Abstract
The increasing availability of complete genome sequences of RNA viruses has the potential to shed new light on fundamental aspects of their biology. Here, I use case studies of 3 RNA viruses to explore the impact of genomic sequence data, with particular emphasis on influenza A virus. Notably, the studies of RNA virus genomics undertaken to date largely focused on issues of evolution and epidemiology, and they have given these disciplines new impetus. However, genomic data have so far made fewer inroads into areas of more direct importance for disease, prevention, and control; thus, harnessing their full potential remains an important goal.
It is a cliché to say that we are now in the midst of the genomic age of biology. As is obvious from even the most cursory trawl through sequence repositories such as GenBank, the number of complete genomes of eukaryotes and their pathogens has increased dramatically in recent years. The advent of new technologies for rapid genome sequencing, in which many millions of nucleotides can be obtained in a single run (1), promises an even richer haul of genomic data in the near future. The gathering pace with which genomic sequence data are being generated is reflected in the case of RNA viruses, the major cause of emerging diseases in humans, and is greatly assisted by their universally small genomes, which have a mean size of approximately 10,000 nucleotides.
The aim of this Review is to document some of the insights that have stemmed from this new wealth of viral genome sequence data, to highlight the current limitations of genomics, and to point to areas where future effort might be directed. In doing so, I make both broad-ranging statements on viral genomics and consider as case studies 3 very different RNA viruses that infect humans: influenza A virus, HIV, and dengue virus (DENV). My major argument is that despite the burgeoning genomic data for RNA viruses, the impact of these data has largely been in the realm of evolution and epidemiology, with relatively little foray into issues of more direct clinical importance, such as studies of pathogenesis and the development of vaccines and antivirals. In short, although these are salad days for population biology, viral genomics has not yet provided a stimulus package to the clinical sciences. In part, this is to be expected, because it is overly simplistic to think that there will always be a simple virological basis to a specific clinical syndrome or that genomic data taken in isolation have much to say about immunogenicity or drug design. More importantly, the power of genomics may be diluted because sequence data are often collected and stored out of context of other key biological variables, particularly clinical manifestation (often because of ethical considerations) and correlates of immunity, so that its full potential has yet to be harnessed.
Influenza A virus
Influenza virus is a major cause of respiratory disease in humans, resulting in annual mortality of 250,000–500,000 globally (2). Influenza viruses possess a segmented negative-sense RNA genome (family Orthomyxoviridae) and cause regular seasonal epidemics in humans, other mammals, and birds. Although 3 types of influenza virus — denoted A, B and C — circulate in humans, the type A viruses are associated with the greatest mortality and morbidity. The genetic (and antigenic) diversity of influenza A virus is described by the specific combination of hemagglutinin (HA) and neuraminidase (NA) surface glycoproteins in each virus. There are 16 known subtypes of HA (H1–H16) and 9 subtypes of NA (N1–N9), all of which are found in wild birds of the orders Anseriformes and Charadriformes (3–5).
The Influenza Genome Sequencing Project (IGSP; http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html) was the obvious call to arms in RNA virus genomics (6). Not only has this remarkable data set allowed new insights into the biology and evolution of this important human pathogen, but an unforeseen spinoff was the enticement of new researchers into the field, which stimulated novel ideas on the fundamentals of viral evolution (7, 8). At the time of this writing, the IGSP has generated more than 3,300 complete genome sequences, with perhaps another 1,600 to come, most of which represent strains of influenza A from infected humans (subtypes A/H1N1 and A/H3N2). There are a growing number of genomes from viruses isolated from birds, reflecting concerns over the potential emergence of highly pathogenic avian influenza A viruses able to infect and spread through human populations (e.g., subtype A/H5N1) (9). Although the focus on avian viruses as the source of human pandemic strains is understandable, the recent emergence in humans of an H1N1 subtype from swine (10) cautions us not to ignore the other mammalian hosts of influenza viruses, particularly as spillover of influenza viruses from pigs to humans seems to occur relatively frequently (11). In addition, one of the most remarkable aspects of this new H1N1 virus is that it is phylogenetically distinct from all other swine H1N1 viruses, highlighting our inadequate surveillance of influenza viruses in this species.
Computational analyses of sequence data generated as part of the IGSP have changed some important concepts in influenza evolution. First, genomic studies have revealed that intrasubtype reassortment — in which recombination occurs between different segments of the viral genome — is a pervasive process, sometimes generating isolates with altered antigenic properties, which in turn may lead to vaccine failure (Figure 1 and refs. 12–14). Although the importance of reassortment in influenza virus was known long before the generation of large-scale genomic sequences (15), and its significance for strain emergence is shown by the appearance of the new strain of H1N1, which was generated by the reassortment of 2 swine lineages (10), the IGSP data provide the only insight into the frequency of this process (12). As a consequence, it is foolish to consider the evolution of the key viral antigens — the HA and NA proteins that sit on the surface of the virion and interact intimately with host cells — outside the context of the rest of the viral genome (16). Indeed, it is clear that the HA protein exhibits complex epistatic interactions with other viral proteins (17). Second, the richness of the genomic sequence data has revealed that specific human populations, such as that of New York state, are characterized by the circulation of multiple and often diverse viral lineages of the same subtype, which provide the raw material for frequent reassortment (12, 14, 18). While this may not sound like a surprise, a reasonable model before the arrival of expansive genome sequence data was that a single viral lineage enters a population at the start of the influenza season (around October in the United States) and gradually spreads through the population over the subsequent 6 months, before dying out the following summer (19). Although there is clearly a major summer die-off of influenza virus in the Northern (and Southern) Hemispheres, for reasons that are still unclear, viral lineages are continually imported during the time course of the influenza season (Figure 2 and refs. 16, 18). Indeed, one of the most interesting epidemiological insights from the IGSP data has been that relatively geographically isolated communities in the United States (such as Hopkinsville, Kentucky) harbor influenza A virus genetic diversity similar to that of major cities with more expansive travel networks (14), highlighting how rapidly this virus is able to spread through populations.
