Summary
Canine parvovirus (CPV), first recognized as an emerging virus of dogs in 1978, resulted from a successful cross-species transmission. CPV emerged from the endemic feline panleukopenia virus (FPV), or from a closely related parvovirus of another host. Here we refine our current understanding of the evolution and population dynamics of FPV and CPV. By analyzing nearly full-length viral sequences we show that the majority of substitutions distinguishing CPV from FPV are located in the capsid protein gene, and that this gene is under positive selection in CPV, resulting in a significantly elevated rate of molecular evolution. This provides strong phylogenetic evidence for a prominent role of the viral capsid in host adaptation. In addition, an analysis of the population dynamics of more recent CPV reveals, on a global scale, a strongly spatially subdivided CPV population with little viral movement among countries and a relatively constant population size. Such limited viral migration contrasts with the global spread of the virus observed during the early phase of the CPV pandemic, but corresponds to the more endemic nature of current CPV infections.
Introduction
Efficient transmission in a new host species is a major obstacle to the successful cross-species transmission and emergence of viruses. Many emergent viruses are likely to be maladapted to the new host species and therefore cause only short transmission chains (Kuiken et al., 2006). Hence, the process of viral emergence will often require multi-stage adaptations to a new host, an idea supported by the observation that RNA viruses, which often exhibit evolutionary plasticity, are the most common agents of emerging disease (Woolhouse et al., 2001). Indeed, there is growing evidence that although replication fidelity is low in RNA viruses (and some ssDNA viruses), it has been fine-tuned in some cases (Sanjuan et al., 2007; Sanjuan et al., 2005; Vignuzzi et al., 2006) and might be environment-specific (Pita et al., 2007).
Canine parvovirus (CPV) represents one of the few examples where the process of cross-species viral transmission has been observed in `real time'. CPV emerged during the 1970s as a host-range variant of an endemic parvovirus of cats, mink, or related hosts within the order Carnivora. This group of closely related viruses includes feline panleukopenia virus (FPV) and mink enteritis virus (MEV), both of which have long been endemic in their natural hosts. These viruses are globally distributed and infect a variety of domestic and feral hosts including cats, raccoons, foxes and mink (Barker et al., 1983; Parrish, 1990). Host range barriers restrict transmission of these viruses among dogs and related canids, in which viral replication is limited to the thymus and probably bone marrow (Truyen & Parrish, 1992).
CPV was first recognized in 1978 when it spread world-wide, but while this virus had probably circulated locally before that time, the exact time of emergence is uncertain. The first CPV identified is referred to as CPV-2 to distinguish it from the previously identified but distantly related parvovirus of dogs, minute virus of canines (canine minute virus). CPV-2 caused a severe pandemic with high mortality in dogs, but was unable to infect cats, raccoon or mink. A variant virus (referred to as CPV-2a) emerged in 1979 and replaced CPV-2 worldwide in approximately one year (Parrish et al., 1988). CPV-2a differed antigenically from CPV-2 and infected both dogs and cats. Another antigenic variant (referred to as CPV-2b) with an equivalent host-range was defined by a single substitution (N426D) in the capsid protein gene and spread globally after 1984. Further mutations arose and spread widely, such as a mutation at residue 297 of the capsid protein. Currently, another mutation at the exposed capsid residue 426 (D426E), with a thus far uncharacterized impact on fitness, is spreading globally. This mutation was first described in Italy in 2000 (Buonavoglia et al., 2001), but has since been observed in other European countries (Decaro et al., 2007; Decaro et al., 2006; Martella et al., 2004), South America (Perez et al., 2007), Asia (Nakamura et al., 2004a) and most recently in the United States (Hong et al., 2007; Kapil et al., 2007).
Parvoviruses are small DNA viruses with a single-stranded linear genome of about 5kb. Their replication is dependent upon the host cell replication machinery, but estimates of rates of nucleotide substitution for both CPV and FPV are more comparable to those observed in RNA viruses than in other DNA viruses (Shackelton et al., 2005). The CPV genome contains two open reading frames (ORFs), one encoding the two non-structural proteins NS1 and NS2, and the other encoding the two structural proteins VP1 and VP2 (Suppl. Fig. 1). The amino terminal (N-terminal) ends of NS1 and NS2 are identical in sequence, but the carboxy-terminal (C-terminal) end of the NS2 protein is derived by differential splicing of the mRNA, being translated in a different reading frame which overlaps that of NS1. VP1 and VP2 are splice variants and are identical in sequence except for a 143 amino acid (aa) long N-terminal stretch unique to VP1. VP2 is the major capsid protein, and accounts for approximately 90% of the viral capsid. The capsid is the major determinant of host range (Hueffer et al., 2003b) and subject to antibody mediated selection (Nelson et al., 2007). To date, most studies have focused on the evolution of the VP2 gene, with limited work on the non-structural genes.
Despite widespread vaccination of domestic carnivores, FPV and CPV have remained important pathogens of domestic and wild carnivores. In addition, both the patterns of CPV dispersal and the selection pressures acting across the viral genome are poorly understood. This non-enveloped virus causes only short infections with no persistent virus remaining after infection, but is highly resistant in the environment. The global spread of the virus could therefore occur through the movement of infected animals or mechanical vectors, although the spatial dynamics of CPV have not been examined in detail. Here we explore these ideas by performing the largest evolutionary analysis of CPV undertaken to date, incorporating 143 sequences from 13 countries sampled over 29 years.
Methods
Isolate collection and sequencing
Viral DNA was amplified by polymerase chain reaction (PCR) from clinical specimens collected at various points before or after the emergence of CPV. The DNA was either amplified directly from the clinical specimen, or after purification using the QIAamp viral DNA purification kit (Qiagen), according to the manufacturer's recommendations. PCR amplification was performed using Phusion hot start polymerase (New England Biolabs) and primers that flanked either the NS1 or VP1 genes, respectively, or that spanned the whole coding region. PCR products were either sequenced directly after purification with the QIAquick PCR purification kit, or after subcloning of the viral DNA into the bacterial vectors pJET 1.2 blunt (Fermentas) or pSMART GC HK (Lucigen Corporation) and each nucleotide was sequenced multiple times. Consensus sequences were constructed using the `Seqman' program in the LaserGene 7.0 software package (DNASTAR). A total of 5 FPV and 6 CPV sequences that cover the complete viral coding region have been generated and were deposited in GenBank (accession numbers EU659111 to EU659121).
