Abstract
Aminoacylase 3 (AA3) deacetylates N-acetyl-aromatic amino acids and mercapturic acids including N-acetyl-1,2-dichlorovinyl-L-cysteine (Ac-DCVC), a metabolite of a xenobiotic trichloroethylene. Previous studies did not demonstrate metal-dependence of AA3 despite a high homology with a Zn2+-metalloenzyme aminoacylase 2 (AA2). A 3D model of mouse AA3 was created based on homology with AA2. The model showed a putative metal binding site formed by His21, Glu24 and His116, and Arg63, Asp68, Asn70, Arg71, Glu177 and Tyr287 potentially involved in catalysis/substrate binding. The mutation of each of these residues to alanine inactivated AA3 except Asn70 and Arg71, therefore the corrected 3D model of mouse AA3 was created. Wild type (wt) mouse AA3 expressed in E. coli contained ~0.35 zinc atoms per monomer. Incubation with Co2+ and Ni2+ activated wt-AA3. In the cobalt-activated AA3 zinc was replaced with cobalt. Metal removal completely inactivated wt-AA3, whereas addition of Zn2+, Mn2+ or Fe2+ restored initial activity. Co2+ and to a lesser extent Ni2+ increased activity several times in comparison with intact wt-AA3. Co2+ drastically increased the rate of deacetylation of Ac-DCVC and significantly increased the toxicity of Ac-DCVC in the HEK293T cells expressing wt-AA3. The results indicate that AA3 is a metalloenzyme significantly activated by Co2+ and Ni2+.
Keywords: Metalloprotein, Cobalt, Zinc, aminoacylase
1. Introduction
The majority of eukaryotic proteins are Nα-terminally acetylated [1,2]. The catabolism of the Nα-acetylated proteins releases N-acetylated amino acids that may be deacetylated by various aminoacylases. Aminoacylase 3 (AA3) deacetylates the N-acetylated aromatic amino acids [3], which are present at relatively low frequencies in eukaryotic proteins [2], and the S-conjugates of N-acetyl-L-cysteine (mercapturic acids). The deacetylation of N-acetyl-S-(1,2-dichlorovinyl)-L-cysteine (Ac-DCVC), a metabolite of the glutathione conjugation pathway of a common industrial contaminant trichloroethylene (TCE), catalyzed by AA3 has been shown to be involved in mediating the cytotoxicity of Ac-DCVC in the mouse proximal tubule cell line mPCT [4]. It is believed that the deacetylation of Ac-DCVC is involved in mediating the nephrotoxicity of TCE in rodents and humans [5–11]. Aminoacylase 1 (AA1; EC 3.5.1.14), similar to AA3 is involved in deacetylation of mercapturic acids and N-acetyl aliphatic amino acids, whereas aminoacylase 2 (AA2; EC 3.5.1.15) strictly deacetylates N-acetyl-L-aspartate (Ac-Asp). AA1, AA2 and AA3 are highly expressed in mammalian renal proximal tubules [4,12–15] where they apparently deacetylate Nα-acetylated amino acids. The AA2 deficiency in humans leads to Canavan disease, which is characterized by dysmyelinization of the white matter in children apparently due to the impaired deacetylation of Ac-Asp, one of the most abundant amino acid derivatives in the vertebrate brain [16–20].
AA1 has been shown to be a Zn2+ -metalloprotein containing stoichiometric amounts of this metal per monomer [21–23]. Co2+ [24] and Mn2+ [25] can substitute for zinc ions in mammalian AA1 without a substantial loss of activity. Nuclear magnetic relaxation studies of the Mn2+ -substituted pig AA1 suggested that Zn2+ stabilizes the active conformation of AA1 without playing any specific role in catalysis [25]. In contrast, another study has shown that zinc plays a catalytic role in human AA1 [26]. The crystal structure of the metal-binding domain of AA1 has revealed two zinc atoms per active site [26]. Zn2+ removal significantly changed the circular dichroism (CD) spectrum of AA1 [27] suggesting that zinc is involved in stabilization of the native conformation of AA1. Zn2+, Mn2+, Mg2+ and Ca2+ have been shown to activate AA2 [28]. Zinc detected in the active site of human AA2 was hypothesized to play an important role in catalysis [17]. Rat and human AA2 expressed in E. coli contained 0.15 and 0.05 zinc atoms per monomer and 0.53 and 0.15 nickel atoms per monomer respectively [17]. In contrast, human AA2 expressed in Pichi pastoris contained ~1.3 zinc atoms per monomer that could not be completely removed after extended dialysis against o-phenanthroline (OP); the content of zinc was proportional to the residual activity of AA2 [18]. The architecture of the hypothetical active sites of AE2 showed close similarity with carboxypeptidases despite a very low sequence identity of these enzymes [17].
The involvement of metal ions in AA3 mediated catalysis has not been documented in previous studies. AA3 has ~40% amino acid identity with AA2 [4], and all three amino acid residues (His21, His116 and Glu24) that have been hypothesized to participate in Zn2+ coordination [17] and which mutations inactivated AA2 [18], are conserved in AA3 [4,14]. Therefore in the present work we determined whether mouse AA3 contains zinc/other metals; whether Zn2+ or other metal ions activate mouse AA3; and whether His21, His116 and Glu24 are necessary for metal binding. In addition, using 3D modeling we determined the amino acid residues of mouse AA3 that may be potentially important for substrate binding/catalysis and performed mutational analysis of them.
2. Materials and methods
2.1. Cloning, expression and purification of wt- and mutant mouse AA3
Mouse wt-AA3 and mutants were expressed in E. coli using the pRSET vector (Invitrogen, Carlsbad, CA, USA) and purified as described before [4,14]. The mutations were made using a QuickChange site-directed mutagenesis kit from Stratagene (La Jolla, CA, USA). Sequences of all constructs were confirmed by a bi-directional sequencing using an ABI 310 sequencer (Perkin Elmer, Foster City, CA, USA). The purity of mouse AA3 preparations was >99% as was estimated by SDS-PAGE. Protein bands were visualized with Coomassie brilliant blue R (Sigma, Milwaukee, WI, USA). The His6-tag was cleaved off using an enterokinase cleavage kit (Novagen), and enterokinase was removed according to the manufacturer protocol.
2.2. AA3 activity assay
AA3 activity was determined with N-acetyl-L-tyrosine (Ac-Tyr) and Ac-DCVC [4,14] by measuring the deacetylated product in fluorescence assay [29]. The calibration curves were created with L-tyrosine and DCVC. The experiments were performed at least in triplicate.
The Km and Vmax values were calculated by fitting data to the Michaelis–Menten equation using the OriginPro 7.5 software (OriginLab Corp., Northampton, MA, USA). All values are means ± S.E. of measurements of at least three separate experiments.
2.3 Metal ion removal from mouse AA3
Metal ions were removed from the mutant and wt-AA3 using dialysis against 5 mM OP in 50 mM Tris-HCl, pH 7.5, for 18 h at 4°C. OP was separated from mouse AA3 by gel-filtration on a PD-10 desalting column equilibrated with 50 mM Tris-HCl, pH 7.5.
2.4. Metal content of mouse AA3
The content of metals in mouse AA3 was determined using inductively coupled plasma mass spectrometry (ICP-MS). To a 50 µl AA3 sample, 100 µl of ultrapure nitric acid (Optima, Fisher, CA, USA) was added. The samples were heated to 90°C on a 48 well digestion block (CPI International, Santa Rosa, CA, USA) with a digital temperature controller and allowed to digest until no particulates or color change was observed. The temperature was then raised to 110°C and the samples were allowed to evaporate to ~50 µl. After cooling, the samples were diluted to 2 ml with ultra pure water, and the final nitric acid concentration was adjusted to 2%. External calibration solutions were diluted from a 100-ppm multi-element stock solution (CPI International) and scandium, gallium, and indium were added to a final concentration of 50 ppb to all samples and calibration standards. Samples were analyzed on an Agilent 7500ce Quadrupole ICP-MS equipped with an H2/He/Xe Octapole Reaction/Collision Cell. The instrument parameters were as follows: 1550 RF power, 1050 ml/min carrier gas, 0.1 rps nebulizer pump, 2°C spray chamber temperature; 4 ml/min and 4.5 ml/min of H2 and He respectively were used when the analysis required interference removal by the collision cell. The means ± S.E. are shown for three independent mouse AA3 isolations.
2.5. Metal induced secondary structure changes in mouse AA3
Changes in the secondary structure of mouse AA3 induced by metal ions were studied using circular dichroism (CD) spectra measurements of 10 µM AA3 solutions in 20 mM Tris-HCl, pH 7.5, with and without metal ions. After 30 min incubation at 20°C with a 0.1 mM metal ion, CD spectra were measured in a JASCO J-715 Circular Dichroism spectrophotometer with a Peltier temperature control (JASCO, Easton, MD, USA) at 20°C.
2.6. Metal induced aggregation of mouse AA3
The aggregation of mouse AA3 induced by metal ions was studied using measurements of the turbidity and dynamic light scattering (DLS) of mouse AA3 solutions. Transmission electron microscopy was used in addition to the above-mentioned methods to characterize the size and shape of mouseAA3 aggregates.
The dynamics of aggregation was recorded by measuring the turbidity of mouse AA3 solutions. In these experiments, the absorbance of 15 µM mouse AA3 solution in 20 mM Tris-HCl, pH 7.5, was measured without or in the presence of 0.1 or 1 mM metal ion on a Genesys 10 UV spectrophotometer (Thermo Fisher Scientific, Pittsburg, PA, USA) at 350 nm.
The DLS measurements were used to characterize the size, shape and polydispersity of mouse AA3 particles formed in the presence of a metal ion. In all experiments Co2+, Zn2+, Fe2+, Ni2+, Cu2+ or Fe3+ was added to a final concentration 0.1 or 1.0 mM into 15 µM solution of mouse AA3 in 20 mM Tris-HCl, pH 7.5, and measurements were performed on a Dynamic Light Scattering Analyzer-N4 Plus (Beckman Coulter, Fullerton, CA, USA) during 2 min at 20°C.
For transmission electron microscopy, Co2+ or Fe2+ was added to a final concentration of 0.1 mM into 15 µM solution of mouse AA3 in 20 mM Tris-HCl, pH 7.5. After incubation at 20°C for 2 min, a 2 µl aliquot was diluted in 98 µl of 20 mM Tris-HCl, pH 7.5, containing 0.1 mM Co2+ or Fe2+ and negatively stained with 1% uranyl acetate as described before [30]. Micrographs were taken on a JEM-1200EX transmission electron microscope (JEOL, Tokyo, Japan) operated at 80 kV. Images were recorded with a BioScan 600W digital camera (Gatan, Pleasanton, CA, USA).