Figure 1. Process of reassortment in influenza A virus.
Reassortment occurs when 2 viruses (denoted here as A and B) coinfect a single host cell. Each virus contains 8 gene segments, shown as open and closed bars. Reassortment among these 2 viruses then produces viruses with a variety of segment combinations.
Figure 2. Genetic diversity of human influenza A/H1N1 virus in the United States during the 2006–2007 influenza season.
The phylogenetic tree depicts the diversity of the gene encoding NA in 284 isolates sampled across the United States. Isolates from different geographical locations are color coded according to the inset map. Note that there is remarkable genetic diversity, manifest as the circulation of multiple lineages likely to be independently imported, and that there is no clear spatial patterning, as reflected in the mix of colors. Figure adapted with permission from PLoS Pathogens (14).
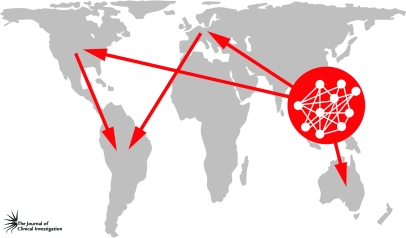
The genomic revolution was also central to the realization that a global source population for human influenza A virus — most likely centered in East and Southeast Asia — annually provides the antigenic variants that ignite the seasonal influenza epidemics in the sink populations of the Northern and Southern Hemispheres (16, 20). Southeast Asia fits the bill for a global influenza source because its large, dense, and well-connected populations provide exactly the conditions that allow natural selection (manifest as ongoing antigenic evolution in the HA and NA proteins) to operate with maximum efficiency (Figure 3). Not only is this observation important from the perspective of basic epidemiology — this is the geographic region that needs to be surveyed most intensively for emergent viruses — but it may also have a major bearing on vaccine design. Vaccines designed to prevent infection with human influenza A viruses tend to fail every few years because of a periodic burst of antigenic evolution, in which a virus emerges with more mutations in the HA protein than predicted (21). Knowing where these antigenic strains are generated every year should speed up the process of choosing the influenza A strain to be incorporated into the vaccine and also yield more effective choices.
Figure 3. Global migration of influenza A virus.
Seasonal epidemics of human influenza A/H3N2 virus are thought to begin in a circulation network located in East and Southeast Asia before being exported to other geographical localities (arrows denote direction of migration). The virus is thought to be exported directly from the circulation network in East and Southeast Asia to Australia, Europe, and North America. It is then exported to South America from both Europe and North America. Figure adapted with permission from Science (20).
The expanse of genomic information about influenza A viruses has also shed new light on the evolution of drug resistance. The most interesting observation from this perspective is that, remarkably, direct drug-selection pressure may not always be responsible for the rise of drug resistance. The first example of what sounds like a highly paradoxical evolutionary process occurred with the adamantanes, a class of first-generation antivirals against which subtype A/H3N2 viruses have shown a dramatic global rise in resistance over the last 4 years (22, 23). Resistance in this case is caused by a single amino acid change — Ser31Asn — in the viral M2 protein, an ion channel that is the target of adamantanes. To the surprise of many, the frequency of the Ser31Asn mutation has risen abruptly in populations where adamantanes are rarely used, such as the United States. The most likely explanation for the global dissemination of Ser31Asn is therefore that it became fortuitously linked to another beneficial mutation — probably conferring immune escape — located elsewhere in the viral genome (24). The population frequency of subtype A/H3N2 viruses carrying the Ser31Asn mutation is therefore not simply a function of the fitness conferred by this mutation, but also that of other advantageous mutations in the viral genome. This again tells us that genome-wide interactions are a critical aspect of viral genetics. Incredibly, exactly the same process of genetic hitchhiking now seems to be taking place with resistance to oseltamivir, a major NA inhibitor. In this case, there has been a dramatic rise in oseltamivir resistance in subtype A/H1N1 viruses circulating in Europe (and somewhat so in the Americas), which was caused by a His275Tyr mutation in the NA protein (25, 26). Because these drugs are not widely used in European populations, linkage to another beneficial mutation seems the most probable explanation for the rise of viruses carrying His275Tyr, although this needs to be formally assessed. More troublesome is that oseltamivir was designed as the frontline defense against highly pathogenic avian A/H5N1 viruses, which have periodically spilled over into human populations. It is therefore possible that subtype A/H5N1 viruses will also readily evolve resistance to oseltamivir, or that an oseltamivir-resistant NA protein from an A/H1N1 virus will incorporate itself into A/H5N1 strains via reassortment.