Sequence data
The sequences of 27 near full-length genomes, 54 NS1 and 199 VP2 genes from CPV, FPV and closely related parvoviruses were either determined here or obtained from GenBank (Supplementary Tables 1-3). Several CPV-2c sequences (GenBank accession numbers AY3880577, AY742942, EF375479, EF375481) covered only smaller parts of the coding region and where therefore excluded from the analysis. Similarly, not all published isolations of CPV-2c strains were apparently accompanied by GenBank submission of the viral sequences (see for example Kapil et al., 2007, Hong et al., 2007 or Decaro et al., 2007). Of the sequences compiled, 23 near full-length sequences, 50 NS1 and 126 VP2 gene sequences contained information on the time of isolation, while the place of isolation was available for 52 NS1 and 178 VP2 gene sequences. The first 4 codons of the VP2 sequences were deleted from the VP2 gene alignment, as some of the sequences were incomplete, resulting in an alignment of 1721 nucleotides (nt). For the analysis of selection pressures acting on the VP1 unique region, a second alignment of 26 VP1 gene sequences was constructed with the intron within the VP1 unique N-terminus removed, resulting in an alignment of 2181 nt. The complete 2001 nt coding region of the NS1 gene was also analyzed, and for the analysis of selection pressures on the NS2 gene, the N-terminal 89 codons were combined with the 76 codon-long C-terminal region in the correct reading frame, resulting in an alignment of 495 nt. The near full-length sequences were trimmed to the complete NS1 and VP1 coding regions, with the introns included, resulting in a 4269 nt alignment. Sequences were aligned by hand in the Se-Al program (http://tree.bio.ed.ac.uk/software/seal/).
Inference of phylogenetic trees
Maximum likelihood (ML) phylogenetic trees were estimated using PAUP* version 4.0 (Swofford, 2003) with the best-fit model of nucleotide substitution determined using Modeltest (Posada and Crandall, 1998). The HKY85 substitution model was the best-fit for the full-length data set and the key parameters (i.e. transition and transversion rates) were estimated from the data. For the VP2 data sets, the GTR+I+G4 substitution model was optimal. To assess support for individual nodes, bootstrap resampling values were estimated with 1000 neighbor-joining trees, again employing PAUP* (Swofford, 2003).
Estimation of substitution rate and population dynamics
Rates of nucleotide substitution per site, per year (subs/site/year) were estimated using the Bayesian Markov chain Monte Carlo (MCMC) method available in the BEAST package (http://beast.bio.ed.ac.uk/). Phylogenetic trees were estimated using the HKY85 substitution model, and either strict (constant) or relaxed (uncorrelated lognormal) molecular clocks were assumed. Four different demographic models were compared, assuming constant population size, exponential or logistic population growth or a Bayesian skyline model, which provides a piecewise depiction of changes in relative genetic diversity through time. Models were compared using Bayes Factors, with uncertainty in each parameter estimate depicted in values of the 95% highest probability density (HPD) interval. All analyses were run for sufficient time to ensure convergence as assessed using the TRACER program (http://tree.bio.ed.ac.uk/software/tracer/), with 10% of runs removed as burn-in. For those models supporting population growth, estimates of the population doubling time (l) were obtained from the BEAST estimates of population growth (r) by using the formula λ = ln (2)/r.
Estimates of site-specific selection pressures
We used the Single Likelihood Ancestor Counting (SLAC), Fixed Effects Likelihood (FEL) and Random Effects Likelihood (REL) algorithms available in the Datamonkey web interface of the HyPhy software package (Kosakovsky Pond & Frost, 2005) to estimate the relative rates of synonymous (dS) and nonsynonymous (dN) substitutions per site (ratio dN/dS) in our sample of CPV and FPV isolates. The HKY85 substitution model was employed in each case.
Analysis of migration patterns
To determine the strength of phylogenetic clustering by country of virus isolation we employed a parsimony character mapping approach (Carrington et al., 2005). Each CPV sequence was first assigned a character state reflecting its country of origin. Given the ML phylogeny for these sequences (as determined above), the minimum number of state changes needed to produce the observed distribution of country character states was estimated using parsimony (excluding ambiguous changes). To determine the expected number of changes under the null hypothesis of complete mixing among countries, the states of all isolates were randomized 1000 times. The difference between the mean number of observed and expected state changes indicates the level of geographic isolation, with statistical significance assessed by comparing the total number of observed state changes to the number expected under random mixing. All analyses were performed using PAUP* (Swofford, 2003).
Results
Phylogenetic analysis of the full-length genome
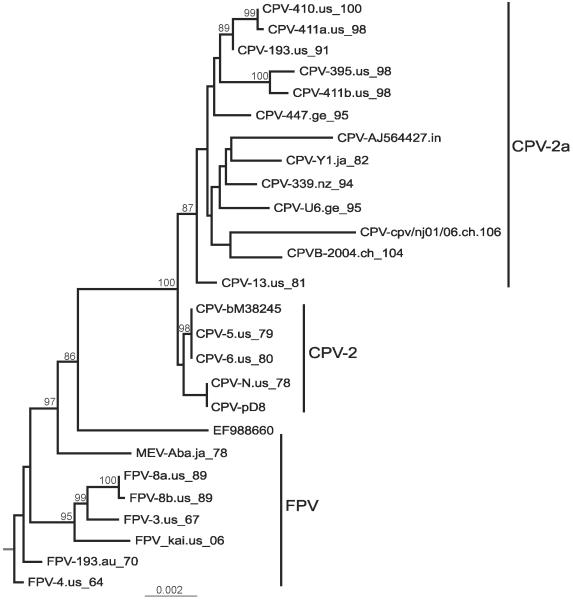
We analyzed 27 near full-length genomes of carnivore parvoviruses, several of which were determined during the course of this study. The FPV sequences cover a time period of 42 years (1964 to 2006) and were collected in various geographical regions. The oldest CPV isolate was collected in 1978, the year in which CPV was first described, and the CPV sequences cover a total of 29 years. The topology of the ML tree of these data (Fig. 1) was similar to that of trees inferred from the VP2 region, with the longest internal branch separating the FPV and CPV clades. 16 nucleotide substitutions separated the FPV and CPV clades, while 7 substitutions separated the CPV-2 from the CPV-2a clade. The majority of these substitutions (11 between FPV and CPV and 5 between CPV-2 and CPV-2a) were located in the capsid protein region (data not shown).
