate (Rochester, NY) for 1 hour, radioactive bands were then visualized with a GE LifeSciences Phosphoimager (Piscataway, NJ). CsrA protein was transferred from the previously visualized gels to a Biorad Immunoblot PVDF membrane (Hercules, CA) by electroblotting in 1× TBE buffer for 45 minutes at 4°C. Blots were blocked with Superblock T20 (cat.# 37536, Pierce Biotech, Rockford, IL) for 1 hour at room temperature according the manufacturer's recommendations. Blots were probed with a CsrA-specific polyclonal primary antibody48 at 1:10,000 in blocking reagent overnight at 4°C with gentle shaking, they were then washed 12 times with 30 ml of Tris-Buffered Saline with Tween (0.05%) (cat # 28360, Pierce Biotech, Rockford, IL) then probed with anti-rabbit IgG secondary antibody conjugated to HRP (cat.# 31460, Peirce Biotech, Rockford, IL) diluted in blocking buffer at 8 ng/ml for 1 hr at room temperature with gentle shaking. The blot was washed again 12 times with Tris-Buffered Saline with Tween and developed with SuperSignal West Femto chemiluminescent substrate (cat.# 34095, Pierce Biotech, Rockford, IL) according to the manufacturer's recommendations. Blots were then placed in a plastic development folder and visualized using a Biorad Versadoc for chemiluminescent detection. Radiolabeled RNA bands and chemiluminescent protein bands were physically matched up for size and migration distance and then quantified with ImageQuant (GE Life Sciences, Piscataway, NJ) and Quantity One Software (Biorad, Hercules, CA) respectively. The HD-CsrA/RNA10-1BS standard complex was assigned a stoichiometry of 1.0 because this combination of protein and RNA was specifically designed to only allow for a 1:1 ratio; all other protein:RNA ratios were compared relative to this standard complex. Stoichiometry values represent the average of triplicate experiments.
In vitro coupled transcription-translation
Effects of WT-CsrA or HD-CsrA proteins on glgC′-‘lacZ (pCZ3-3) and glgC′ΔGGA-‘lacZ (pCSB25) expression were examined by S30-driven transcription-translation using an S30 extract from a csrA mutant strain (TRBW3414), as previously described.27; 49 Reaction products were separated by 7.5% SDS-PAGE and radiolabeled proteins were detected by fluorography using sodium salicylate.70 35S-methionine incorporation was measured by densitometry as well as by dissolving excised gel slices in hydrogen peroxide and liquid scintillation counting.49
Boundary Analysis
A 3′-boundary analysis was carried out by modifying previously published procedures.21; 71 Plasmid pAltC4 contains portions of the WT glgCAP operon leader and glgC coding regions (+46 to +179 relative to the start of transcription) cloned downstream of a T7 RNA polymerase promoter.72 RNA was synthesized in vitro using the Ambion MEGAscript kit and plasmid pAltC4 that had been linearized with BamH1 as template. Gel-purified RNA was dephosphorylated with calf intestinal alkaline phosphatase and subsequently 5′-end-labeled using [γ-32P] ATP and polynucleotide kinase. Labeled RNA was gel purified, suspended in TE (10 mM Tris-HCl, pH 8.0, 1 mM EDTA) and renatured by heating to 85 °C followed by slow cooling to room temperature. To generate 5′-labeled alkaline hydrolysis ladders, 60-μl RNA samples (200 nM) were incubated for 5 min at 95 °C in alkaline hydrolysis buffer (100 mM NaHCO3/Na2CO3, pH 9.0, 2 mM EDTA) and then recovered by ethanol precipitation. Hydrolyzed RNAs were mixed with 1 μM CsrA (20 μl reaction volume) in binding buffer (10 mM Tris-HCl, pH 7.5, 100 mM KCl, 10 mM MgCl2, 10% glycerol, 20 mM DTT). Reaction mixtures were incubated for 30 min at 37 °C to allow CsrA-glgC RNA complex formation. Samples were fractionated through 10% native polyacrylamide gels. Bound and unbound transcripts were visualized by autoradiography, excised and eluted from the gel. RNAs were ethanol precipitated and fractionated through 10% denaturing polyacrylamide gels. RNase T1 and alkali digestion ladders of the same transcript were prepared as described previously and used as molecular size standards.73
Acknowledgments