2.7. Cytotoxicity experiments
Cytotoxicity experiments were performed using HEK293T cells. These cells did not express human AA3 and the extracts from these cells did not significantly deacetylate Ac-DCVC. To express mouse AA3 in HEK293T cells, the coding sequence of mouse AA3 inserted into the multiple cloning site of an Invitrogen pcDNA3.1(+)/Zeocin vector [4] was used. The cells were transfected with the plasmid using LipofectAMINE 2000 (Invitrogen) per the matufactorer’s protocol. Mock-transfected HEK293T cells were generated by transforming the cells with the vector. The cells (0.5–10·106 cells/ml) were seeded into 96-well plates with Dulbecco’s modified Eagle’medium containing 5% fetal bovine serum, 1 mM Ac-DCVC and different concentrations of Co2+ for 2 h at 37°C. The mock-transfected cells were used as a control. Cell viability was estimated using a CytoTox96 cytotoxicity assay (Promega, Madison, WI) as was described before [4].
2.8. Expression of human AA3 in HEK293T cells
The coding region of human AA3 was inserted into the BamHI-XhoI site of a pcDNA3.1-His vector (Invitrogen, Carlsbad, CA) was expressed in HEK293T cells. The cell extracts were analyzed by SDS-PAGE and immunoblotting using our rabbit anti-human AA3 antibody HR-C1 specific for ten C-terminal amino acids of human AA3.
2.9. 3D modeling of mouse AA3
The 3D model of mouse AA3 was generated using the structure of the highly homologous protein rat AA2 (PDB accession code 2GU2) by the program SWISS-MODEL [31]. A cobalt atom was substituted for the zinc present in the rat AA2 structure based on biochemical observations in this manuscript. The substrate, Ac-Tyr, was generated using the program Discovery Studio ViewerPro 5.0 from Accelrys [www.accelrys.com]. The amino acid side chains were remodeled by hand using the program Coot [32] based on the biochemical observations in this manuscript. The model was energy minimized using Refmac [33].
2.10. Reagents
If not specifically indicated, the reagents of the highest purity were purchased from Sigma. N-acetyl-S-(1,2-dichlorovinyl)-L-cysteine (Ac-DCVC) and S-(1,2-dichlorovinyl)-L-cysteine (DCVC) were synthesized as described previously [4,34], purified using HPLC and their purity (>95%) was confirmed by mass spectrometry (MS).
3. Results and discussion
3.1. The initial 3D model of mouse AA3
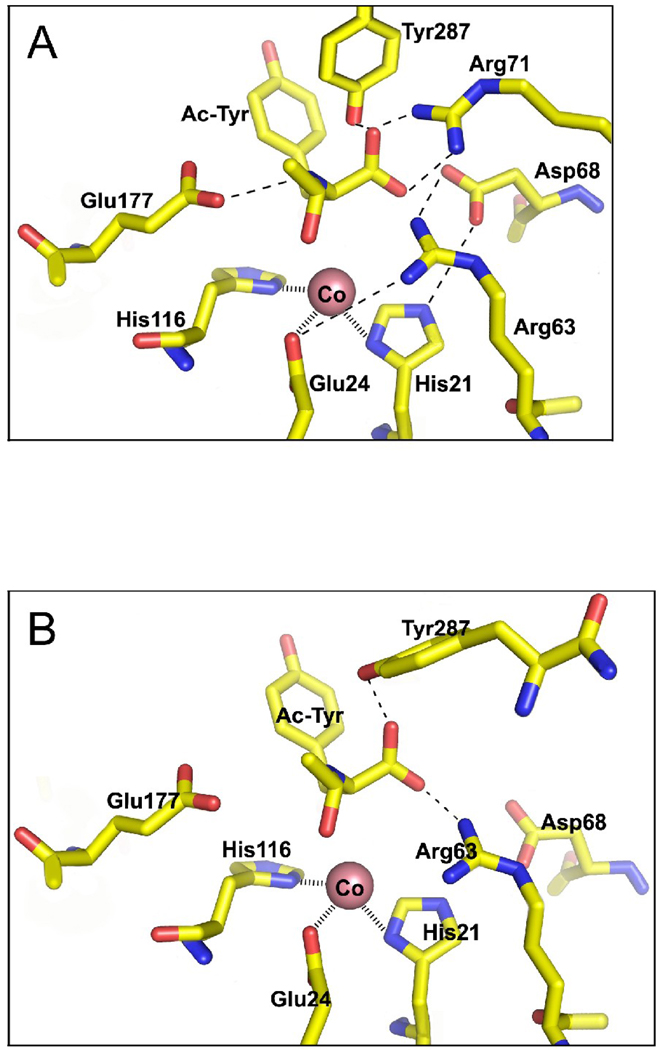
3D modeling of mouse AA3 using the 3D structure of rat AA2 as a template showed structural similarity of these proteins. A putative metal binding site formed by His21, Glu24 and His116 (Fig. 1A) was identified. The identical amino acids formed a metal binding site in the putative active site of rat and human AA2 [17]. Therefore it is likely that these conserved amino acids might be involved in metal coordination in mouse AA3. Several amino acid residues have been hypothesized to participate in N-acetyl-aspartate binding/catalysis of rat and human AA2 [17,35,36]. These amino acids are conserved not only in AA2 but also in AA3 (Arg63, Asp68, Asn70, Arg71, Asp114, Glu177 and Tyr287 in mouse AA3). Contrary to AA2, which specifically deacetylates Ac-Asp [3,18], AA3 uses a wide range of substrates but not Ac-Asp [3,4,37–39]. Therefore these common amino acid residues cannot be responsible for different substrate specificity of AA2 and AA3 but rather may be involved in binding of common α-carboxyl and N-acetyl groups and catalysis. Their location in the mouse AA3 model (Fig. 1A) supports this hypothesis.
Fig. 1.
3D modeling of mouse AA3. (A) The preliminary model of mouse AA3 created on base of the homology with rat AA2 (PBD ID code 2GU2). (B) The corrected model of mouse AA3 based on the mutational analysis of mouse AA3.
3.2. Effect of metal ions and chelating agents and metal content in mouse wt-AA3 activity
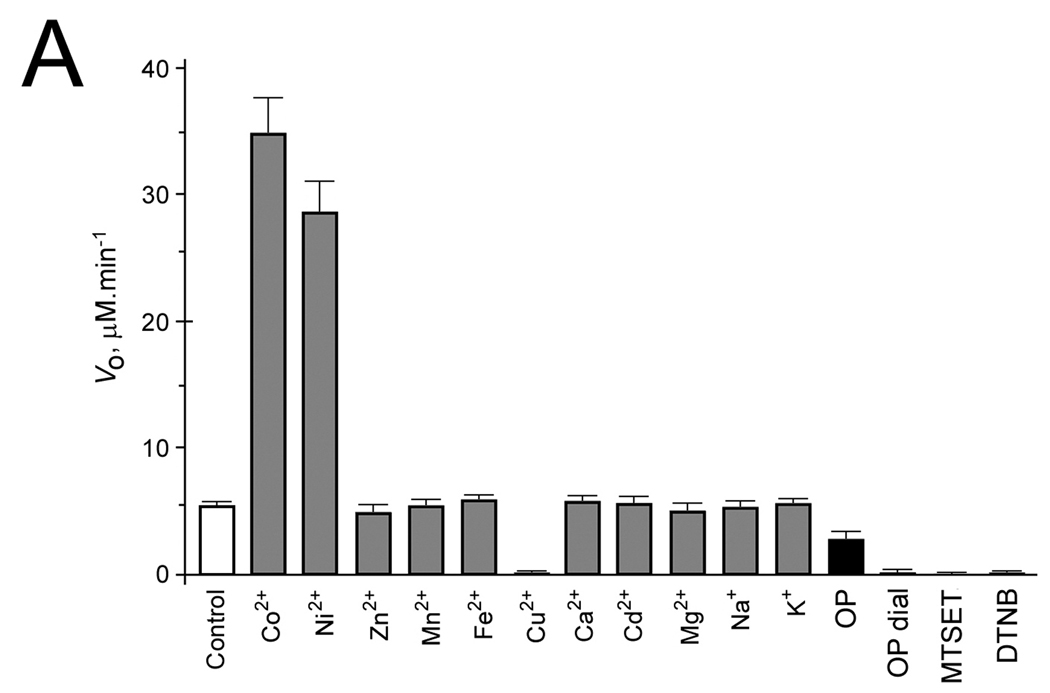
Zn2+, Mn2+, Fe2+, Cd2+, Mg2+, Ca2+, K+ and Na+ did not significantly alter activity whereas Co2+ and Ni2+ stimulated activity of mouse wt-AA3 several times (Fig. 2A). 5 mM OP added into AA3 reaction assay partially inhibited mouse AA3 (Fig. 2A) whereas even 30 mM EDTA has been previously shown not to inhibit mouse AA3 [4]. Overnight dialysis against 5 mM OP completely inactivated mouse AA3 (Fig. 2A). Zn2+, and Fe2+ added to apoenzyme restored catalytic activity to the level of holoenzyme; Co2+ and Ni2+ increased activity several times above the holoenzyme level (Fig. 2B).
Fig. 2.
(A) The effect of metal ions, OP, MTSET and DTNB on the rate of deacetylation of Ac-Tyr mediated by mouse wt-AA3. The reaction was started by adding AA3 (1.2 µM) in the assay of a total volume 0.1 ml, which contained 50 mM Tris-HCl, pH 7.5, 2.5 mM Ac-Tyr and 1 mM metal ion, or 1 mM MTSET, 1 mM DTNB or 5 mM OP. (OP dial) means mouse wt-AA3 dialyzed for 18 h against 5 mM OP. (Control) means mouse wt-AA3 without any treatment. (B) The effect of metal ions on the rate of deacetylation of Ac-Tyr by the mouse wt-AA3 dialyzed against 5 mM OP; Co2+ (■), Ni2+ (●), Fe2+ (▼), and Zn2+ (▲) were added into the reaction assay (see above).
The results suggested that mouse AA3 requires for catalysis and may contain metal ions in agreement with the hypothesized model (Fig. 1A). To test this hypothesis, we measured Zn, Fe, Mn, Co, Ni and Cu content in mouse wt-AA3 before and after dialysis against 5 mM OP (Table 1) using ICP-MS. Before dialysis, mouse AA3 contained ~0.35 atoms of Zn, ~0.1 atom of Ni per monomer and 0.03–0.05 atoms of Fe, Mn and Cu. Therefore Zn was the predominant metal present in mouse wt-AA3, but its amount was not equimolar to the number of monomers. Incubation of wt-AA3 with 0.1 mM Zn2+ increased the content of bound zinc to ~4 atoms per monomer (Table 1) but did not affect activity (Fig. 1A). Rat and human AA2 expressed in E. coli also contained less than equimolar amounts of zinc, 0.15 and 0.05 atoms per monomer respectively [17], whereas human AA2 expressed in P. pastoris contained ~1.3 atoms of zinc per monomer [18]. It is possible that not all mouse AA3 molecules expressed in bacteria are properly folded and active, and only the properly folded mouse AA3 contains metal. Dialysis against OP completely removed metal ions from mouse wt-AA3 (Table 1) and inactivated mouse AA3 (Fig. 2A). Therefore only the metal (zinc) containing mouse wt-AA3 is catalytically active.