Figure 1.
Phylogenetic tree of 27 full-length carnivore parvovirus sequences, rooted with the oldest FPV sequence (FPV-4.us.64). Horizontal branch lengths are drawn to scale (nucleotide substitutions per site), and bootstrap support (>85%) is indicated above the respective branches. The three recognized clades, FPV and related viruses (red), CPV-2 (blue) and CPV-2a and derived viruses (green), are indicated.
Substitution rates and population dynamics
Bayesian MCMC estimates of rates of nucleotide substitution were inferred for the full-length genome, and separately for the NS1 and VP2 genes, either analyzing all carnivore parvoviruses simultaneously or subdividing the sequences into FPV and CPV (i.e. all CPV-2, CPV-2a and derived viruses). For the VP2 sequence, two additional data sets were analyzed, one consisting of the CPV-2a sequences, and the other comprising all FPV and most CPV sequences but excluding the original CPV-2 sequences. Very similar substitution rates were obtained for individual data sets under all molecular clock and demographic models, reflecting the robustness of this analysis (all results available from the authors on request). However, estimates of the substitution rate from the capsid protein gene were strikingly different between FPV and CPV (Table 1). The mean substitution rate for FPV was 8.2 × 10-5 subs/site/year (95% HPD = 3.6 × 10-5 - 1.3 × 10-4 subs/site/year), while that of CPV was significantly higher at 2.2 × 10-4 subs/site/year (95% HPD = 1.7 × 10-4 - 2.7 × 10-4 subs/site/year). Estimates of substitution rate for both FPV and CPV based on the NS1 gene were similar (6.6 × 10-5 subs/site/year and 1.0 × 10-4 subs/site/year, respectively), with overlapping HPD values.
Table 1.
Nucleotide substitution rates, population dynamics and times to common ancestry for CPV and FPV.
| VP2 | NS1 | full-length | |||||||
|---|---|---|---|---|---|---|---|---|---|
| FPV | CPV | CPV-2a | no CPV-2 | all | FPV | CPV | all | ||
| demographic model | const.* | log.† | log.† | const.* | BSP‡ | const.* | const.* | const.* | const.* |
| sequence length (bp) | 1721 | 1721 | 1721 | 1721 | 1721 | 2007 | 2007 | 2007 | 4269 |
| number of sequences | 30 | 96 | 90 | 120 | 126 | 22 | 27 | 49 | 23 |
| date range of sequences | 1962-2006 | 1978-2004 | 1980-2004 | 1962-2006 | 1962-2006 | 1963-2006 | 1978-2004 | 1963-2006 | 1964-2006 |
| Mean substitution rate§ | 8.2 × 10-5 | 2.2 × 10-4 | 2.1 × 10-4 | 1.6 × 10-4 | 1.4 × 10-4 | 6.4 × 10-5 | 1.0 × 10-4 | 6.2 × 10-5 | 1.2 × 10-4 |
| HPD substitution rate ‖ | 3.6× 10-5-1.3× 10-4 | 1.7 × 10-4-2.7 × 10-4 | 1.6 × 10-4-2.7 × 10-4 | 1.1 × 10-4-2.0 × 10-4 | 1.1 × 10-4-1.7 × 10-4 | 1.7 × 10-5-1.2 × 10-4 | 2.2 × 10-5-2.0 × 10-4 | 3.4 × 10-5-9.3 × 10-5 | 7.1 × 10-5-1.7 × 10-5 |
| Time to Most Recent Common Ancestor (years)¶ | 98 | 32 | 28 | 71 | 105 | 128 | 85 | 127 | 71 |
| HPD age | 53-169 | 29-37 | 26-32 | 50-100 | 100-117 | 50-260 | 30-192 | 65-215 | 47-103 |
| mean growth rate (1/years) | - | 0.2 | 0.4 | - | - | - | - | - | - |
| HPD growth rate (years) | - | 0.1 | 0.1 | - | - | - | - | - | - |
| - | 0.3 | 0.4 | - | - | - | - | - | - | |
| Mean epidemic doubling time (λ) | - | 3.7 | 1.8 | - | - | - | - | - | - |
not applicable
const. = constant population size
log. = logistic population growth
BSP = Bayesian skyline model
Mean substitution rate = subs/site/year
HPD = 95% highest probability density interval
Time to Most Recent Common Ancestor (TMRCA) = age since most recent sequence included in analysis (i.e. 2006)
Using the same approach we were able to estimate the Time to the Most Recent Common Ancestor (TMRCA) of both FPV and CPV. The estimated TMRCA for FPV was 98 years from the most recent analyzed isolate collected in 2006 (95% HPD = 53 - 168 years) while that for CPV was 32 years (95% HPD = 29 - 37 years) which closely corresponds with the first CPV case recognized in 1978. The mean TMRCA estimate for CPV-2a (28 years) is lower than that for all CPVs, again matching the first observed CPV-2a collected in 1979. The TMRCA estimates based on the NS1 gene place the emergence of FPV, and particularly CPV, considerably earlier than those based on the capsid protein gene (128 years for FPV and 85 years for CPV). However, the HPD values are very wide (and overlap with those inferred from VP2), revealing the inherent statistical uncertainty in this estimate (although more recent dates are observed under other demographic models tested - results not shown).
The population dynamics of all CPVs and the CPV-2a subset were best described by logistic population growth in which a rapid growth phase is followed by a phase of increasingly slower growth, while a constant population size provided a better description of both the FPV data set and the data set containing all sequences except the CPV-2 strain. The complete data set, comprising all VP2 sequences, was best described by the more complex Bayesian skyline model, reflecting the inclusion of viruses with differing epidemiological dynamics. Notably, the mean epidemic doubling time for CPV-2a and derived viruses (1.8 years) is approximately half that for all CPVs combined (3.7 years), revealing the initially rapid spread of the CPV-2a variant (although, again, the 95% HPD values overlap). However, these estimates of growth rate should be treated with caution due to the potentially confounding effect of population subdivision. Both FPV and CPV NS1 sequences were best described by models of constant population size.