What the IGSP has not provided is clear information on whether there is a viral genetic basis to the variance in clinical presentation of influenza in humans (i.e., from subclinical to severe respiratory illness). This kind of analysis is clearly a major goal of second-generation studies in RNA virus genomics. In my opinion, there are a number of reasons, all of which are surmountable, why genomic data from influenza A virus have thus far been of limited use for clinical studies. First, the available sample of sequences, although huge from a genomics perspective, is still small relative to those normally used in clinical epidemiology. Thankfully, advances in sequencing technology mean that this major barrier will soon be breached (1). Second, the isolates of influenza A virus chosen for sequencing evidently do not amass a representative sample of those that circulate in human populations, as they were taken from patients who were ill enough to seek medical attention. Hence, there is necessarily a profound sampling bias against low virulence — even asymptomatic — strains of influenza A virus. Last, it is clear that most mortality among patients with influenza disease is due to secondary bacterial pneumonia (caused by Streptococcus pneumoniae; ref. 27). Hence, it is impossible to discuss the epidemiology of influenza virus out of context of the other pathogens that cocirculate during the influenza season (28).
One of the few observations made using data generated as part of the IGSP that might inform on aspects of clinical disease is that the influenza epidemics of 1947 and 1951, both of which were characterized by high case numbers in specific geographical localities (including parts of the United States), were seemingly associated with A/H1N1 influenza viruses that were produced by large-scale reassortment events (29). This is particularly noteworthy in the case of the 1947 epidemic, as previous studies indicated that the virus associated with this outbreak— during which there was a widespread vaccine failure — was antigenically distinct from previously circulating viruses (although still classified as a subtype A/H1N1 virus; ref. 30). Modern genomic data now tells us that this antigenically distinct variant of HA was acquired by reassortment (from an as-yet unidentified source), further emphasizing how processes of genetic exchange can alter viral phenotype (29).
Finally, despite the genomic revolution, there are still aspects of the evolution and epidemiology of influenza A virus that remain uncertain but may be central to its future control. First, it is unclear why the A/H1N1 and A/H3N2 subtypes that currently cocirculate in human populations have such different epidemiological dynamics: the former is characterized by greater circulating genetic diversity but weaker antigenic evolution (i.e., accumulation of mutations at antigenic sites), while the latter is characterized by lower levels of circulating genetic diversity (i.e., seasonal population crashes in temperate regions) but more rapid antigenic evolution (16). Second, and of more clinical importance, it is essential that we understand the rules by which antigenic evolution occurs in influenza A virus (31, 32). Specifically, if it were possible to determine how and why large antigenic changes occur periodically — how the virus moves from antigenic type to antigenic type — it would doubtless be possible to design vaccines that are far more effective vaccines, perhaps even evolution proof. It is therefore essential that future genome sequence data be gathered with an associated measure of viral antigenicity, such as hemagglutinin-inhibition distance, which depicts the differing ability of isolates of influenza virus to agglutinate red blood cells and the corresponding ability of antisera raised against these isolates to inhibit agglutination (21).
HIV
Not only is HIV/AIDS the most severe infectious disease inflicting human populations today, and arguably the most serious that we have ever faced, but it is also the pandemic that kick started the current interest in emerging disease. Since the discovery of the virus that causes AIDS in the early 1980s (33), the various research themes directed toward HIV/AIDS have experienced remarkably mixed fortunes. There has been depressingly little progress toward an HIV vaccine, reflecting a marked lack of protective immunity in both human and animal trials. Although there is more genomic sequence data for HIV than for any other virus, and this has told us a great deal about its biology, genomics has yet to contribute much to HIV vaccine development. Given the current impasse in vaccine development, debates over how phylogenetics should assist in deciding which HIV strain to use are perhaps overly optimistic (34, 35). In stark contrast, the study of the origins, evolution, and molecular epidemiology of HIV has been wildly successful: we now know approximately when both types of AIDS virus (HIV-1 and HIV-2) first appeared in humans; we now know roughly where these emergence events took place; and we can make more than an educated guess about the precise mechanisms of cross-species transmission (36–39). Hence, for HIV-1, the most prevalent and clinically relevant type of HIV, analysis of genomic sequence data has told us that this virus most likely jumped from common chimpanzees (Pan troglodytes troglodytes) to humans on multiple occasions during the early years of the twentieth century, and that this occurred somewhere in West and Central Africa, probably through the preparation and consumption of virally infected bushmeat. These early epidemic embers were fanned by an expanding logging industry as well as the urbanization of West and Central Africa (39–41). Recent dating studies using molecular clocks, in which it is assumed that viral mutations accumulate through time at a fairly constant rate so that divergence times can be estimated across phylogenetic trees (42), have also suggested that HIV-1 first entered the Western Hemisphere during the 1960s, although it was not detected as a clinical disease (AIDS) until some years later (43).
The other major insight stemming from genomic approaches to studying HIV-1 is that this virus is highly genetically diverse, both within an individual host and on a global scale (44). This diversity is a major impediment to successful vaccination, and at the very least means that different vaccine strains will be required in particular geographic areas and/or that vaccines will need to be continually updated. Currently, there are 9 recognized genetic subtypes of HIV-1 (A–K, excluding E and I) as well as 35 circulating recombinant forms (CRFs), which are created by frequent intersubtype recombination (i.e., genetic exchange among subtypes). Some of these CRFs are complex, which indicates that individual hosts were infected by multiple viruses — again highlighting the lack of cross-protective immunity — followed by frequent recombination. Indeed, intersubtype recombination is so common in HIV-1, it is arguable that the subtypes — originally described as discrete phylogenetic entities — do not exist at all. Rather, what are thought of as HIV-1 subtypes are merely clusters of sequences that were generated by a series of localized outbreaks in specific populations (45). The ultimate demise of the subtype concept also confounds attempts to associate particular viral lineages with specific disease manifestations. Indeed, it is notable that few studies have proposed associations between different HIV subtypes and specific phenotypic features (46, 47), some of which have not withstood further scrutiny (48).