We thank Kazushi Suzuki for construction of the CsrA-intein gene fusion. These studies were supported by the National Institutes of Health (R01 GM059969).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–21. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 2.Abaza I, Gebauer F. Trading translation with RNA-binding proteins. RNA. 2008;14:404–9. doi: 10.1261/rna.848208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolognani F, Perrone-Bizzozero NI. RNA-protein interactions and control of mRNA stability in neurons. J Neurosci Res. 2008;86:481–9. doi: 10.1002/jnr.21473. [DOI] [PubMed] [Google Scholar]
- 4.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–86. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chmiel NH, Rio DC, Doudna JA. Distinct contributions of KH domains to substrate binding affinity of Drosophila P-element somatic inhibitor protein. RNA. 2006;12:283–91. doi: 10.1261/rna.2175706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nogueira T, Springer M. Post-transcriptional control by global regulators of gene expression in bacteria. Curr Opin Microbiol. 2000;3:154–8. doi: 10.1016/s1369-5274(00)00068-0. [DOI] [PubMed] [Google Scholar]
- 7.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–8. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–90. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Babitzke P, Baker CS, Romeo T. Regulation of Translation Initiation by RNA Binding Proteins. Annu Rev Microbiol. 2009 doi: 10.1146/annurev.micro.091208.073514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gopal B, Haire LF, Gamblin SJ, Dodson EJ, Lane AN, Papavinasasundaram KG, Colston MJ, Dodson G. Crystal structure of the transcription elongation/anti-termination factor NusA from Mycobacterium tuberculosis at 1.7 A resolution. J Mol Biol. 2001;314:1087–95. doi: 10.1006/jmbi.2000.5144. [DOI] [PubMed] [Google Scholar]
- 11.Beuth B, Pennell S, Arnvig KB, Martin SR, Taylor IA. Structure of a Mycobacterium tuberculosis NusA-RNA complex. EMBO J. 2005;24:3576–87. doi: 10.1038/sj.emboj.7600829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antson AA, Dodson EJ, Dodson G, Greaves RB, Chen X, Gollnick P. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 1999;401:235–42. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
- 13.Babitzke P, Bear DG, Yanofsky C. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a toroid-shaped molecule that binds transcripts containing GAG or UAG repeats separated by two nucleotides. Proc Natl Acad Sci U S A. 1995;92:7916–20. doi: 10.1073/pnas.92.17.7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori D, Sasagawa N, Kino Y, Ishiura S. Quantitative analysis of CUG-BP1 binding to RNA repeats. J Biochem. 2008;143:377–83. doi: 10.1093/jb/mvm230. [DOI] [PubMed] [Google Scholar]
- 15.Sabnis NA, Yang H, Romeo T. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J Biol Chem. 1995;270:29096–104. doi: 10.1074/jbc.270.49.29096. [DOI] [PubMed] [Google Scholar]
- 16.Liu MY, Yang H, Romeo T. The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J Bacteriol. 1995;177:2663–72. doi: 10.1128/jb.177.10.2663-2672.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol. 2005;56:1648–63. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- 18.Jackson DW, Suzuki K, Oakford L, Simecka JW, Hart ME, Romeo T. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J Bacteriol. 2002;184:290–301. doi: 10.1128/JB.184.1.290-301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei BL, Brun-Zinkernagel AM, Simecka JW, Pruss BM, Babitzke P, Romeo T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol. 2001;40:245–56. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- 20.Ang S, Horng YT, Shu JC, Soo PC, Liu JH, Yi WC, Lai HC, Luh KT, Ho SW, Swift S. The role of RsmA in the regulation of swarming motility in Serratia marcescens. J Biomed Sci. 2001;8:160–9. doi: 10.1007/BF02256408. [DOI] [PubMed] [Google Scholar]