Selection pressures in CPV and FPV
An analysis of selection pressures, manifest as dN/dS ratios, across both viruses and genes revealed the sporadic action of positive selection, with mean dN/dS values of 0.15 for NS1, 0.53 for NS2, 0.15 for the VP1 unique region and 0.12 for VP2 (Table 2). Notably, the short NS2 protein contained four positively selected sites, all within the 76 aa long C-terminal region where the encoded sequence overlaps that of NS1 in a different reading frame. Hence, the `positively selected' sites in this region are difficult to interpret. While the possibility of positive selection cannot be ruled out, the results might represent false-positives caused by synonymous mutations in the alternate reading frame. In contrast, only a single NS1 site, at codon 597, appeared to be under positive selection. This amino acid is located in the C-terminal region of NS1, in the region where NS2 is encoded in the different reading frame, and overlaps with residues 106 and 107 of NS2, again suggesting that this result might be false-positive. No positively selected sites were detected in the VP1 unique region. As expected given the capsid's known role in host range control, four capsid residues, at VP2 positions 101 (dN/dS= infinity; dN= 2.24), 300 (dN/dS = infinity; dN= 3.0), 324 (dN/dS = infinity; dN= 4.43) and 426 (dN/dS = infinity; dN= 4.56), were under positive selection across all sequences. Among the FPV sequences, only VP2 codon 562 in the VP2 gene appears to be under positive selection (dN/dS = infinity; dN = 13.05). Among the CPV sequences, VP2 residues 87 (dN/dS = infinity; dN= 5.03) and 426 (dN/dS = infinity; dN= 4.55) were under positive selection.
Table 2.
Analysis of selection pressures in each genomic region. P-values or, in the case of REL, Bayes factors, are reported. P-values < 0.1 and Bayes factors > 100 are shown in bold.
| gene | mean dN/dS | mutation | dN | dS | dN/dS | SLAC | FEL | IFEL | REL | location‡ | potential function‡ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NS1* | 0.15 | L597H | 10.47 | 0 | ∞ | 0.23 | 0.089 | 0.114 | 77 | C-terminus | P38 promoter transactivation | |
| NS2* | 0.53 | T94R | 2.42 | 0.91 | 2.66 | 0.445 | 0.25 | 0.195 | 331 | C-terminus | CRM1 interaction | |
| N107R | 2.42 | 2.12 | 1.14 | 0.602 | 0.76 | 0.26 | 136 | C-terminus | CRM1 interaction | |||
| D151N | 2.42 | 0.92 | 2.63 | 0.615 | 0.296 | 0.09 | 301 | C-terminus | 14-3-3 family interaction | |||
| E160K | 2.42 | 0.87 | 2.78 | 0.52 | 0.38 | 1 | 325 | C-terminus | unclear | |||
| VP1 | 0.15 | - | - | - | - | - | - | - | - | - | - | |
| VP2 | all | 0.12 | I101T† | 2.24 | 0 | ∞ | 0.35 | 0.10 | 0.34 | 5617† | 3fold spike | possibly TfR binding§ |
| A300G | 3.0 | 0 | ∞ | 0.089 | 0.189 | 0.361 | 6† | 3fold spike | TfR binding, antigenic change§ | |||
| Y324I† | 4.43 | 0 | ∞ | 0.506 | 0.54 | 0.286 | 504 † | 3fold spike | likely TfR binding | |||
| N426D/E | 4.56 | 0 | ∞ | 0.043 | 0.255 | 0.124 | 6† | capsid surface | antigenic escape | |||
| FPV | 0.11 | V562L | 13.05 | 0 | ∞ | 0.408 | 0.33 | 0.097 | 4704 | capsid surface | unclear | |
| CPV | 0.11 | M87L | 5.03 | 0 | ∞ | 0.465 | 0.219 | 0.069 | # | shoulder | TfR binding; possibly antigenic§ | |
| N426D/E | 4.55 | 0 | ∞ | 0.045 | 0.251 | 0.104 | # | capsid surface | antigenic escape | |||
none detected
“infinity”
residues were mapped to the NS2 gene of minute virus of mice using a previously published alignment (Wang et al., 1998) and the functions listed correspond to those known in that virus.
only detected in the full-length data set
location and function of the NS1 and NS2 mutations were inferred from the known functions of homologous regions in minute virus of mice.
TfR = Transferrin receptor
contained too many sequences for analysis
Migration patterns of CPV
The differences between the observed and expected numbers of migration events involving CPV isolates sampled from different countries are shown in Table 3, with negative values indicative of population subdivision and positive values characterizing directional migration between the specified geographic regions. Overall, we detected a strong signal of population subdivision, with only a few instances of migration between specific localities. Indeed, only 12 of the 182 possible directional migration pathways showed signs of CPV migration, and none had strong signals, suggesting only sporadic geographical movement of these viruses. Conversely, 95 of the 182 possible pathways were negative, indicative of population subdivision, sometimes strongly so. Interestingly, China represents a far weaker source of viruses for other regions than expected given the number of available sequences from this country. Similarly, Brazil is also a weaker source population than expected by chance (especially for movements to China and Taiwan), but appears to be a source for movement to Europe (Italy and Poland) and potentially the United States. Overall, the strongest evidence of viral movement was from the United States to Vietnam (observed-expected = 1.54) and from Vietnam to China (observed-expected = 1.36), but the signal was weak.
Table 3.
Migration analysis of CPV, based on VP2 gene. Observed minus expected migration pathways are shown below, with directional movements from country indicated in row to country shown in column. Likely migration events (positive values) are indicated by shading.