Although it is often stated that the success of HIV-1 is due to its capacity for rapid mutation, this idea lacks some important context. Estimates of the intrinsic mutation rate for HIV-1, reflecting the process of replication with a highly error-prone reverse transcriptase enzyme, place this in the realm of 0.1–0.3 mutations per genome, per replication (49, 50). While this mutation rate is evidently far higher than those observed in organisms with double-strand DNA, it may still be 5-fold lower than that in RNA viruses that replicate using the even more error-prone RNA-dependent RNA polymerase, in which mutation rates frequently reach approximately 1.0 mutation per genome, per replication (51, 52). As a consequence, while it is certainly the case that the raw material for genetic variation in HIV-1 is produced by mutation, the subsequent processes of recombination and natural selection for immune escape are equally responsible for the complex and abundant genetic diversity in this virus (53). In fact, as many as 2–3 recombination events may occur during each replication cycle in HIV-1, making it one of the highest recombination rates recorded (54, 55).
DENV
Dengue is the most common arbovirus infection of humans, responsible for perhaps 50 million dengue fever cases annually throughout the tropical and subtropical world and an extremely serious pediatric disease in Southeast Asia (56). In a minority of cases, infection with DENV results in more serious disease syndromes, usually referred to as dengue hemorrhagic fever and dengue shock syndrome, with some 500,000 patients hospitalized globally on an annual basis (90% of whom are children). DENV is a single-strand positive-sense RNA virus of the family Flaviviridae that is transmitted among primate hosts by the Aedes spp. mosquito. Importantly, it is organized as 4 serotypes (DENV-1–DENV-4) that diverge at approximately 30% of nucleotide sites across their genome.
The gradual increase in the frequency of DENV disease over the last 30 years also makes it an archetypal emerging virus, one that may expand its geographical range as a result of global warming, particularly because of more widespread vector distributions (57). It is therefore no surprise that there is now a concerted effort to obtain the complete genome sequences of many thousands of DENV isolates, an effort directed by the Broad Institute of MIT/Harvard (http://www.broad.mit.edu/annotation/viral/Dengue/).
Analysis of the genome data already available has been the centerpiece of studies on DENV evolution and epidemiology. In particular, each of the 4 serotypes of DENV harbors extensive genetic diversity in the form of phylogenetically distinct clusters termed genotypes (58, 59), although there is no consensus on how genotypes should be defined. Unlike HIV-1, this population structure has not been broken down by frequent recombination. As is now well documented (reviewed in ref. 59), the most obvious biological feature of the DENV genotypes is their differing geographical distributions (Figure 4). Interestingly, some genotypes have proven more cosmopolitan than others by infecting humans on several continents (Africa, Asia, Oceania, and Latin America), while other genotypes are more geographically restricted. For example, DENV-1 genotype III has spread to Africa, the Americas, India, and Southeast Asia, while genotype I is restricted to Southeast Asia. It is unclear whether this variation in geographic range is simply a result of the chance exportation of specific lineages or reflects intrinsic differences in fitness (such that some genotypes have more epidemic potential than others), although this issue should be addressed in more detail when additional genomic data are processed.
Figure 4. Global distribution of DENV.
The broad-scale geographic locations of the different serotypes and genotypes of DENV are indicated: (I)–(IV), genotypes I–IV; (Am), American; (Am/As), American/Asian; (As I), (As II); Asian I and II; (Cos), cosmopolitan. Note the abundance of viral genetic diversity in Southeast Asia and the relative lack of information about DENV in Africa. Figure adapted with permission from The evolution and emergence of RNA viruses (90).
There is also some evidence that DENV genotypes differ in both fitness and virulence and that this may have a major bearing on the structure of viral populations. This is most evident in cases of lineage (or clade) replacement, in which there is a relatively abrupt change in the genetic composition of the infecting viruses in a specific population — that is, one viral lineage displaces another (60–64). There are a number of documented replacement events, and, in some of these cases, the evidence that the viruses also differ in fitness is compelling. The best-documented of these cases is the replacement of low-virulence American genotype DENV-2 by high-virulence Southeast Asian genotype DENV-2 in Latin America (65–67). Not only is this pattern apparent in comparative phylogenies, but there is now good experimental evidence that Southeast Asian DENV-2 outcompetes American strains in cell culture assays involving both human and Aedes spp. mosquito cells (68, 69).