Table 1.
Metal content of wt-AA3 and the H21A mutant of mouse AA3. Protein samples were desalted before metal measurements on PD-10 columns (GE HealthCare) equilibrated with 50 mM Tris-HCl, pH 7.5. (No) means no treatment, ( + Co2+) incubated with 0.1 mM Co2+, (OP) means after dialysis against 5 mM OP in 50 mM Tris-HCl, and (OP + Co2+), (OP + Zn2+) and (OP + M2+) means after incubation of dialyzed proteins with respectively 0.1 mM Co2+, 0.1 mM Zn2+, and a mixture of 0.1 mM Zn2+, Mn2+, Fe2+, Cu2+, Ni2+ and Co2+. The measurements were performed on an Agilent 7500ce Quadrupole ICP-MS equipped with an H2/He/Xe Octapole Reaction/Collision Cell. Mean values of at least 3 independent isolations in atoms per monomer (35 kDa) ± S.E. are shown.
| wt/mutant | Treatment | Metal content (atom/monomer) | ||||||
|---|---|---|---|---|---|---|---|---|
| Zn | Mn | Fe | Cu | Ni | Co | Total | ||
| wt | No | 0.351±0.036 | 0.031±0.003 | 0.048±0.009 | 0.043±0.012 | 0.101±0.002 | 0.000±0.001 | 0.574 |
| + Co2+ | 0.040±0.008 | 0.005±0.003 | 0.029±0.009 | 0.000±0.000 | 0.002±0.001 | 0.891±0.250 | 0.967 | |
| OP | 0.015±0.002 | 0.004±0.002 | 0.001±0.001 | 0.013±0.002 | 0.000±0.000 | 0.002±0.001 | 0.035 | |
| OP + Co2+ | 0.026±0.020 | 0.006±0.003 | 0.002±0.001 | 0.001±0.001 | 0.000±0.000 | 1.582±0.413 | 1.617 | |
| OP + Zn2+ | 4.101±0.267 | 0.006±0.002 | 0.003±0.001 | 0.007±0.004 | 0.010±0.004 | 0.001±0.001 | 4.128 | |
| OP + M2+ | 0.284±0.037 | 0.704±0.025 | 0.164±0.016 | 0.609±0.026 | 0.247±0.196 | 0.627±0.045 | 2.635 | |
| H21D | No | 1.025±0.027 | 0.006±0.001 | 0.022±0.005 | 0.012±0.003 | 0.340±0.044 | 0.000±0.000 | 1.405 |
| OP | 0.012±0.005 | 0.001±0.001 | 0.003±0.002 | 0.001±0.001 | 0.008±0.005 | 0.000±0.000 | 0.025 | |
| OP + Co2+ | 0.015±0.011 | 0.001±0.000 | 0.002±0.001 | 0.003±0.002 | 0.012±0.008 | 0.538±0.034 | 0.571 | |
| OP + Zn2+ | 3.212±0.394 | 0.000±0.000 | 0.005±0.003 | 0.004±0.002 | 0.000±0.000 | 0.002±0.000 | 3.223 | |
| H21A | No | 0.398±0.055 | 0.001±0.001 | 0.048±0.007 | 0.033±0.004 | 0.034±0.044 | 0.005±0.002 | 0.456 |
| E24A | No | 0.443±0.100 | 0.024±0.003 | 0.021±0.006 | 0.096±0.037 | 0.048±0.032 | 0.050±0.011 | 0.682 |
| H116A | No | 0.497±0.131 | 0.009±0.004 | 0.031±0.020 | 0.065±0.024 | 0.082±0.016 | 0.015±0.011 | 0.699 |
Surprisingly mouse wt-AA3 expressed in E. coli did not contain cobalt (Table 1), the most significant metal-activator of this enzyme. To determine whether mouse AA3 can bind Co2+, we incubated the apo- and holoenzyme with 0.1 mM Co2+. After separation of the unbound metal, the enzyme contained ~0.9–1.5 atoms of cobalt per monomer. No zinc was detected in the holoenzyme after incubation with 0.1 mM Co2+ indicating that zinc (0.35 atoms per monomer) was replaced by cobalt. The results indicated that Co2+ and Zn2+ might bind to the same site(s) in mouse AA3 and suggested that Co2+ might have a higher affinity than Zn2+ to mouse AA3. Given that mouse AA3 binds more than 1 metal atom per monomer and only 1 metal binding site was identified using our mouse AA3 model (Fig. 1A), we compared mean relative affinities mouse AA3 for zinc, cobalt, nickel, iron, manganese and copper. In this experiment the apoenzyme was incubated with the solution containing a mixture of Zn2+, Co2+, Ni2+, Fe2+, Cu2+ and Mn2+ each used at 0.1 mM concentration. The amount of bound metals were ~0.3, 0.6, 0.25, 0.15, 0.6 and 0.7 atoms of respectively Zn, Co, Ni, Fe, Cu and Mn bound to one wt-AA3 monomer (Table 1). Therefore mouse AA3 has similar affinities for these divalent metal ions (Table 1). Given that only 1 site is necessary for catalysis, similar binding properties of these divalent cations to mouse AA3 apparently represent their binding to the sites different than the active site. Although Co2+ and Ni2+ are the most potent activators of mouse AA3, no cobalt was detected in the intact AA3 expressed in E. coli. It is likely that the amount of metal ions bound to AA3 is determined by their relative contents in growth and purification media. Indeed a very low concentration of cobalt was detected in the growth media whereas zinc content was relatively high (data not shown).
3.3. Metal content and activity of H21D, H21A, E24A and H116A mutants of mouse AA3
H21A, E24A and H116A mutants of mouse AA3 were inactive with both Ac-Tyr and Ac-DCVC (Table 2). Incubation with cobalt or other divalent metal ions did not restore activity suggesting that the metal coordination mediated by His21, Glu24 and His116 is necessary for catalysis. We expected that these mutants should have impaired metal binding properties. Surprisingly, Zn content in the mutants was similar or even higher (H21D) than in wt-AA3 (Table 1). Given that ~4 and ~3 zinc atoms can be bound to the monomer of wt-AA3 and H21D mutant respectively, the mutant can bind ~1 atom of zinc less than wt-AA3 (Table 1) that correlates with the proposed impaired metal binding in the active site. Then different level of zinc in H21D, H21A, E24A and H116A mutants and wt-AA3 just apparently reflects different availability of zinc for active AA3 biosynthesis due to variations of zinc and other metal ions levels in growth media. Similar changes in metal binding properties of H21E mutant of human AA2 have been described by Hershfield et al. [40]. It is interesting to note that the incubation of either intact mouse AA3 or apoenzyme with zinc is accompanied with binding of ~ 4 zinc atoms per monomer but does not affect enzymatic activity. This fact suggests that 1) not all mouse AA3 molecules expressed in E. coli are catalytically active, and 2) only those metal ions that bound to the active site are important for catalytic activity. Contrary to Zn2+, Co2+ and Ni2+ significantly activate mouse AA3. The Co2+ -activated enzyme contains ~1 cobalt atom per monomer and trace amounts of zinc and other metals indicating that cobalt completely replaces them from the enzyme.
Table 2.
Kinetic characteristics of mouse mutant and wt-AA3. The results are means of at least 3 experiments ±S.E. ND means not detected, NM means not measured.