- 21.Dubey AK, Baker CS, Suzuki K, Jones AD, Pandit P, Romeo T, Babitzke P. CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J Bacteriol. 2003;185:4450–60. doi: 10.1128/JB.185.15.4450-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenz DH, Miller MB, Zhu J, Kulkarni RV, Bassler BL. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol Microbiol. 2005;58:1186–202. doi: 10.1111/j.1365-2958.2005.04902.x. [DOI] [PubMed] [Google Scholar]
- 23.Heurlier K, Williams F, Heeb S, Dormond C, Pessi G, Singer D, Camara M, Williams P, Haas D. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J Bacteriol. 2004;186:2936–45. doi: 10.1128/JB.186.10.2936-2945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao NX, Wei K, Chen Q, Meng QL, Tang DJ, He YQ, Lu GT, Jiang BL, Liang XX, Feng JX, Chen B, Tang JL. The rsmA-like gene rsmA(Xcc) of Xanthomonas campestris pv. campestris is involved in the control of various cellular processes, including pathogenesis. Mol Plant Microbe Interact. 2008;21:411–23. doi: 10.1094/MPMI-21-4-0411. [DOI] [PubMed] [Google Scholar]
- 25.Mulcahy H, O'Callaghan J, O'Grady EP, Macia MD, Borrell N, Gomez C, Casey PG, Hill C, Adams C, Gahan CG, Oliver A, O'Gara F. Pseudomonas aeruginosa RsmA plays an important role during murine infection by influencing colonization, virulence, persistence, and pulmonary inflammation. Infect Immun. 2008;76:632–8. doi: 10.1128/IAI.01132-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fortune DR, Suyemoto M, Altier C. Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar Typhimurium. Infect Immun. 2006;74:331–9. doi: 10.1128/IAI.74.1.331-339.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu MY, Gui G, Wei B, Preston JF, 3rd, Oakford L, Yuksel U, Giedroc DP, Romeo T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem. 1997;272:17502–10. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 28.Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I, Baker CS, Georgellis D, Babitzke P, Romeo T. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol. 2003;48:657–70. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- 29.Gutierrez P, Li Y, Osborne MJ, Pomerantseva E, Liu Q, Gehring K. Solution structure of the carbon storage regulator protein CsrA from Escherichia coli. J Bacteriol. 2005;187:3496–501. doi: 10.1128/JB.187.10.3496-3501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heeb S, Kuehne SA, Bycroft M, Crivii S, Allen MD, Haas D, Camara M, Williams P. Functional analysis of the post-transcriptional regulator RsmA reveals a novel RNA-binding site. J Mol Biol. 2006;355:1026–36. doi: 10.1016/j.jmb.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 31.Rife C, Schwarzenbacher R, McMullan D, Abdubek P, Ambing E, Axelrod H, Biorac T, Canaves JM, Chiu HJ, Deacon AM, DiDonato M, Elsliger MA, Godzik A, Grittini C, Grzechnik SK, Hale J, Hampton E, Han GW, Haugen J, Hornsby M, Jaroszewski L, Klock HE, Koesema E, Kreusch A, Kuhn P, Lesley SA, Miller MD, Moy K, Nigoghossian E, Paulsen J, Quijano K, Reyes R, Sims E, Spraggon G, Stevens RC, van den Bedem H, Velasquez J, Vincent J, White A, Wolf G, Xu Q, Hodgson KO, Wooley J, Wilson IA. Crystal structure of the global regulatory protein CsrA from Pseudomonas putida at 2.05 A resolution reveals a new fold. Proteins. 2005;61:449–53. doi: 10.1002/prot.20502. [DOI] [PubMed] [Google Scholar]
- 32.Mercante J, Suzuki K, Cheng X, Babitzke P, Romeo T. Comprehensive alanine-scanning mutagenesis of Escherichia coli CsrA defines two subdomains of critical functional importance. J Biol Chem. 2006;281:31832–42. doi: 10.1074/jbc.M606057200. [DOI] [PubMed] [Google Scholar]
- 33.Schubert M, Lapouge K, Duss O, Oberstrass FC, Jelesarov I, Haas D, Allain FH. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat Struct Mol Biol. 2007;14:807–13. doi: 10.1038/nsmb1285. [DOI] [PubMed] [Google Scholar]
- 34.Gollnick P, Babitzke P, Antson A, Yanofsky C. Complexity in regulation of tryptophan biosynthesis in Bacillus subtilis. Annu Rev Genet. 2005;39:47–68. doi: 10.1146/annurev.genet.39.073003.093745. [DOI] [PubMed] [Google Scholar]
- 35.Hopcroft NH, Manfredo A, Wendt AL, Brzozowski AM, Gollnick P, Antson AA. The interaction of RNA with TRAP: the role of triplet repeats and separating spacer nucleotides. J Mol Biol. 2004;338:43–53. doi: 10.1016/j.jmb.2004.02.038. [DOI] [PubMed] [Google Scholar]