Although it has yet to be experimentally verified, another interesting example of lineage replacement occurred in Bangkok, Thailand. Here, 2 lineages of DENV-1 genotype I cocirculated among patients admitted to a children’s hospital during the 1990s, until one lineage eventually took over (70). Interestingly, this switch occurred alongside a decline in the overall frequency of DENV-1 in the population and the rise of DENV-4 (because these 2 serotypes exhibit out-of-phase epidemiological dynamics; ref. 71). This in turn suggests that the success of the winning lineage of DENV-1 may be the result of immune-mediated natural selection — that is, fitness was a function of how this viral lineage was better able to evade the cross-immunity generated by DENV-4 (71). More generally, this example reminds us that fitness is always context dependent, and that as the immunological landscape changes, so may different viral lineages be favored. This has major implications for the future use of tetravalent vaccines that are designed to induce immunity to all 4 DENV serotypes but which are unlikely to be completely cross-protective (72), as viral lineages that evade cross-immunity will be at a selective advantage. Indeed, imperfect vaccination may have profound implications for viral evolution, perhaps even increasing virulence (73), although this has been the subject of considerable debate (74). I believe that the priority for future studies in DENV genomics should be to obtain sequence data from a more diverse range of geographical localities — Africa is notoriously undersampled — and a broader set of clinical syndromes, in particular from patients who experience mild clinical symptoms after DENV infection, as they are normally rarely sampled.
The importance of clonal sequence data
Most of the epidemiological inferences made to date from genomic sequence analysis of RNA viruses have used the consensus sequences of populations of RNA viruses; from each patient, a single sequence is generated that represents the most common nucleotide in every position in this particular viral sample. Although the use of consensus sequences is a valid (and cheap) way to approach many questions in molecular epidemiology, it is also a poor way to reveal many of the intricate patterns of RNA virus evolution. In particular, there is growing evidence that the use of consensus sequences hides a great deal of the genotypic and phenotypic diversity that resides within an individual host. This intrahost diversity may ultimately have major bearing on how viruses respond to treatment: more preexisting variation in the population provides more raw material for natural selection. While the importance of intrahost genetic variation has long been known to those working on chronic viral infections such as HIV and HCV, where the cloning of individual RNA molecules is routine (75, 76), its relevance for acute infections only recently became apparent. For example, in the case of influenza A virus, individuals have been observed to harbor both adamantane-sensitive and -resistant strains (77), and both functional and truncated strains of DENV have been found within single patients (78).
One important facet of the demonstration of abundant genetic variation within individual hosts is that the viral lineages sampled are often so phylogenetically divergent that they are unlikely to have been generated within hosts by de novo mutation. This has 2 immediate potential implications. First, there is not, as is often assumed, a severe population bottleneck at interhost transmission (79), such that multiple viral lineages are able to pass among hosts. Second, the immune response may be less effective than is often imagined or may develop with insufficient speed, such that hosts can be rapidly superinfected with multiple viral lineages. Irrespective of which (or both) of these models is correct, it is clear that single individuals can carry both genetically and phenotypically diverse viral lineages, highlighting the potential for RNA viruses to quickly respond to intervention strategies.
Virus discovery and metagenomics: the future of genomic analysis of RNA viruses
One of the most important recent developments in microbiology is the rise of metagenomics approaches to explore the biodiversity of viruses directly in different environments, including those that potentially cause disease in humans. For example, this approach has been essential to revealing the remarkable diversity of viruses in aquatic environments, even enabling the researchers to identify new viral families (80–82), and documenting the virosphere in human fecal samples (83, 84). Metagenomics may also have identified the agent of the colony collapse disorder that is ravaging populations of honey bees in North America (85), although this awaits confirmation.
Although virus discovery may sound like an intellectually vacuous fishing exercise, I believe that, if done properly, it should be a central component of the biomedical sciences. Not simply stamp collecting for the twenty-first century, virus discovery through metagenomics has the potential to rapidly survey and identify new human diseases. The ecology of modern human populations facilitates the emergence of potentially devastating diseases — our populations are increasingly large, urbanized, and well connected through global travel, which may allow for the rapid spread of new viruses; there are profound changes in land use, including deforestation, so that humans come into contact with previously isolated species; and the HIV/AIDS epidemic has ignited widespread immunodeficiency, which may allow new viruses to gain a foothold in immunocompromised patients (86). It is therefore crucial that we undertake active surveillance to identify both new viruses as they initially invade human populations and potentially emergent viruses that currently reside in animal populations, but may eventually jump our species boundary. For example, although dengue is now very much a disease of humans, it also exists in a sylvatic (i.e., jungle) cycle involving nonhuman primates (and perhaps other mammals; ref. 87) and various Aedes spp. mosquitoes. The sylvatic cycle is rarely studied, but given the increasing encroachment of human populations into areas of virgin forest, viruses within this setting may pose a threat to humans in the future. As each of the 4 serotypes of DENV currently circulating in humans almost certainly have their origins in those viruses found in nonhuman primates (88), it seems a safe bet that there is a far greater diversity of potentially emergent DENV lurking in tropical forests. Ominously, jumping species boundaries from monkeys to humans has not posed much of an adaptive challenge to DENV in the past (89).
Genomic analyses will have a profound impact on the biomedical sciences over the next decade. For the first time, many of the biggest problems in viral biology are no longer limited by the ability to generate sequence data. As a consequence, viral genomics may soon become an important clinical tool. The true challenge of genomic biology therefore lies elsewhere. First, so much sequence data is now produced so rapidly that computational analysis of the sort described here is increasingly difficult. I hope that the new wealth of genome sequence data will entice computer scientists to upgrade their own technology in the near future. Second, as discussed above, the true power of viral genome sequences will not be manifest unless they are linked to additional epidemiological, clinical, and immunological data. At the very least, we require databases that integrate all these variables. Such a happy marriage will truly usher in the golden age of genomic biology.