| wt/mutant | Km (mM) | Vmax (nmol·mg−1·min−1) | ||||||
|---|---|---|---|---|---|---|---|---|
| Ac-Tyr | Ac-DCVC | Ac-Tyr | Ac-DCVC | |||||
| −Co | +Co | −Co | +Co | −Co | +Co | −Co | +Co | |
| wt-AA3 | 1.30±0.20 | 0.56±0.17 | 4.51±0.56 | 3.02±0.61 | 226±54 | 1890±228 | 483±123 | 3950±776 |
| H21A | ND | ND | ND | ND | 0 | 0 | 0 | 0 |
| E24A | ND | ND | ND | ND | 0 | 0 | 0 | 0 |
| R63A | ND | ND | ND | ND | 0 | 0 | 0 | 0 |
| R67D | 0.26±0.12 | 0.53±0.13 | NM | NM | 49±5 | 424±24 | NM | NM |
| D68A | ND | ND | ND | ND | 0 | 0 | 0 | 0 |
| N70A | 2.80±0.22 | 2.83±0.34 | 2.48±0.57 | 3.49±0.45 | 524±48 | 789±143 | 501±101 | 1823±90 |
| R71A | 0.71±0.09 | 2.02±0.38 | 7.00±1.88 | 12.40±5.11 | 784±82 | 3780±549 | 595±97 | 2170±443 |
| D114A | 0.25±0.05 | 0.18±0.04 | 2.46±0.77 | 5.14±2.09 | 63±33 | 510±62 | 70±10 | 558±84 |
| H116A | ND | ND | ND | ND | 0 | 0 | 0 | 0 |
| Y156A | 0.79±0.29 | 0.22±0.14 | NM | NM | 473±69 | 1495±78 | NM | NM |
| E167A | 0.64±0.11 | 0.83±0.26 | NM | NM | 317±37 | 3335±302 | NM | NM |
| E167R | 0.61±0.07 | 1.30±0.11 | NM | NM | 400±56 | 1040±99 | NM | NM |
| E177A | ND | ND | ND | ND | 0 | 0 | 0 | 0 |
| D235R | 1.30±0.22 | 0.55±0.31 | NM | NM | 247±66 | 2392±599 | NM | NM |
| Y287A | 0.23±0.11 | 0.23±0.05 | 13.61±4.57 | 4.21±1.43 | 5±2 | 152±5 | 1±1 | 99±48 |
| Y288A | 5.20±0.86 | 3.71±0.28 | 6.76±2.74 | 17.73±7.84 | 450±37 | 7030±1529 | 45±10 | 7936±1334 |
| E289A | 0.25±0.05 | 0.25±0.06 | 3.40±1.5 | 11.03±6.92 | 221±22 | 1526±98 | 439±45 | 2925±505 |
| K290A | 0.27±0.05 | 0.26±0.03 | 1.11±0.37 | 11.00±5.66 | 325±65 | 1607±111 | 1123±125 | 7560±987 |
3.4. Metal induced aggregation of mouse AA3
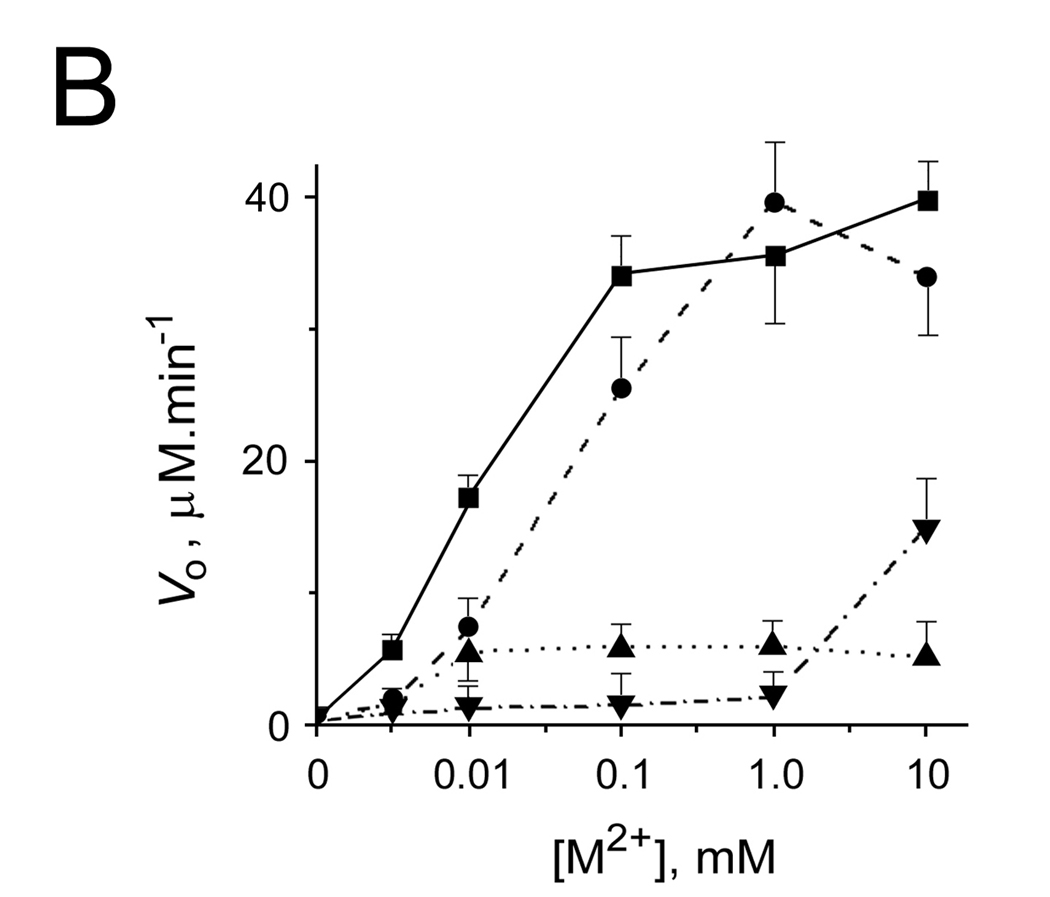
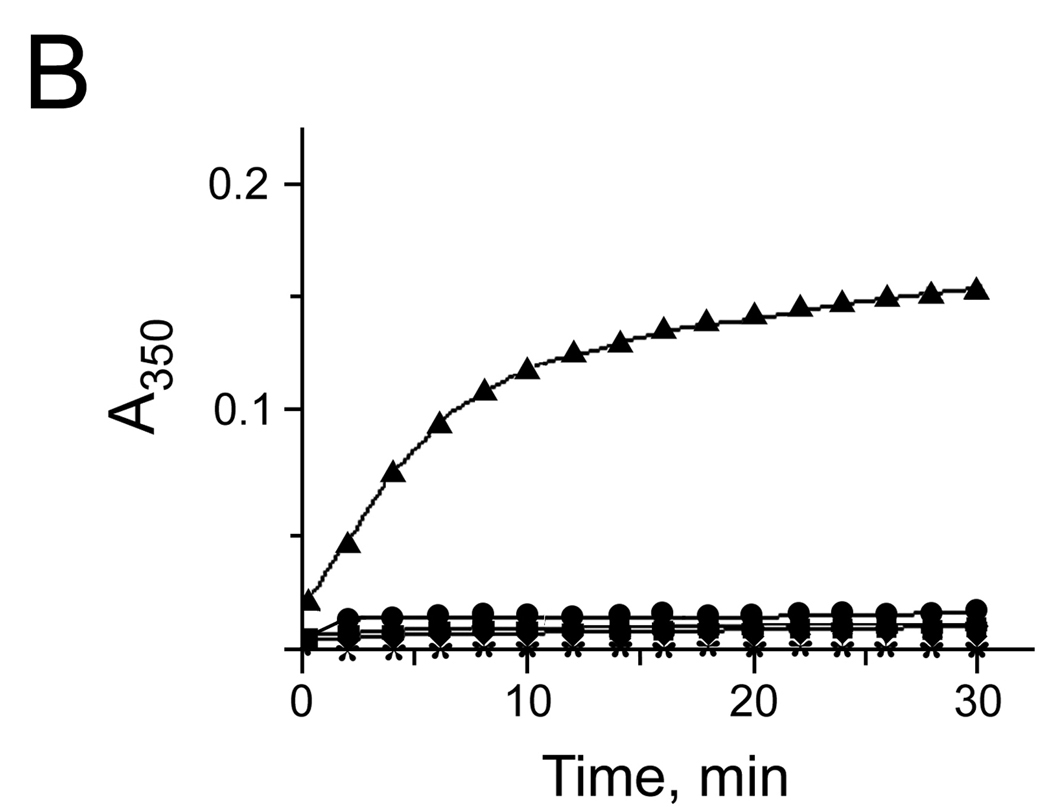
Our data showed that more than 1 metal atom could be bound per AA3 monomer. The location of the metal binding sites, except the putative active site, cannot be exactly predicted using the 3D model of mouse AA3 although it is likely that metal ions bind to AA3 periphery. In agreement with this suggestion, further studies showed that metal ions might induce mouse AA3 aggregation. In the absence of substrate, 1 mM Zn2+, Cu2+, Fe2+, Co2+ or Ni2+ induced fast aggregation of wt-AA3 (Fig. 3A). At a lower concentration (0.1 mM), only Fe2+ caused significant aggregation (Fig. 3B). The Zn2+, Co2+ -, Fe2+ - and Ni2+ -aggregated enzyme retained catalytic activity (data not shown).
Fig. 3.
(A, B) The aggregation of mouse wt-AA3 induced by Fe2+ (▲), Co2+ (♦), Ni2+ (■), Cu2+ (●), and Zn2+ (*). A 1 mM (A) and 0.1 mM (B) concentration of the metal ions was used. Typical curves are shown. The aggregation of AA3 was determined by measuring the turbidity of AA3 solutions (A350)
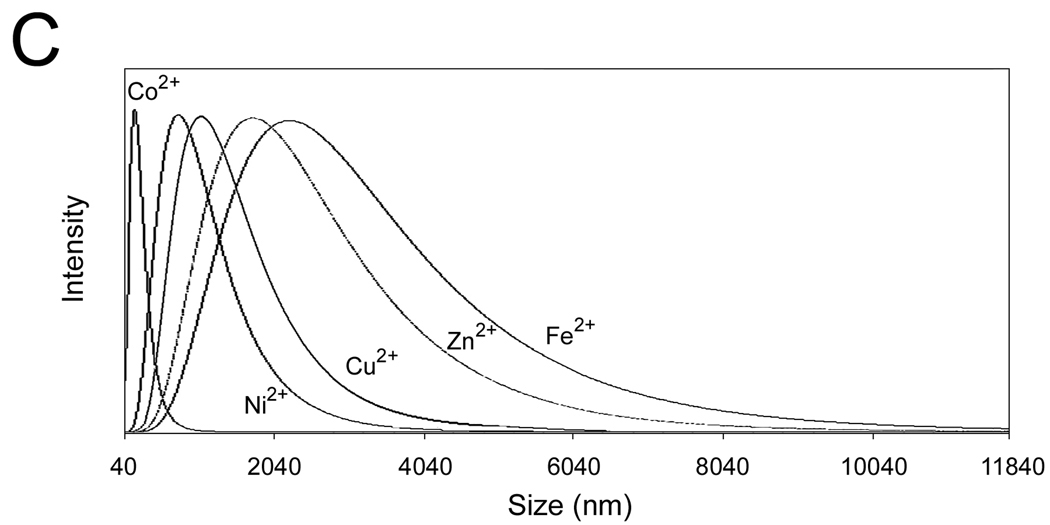
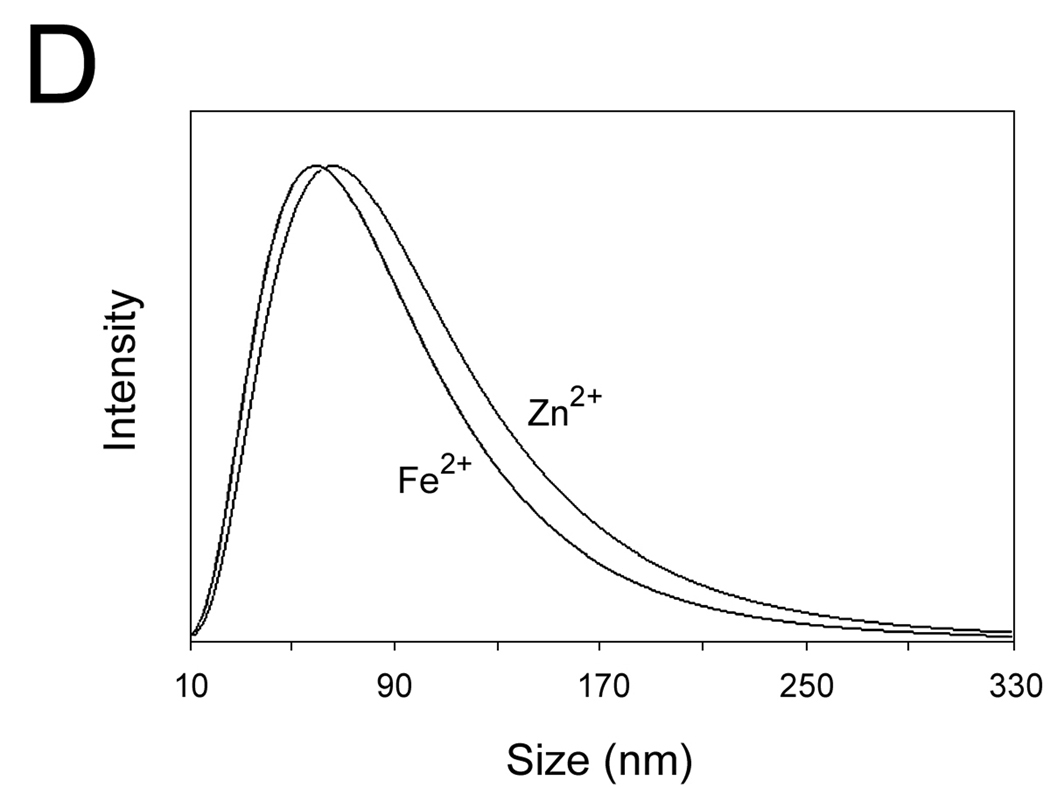
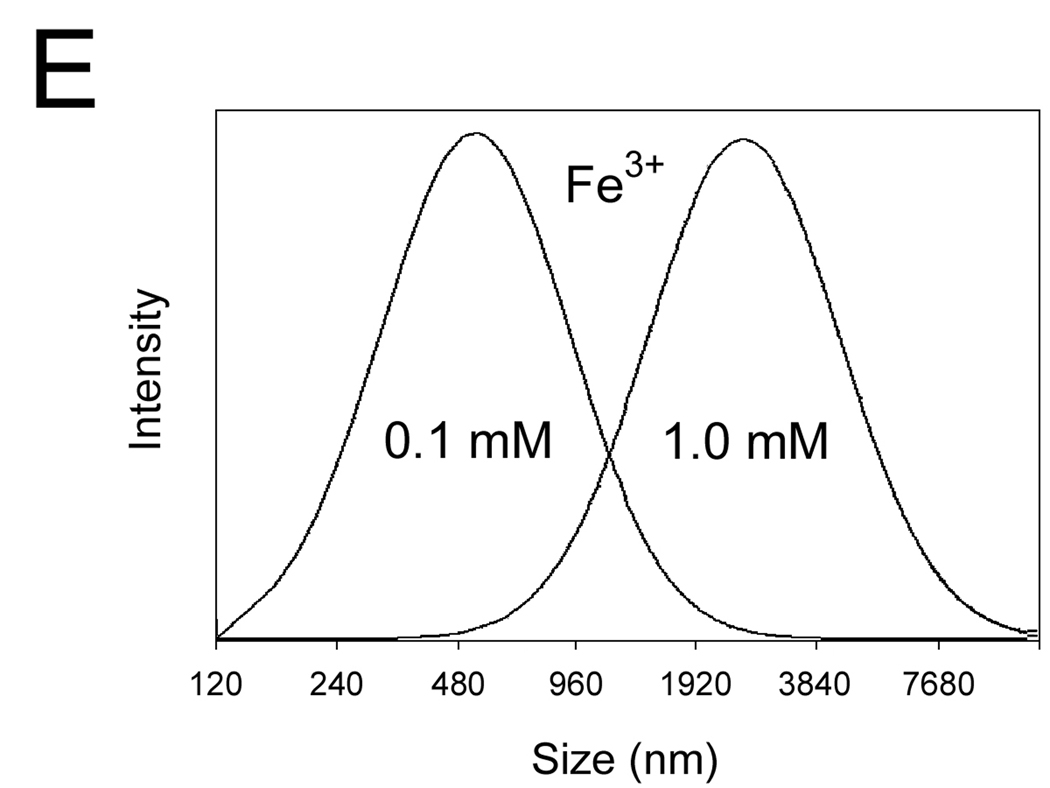
(C–E) The DLS spectra of the wt-AA3 solutions in the presence of 1.0 mM (C) and 0.1 mM (D) Co2+, Ni2+, Cu2+, Zn2+ or Fe2+, and in the presence of 0.1 and 1.0 mM Fe3+ (E).
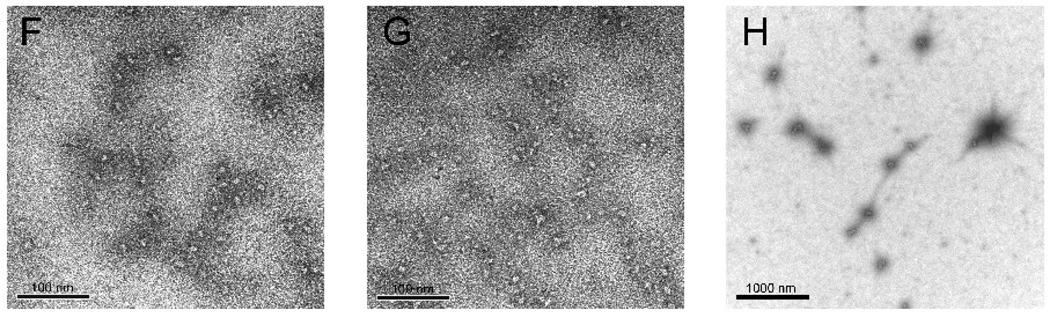
(F–H) Electron micrographs of wt-AA3 without metal ions (F), or with 0.1 mM Co2+ (G) and 0.1 mM Fe2+ (H) negatively stained with 1% uranyl acetate.