- 36.He F, Dang W, Abe C, Tsuda K, Inoue M, Watanabe S, Kobayashi N, Kigawa T, Matsuda T, Yabuki T, Aoki M, Seki E, Harada T, Tomabechi Y, Terada T, Shirouzu M, Tanaka A, Guntert P, Muto Y, Yokoyama S. Solution structure of the RNA binding domain in the human muscleblind-like protein 2. Protein Sci. 2009;18:80–91. doi: 10.1002/pro.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker CS, Morozov I, Suzuki K, Romeo T, Babitzke P. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol Microbiol. 2002;44:1599–610. doi: 10.1046/j.1365-2958.2002.02982.x. [DOI] [PubMed] [Google Scholar]
- 38.Rippe K. Making contacts on a nucleic acid polymer. Trends Biochem Sci. 2001;26:733–40. doi: 10.1016/s0968-0004(01)01978-8. [DOI] [PubMed] [Google Scholar]
- 39.Dubey AK, Baker CS, Romeo T, Babitzke P. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. Rna. 2005;11:1579–87. doi: 10.1261/rna.2990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yakhnin H, Pandit P, Petty TJ, Baker CS, Romeo T, Babitzke P. CsrA of Bacillus subtilis regulates translation initiation of the gene encoding the flagellin protein (hag) by blocking ribosome binding. Mol Microbiol. 2007;64:1605–20. doi: 10.1111/j.1365-2958.2007.05765.x. [DOI] [PubMed] [Google Scholar]
- 41.Sorger-Domenigg T, Sonnleitner E, Kaberdin VR, Blasi U. Distinct and overlapping binding sites of Pseudomonas aeruginosa Hfq and RsmA proteins on the non-coding RNA RsmY. Biochem Biophys Res Commun. 2007;352:769–73. doi: 10.1016/j.bbrc.2006.11.084. [DOI] [PubMed] [Google Scholar]
- 42.Valverde C, Lindell M, Wagner EG, Haas D. A repeated GGA motif is critical for the activity and stability of the riboregulator RsmY of Pseudomonas fluorescens. J Biol Chem. 2004;279:25066–74. doi: 10.1074/jbc.M401870200. [DOI] [PubMed] [Google Scholar]
- 43.Burrowes E, Abbas A, O'Neill A, Adams C, O'Gara F. Characterisation of the regulatory RNA RsmB from Pseudomonas aeruginosa PAO1. Res Microbiol. 2005;156:7–16. doi: 10.1016/j.resmic.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 44.Lapouge K, Sineva E, Lindell M, Starke K, Baker CS, Babitzke P, Haas D. Mechanism of hcnA mRNA recognition in the Gac/Rsm signal transduction pathway of Pseudomonas fluorescens. Mol Microbiol. 2007;66:341–56. doi: 10.1111/j.1365-2958.2007.05909.x. [DOI] [PubMed] [Google Scholar]
- 45.Baker CS, Eory LA, Yakhnin H, Mercante J, Romeo T, Babitzke P. CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J Bacteriol. 2007;189:5472–81. doi: 10.1128/JB.00529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci U S A. 1998;95:1460–5. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 48.Gudapaty S, Suzuki K, Wang X, Babitzke P, Romeo T. Regulatory interactions of Csr components: the RNA binding protein CsrA activates csrB transcription in Escherichia coli. J Bacteriol. 2001;183:6017–27. doi: 10.1128/JB.183.20.6017-6027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romeo T, Preiss J. Genetic regulation of glycogen biosynthesis in Escherichia coli: in vitro effects of cyclic AMP and guanosine 5′-diphosphate 3′-diphosphate and analysis of in vivo transcripts. J Bacteriol. 1989;171:2773–82. doi: 10.1128/jb.171.5.2773-2782.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–81. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki K, Babitzke P, Kushner SR, Romeo T. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 2006;20:2605–17. doi: 10.1101/gad.1461606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jonas K, Edwards AN, Simm R, Romeo T, Romling U, Melefors O. The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol. 2008;70:236–57. doi: 10.1111/j.1365-2958.2008.06411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kushner SR. mRNA decay in Escherichia coli comes of age. J Bacteriol. 2002;184:4658–65. doi: 10.1128/JB.184.17.4658-4665.2002. discussion 4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coburn GA, Mackie GA. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog Nucleic Acid Res Mol Biol. 1999;62:55–108. doi: 10.1016/s0079-6603(08)60505-x. [DOI] [PubMed] [Google Scholar]