Footnotes
Conflict of interest: The author has declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 119:2488–2495 (2009). doi:10.1172/JCI38050
References
- 1.Margulies M., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. 2003. Fact sheet N°211. Influenza. http://www.who.int/mediacentre/factsheets/fs211/.
- 3.Dugan V.G., et al. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 2008;4:e1000076. doi: 10.1371/journal.ppat.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olsen B., et al. Global patterns of influenza a virus in wild birds. Science. 2006;312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 5.Webster R.G., Bean W.J., Gorman O.T., Chambers T.M., Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghedin E., et al. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature. 2005;437:1162–1166. doi: 10.1038/nature04239. [DOI] [PubMed] [Google Scholar]
- 7.Rabadan R., Levine A.J., Robins H. Comparison of avian and human influenza A viruses reveals a mutational bias on the viral genomes. . J. Virol. 2006;80:11887–11891. doi: 10.1128/JVI.01414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenbaum B.D., Levine A.J., Bhanot G., Rabadan R. Patterns of evolution and host gene mimicry in influenza and other RNA viruses. PLoS Pathog. 2008;4:e1000079. doi: 10.1371/journal.ppat.1000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kandun I.N., et al. Three Indonesian clusters of H5N1 virus infection in 2005. N. Engl. J. Med. 2006;355:2186–2194. doi: 10.1056/NEJMoa060930. [DOI] [PubMed] [Google Scholar]
- 10.Novel Swine-Origin Influenza A (H1N1) Virus Investigating Team et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 11.Shinde V., et al. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005–2009. N. Engl. J. Med. 2009;360:2616–2625. doi: 10.1056/NEJMoa0903812. [DOI] [PubMed] [Google Scholar]
- 12.Holmes E.C., et al. Whole genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 2005;3:e300. doi: 10.1371/journal.pbio.0030300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson M.I., et al. Stochastic processes are key determinants of the short-term evolution of influenza A virus. PLoS Pathog. 2006;2:e125. doi: 10.1371/journal.ppat.0020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson M.I., et al. Molecular epidemiology of A/H3N2 and A/H1N1 influenza virus during a single epidemic season in the United States. PLoS Pathog. 2008;4:e1000133. doi: 10.1371/journal.ppat.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bean W.J., Jr., Cox N.J., Kendal A.P. Recombination of human influenza A viruses in nature. Nature. 1980;284:638–640. doi: 10.1038/284638a0. [DOI] [PubMed] [Google Scholar]
- 16.Rambaut A., et al. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitnaul L.J., et al. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J. Virol. 2000;74:6015–6020. doi: 10.1128/JVI.74.13.6015-6020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson M.I., Simonsen L., Viboud C., Miller M.A., Holmes E.C. Phylogenetic analysis reveals the global migration of seasonal influenza A viruses. PLoS Pathog. . 2007;3:1220–1228. doi: 10.1371/journal.ppat.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fitch W.M., Leiter J.M.E., Li X., Palese P. Positive Darwinian evolution in human influenza A viruses. Proc. Natl. Acad. Sci. U. S. A. 1991;88:4270–4274. doi: 10.1073/pnas.88.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell C.A., et al. The global circulation of seasonal influenza A (H3N2) viruses. Science. 2008;320:340–346. doi: 10.1126/science.1154137. [DOI] [PubMed] [Google Scholar]
- 21.Smith D.J., et al. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 22.Bright R.A., et al. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet. 2005;366:1175–1181. doi: 10.1016/S0140-6736(05)67338-2. [DOI] [PubMed] [Google Scholar]
- 23.Bright R.A., Shay D.K., Shu B., Cox N.J., Klimov A.I. Adamantane resistance among influenza A viruses isolated early during the 2005-2006 influenza season in the United States. JAMA. 2006;295:891–894. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- 24.Simonsen L., et al. The rapid global spread of reassortant human influenza A/H3N2 viruses conferring adamantane-resistant. Mol. Biol. Evol. . 2007;24:1811–1820. doi: 10.1093/molbev/msm103. [DOI] [PubMed] [Google Scholar]
- 25.Nicoll A., Ciancio B., Kramarz P., Influenza Project Team. Observed oseltamivir resistance in seasonal influenza viruses in Europe interpretation and potential implications. Euro Surveill. 2008;13:8025. doi: 10.2807/ese.13.05.08025-en. [DOI] [PubMed] [Google Scholar]
- 26.Rameix-Welti M.A., Enouf V., Cuvelier F., Jeannin P., van der Werf S. Enzymatic properties of the neuraminidase of seasonal H1N1 influenza viruses provide insights for the emergence of natural resistance to oseltamivir. PLoS Pathog. 2008;4:e1000103. doi: 10.1371/journal.ppat.1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCullers J.A. Insights into the interaction between influenza virus and pneumococcus. Clin. Microbiol. Rev. . 2006;19:571–582. doi: 10.1128/CMR.00058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmes E.C. Viral evolution in the genomic age. PLoS Biol. 2007;5:e278. doi: 10.1371/journal.pbio.0050278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson M.I., et al. Multiple Reassortment events in the evolutionary history of H1N1 influenza A virus since 1918. PLoS Pathog. . 2008;4:e1000012. doi: 10.1371/journal.ppat.1000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kilbourne E.D., et al. The total influenza vaccine failure of 1947 revisited: major intrasubtypic antigenic change can explain failure of vaccine in a post-World War II epidemic. Proc. Natl. Acad. Sci. U. S. A. 2002;99:10748–10752. doi: 10.1073/pnas.162366899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson M.I., Holmes E.C. The evolution of epidemic influenza. Nat. Rev. Genet. 2007;8:196–205. doi: 10.1038/nrg2053. [DOI] [PubMed] [Google Scholar]