| to from | A | B | C | F | G | I | J | N | O | P | S | T | U | V | Σ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | - | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| B | -0.15 | - | -2.71 | -0.13 | -0.31 | 1.10 | -0.46 | -0.14 | -0.15 | 0.86 | -1.25 | -1.04 | 0.07 | -2.08 | -6.37 |
| C | -0.26 | -3.79 | - | -0.27 | -0.48 | -1.68 | 0.24 | -0.28 | -0.25 | -0.24 | -2.29 | -1.85 | -3.49 | -3.67 | -18.3 |
| F | 0 | 0 | 0 | - | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| G | 0 | 0 | 0 | 0 | - | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| I | -0.01 | -0.08 | -0.06 | 0 | -0.01 | - | -0.02 | -0.01 | -0.01 | 0 | -0.04 | -0.04 | -0.06 | -0.06 | -0.39 |
| J | 0 | 0 | -0.01 | 0 | 0 | 0 | - | 0 | 0 | 0 | 0 | 0 | 0 | 0 | -0.01 |
| N | 0 | 0 | 0 | 0 | 0 | 0 | 0 | - | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | - | 0 | 0 | 0 | 0 | 0 | 1 |
| P | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | - | 0 | 0 | 0 | 0 | 0 |
| S | -0.01 | -0.13 | -0.12 | -0.01 | -0.02 | -0.05 | 0.02 | -0.01 | -0.01 | -0.01 | - | -0.07 | -0.1 | -0.12 | -0.67 |
| T | 0 | -0.08 | 0.92 | -0.01 | -0.01 | -0.04 | 0.98 | -0.01 | -0.01 | -0.04 | -0.05 | - | -0.08 | -0.05 | 1.58 |
| U | -0.03 | -0.52 | -0.48 | -0.04 | 0.95 | 0.80 | -0.09 | -0.03 | -0.03 | -0.02 | -0.29 | -0.24 | - | 1.54 | 1.53 |
| V | -0.04 | -0.65 | 1.36 | -0.03 | -0.09 | -0.27 | -0.1 | -0.05 | -0.04 | -0.04 | -0.33 | 0.73 | -0.53 | - | -0.08 |
| Σ | -0.49 | -4.25 | -1.11 | -0.49 | 0.03 | -0.14 | 0.54 | -0.51 | -0.48 | 0.54 | -4.25 | -2.5 | -4.20 | -4.45 | |
not applicable
Country codes: A: India; B: Brazil; C: China; F: Finland; G: Germany; I: Italy; J: Japan; N: New Zealand; O: outgroup (1964 FPV isolate, UK); P: Poland; S: South Korea; T: Taiwan; U: USA; V: Vietnam;
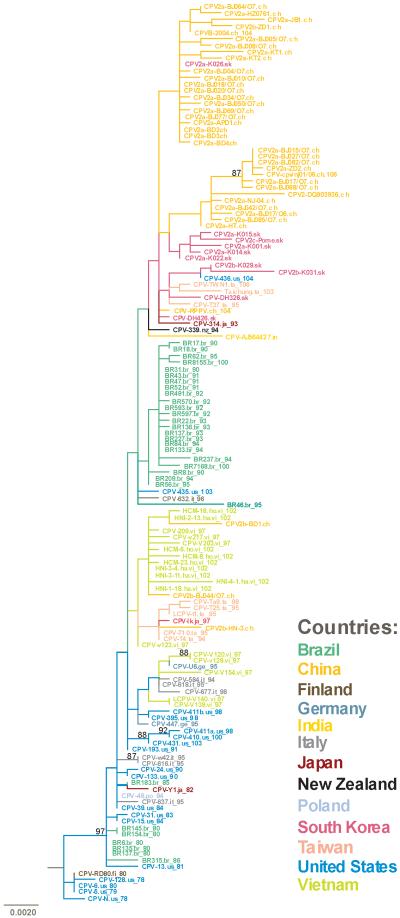
When we mapped the positively selected sites onto the phylogenetic tree depicted in Figure 2 (data not shown), the VP2 N426D mutation appeared to be co-circulating in several countries, such as South Korea, the United States and Taiwan, but was absent in all but four sequences collected in China (CPV2b-BD1.ch, CPV2b-BK044/07.ch,CPV2b-HN-3.ch and CPV2b-ZD1.ch), of which the first three clustered with sequences from Vietnam. Interestingly the N426D mutation was not monophyletic, indicating that it has arisen independently in these countries. Only a single Brazilian sequence (BR185.br_85) had the N426D mutation, and this sequence had signatures of migration, since it clustered with the European and U.S. isolates instead of the other Brazilian isolates. Geographic structure was even more striking in the case of the Y324I mutation. This was only present in sequences from China and South Korea and again was not monophyletic. Conversely, the VP2 mutations at codons 87, 101 and 300 were present in all but five CPV-2a sequences. Three of the Vietnamese sequences collected in 1997 from feral cats (CPV-V139.vi_97, CPV-V203.vi_97 and LCPV_V140.vi_97) and one sequence from South Korea (CPV-2c-Pome) harbored a G300D mutation (Ikeda et al., 2000) and one Chinese sequence isolated from a red panda in 2004 (CPV-RPPV.ch_104) possessed a G300V mutation (Qin et al., 2006). Finally, only one sequence in the data set (HNI-4-1.ha.vi_102) contained the VP2 D426E mutation. As such, no inference can be made from this dataset regarding the global movement of this mutation.
Figure 2.
Phylogenetic tree of 152 CPV VP2 sequences used for the analysis of migration patterns. Horizontal branch lengths are drawn to scale (nucleotide substitutions per site), and bootstrap support (>85%) is indicated above their respective branches. The country of isolation is indicated by color-coding, as indicated in the key.
Discussion
This study provides novel insights into the population dynamics of the emerging CPV virus, contrasting the evolutionary pressures acting on CPV and FPV, and among the genes of CPV. The emergence of CPV was characterized by strong selection for specific mutations within VP2 but not NS1. Optimal receptor binding and antigenic escape were the likely driving forces, as indicated by the location and function of the positively selected sites in the capsid protein (located at VP2 codons 101, 300, 324, 426).
This study is the first to systematically explore CPV dynamics on a global scale. Notably, we found the CPV population to be spatially structured, with very limited gene flow between clusters. This is in contrast with a previous study which reported strong temporal, but mild or absent spatial clustering, among CPV isolates from Brazil, the U.S. and Europe (Pereira et al., 2000), but is consistent with other reports of geographic clustering of isolates from Vietnam, Japan/Taiwan and the United States (Doki et al., 2006). Three of the mutations at positively selected sites (at VP2 residues 87,101 and 300) arose soon after CPV emerged. The mutations at residues 87 and 101 have arisen once and all but five of the CPV-2a viruses harbor the A300G mutation. The remaining CPV-2a viruses were isolated from feral feline hosts and show alternative substitutions of residue 300, suggesting a role of this residue in the feline adaptation of CPV. We have previously shown that an Asp at residue 300 is selected during passage of CPV-2 in feline cells in culture, and also that this prevents the infection of dog cells (Parrish and Carmichael, 1986; Parker and Parrish, 1997). The two positively selected mutations at VP2 residues 324 and 426 arose later, and apparently independently, in different geographic locations. Thus the early period of CPV emergence was characterized by global viral spread and global strain replacement, while later mutations likely arose through parallel emergence of the same mutations in separate geographic regions. Indeed, by far the strongest phylogeographic signal in these data is that of population subdivision, with relatively little movement of viruses between geographic areas. In addition, the majority of migrations that do occur are between countries in close geographic proximity, such as Taiwan or Vietnam and China, or between countries with close economic connections such as the United States and Germany or Italy, indicating that CPV is likely spread among countries through the movement of mechanical vectors or, in some cases, infected animals. The movement of viruses from the United States to Vietnam appears surprising, but all of these sequences were isolated in 1997, only 3 years after the embargo against Vietnam was lifted, indicating a potential role of the developing trade relations between the countries.