We further used dynamic light scattering (DLS) to determine the size and polydispersity of mouse AA3 aggregates. The mean size of the aggregates after 2 min incubation with 1 mM Co2+, Ni2+, Cu2+, Zn2+ or Fe2+ was 200, 845, 1100, 1950 and 2500 nm respectively (Fig. 3C). No aggregates were formed in the presence of 0.1 mM Co2+, Ni2+ and Cu2+, whereas in the presence of 0.1 mM Zn2+ and Fe2+ the aggregates of 74 and 66 nm were detected (Fig. 3D). Iron (III) demonstrated even higher aggregating effect than Fe2+ inducing the formation of the 590 and 2820 nm aggregates at 0.1 and 1 mM concentrations respectively (Fig. 3E).
To further characterize mouse AA3 aggregation in the presence of 0.1 mM Fe2+ and to confirm that 0.1 mM Co2+ does not induce aggregation, we used transmission electron microscopy of negatively stained enzyme particles. Fig. 3F shows a representative micrograph of mouse AA3 without metal ions. The particles of 10–12 × 6–7 nm are seen corresponding to the mouse AA3 dimers described before [41]. No aggregation of mouse AA3 dimers was detected after the incubation with 0.1 mM Co2+ (Fig. 3G). The aggregates of 60–80 nm were formed in the presence of 0.1 mM Fe2+ (Fig. 3H). Their size was in a good agreement with the size determined by DLS.
The metal binding site in the putative active center was not involved in the metal-induced aggregation of mouse AA3 because H21A, E24A or H116A mutants formed aggregates of similar size and shape as mouse wt-AA3 (data not shown). The metal induced aggregation was not restricted to mouse AA3 since similar metal induced aggregation was attributed to human AA3 (data not shown).
Although it is unlikely that cobalt, nickel or zinc may induce aggregation of human AA3 in vivo given their low physiological levels, the iron-induced aggregation may play a role in mediating of the iron-induced toxicity in the liver, kidney and other organs/tissues where AA3 is expressed. The concentration of total iron in normal human liver is 20–40 µmol/g [42], which is equal to ~20–40 mM. Iron is mostly bound to proteins and therefore it cannot induce aggregation of human AA3 [43]. Pathological conditions that increase a free iron level to 100 µM, may lead to human AA3 aggregation, which in turn may induce cellular damage similar to the amyloid diseases [44,45].
It is necessary to note that Co2+, Zn2+, Fe2+, Ni2+ and Cu2+ did not affect the CD spectra of wt-AA3 and the mutants (data not shown) indicating that metal induced aggregation of mouse AA3 does not involve changes in the secondary structure of AA3.
3.5. Effect of Cu2+
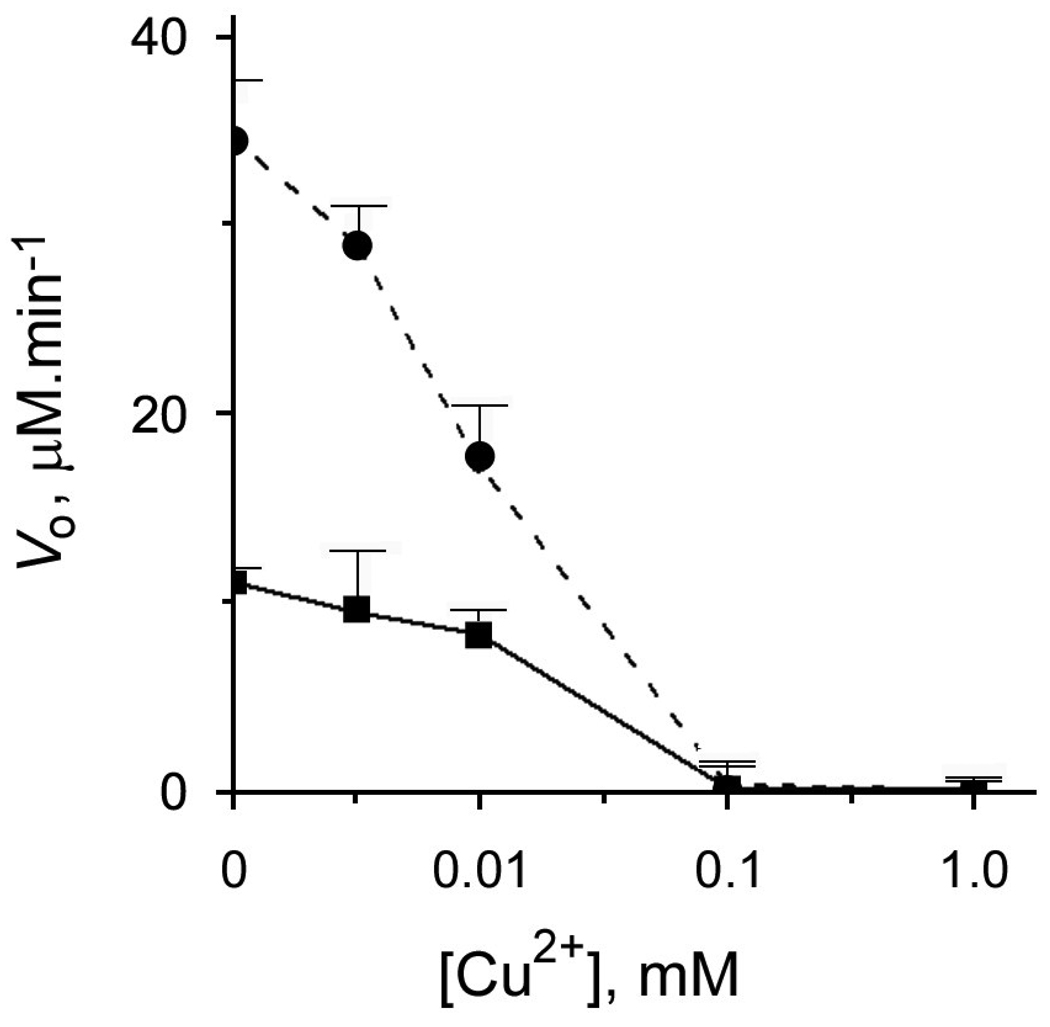
In contrast to Co2+, Ni2+, Zn2+, Mn2+ or Fe2+, 0.1 mM Cu2+ completely inhibited both the intact and Co2+ -activated mouse AA3 even when was used at an equimolar concentration with Co2+ (Fig. 4). Cu2+ is known to bind both N and S ligands [30,46,47]. Therefore its binding for example to a cysteine residue(s) may affect the catalysis for example via affecting metal coordination in the active site or by inducing unfavorable structural changes of mouse AA3, which has 8 cysteine residues. Alternatively mouse AA3 may contain a cysteine residue(s) important for catalysis. This hypothesis is supported by our previous finding that p-chloromercuribenzoate completely inhibits mouse AA3 [4]. In the present study other thiol-reactive compounds, [2-(trimethylammonium)ethyl]methanethiosulfonate bromide and 5,5’-dithio-bis(2-nitrobenzoate) (DTNB) also completely inhibited mouse AA3 (Fig. 2A). Similar effect has been shown for AA1 [48]. Nevertheless the hypothetical involvement of a cysteine residue in catalysis is not supported by the 3D model of mouse AA3 (Fig. 1A).
