Abstract
Six1 is a developmentally regulated homeoprotein with limited expression in most normal adult tissues and frequent misexpression in a variety of malignancies. Here we demonstrate, using a bitransgenic mouse model, that misexpression of human Six1 in adult mouse mammary gland epithelium induces tumors of multiple histological subtypes in a dose-dependent manner. The neoplastic lesions induced by Six1 had an in situ origin, showed diverse differentiation, and exhibited progression to aggressive malignant neoplasms, as is often observed in human carcinoma of the breast. Strikingly, the vast majority of Six1-induced tumors underwent an epithelial-mesenchymal transition (EMT) and expressed multiple targets of activated Wnt signaling, including cyclin D1. Interestingly, Six1 and cyclin D1 coexpression was found to frequently occur in human breast cancers and was strongly predictive of poor prognosis. We further show that Six1 promoted a stem/progenitor cell phenotype in the mouse mammary gland and in Six1-driven mammary tumors. Our data thus provide genetic evidence for a potent oncogenic role for Six1 in mammary epithelial neoplasia, including promotion of EMT and stem cell–like features.
Introduction
Normal embryogenesis and neoplasia share many of the same basic processes and molecular pathways, suggesting that tumor development is an aberrant form of morphogenesis (1). Indeed, there is now overwhelming evidence that developmental genes are often misexpressed in human cancers and that this misexpression can impact neoplastic disease through the re-initiation of developmental programs (2).
Recently, much attention has focused on a process typically associated with normal development, the epithelial-mesenchymal transition (EMT), as an important mechanism during tumor progression. In normal development, epithelial cells lose adhesion and polarity, delaminate, and acquire an invasive, so-called “mesenchymal” phenotype, allowing migration to a site appropriate for organ formation (3). In neoplasia, a similar process is thought to occur at the tumor front, allowing for cellular invasion and eventual metastatic dissemination of cancer cells (4–6). Multiple signaling pathways have been implicated in both developmental and oncogenic EMT, including the Notch, TGF-β, and Wnt signaling pathways (7–13). Recent evidence demonstrates that cells undergoing EMT take on stem cell characteristics (14), implicating developmental regulators of EMT as potential factors involved in stem cell maintenance. Additionally, cells that take on EMT and stem cell characteristics have increased tumorigenic and metastatic potential, underscoring the critical link between developmental processes and cancer (4–6, 14).
Homeobox genes encode transcription factors that are “master regulators” of normal development and control processes such as proliferation, apoptosis, migration, and invasion. In particular, the processes of migration and invasion are associated with an EMT, and several homeoproteins have been implicated in EMT and stem cell maintenance (15–18). Our laboratory focuses on the sine oculis-related homeobox 1 homolog (Six1) homeoprotein that is expressed during early embryogenesis but lost in most adult tissues (19). It is essential for the development of numerous organs, in which it is involved in the expansion of progenitor cell populations through its ability to increase cellular proliferation and survival (19–26). In addition, recent evidence demonstrates that Six1 plays a role in cellular migration and invasion during embryogenesis (20–24) through a mechanism that may involve an EMT. Interestingly, the closely related family member, Six2, regulates both a mesenchymal and stem cell population in the kidney, suggesting that Six family members may play important roles in both EMT and stem cell regulation (27), 2 processes that are now believed to be intimately related (14).
Overexpression of Six1 is observed in numerous cancers, including breast (19, 28, 29), ovarian (26), cervical (30), and hepatocellular carcinomas (31) as well as rhabdomyosarcomas (32–34) and Wilms tumors (35). In several of the aforementioned cancers, Six1 enhances cancer cell proliferation and survival (19, 25, 26, 28, 33), and its overexpression in immortalized mammary epithelial cells induces transformation, leading to highly aggressive and invasive tumors when transplanted into nude mice (25). Although Six1 expression is strongly correlated with neoplasia, its ability to initiate aggressive tumors from normal mammary epithelial cells, or any other normal cells, has not previously been examined.