While recombination can potentially impact phylogenetic relationships, we found no clear signal of recombination among the FPV or CPV isolates studied here (unpublished data; available from the authors on request). Indeed, the presence and role of recombination among FPV or CPV sequences is uncertain. While phylogenetic evidence of recombination has been detected for related parvoviruses of mink (Aleutian mink disease virus), swine (porcine parvovirus) and rodents (several rodent parvoviruses including LuIII) (Shackelton et al, 2007), there is no clear evidence for the process among the FPV- and CPV-related parvoviruses studied to date. It is therefore unlikely that the analyses reported here were strongly impacted by recombination.
Our molecular clock estimates of emergence times for CPV-2 and CPV-2a are notable in that they are very close to the dates when the viruses were first detected in nature. In particular, we estimate that CPV-2 emerged around 1974 (i.e. 32 years before the most recent analyzed sequence collected in 2006), only four years before it spread globally in 1978. This is compatible with serological evidence that the virus circulated in Europe a few years before the CPV pandemic (Truyen et al., 1994b). Similarly, our estimate for the emergence date of CPV-2a is approximately four years later than CPV-2, which corresponds closely with the first detection of CPV-2a in nature in 1979. Notably, the pattern of logistic population growth for CPV-2 and CPV-2a indicates the initially rapid spread of the virus among animals. This corresponds to the expected population dynamics of a newly emerging pathogen, and has also been reported by previous studies (Pereira et al., 2006; Shackelton et al., 2005). In marked contrast, FPV evolution is characterized by constant population sizes, as expected for an endemic pathogen.
The change in evolutionary dynamics associated with the process of emergence is also apparent in the significantly higher rate of substitution in the capsid protein gene of CPV compared to the FPV sequences, with mean rates of 1.2 and 0.4 substitutions per genome per year, respectively. Although these rates are comparable to those inferred previously (Horiuchi et al., 1998; Pereira et al., 2006; Shackelton et al., 2005), this is the first time a statistically significant difference between FPV and CPV has been revealed. In theory, the elevated rate of evolutionary change in CPV might be due to either lower replication fidelity in dogs or increased positive selection after the transmission to the new host. Since these viruses are all replicated by host cell DNA polymerases, significantly different fidelities appear unlikely. Moreover, substitution rates inferred from the NS1 gene were comparable between FPV and CPV sequences, and similar to that inferred for FPV based on the capsid gene, indicating that the elevated substitution rate observed among the CPV sequences was most likely due to strong positive selection, probably driven by host adaptation and immune escape.
There is clear evidence for strong positive selection on the capsid protein gene, and differences among the host species again appear to be of importance in this regard. Four VP2 residues - 101, 300, 324 and 426 - are subject to positive selection among all carnivore parvoviruses. Residues 101 and 300 are located on a surface-exposed region of the viral capsid (Suppl. Fig. 2), interact with the transferrin receptor (which is the receptor mediating viral entry), and partially determine the re-acquired feline host range of CPV-2a. The substitution at residue 324 has not been previously characterized, but is adjacent to residue 323, a known host-range mutation, which together with residue 93 controls canine TfR binding and the canine host range (Hueffer et al., 2003a). The 324 mutation appears to be circulating only in China and South Korea (data not shown), and has appeared a number of times independently. Although clear information on time of sampling is available for only a few of the sequences containing this mutation, most appear to have been collected in 2006 or 2007, indicating that this mutation probably arose recently. Mutations at aa 426 change the antigenic profile of the virus (Nakamura et al., 2004b; Parrish et al., 1991). This codon has undergone two mutations since CPV emerged, first from asparagine to aspartate, and more recently to glutamate, indicating complicated selection dynamics acting on this residue. The exact phenotypic consequences of the mutations are not clear, and the N426D mutation has been present in some CPV populations for decades without replacing the wild-type (Parrish et al., 1991).
Mapping the codon 426 variants onto the phylogenetic tree in Figure 2 revealed both the Asn and Asp amino acids were co-circulating in the United States, Taiwan, Vietnam and Europe, while the Asp mutation was at very low frequency in China and Brazil (data not shown; available from the authors on request). Interestingly, the Asp codon-containing sequences from those countries showed signatures of migration and clustered with sequences from other countries that carried the respective mutation. The D426E mutation has recently been isolated from several countries around the world, but only a single VP2 sequence of known origin that harbored this mutation was available, preventing more detailed analysis. One additional sequence containing the D426E mutation (GenBank accession number AF401519) was excluded from the analysis since the place of isolation could not be clearly determined. Among the CPV sequences, residues 87 and 426 are under strong positive selection. Residue 87 is also surface exposed and, although its specific functions are unknown, appears to be associated with the re-gained feline host range of CPV-2a, together with residue 300.
Less positive selection was observed in FPV, and only VP2 codon 562 appeared to be under positive selection, although relatively strongly. Adaptive evolution at this site has not been described previously. However, two residues in close spatial proximity, 564 and 568, partially determine the in vivo host range of FPV (Truyen et. al, 1994a).
Finally, our study is notable in that we detected putatively positively selected sites in the NS1 (codon 597, overlapping codon 107 in NS2) and NS2 (codons 94, 107,151 and 160) genes, although these are potentially false positives due to the overlapping reading frames. The role of NS2 in the CPV life-cycle is unclear and knock-out mutants appear to behave similarly to wild-type both in tissue culture and in dog infections (Wang et al., 1998). However, in the related rodent parvoviruses, NS2 is required for efficient translation and capsid assembly in cells from natural host species but not in cells from other hosts (Choi et al., 2005; Cotmore et al., 1997; D'Abramo et al., 2005; Eichwald et al., 2002; Li, 1991; Naeger et al., 1993). A possible role for NS2 in host adaptation therefore merits further study.
Supplementary Material
Acknowledgements
Leland E. Carmichael and Edward Dubovi provided clinical samples, and Wendy S. Weichert and Nell Bond provided excellent technical support. Christian Nelson provided valuable help with the three-dimensional representation of the positively selected capsid residues. This work was supported by NIH grants GM080533-01 to E.C.H and AI028385 to CRP.