Fig. 4.
The effect of Cu2+ on the rate of deacetylation of Ac-Tyr mediated by mouse wt-AA3. Copper was added into AA3 reaction assay (see above) without (■) and with 100 µM Co2+ (●).
3.6. Amino acids involved in substrate binding/catalysis
We performed a mutational analysis of the amino acid residues in mouse AA3 that may potentially be involved in substrate binding/catalysis based on the homology with AA2 and the 3D model of mouse AA3. The first model of mouse AA3 (Fig. 1A) suggested that Arg63, Asp68, Asn70, Arg71, Glu177 and Tyr287 might be involved in the active site formation. These residues conserved in mammalian AA2 and AA3, have been shown to be important for catalysis/substrate binding in AA2 [17,18,40]. The results of the mutational analysis did not completely confirm this model (Table 2 and Table 3): Arg63, Asp68, Glu177 and Tyr287 were apparently involved in substrate binding/catalysis since the mutation of these amino acids to alanine completely inactivated mouse AA3. The kinetics parameters of N70A and R71A were very similar to wt- AA3. Asn114 conserved in AA2 and AA3 formed in our model an H-H bond with His24. D114A mutant retained a partial activity with both Ac-Tyr and Ac-DCVC suggesting that this H-H bond is important for the architecture of the active site of mouse AA3. Nevertheless D114A as well as N70A and R71A were significantly activated by Co2+ similar to wt-AA3. Arg71 has been proposed to be involved in binding of α-carboxyl group of Ac-Asp in the rat and human AA2 models [17,36] and was suggested to play a similar role in our first 3D model of AA3 (Fig. 1A).
Table 3.
Vmax / Km values of mouse mutant and wt-AA3 with Ac-Tyr and Ac-DCVC. ND means not detected, NM means not measured.
| wt/mutant | Vmax / Km | |||||||
|---|---|---|---|---|---|---|---|---|
| Ac-Tyr | Ac-Tyr | |||||||
| −Co2+ | +Co2+ | −Co2+ | +Co2+ | |||||
| nmol·mg−1·min−1∙mM−1 | % | nmol·mg−1·min−1·mM−1 | % | nmol·mg−1·min−1·mM−1 | % | nmol·mg−1·min−1·mM−1 | % | |
| wt-AA3 | 174 | 100 | 3375 | 100 | 107 | 100 | 1317 | 100 |
| H21A | ND | ND | ND | ND | ND | ND | ND | ND |
| E24A | ND | ND | ND | ND | ND | ND | ND | ND |
| R63A | ND | ND | ND | ND | ND | ND | ND | ND |
| R67D | 188 | 108 | 800 | 24 | NM | NM | NM | NM |
| D68A | ND | ND | ND | ND | ND | ND | ND | ND |
| N70A | 187 | 107 | 226 | 8 | 200 | 187 | 492 | 37 |
| R71A | 1104 | 635 | 1890 | 56 | 85 | 79 | 175 | 13 |
| D114A | 252 | 145 | 2833 | 84 | 28 | 27 | 109 | 8 |
| H116A | ND | ND | ND | ND | ND | ND | ND | ND |
| Y156A | 599 | 344 | 6795 | 201 | NM | NM | NM | NM |
| E167A | 495 | 284 | 4018 | 119 | NM | NM | NM | NM |
| E167R | 656 | 491 | 800 | 24 | NM | NM | NM | NM |
| E177A | ND | ND | ND | ND | ND | ND | ND | ND |
| D235R | 190 | 109 | 4349 | 129 | NM | NM | NM | NM |
| Y287A | 22 | 12 | 661 | 20 | 0.07 | 0.06 | 24 | 2 |
| Y288A | 87 | 50 | 1900 | 56 | 7 | 6 | 448 | 34 |
| E289A | 884 | 506 | 6104 | 181 | 129 | 121 | 265 | 20 |
| K290A | 1203 | 691 | 6181 | 183 | 1012 | 121 | 687 | 52 |
Based on our model we performed mutational analysis of several conserved amino acids in mouse AA3. Without cobalt the mutation of Tyr288 to alanine did not affect the deacetylation rate of Ac-Tyr but severely decreased the deacetylation rate of Ac-DCVC (Table 2). In the presence of cobalt, the mutant enzyme restored Ac-DCVC deacetylating activity. The mutation of another conserved residue in this area, Glu289 to an alanine also did not have a significant effect on mouse AA3. We expected that another conserved residue in this area, Lys290, might be involved in substrate binding instead of Arg71. The results (Table 2 and Table 3) also did not confirm our expectation.
It has been suggested that Arg168 in human AA2 provides specificity for the binding of the β-carboxyl group of Ac-Asp [17]. The mutations of Arg168 to cysteine, histidine and glutamate were found in patients with Canavan disease [17,45]. Mouse AA3 has in this position a glutamate, therefore we determined 1) How E167R and E167A mutations affect the activity with Ac-Tyr and Ac-DCVC; 2) Whether wt-AA3 and E167R mutant deacetylate Ac-Asp. In the deacetylation of Ac-Tyr, E167R demonstrated an increased activity without Co2+ and decreased activity in the presence of Co2+ in comparison with wt-AA3. Wt-AA3 showed no Ac-Asp deacetylating activity with or without Co2+, whereas E167R mutant weakly deacetylated Ac-Asp. Without and with Co2+, Km was 2.46± 0.51 and 9.71±2.33 mM, and Vmax without and with Co2+ was 22.1±6.17 and 241±34 nmol·mg−1·min−1 respectively. In comparison with the deacetylation rate of Ac-Tyr, the efficiency of the deacetylation of Ac-Asp mediated by E167R mutant without and with Co2+ was 1.05 and 3.01% respectively. The data indicated that Ac-Asp is not an efficient substrate of E167R mutant, and Glu167 is not as important for substrate binding/catalysis in mouse AA3 as in AA2. It is possible that in mouse AA3 the substrate binding is basically mediated by Arg63 and Tyr287 whereas no specific interaction of the side groups of a substrate with the enzyme takes place. This hypothesis could explain a wider substrate specificity of AA3 in comparison with AA2. Nevertheless the architecture of the area near the substrate binding site restricts the use of certain N-acetylated substrates that are used by AA1 but practically not used by AA3 [4].
The results indicate that almost all the conserved amino acid residues important for AA2, also play important roles in the catalysis mediated by mouse AA3 (Arg63, Asp68, Glu177 and Tyr287). Contrary to AA2, Arg71 and Asn70 do not play important roles in substrate binding/catalysis in mouse AA3. Based on this data we revised correspondingly our first 3D mouse AA3 model. In this model (Fig. 1B), Arg71 and Asn70 are not involved in substrate binding/catalysis. Instead Arg63 and Tyr287 play key roles in the binding of α-carboxyl group of the substrate in the revised model. A very high sequence and 3D homology of AA3 with AA2 suggests that AA3 utilizes the same catalytic mechanism as AA2. Nevertheless only a high-resolution atomic structure of AA3 may prove or disprove this speculation.
3.7. Effect of Co2+ on the kinetic characteristics of mouse AA3 and the Ac-DCVC-induced cytotoxicity in HE293T cells expressing mouse AA3
With both Ac-Tyr and Ac-DCVC, the addition of cobalt increased the Vmax several times and decreased the Km of mouse wt-AA3 (Table 2 and Table 3). In general, Co2+ increases Vmax as well as Km in the deacetylation of Ac-DCVC, whereas in the deacetylation of Ac-Tyr the increase of Vmax in the presence of Co2+ was not accompanied by the increase of Km. As a result, the efficiency of deacetylation of Ac-Tyr and Ac-DCVC measured as the Vmax/Km value was significantly increased in the presence of cobalt (Table 2 and Table 3). The results demonstrated that Co2+ plays a crucial role in the activation of mouse AA3.