- 32.Smith D.J. Predictability and preparedness in influenza control. Science. 2006;312:392–394. doi: 10.1126/science.1122665. [DOI] [PubMed] [Google Scholar]
- 33.Barré-Sinoussi F., et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 34.Gaschen B., et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 35.Nickle D.C., et al. Consensus and ancestral state HIV vaccines. Science. 2003;299:1515–1518. doi: 10.1126/science.299.5612.1515c. [DOI] [PubMed] [Google Scholar]
- 36.Gao F., et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. . Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 37.Keele B.F., et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemey P., et al. Tracing the origin and history of the HIV-2 epidemic. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6588–6592. doi: 10.1073/pnas.0936469100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Worobey M., et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature. 2008;455:661–664. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolfe N.D., Daszak P., Kilpatrick A.M., Burke D.S. Bushmeat hunting, deforestation, and prediction of zoonoses emergence. Emerg. Infect. Dis. 2005;11:1822–1827. doi: 10.3201/eid1112.040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hahn B.H., Shaw G.M., de Cock K.M., Sharp P.M. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 42.Drummond A.J., Ho S.Y.W., Phillips M.J., Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gilbert M.T., et al. The emergence of HIV/AIDS in the Americas and beyond. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18566–18570. doi: 10.1073/pnas.0705329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rambaut A., Crandall K.A., Posada D., Holmes E.C. The causes and consequences of HIV evolution. Nat. Rev. Genet. 2004;5:52–61. doi: 10.1038/nrg1246. [DOI] [PubMed] [Google Scholar]
- 45.Rambaut A., Robertson D.L., Pybus O.G., Peeters M., Holmes E.C. Phylogeny and the origin of HIV-1. Nature. 2001;410:1047–1048. doi: 10.1038/35074179. [DOI] [PubMed] [Google Scholar]
- 46.Iversen A.K., et al. Preferential detection of HIV subtype C’ over subtype A in cervical cells from a dually infected woman. AIDS. 2005;19:990–993. doi: 10.1097/01.aids.0000171418.91786.ad. [DOI] [PubMed] [Google Scholar]
- 47.John-Stewart G.C., et al. Subtype C is associated with increased vaginal shedding of HIV-1. . J. Infect. Dis. 2005;192:492–496. doi: 10.1086/431514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soto-Ramirez L.E., et al. HIV-1 Langerhans’ cell tropism associated with heterosexual transmission of HIV. Science. 1996;271:1291–1293. doi: 10.1126/science.271.5253.1291. [DOI] [PubMed] [Google Scholar]
- 49.Mansky L.M. Retrovirus mutation rates and their role in genetic variation. J. Gen. Virol. 1998;79:1337–1345. doi: 10.1099/0022-1317-79-6-1337. [DOI] [PubMed] [Google Scholar]
- 50.Mansky L.M., Temin H.M. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. . 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drake J.W., Charlesworth B., Charlesworth D., Crow J.F. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duffy S., Shackelton L.A., Holmes E.C. Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 53.Lemey P., Rambaut A., Pybus O.G. HIV dynamics within and among hosts. AIDS Rev. 2006;8:125–140. [PubMed] [Google Scholar]
- 54.Jetzt A.E., et al. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 2000;74:1234–1240. doi: 10.1128/JVI.74.3.1234-1240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jung A., et al. Recombination - Multiply infected spleen cells in HIV patients. Nature. 2002;418:144. doi: 10.1038/418144a. [DOI] [PubMed] [Google Scholar]
- 56.Gubler D.J. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 2002;10:100–103. doi: 10.1016/S0966-842X(01)02288-0. [DOI] [PubMed] [Google Scholar]
- 57.Jetten T.H., Focks D.A. Potential changes in the distribution of dengue transmission under climate warming. Am. J. Trop. Med. Hyg. 1997;57:285–297. doi: 10.4269/ajtmh.1997.57.285. [DOI] [PubMed] [Google Scholar]
- 58.Rico-Hesse R. Microevolution and virulence of dengue viruses. Adv. Virus Res. 2003;59:315–341. doi: 10.1016/S0065-3527(03)59009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holmes E.C., Twiddy S.S. The origin, emergence and evolutionary genetics of dengue virus. Infect. Genet. Evol. 2003;3:19–28. doi: 10.1016/S1567-1348(03)00004-2. [DOI] [PubMed] [Google Scholar]
- 60.Bennett S.N., et al. Selection-driven evolution of emergent dengue virus. Mol. Biol. Evol. 2003;20:1650–1658. doi: 10.1093/molbev/msg182. [DOI] [PubMed] [Google Scholar]
- 61.Klungthong C., Zhang C., Mammen M.P., Jr., Ubol S., Holmes E.C. The molecular epidemiology of dengue virus serotype 4 in Bangkok, Thailand. Virology. 2004;329:168–179. doi: 10.1016/j.virol.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 62.Sittisombut N., et al. Possible occurrence of a genetic bottleneck in dengue serotype 2 viruses between the 1980 and 1987 epidemic seasons in Bangkok, Thailand. Am. J. Trop. Med. Hyg. 1997;57:100–108. doi: 10.4269/ajtmh.1997.57.100. [DOI] [PubMed] [Google Scholar]