Footnotes
Publisher's Disclaimer: This is an author manuscript that has been accepted for publication in Journal of General Virology, copyright Society for General Microbiology, but has not been copy-edited, formatted or proofed. Cite this article as appearing in Journal of General Virology. This version of the manuscript may not be duplicated or reproduced, other than for personal use or within the rule of `Fair Use of Copyrighted Materials' (section 17, Title 17, US Code), without permission from the copyright owner, Society for General Microbiology. The Society for General Microbiology disclaims any responsibility or liability for errors or omissions in this version of the manuscript or in any version derived from it by any other parties. The final copy-edited, published article, which is the version of record, can be found at http://vir.sgmjournals.org, and is freely available without a subscription.
References
- Barker IK, Povey RC, Voigt DR. Response of mink, skunk, red fox and raccoon to inoculation with mink virus enteritis, feline panleukopenia and canine parvovirus and prevalence of antibody in wild carnivores in Ontario. Can J Comp Med. 1983;47 [PMC free article] [PubMed] [Google Scholar]
- Buonavoglia C, Martella V, Pratelli A, Tempesta M, Cavalli A, Buonavoglia D, Bozzo G, Elia G, Decaro N, Carmichael L. Evidence for evolution of canine parvovirus type 2 in Italy. J Gen Virol. 2001;82:3021–3025. doi: 10.1099/0022-1317-82-12-3021. [DOI] [PubMed] [Google Scholar]
- Carrington CV, Foster JE, Pybus OG, Bennett SN, Holmes EC. Invasion and maintenance of dengue virus type 2 and type 4 in the Americas. J Virol. 2005;79:14680–14687. doi: 10.1128/JVI.79.23.14680-14687.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi EY, Newman AE, Burger L, Pintel D. Replication of minute virus of mice DNA is critically dependent on accumulated levels of NS2. J Virol. 2005;79:12375–12381. doi: 10.1128/JVI.79.19.12375-12381.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore SF, D'Abramo AM, Jr., Carbonell LF, Bratton J, Tattersall P. The NS2 polypeptide of parvovirus MVM is required for capsid assembly in murine cells. Virology. 1997;231:267–280. doi: 10.1006/viro.1997.8545. [DOI] [PubMed] [Google Scholar]
- D'Abramo AM, Jr., Ali AA, Wang F, Cotmore SF, Tattersall P. Host range mutants of minute virus of mice with a single VP2 amino acid change require additional silent mutations that regulate NS2 accumulation. Virology. 2005;340:143–154. doi: 10.1016/j.virol.2005.06.019. [DOI] [PubMed] [Google Scholar]
- Decaro N, Desario C, Addie DD, Martella V, Vieira MJ, Elia G, Zicola A, Davis C, Thompson G, Thiry E, Truyen U, Buonavoglia C. The study molecular epidemiology of canine parvovirus, Europe. Emerg Infect Dis. 2007;13:1222–1224. doi: 10.3201/eid1308.070505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N, Martella V, Desario C, Bellacicco AL, Camero M, Manna L, d'Aloja D, Buonavoglia C. First detection of canine parvovirus type 2c in pups with haemorrhagic enteritis in Spain. J Vet Med. 2006;53:468–472. doi: 10.1111/j.1439-0450.2006.00974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doki M, Fujita K, Miura R, Yoneda M, Ishikawa Y, Taneno A, Kai C. Sequence analysis of VP2 gene of canine parvovirus isolated from domestic dogs in Japan in 1999 and 2000. Comp Immunol Microbiol Infect Dis. 2006;29:199–206. doi: 10.1016/j.cimid.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Eichwald V, Daeffler L, Klein M, Rommelaere J, Salome N. The NS2 proteins of parvovirus minute virus of mice are required for efficient nuclear egress of progeny virions in mouse cells. J Virol. 2002;76:10307–10319. doi: 10.1128/JVI.76.20.10307-10319.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C, Decaro N, Desario C, Tanner P, Pardo MC, Sanchez S, Buonavoglia C, Saliki JT. Occurrence of canine parvovirus type 2c in the United States. J Vet Diagn Invest. 2007;19:535–539. doi: 10.1177/104063870701900512. [DOI] [PubMed] [Google Scholar]
- Horiuchi M, Yamaguchi Y, Gojobori T, Mochizuki M, Nagasawa H, Toyoda Y, Ishiguro N, Shinagawa M. Differences in the evolutionary pattern of feline panleukopenia virus and canine parvovirus. Virology. 1998;249:440–452. doi: 10.1006/viro.1998.9335. [DOI] [PubMed] [Google Scholar]
- Hueffer K, Govindasamy L, Agbandje-McKenna M, Parrish CR. Combinations of two capsid regions controlling canine host range determine canine transferrin receptor binding by canine and feline parvoviruses. J Virol. 2003a;77:10099–10105. doi: 10.1128/JVI.77.18.10099-10105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueffer K, Parker JS, Weichert WS, Geisel RE, Sgro JY, Parrish CR. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J Virol. 2003b;77:1718–1726. doi: 10.1128/JVI.77.3.1718-1726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Mochizuki M, Naito R, Nakamura K, Miyazawa T, Mikami T, Takahashi E. Predominance of canine parvovirus (CPV) in unvaccinated cat populations and emergence of new antigenic types of CPVs in cats. Virology. 2000;278:13–19. doi: 10.1006/viro.2000.0653. [DOI] [PubMed] [Google Scholar]
- Kapil S, Cooper E, Lamm C, Murray B, Rezabek G, Johnston L, 3rd, Campbell G, Johnson B. Canine parvovirus types 2c and 2b circulating in North American dogs in 2006 and 2007. J Clin Microbiol. 2007;45:4044–4047. doi: 10.1128/JCM.01300-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. 2005. [DOI] [PubMed] [Google Scholar]
- Kuiken T, Holmes EC, McCauley J, Rimmelzwaan GF, Williams CS, Grenfell BT. Science. Vol. 312. New York, NY: 2006. Host species barriers to influenza virus infections; pp. 394–397. [DOI] [PubMed] [Google Scholar]
- Li X, Rhode SL. Nonstructural protein NS2 of parvovirus H-1 is required for efficient viral protein synthesis and virus production in rat cells in vivo and in vitro. Virology. 1991;184:117–130. doi: 10.1016/0042-6822(91)90828-y. [DOI] [PubMed] [Google Scholar]