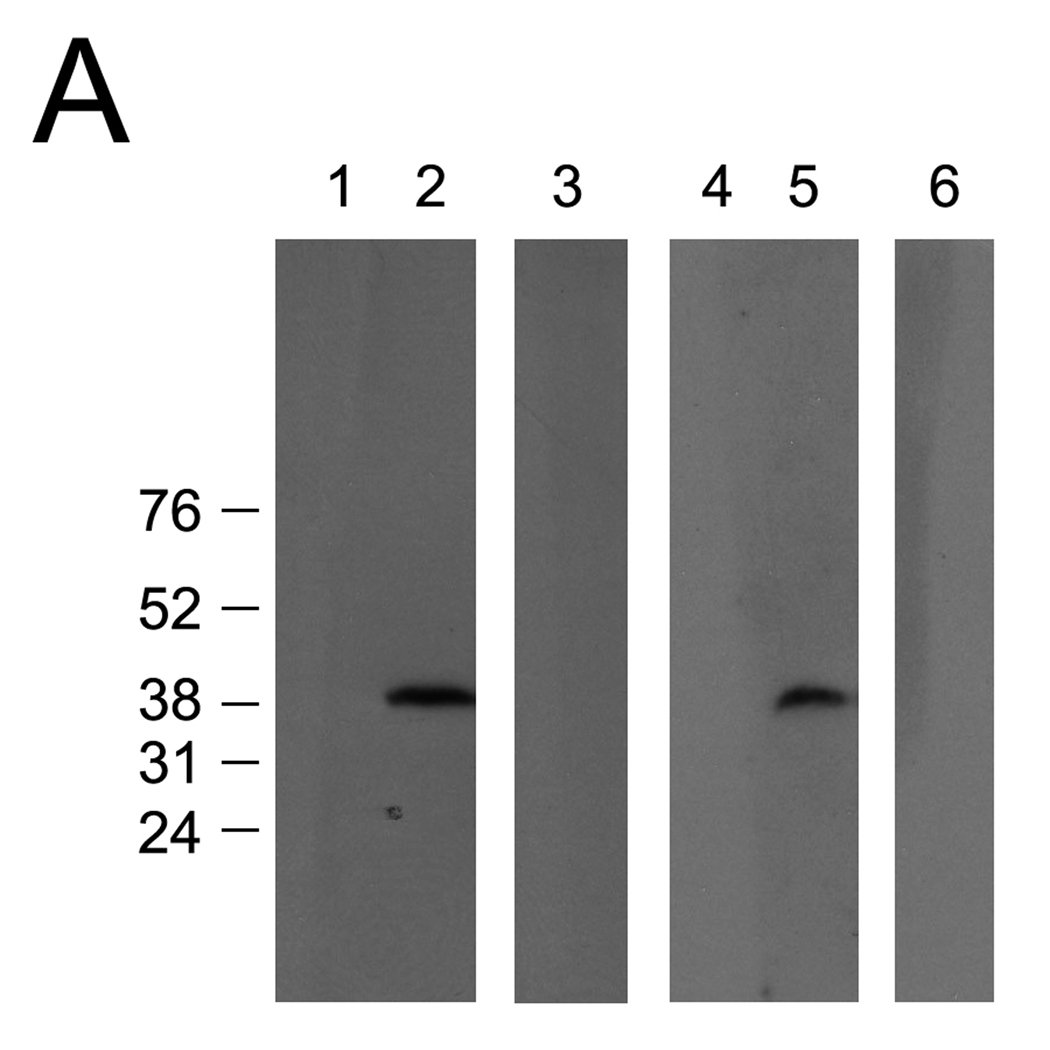
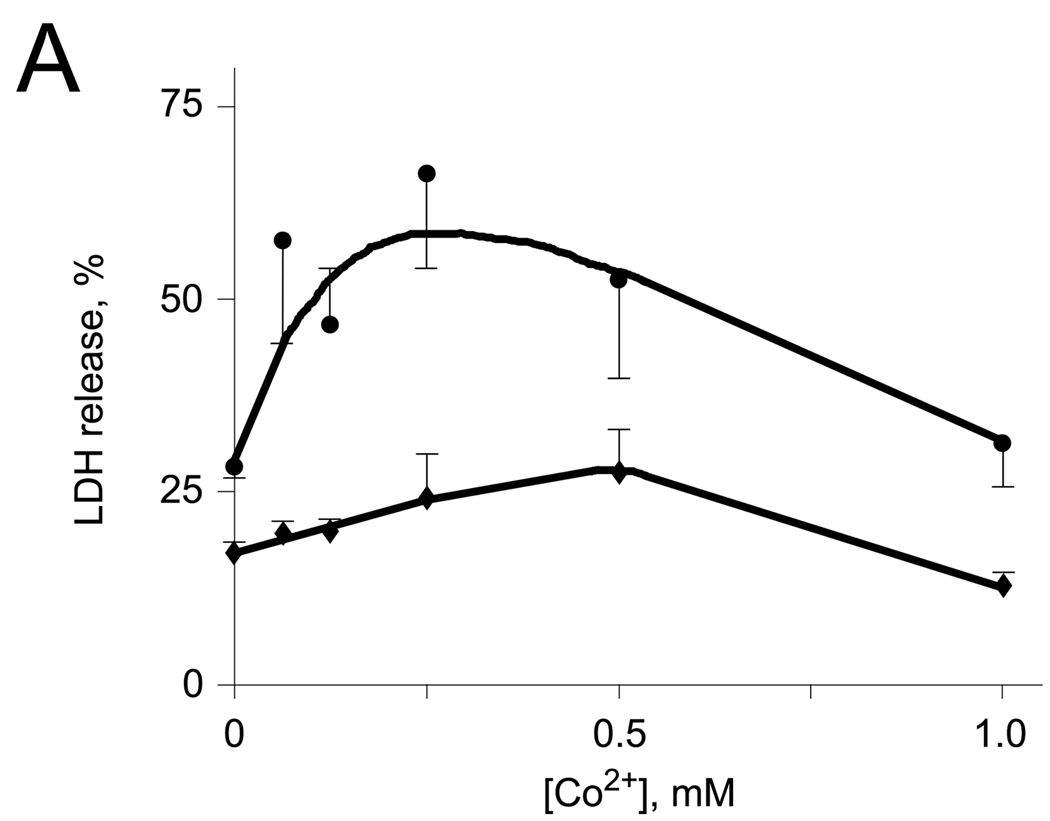
In order to determine whether Co2+ may accelerate the rate of the deacetylation of Ac-DCVC in the cells expressing AA3 and therefore increase the toxicity of Ac-DCVC, we studied the effect of cobalt ions on the toxicity of Ac-DCVC in the HEK293T cells exogenously expressing mouse wt-AA3. No human AA3 protein was detected in these cells (Fig. 5A) and no deacetylation of Ac-DCVC was catalyzed by the extract from these cells (data not shown). The toxic effect of Ac-DCVC in the cells expressing mouse wt-AA3 was significantly increased in the presence of Co2+ in comparison with the same cells not treated with cobalt (Fig. 5B). The maximum effect was induced by 0.25 mM Co2+ and then it was decreased. The decrease of the toxic effect of Ac-DCVC present at higher concentrations (0.5–1 mM) of cobalt was probably not resembled a real decrease and but rather reflected the Co2+-induced inhibition of LDH. The results suggested that Co2+ increases the toxicity of Ac-DCVC in HEK293T cells probably via activation of mouse AA3. Via affecting the rate of Ac-DCVC deacetylation, Co2+ and other metal ions may play a role in the glutathione conjugation pathway of the TCE metabolism in kidney and liver and therefore may play an important role in mediating the cytotoxicity of Ac-DCVC.
Fig. 5.
(A) Immunoblotting of the extracts from mock-transfected (1,4) and transfected with His6-tagged human AA3 (2,3) or mouse AA3 (5,6). The C-terminal human AA3 specific antibody HR-C1 (1,2) or the same antibody pre-incubated with the immunized peptide (3) were used. The C-terminal mouse AA3 specific antibody MR-C1 (4,5) or the same antibody pre-incubated with the immunized peptide (6) were used. (B) The effect of cobalt ions on the toxicity of Ac-DCVC in the HEK293T cells transiently expressing mouse wt-AA3 (●). Mock-transfected HEK293T cells (♦) were used as a control.
Acknowledgements
The authors thank Jeff Abramson for help with 3D modeling of mouse AA3. This work was supported by the National Institutes of Health grant R01 ES012935.
Abbreviations
- Ac-DCVC
N-acetyl-S-(1,2-dichlorovinyl)-L-cysteine
- Ac-Tyr
N-acetyl-L-tyrosine
- AA3
aminoacylase AA3
- AA2
aminoacylase 2, aspartoacylase
- AA1
aminoacylase 1
- CD
circular dichroism
- DEAE-cellulose
diethylaminoethyl cellulose
- DLS
dynamic light scattering
- DTNB
Ellman’s reagent, 5,5’-dithio-bis(2-nitrobenzoate)
- EDTA
ethylenediaminetetraacetic acid
- HPLC
high performance liquid chromatography
- ICP-MS
inductively coupled plasma mass spectrometry
- LC
liquid chromatography
- MTSET
[2-(trimethylammonium)ethyl]methanethiosulfonate bromide
- MS
mass spectrometry
- OP
o-phenanthroline
- PBS
phosphate buffered saline
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TCE
trichloroethylene.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Perrson B, Flinta C, von Heijne G, Jörnvall H. Structures of N-terminally acetylated proteins. Eur. J. Biochem. 1985;152:523–527. doi: 10.1111/j.1432-1033.1985.tb09227.x. [DOI] [PubMed] [Google Scholar]
- 2.Driesen HPC, De Jong WW, Tesser GI, Bloemendal H. The mechanism of N-terminal acetylation of proteins. Crit. Rev. Biochem. Mol. Biol. 1985;18:281–325. doi: 10.3109/10409238509086784. [DOI] [PubMed] [Google Scholar]
- 3.Anders MW, Dekant W. Aminoacylases. Adv. Pharmacol. 1994;27:431–448. doi: 10.1016/s1054-3589(08)61042-x. [DOI] [PubMed] [Google Scholar]
- 4.Newman D, Abuladze N, Scholz K, Dekant W, Tsuprun V, Ryazantsev S, Bondar G, Sassani P, Kurtz I, Pushkin A. Specificity of aminoacylase III-mediated deacetylation of mercapturic acids. Drug Metabol. Disposit. 2007;35:43–50. doi: 10.1124/dmd.106.012062. [DOI] [PubMed] [Google Scholar]
- 5.Anders MW, Dekant W, Vamvakas S. Formation and fate of nephrotoxic and cytotoxic glutathione S-conjugates: cysteine conjugate beta-lyase pathway. Adv. Pharmacol. 1994;27:115–162. doi: 10.1016/s1054-3589(08)61031-5. [DOI] [PubMed] [Google Scholar]
- 6.Hayden PJ, Stevens JL. Cysteine conjugate toxicity, metabolism and binding to macromolecules in isolated rat kidney mitochondria. Mol. Pharmacol. 1990;37:468–476. [PubMed] [Google Scholar]
- 7.Commandeur JNM, Stijntjes GJ, Vermeulen NPE. Enzymes and transport systems involved in the formation and disposition of glutathione S-conjugates. Role of bioactivation and detoxification mechanisms of xenobiotics. Pharmacol. Rev. 1995;47:271–330. [PubMed] [Google Scholar]
- 8.Birner G, Bernauer U, Werner M, Dekant W. Biotransformation, excretion, and nephrotoxicity of haloalkene-derived cysteine S-conjugates. Arch. Toxicol. 1997;72:1–8. doi: 10.1007/s002040050461. [DOI] [PubMed] [Google Scholar]
- 9.Boogaard PJ, Commandeur JNM, Mulder GJ, Vermeulen NPE, Nagelkerke JF. Toxicity of the cysteine-S-conjugates and mercapturic acids of four structurally related difluoroethylenes in isolated proximal tubular cells from rat kidney: uptake of the conjugates and activation to toxic metabolites. Biochem. Pharmacol. 1989;38:3731–3741. doi: 10.1016/0006-2952(89)90579-0. [DOI] [PubMed] [Google Scholar]
- 10.Commandeur JN, Boogaard PJ, Mulder GJ, Vermeulen NP. Mutagenicity and cytotoxicity of two regioisomeric mercapturic acids and cysteine S-conjugates of trichloroethylene. Arch. Toxicol. 1991;65:373–380. doi: 10.1007/BF02284259. [DOI] [PubMed] [Google Scholar]
- 11.Lash LH, Hueni SE, Putt DA. Apoptosis, necrosis and cell proliferation induced by S-(1,2-dichlorovinyl)-L-cysteine in primary cultures of human proximal tubular cells. Toxicol. Appl. Pharmacol. 2001;177:1–16. doi: 10.1006/taap.2001.9295. [DOI] [PubMed] [Google Scholar]
- 12.Löffler HG, Schneider F, Aumuller G, Unsicker K. Immunocytochemical studies of aminoacylase I (EC 3.5.1.14) localization in the swine kidney. Acta Histochem. 1982 Suppl. 25:57–60. [PubMed] [Google Scholar]
- 13.Lindner H, Hopfner S, Tafler-Naumann M, Miko M, Konrad L, Röhm KH. The distribution of aminoacylase I among mammalian species and localization of the enzyme in porcine kidney. Biochimie. 2000;82:129–137. doi: 10.1016/s0300-9084(00)00191-7. [DOI] [PubMed] [Google Scholar]
- 14.Pushkin A, Carpenito G, Abuladze N, Newman D, Tsuprun V, Ryazantsev S, Motemoturu S, Sassani P, Solovieva N, Dukkipati R, Kurtz I. (Structural characterization, tissue distribution, and functional expression of murine aminoacylase III. Am. J. Physiol. 2004;286:C848–C856. doi: 10.1152/ajpcell.00192.2003. [DOI] [PubMed] [Google Scholar]
- 15.Uttamsingh V, Baggs RB, Krenitsky DM, Anders MW. Immunohistochemical localization of the acylases that catalyze the deacetylation of N-acetyl-L-cysteine and haloalkene-derived mercapturates. Drug Metab. Dispos. 2000;28:625–632. [PubMed] [Google Scholar]
- 16.Matalon R, Michals K, Sebesta D, Deanching M, Gashkoff P, Casanova J. Aspartoacylase deficiency and N-acetylaspartic aciduria in patients with Canavan disease. Am. J. Med. Genet. 1988;29:463–471. doi: 10.1002/ajmg.1320290234. [DOI] [PubMed] [Google Scholar]