- 56.Majumdar A, Adhya S. Demonstration of two operator elements in gal: in vitro repressor binding studies. Proc Natl Acad Sci U S A. 1984;81:6100–4. doi: 10.1073/pnas.81.19.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Irani MH, Orosz L, Adhya S. A control element within a structural gene: the gal operon of Escherichia coli. Cell. 1983;32:783–8. doi: 10.1016/0092-8674(83)90064-8. [DOI] [PubMed] [Google Scholar]
- 58.Kramer H, Niemoller M, Amouyal M, Revet B, von Wilcken-Bergmann B, Muller-Hill B. lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J. 1987;6:1481–91. doi: 10.1002/j.1460-2075.1987.tb02390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin K, Huo L, Schleif RF. The DNA loop model for ara repression: AraC protein occupies the proposed loop sites in vivo and repression-negative mutations lie in these same sites. Proc Natl Acad Sci U S A. 1986;83:3654–8. doi: 10.1073/pnas.83.11.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee DH, Schleif RF. In vivo DNA loops in araCBAD: size limits and helical repeat. Proc Natl Acad Sci U S A. 1989;86:476–80. doi: 10.1073/pnas.86.2.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dodd IB, Shearwin KE, Perkins AJ, Burr T, Hochschild A, Egan JB. Cooperativity in long-range gene regulation by the lambda CI repressor. Genes Dev. 2004;18:344–54. doi: 10.1101/gad.1167904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Svenningsen SL, Costantino N, Court DL, Adhya S. On the role of Cro in lambda prophage induction. Proc Natl Acad Sci U S A. 2005;102:4465–9. doi: 10.1073/pnas.0409839102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci U S A. 1998;95:12462–7. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Struct Mol Biol. 2004;11:1206–14. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y: 1972. [Google Scholar]
- 66.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–30. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haldimann A, Wanner BL. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol. 2001;183:6384–93. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 69.Yakhnin AV, Trimble JJ, Chiaro CR, Babitzke P. Effects of mutations in the L-tryptophan binding pocket of the Trp RNA-binding attenuation protein of Bacillus subtilis. J Biol Chem. 2000;275:4519–24. doi: 10.1074/jbc.275.6.4519. [DOI] [PubMed] [Google Scholar]
- 70.Chamberlain JP. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979;98:132–5. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- 71.Bevilacqua PC, George CX, Samuel CE, Cech TR. Binding of the protein kinase PKR to RNAs with secondary structure defects: role of the tandem A-G mismatch and noncontiguous helixes. Biochemistry. 1998;37:6303–16. doi: 10.1021/bi980113j. [DOI] [PubMed] [Google Scholar]
- 72.Liu MY, Romeo T. The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. J Bacteriol. 1997;179:4639–42. doi: 10.1128/jb.179.14.4639-4642.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bevilacqua JM, Bevilacqua PC. Thermodynamic analysis of an RNA combinatorial library contained in a short hairpin. Biochemistry. 1998;37:15877–84. doi: 10.1021/bi981732v. [DOI] [PubMed] [Google Scholar]
- 74.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Romeo T, Gong M, Liu MY, Brun-Zinkernagel AM. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J Bacteriol. 1993;175:4744–55. doi: 10.1128/jb.175.15.4744-4755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Silhavy TJ, Berman ML, Enquist LW Cold Spring Harbor Laboratory. Experiments with gene fusions. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y: 1984. [Google Scholar]
- 77.Kay E, Dubuis C, Haas D. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc Natl Acad Sci U S A. 2005;102:17136–41. doi: 10.1073/pnas.0505673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heeb S, Blumer C, Haas D. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J Bacteriol. 2002;184:1046–56. doi: 10.1128/jb.184.4.1046-1056.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