- 63.Thu H.M., et al. Myanmar dengue outbreak associated with displacement of serotypes 2, 3, and 4 by dengue 1. Emerg. Infect. Dis. . 2004;10:593–597. doi: 10.3201/eid1004.030216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wittke V., et al. Extinction and rapid emergence of strains of dengue 3 virus during an interepidemic period. Virology. 2002;301:148–156. doi: 10.1006/viro.2002.1549. [DOI] [PubMed] [Google Scholar]
- 65.Cologna R., Armstrong P.M., Rico-Hesse R. Selection for virulent dengue viruses occurs in humans and mosquitoes. J. Virol. 2005;79:853–859. doi: 10.1128/JVI.79.2.853-859.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leitmeyer K.C., et al. Dengue virus structural differences that correlate with pathogenesis. J. Virol. 1999;73:4738–4747. doi: 10.1128/jvi.73.6.4738-4747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rico-Hesse R., et al. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology. 1997;230:244–251. doi: 10.1006/viro.1997.8504. [DOI] [PubMed] [Google Scholar]
- 68.Armstrong P.M., Rico-Hesse R. Differential susceptibility of Aedes aegypti to infection by the American and Southeast Asian genotypes of dengue type 2 virus. . Vector Borne Zoonotic Dis. 2001;1:159–168. doi: 10.1089/153036601316977769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cologna R., Rico-Hesse R. American genotype structures decrease dengue virus output from human monocytes and dendritic cells. J. Virol. 2003;77:3929–3938. doi: 10.1128/JVI.77.7.3929-3938.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang C., et al. Clade replacements in dengue virus serotypes 1 and 3 are associated with changing serotype prevalence. J. Virol. 2005;79:15123–15130. doi: 10.1128/JVI.79.24.15123-15130.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Adams B., et al. Cross-protective immunity can account for the alternating epidemic pattern of dengue virus serotypes circulating in Bangkok. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14234–14239. doi: 10.1073/pnas.0602768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Whitehead S.S., Blaney J.E., Durbin A.P., Murphy B.R. Prospects for a dengue virus vaccine. Nat. Rev. Microbiol. 2007;5:518–528. doi: 10.1038/nrmicro1690. [DOI] [PubMed] [Google Scholar]
- 73.Gandon S., Mackinnon M.J., Nee S., Read A.F. Imperfect vaccines and the evolution of pathogen virulence. Nature. 2001;414:751–756. doi: 10.1038/414751a. [DOI] [PubMed] [Google Scholar]
- 74.Soubeyrand B., Plotkin S.A. Microbial evolution: antitoxin vaccines and pathogen virulence. Nature. 2002;417:609–610. doi: 10.1038/417609b. [DOI] [PubMed] [Google Scholar]
- 75.Farci P., et al. The outcome of acute Hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288:339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 76.Shankarappa R., et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. . J. Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klimov A.I., et al. Prolonged shedding of amantadine-resistant influenza A viruses by immunodeficient patients: detection by polymerase chain reaction-restriction analysis. J. Infect. Dis. 1995;172:1352–1355. doi: 10.1093/infdis/172.5.1352. [DOI] [PubMed] [Google Scholar]
- 78.Aaskov J., Buzacott K., Thu H.M., Lowry K., Holmes E.C. Long–term transmission of defective RNA viruses in humans and Aedes mosquitoes. . Science. 2006;311:236–238. doi: 10.1126/science.1115030. [DOI] [PubMed] [Google Scholar]
- 79.Keele B.F., et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Angly F.E., et al. The marine virospheres of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Culley A.I., Lang A.S., Suttle C.A. High diversity of unknown picorna-like viruses in the sea. Nature. 2003;424:1054–1057. doi: 10.1038/nature01886. [DOI] [PubMed] [Google Scholar]
- 82.Culley A.I., Lang A.S., Suttle C.A. Metagenomic analysis of coastal RNA virus communities. Science. 2006;312:1795–1798. doi: 10.1126/science.1127404. [DOI] [PubMed] [Google Scholar]
- 83.Finkbeiner S.R., et al. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008;4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang T., et al. RNA viral community in human feces: Prevalence of plant pathogenic viruses. PLoS Biol. 2005;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cox-Foster D.L., et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 86.Weiss R.A. The Leeuwenhoek Lecture 2001. Animal origins of human infectious disease. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2001;356:957–977. doi: 10.1098/rstb.2001.0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de Thoisy B., Dussart P., Kazanji M. Wild terrestrial rainforest mammals as potential reservoirs for flaviviruses (yellow fever, dengue 2 and St. Louis encephalitis viruses) in French Guiana. Trans. R. Soc. Trop. Med. Hyg. 2004;98:409–412. doi: 10.1016/j.trstmh.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 88.Wang E., et al. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. . J. Virol. 2000;74:3227–3234. doi: 10.1128/JVI.74.7.3227-3234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vasilakis N., et al. Potential of ancestral sylvatic dengue-2 viruses to re-emerge. Virology. 2007;358:402–412. doi: 10.1016/j.virol.2006.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Holmes, E.C. 2009.The evolution and emergence of RNA viruses. Oxford Series in Ecology and Evolution. P.H. Harvey, and R.M. May, editors. Oxford University Press, Oxford, United Kingdom. In press. [Google Scholar]