- Martella V, Cavalli A, Pratelli A, Bozzo G, Camero M, Buonavoglia D, Narcisi D, Tempesta M, Buonavoglia C. A canine parvovirus mutant is spreading in Italy. J Clin Microbiol. 2004;42:1333–1336. doi: 10.1128/JCM.42.3.1333-1336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeger LK, Salome N, Pintel DJ. NS2 is required for efficient translation of viral mRNA in minute virus of mice-infected murine cells. J Virol. 1993;67:1034–1043. doi: 10.1128/jvi.67.2.1034-1043.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Tohya Y, Miyazawa T, Mochizuki M, Phung HT, Nguyen NH, Huynh LM, Nguyen LT, Nguyen PN, Nguyen PV, Nguyen NP, Akashi H. A novel antigenic variant of canine parvovirus from a Vietnamese dog. ArchVirol. 2004a;149:2261–2269. doi: 10.1007/s00705-004-0367-y. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Tohya Y, Miyazawa T, Mochizuki M, Phung HTT, Nguyen NH, Huynh LMT, Nguyen LT, Nguyen PN, Nguyen PV, Nguyen NPT, Akashi H. A novel antigenic variant of Canine parvovirus from a Vietnamese dog. Arch Virol. 2004b;149:2261–2269. doi: 10.1007/s00705-004-0367-y. [DOI] [PubMed] [Google Scholar]
- Nelson CD, Palermo LM, Hafenstein SL, Parrish CR. Different mechanisms of antibody-mediated neutralization of parvoviruses revealed using the Fab fragments of monoclonal antibodies. Virology. 2007;361:283–293. doi: 10.1016/j.virol.2006.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JSL, Parrish CR. Canine parvovirus host range is determined by the specific conformation of an additional region of the capsid. J Virol. 1997;77:9214–9222. doi: 10.1128/jvi.71.12.9214-9222.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish CR. Emergence, natural history, and variation of canine, mink, and feline parvoviruses. Adv Virus Res. 1990;38:403–450. doi: 10.1016/S0065-3527(08)60867-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish CR, Aquadro CF, Strassheim ML, Evermann JF, Sgro JY, Mohammed HO. Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J Virol. 1991;65:6544–6552. doi: 10.1128/jvi.65.12.6544-6552.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish CR, Have P, Foreyt WJ, Evermann JF, Senda M, Carmichael LE. The global spread and replacement of canine parvovirus strains. J Gen Virol. 1988;69(Pt 5):1111–1116. doi: 10.1099/0022-1317-69-5-1111. [DOI] [PubMed] [Google Scholar]
- Parrish CR, Carmichael LE. Characterization and recombination mapping of an antigenic and hostrange mutation of canine parvovirus. Virology. 1986;148:121–132. doi: 10.1016/0042-6822(86)90408-3. [DOI] [PubMed] [Google Scholar]
- Pereira CA, Leal ES, Durigon EL. Selective regimen shift and demographic growth increase associated with the emergence of high-fitness variants of canine parvovirus. Infect Genet Evol. 2006;7:399–409. doi: 10.1016/j.meegid.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Pereira CA, Monezi TA, Mehnert DU, D'Angelo M, Durigon EL. Molecular characterization of canine parvovirus in Brazil by polymerase chain reaction assay. Vet Microbiol. 2000;75 doi: 10.1016/s0378-1135(00)00214-5. [DOI] [PubMed] [Google Scholar]
- Perez R, Francia L, Romero V, Maya L, Lopez I, Hernandez M. First detection of canine parvovirus type 2c in South America. Vet Microbiol. 2007;124:147–152. doi: 10.1016/j.vetmic.2007.04.028. [DOI] [PubMed] [Google Scholar]
- Pita JS, de Miranda JR, Schneider WL, Roossinck MJ. Environment determines fidelity for an RNA virus replicase. J Virol. 2007;81:9072–9077. doi: 10.1128/JVI.00587-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Qin Q, Loeffler IK, Li M, Tian K, Wei F. Sequence analysis of a canine parvovirus isolated from a red panda (Ailurus fulgens) in China. Virus genes. 2006;34:299–302. doi: 10.1007/s11262-006-0023-6. [DOI] [PubMed] [Google Scholar]
- Sanjuan R, Cuevas JM, Furio V, Holmes EC, Moya A. Selection for robustness in mutagenized RNA viruses. PLoS genetics. 2007;3:e93. doi: 10.1371/journal.pgen.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan R, Cuevas JM, Moya A, Elena SF. Epistasis and the adaptability of an RNA virus. Genetics. 2005;170:1001–1008. doi: 10.1534/genetics.105.040741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelton LA, Hoelzer K, Parrish CR, Holmes EC. Comparative analysis reveals frequent recombination in the parvoviruses. J Gen Virol. 2007;88:3294–3301. doi: 10.1099/vir.0.83255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelton LA, Parrish CR, Truyen U, Holmes EC. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc Natl Acad Sci U S A. 2005;102:379–384. doi: 10.1073/pnas.0406765102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4 Sinauer Associates; Sunderland, Massachusetts: 2003. [Google Scholar]
- Truyen U, Parrish CR. Canine and feline host ranges of canine parvovirus and feline panleukopenia virus: distinct host cell tropisms of each virus in vitro and in vivo. J Virol. 1992;66:5399–5408. doi: 10.1128/jvi.66.9.5399-5408.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truyen U, Agbandje M, Parrish CR. Characterization of the feline host range and a specific epitope of feline panleukopenia virus. Virology. 1994a;200:494–503. doi: 10.1006/viro.1994.1212. [DOI] [PubMed] [Google Scholar]
- Truyen U, Platzer G, Parrish CR, Hanichen T, Hermanns W, Kaaden OR. Detection of canine parvovirus DNA in paraffin-embedded tissues by polymerase chain reaction. Zentralbl Veterinarmed [B] 1994b;41:148–152. doi: 10.1111/j.1439-0450.1994.tb00218.x. [DOI] [PubMed] [Google Scholar]
- Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Yuan W, Davis I, Parrish CR. Nonstructural protein-2 and the replication of canine parvovirus. Virology. 1998;240:273–281. doi: 10.1006/viro.1997.8946. [DOI] [PubMed] [Google Scholar]
- Woolhouse ME, Taylor LH, Haydon DT. Science. Vol. 292. New York, NY: 2001. Population biology of multihost pathogens; pp. 1109–1112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.