- 17.Bitto E, Bingman CA, Wesenberg GE, McCoy JG, Phillips GN., Jr (2007) Structure of aspartoacylase, the brain enzyme impaired in Canavan disease. Proc. Natl. Acad. Sci. USA. 2007;104:456–461. doi: 10.1073/pnas.0607817104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Coq J, An HJ, Lebrilla C, Viola RE. Characterization of human aspartoacylase: the brain enzyme responsible for Canavan disease. Biochemistry. 2006;45:5878–5884. doi: 10.1021/bi052608w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaul R, Gao GP, Balamurugan K, Matalon R. Cloning of the human aspartoacylase cDNA and a common missence mutation in Canavan disease. Nat. Genet. 1993;5:118–123. doi: 10.1038/ng1093-118. [DOI] [PubMed] [Google Scholar]
- 20.Herga S, Berrin JG, Perrier J, Puigserver A, Giardina AT. Identification of the zinc binding ligands and the catalytic residue in human aspartoacylase, an enzyme involved in Canavan disease. FEBS Lett. 2006;580:5899–5904. doi: 10.1016/j.febslet.2006.09.056. [DOI] [PubMed] [Google Scholar]
- 21.Kördel W, Schneider F. Chemical investigations on pig kidney aminoacylase. Biochim. Biophys. Acta. 1976;445:446–457. doi: 10.1016/0005-2744(76)90098-x. [DOI] [PubMed] [Google Scholar]
- 22.Kördel W, Schneider F. Renal aminoacylase, a zinc enzyme. Z. Naturforsch. 1977;32C:342–344. doi: 10.1515/znc-1977-5-605. [DOI] [PubMed] [Google Scholar]
- 23.Palm CJ, Röhm KH. Aminoacylase I from porcine kidney: identification and characterization of two major protein domains. J. Protein Chem. 1995;14:233–240. doi: 10.1007/BF01886764. [DOI] [PubMed] [Google Scholar]
- 24.Wu HB, Tsou CL. A comparison of Zn(II) and Co(II) in the kinetics of inactivation of aminoacylase by 1,10-phenanthroline and reconstitution of the apoenzyme. Biochem. J. 1993;296:435–441. doi: 10.1042/bj2960435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heese D, Berger S, Röhm K-H. Nuclear magnetic relaxation studies of the role of the metal ion in Mn2+-substituted aminoacylase I. Eur. J. Biochem. 1990;188:175–180. doi: 10.1111/j.1432-1033.1990.tb15385.x. [DOI] [PubMed] [Google Scholar]
- 26.Lindner HA, Lunin VV, Alary A, Hecker R, Cygler M, Menard R. R. Essential roles of zinc ligation and enzyme dimerization for catalysis in the aminoacylase-1/M20 family. J. Biol. Chem. 2003;278:44496–44504. doi: 10.1074/jbc.M304233200. [DOI] [PubMed] [Google Scholar]
- 27.Tang ZY, Yu JY, Zhou Q, He B, Wang ZF, Zhou HM. Secondary structure of holo- and apo-aminoacylase from prediction, circular dichroism, and FT-Raman spectroscopy. J. Biochem. 1995;118:706–709. doi: 10.1093/oxfordjournals.jbchem.a124969. [DOI] [PubMed] [Google Scholar]
- 28.Kaul R, Casanova J, Johnson AB, Tang P, Matalon R. Purification, characterization, and localization of aspartoacylase from bovine brain. J. Neurochem. 1991;56:129–135. doi: 10.1111/j.1471-4159.1991.tb02571.x. [DOI] [PubMed] [Google Scholar]
- 29.Udenfriend S, Stein S, Bohlen P, Dairman W, Leimgruber W, Weigele M. Fluorescamine: a reagent for assay of amino acids, Peptides, proteins, and primary amines in the picomole range. Science. 1972;178:871–872. doi: 10.1126/science.178.4063.871. [DOI] [PubMed] [Google Scholar]
- 30.Valentine RC, Green NM. Electron microscopy of an antibody-hapten complex. J. Mol. Biol. 1967;27:615–617. doi: 10.1016/0022-2836(67)90063-0. [DOI] [PubMed] [Google Scholar]
- 31.Kopp J, Schwede T. The SWISS-MODEL Repository of annotated three-dimensional protein structure homology models. Nuc. Acid. Res. 2004;32:D230–D234. doi: 10.1093/nar/gkh008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K, Coot K. model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 34.Elfarra AA, Jakobson I, Anders IMW. Mechanism of S-(1,2-dichlorovinyl)glutathione-induced nephrotoxicity. Biochem. Pharmacol. 1986;35:283–288. doi: 10.1016/0006-2952(86)90527-7. [DOI] [PubMed] [Google Scholar]
- 35.Hershfield JR, Madhavarao CN, Moffett JR, Benjamins JA, Garbern JY, Narboodiri A. (2006) Aspartoacylase is a regulated nuclear-cytoplasmic enzyme. FASEB J. 2006;20:2139–2141. doi: 10.1096/fj.05-5358fje. [DOI] [PubMed] [Google Scholar]
- 36.Le Coq J, Pavlovsky A, Malik R, Sanishvili R, Xu C, Viola RE. Examination of the mechanism of human brain aspartoacylase through the binding of an intermediate analogue. Biochemistry. 2008;47:3484–3492. doi: 10.1021/bi702400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki S, Tateishi M. M. Purification and characterization of a rat liver enzyme catalyzing N-deacetylation of mercapturic acid conjugates. Drug Metabol. Disposit. 1981;9:573–577. [PubMed] [Google Scholar]
- 38.Uttamsingh V, Anders MW. Acylase-catalyzed deacetylation of haloalkene-derived mercapturates. Chem. Res. Toxicol. 1999;12:937–942. doi: 10.1021/tx990090p. [DOI] [PubMed] [Google Scholar]
- 39.Endo Y. N-acetyl-aromatic amino acid deacylase in animal tissues. Biochim. Biophys. Acta. 1978;523:207–214. doi: 10.1016/0005-2744(78)90023-2. [DOI] [PubMed] [Google Scholar]
- 40.Hershfield JR, Pattabiraman N, Madhavarao CN, Namboodiri MAA. Mutational analysis of aspartoacylase: implications for Canavan disease. Brain Res. 2007;1148:1–14. doi: 10.1016/j.brainres.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryazantsev S, Abuladze N, Newman D, Bondar G, Kurtz I, Pushkin A. Structural characterization of dimeric murine aminoacylase III. FEBS Lett. 2007;581:1898–1902. doi: 10.1016/j.febslet.2007.03.088. [DOI] [PubMed] [Google Scholar]
- 42.Boucher E, Bourienne A, Adams P, Turlin B, Brissot P, Deugnier Y. Liver iron concentration and distribution in chronic hepatitis C before and after interferon treatment. Gut. 1997;41:115–120. doi: 10.1136/gut.41.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cabantchik ZI, Breuer W, Zaninelli G, Cianciulli P. LPI-labile iron in iron overload. Best Pract. Res. Clin. Haematol. 2005;18:277–287. doi: 10.1016/j.beha.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Frieden C. Protein aggregation processes: In search of the mechanism. Protein Sci. 2007;16:2334–2344. doi: 10.1110/ps.073164107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fändrich M. On the structural definition of amyloid fibrils and other polypeptide aggregates. Cell. Mol. Life Sci. 2007;64:2066–2078. doi: 10.1007/s00018-007-7110-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castagnetto JM, Hennessy SW, Roberts VA, Getzoff ED, Tainer JA, Pique ME. MDB: the metalloprotein database and Browser at the Scripps research institute. Nucl. Acid. Res. 2002;30:379–382. doi: 10.1093/nar/30.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karlin S, Zhu ZY, Karlin KD. The extended environment of mononuclear metal centers in protein structures. Proc. Natl. Acad. Sci. USA. 1997;94:14225–14230. doi: 10.1073/pnas.94.26.14225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heese D, Röhm KH. Reactivities of sulfhydryl groups in native and metal-free amimacylase I. Biol. Chem. Hoppe Seyler. 1989;370:607–612. doi: 10.1515/bchm3.1989.370.1.607. [DOI] [PubMed] [Google Scholar]