In this paper, we test the hypothesis that Six1 overexpression in the adult mammary gland leads to activation of developmental pathways out of context, resulting in breast tumor formation. Using a mammary-specific, inducible mouse model of Six1 overexpression, we show that Six1, when misexpressed in the adult mammary epithelium, does indeed induce mammary hyperplasia and aggressive tumor formation. Mammary tumors formed in Six1-overexpressing mice manifest diverse histologic features, including apparent dedifferentiation into high-grade solid and sarcomatoid tumors. The sarcomatoid tumors display full oncogenic EMT, including complete loss of E-cadherin and gain of mesenchymal markers. Furthermore, the majority of nonsarcomatoid tumors (80%) display a partial EMT, including regional loss of membranous E-cadherin, with concomitant gain of nuclear β-catenin and its downstream targets cyclin D1 (Ccnd1), transcription factor 7, T cell specific (Tcf7), Axin2, and myelocytomatosis oncogene (c-myc), suggesting that activation of the Wnt signaling pathway occurs with Six1 overexpression in mouse mammary tumorigenesis. As might be expected of a gene that induces EMT and is associated with Wnt signaling, mammary epithelial cells from Six1-overexpressing mice are enriched for stem cells and have increased mammosphere-forming capability. Additionally, Six1-driven tumors contain cells of multiple mammary lineages and express the stem/progenitor cell marker Sca-1, suggesting that Six1 promotes a stem cell phenotype in the mammary gland. Thus, Six1 is able to induce both EMT and stem cell features in vivo. Consistent with our mouse model, Six1 and cyclin D1 are coexpressed in human breast cancer, and while Six1 alone correlates with poor prognosis, this correlation is significantly enhanced by coexpression of Ccnd1. Together, these data demonstrate that Six1 is a powerful inducer of mammary tumorigenesis and suggest that it may predict poor prognosis in human tumors through its ability to cooperatively interact with cyclin D1 and to enable tumors to acquire both EMT and stem cell–like features.
Results
Development of an inducible, mammary-specific Six1 transgenic mouse model.
Six1 is dynamically expressed in the developing mouse mammary gland, with the highest levels of Six1 transcript present in the late embryonic and young virgin mammary gland. In contrast, levels of Six1 significantly decrease as the mammary gland differentiates, with very low to no expression observed during pregnancy, lactation, and involution (28). Because Six1 overexpression has been detected in human breast cancer, aberrant expression of Six1 in the adult mammary gland may contribute to breast tumorigenesis. To directly test the role of aberrant Six1 expression in the induction of mammary tumorigenesis, and to best mimic the reexpression of Six1 that may occur in the adult human to initiate breast cancer, we developed a (doxycycline-inducible) dox-inducible, bitransgenic mouse model. To this end, we crossed the MTB transgenic line developed by Chodosh and colleagues (36) with transgenic mice containing an HA-tagged Six1 construct, in which Six1 was placed downstream of tet operator sequences (TetSix line). It should be noted that the affect of the HA-tag on Six1 transcriptional activity was previously tested in a cell culture model to ensure that it did not interfere with the normal function of Six1 (data not shown). The MTB line expresses a mouse mammary tumor virus long terminal repeat region–driven (MMTV-LTR–driven) reverse tetracycline transcriptional activator (rtTA), so that treatment of bitransgenic animals (the TetSix line crossed with the MTB line, referred to herein as the TOSix line) with dox activates rtTA, which binds the tet promoter and initiates transcription of Six1 (Figure 1A). Five TetSix founder lines were established, of which 4 transmitted through the germ line and contained variable copy numbers of the transgene (Figure 1B). In our model, Six1 was highly expressed in the mammary gland, with some expression in the salivary gland but not in other organs, as expected when using the MMTV promoter (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI37691DS1).
Figure 1. Characterization of the inducible, mammary-specific Six1 transgenic mouse model.
(A) Schematic representation of the inducible, bitransgenic mouse model system. Tet-O, tet operator. (B) Southern blot analysis shows varying copy numbers in TetSix animal lines (4910, 4922, 6239, and 6245) as compared with spiked-plasmid control. (C) qPCR, using transgene-specific primers and probe, reveals that HASix1 is not expressed in the MTB +dox control animals but is expressed at low levels in the uninduced TOSix mammary glands and at high levels in the induced TOSix mammary glands. Differences in expression between –dox and +dox mammary glands are much greater than differences between transgenic lines (4922 and 6239). Values were transformed using log10(value+1) equation and plotted using a linear axis. Each point represents the value for 1 mammary gland. The middle horizontal lines represent the mean, and error bars represent mean ± SEM. Analysis was performed on multiparous animals. (D) Immunohistochemistry using Six1 antibody reveals no Six1 protein in the control MTB +dox mammary glands (No Six1), low levels of protein in the TOSix –dox mammary glands (Low Six1), and higher levels of Six1 protein in the TOSix +dox mammary glands (High Six1) (scale bar: 100 μm; original magnification, ×40). Clear nuclear staining is shown in insets at higher magnification (arrows) (original magnification, ×100).
The inducible model generates mice with no, low, or high Six1 expression.
After establishing the TOSix transgenic model, we determined levels of expression of Six1 in the absence and presence dox. Experimental female and male TOSix animals from both the 4922 and 6239 lines, as well as control animals (MTB littermates), were continuously treated with 2 mg/ml dox in the drinking water at 12 weeks of age, well after the mice had reached sexual maturity. TOSix animals from both lines were continuously treated with sucrose-only water after 12 weeks of age, to establish levels of HA-Six1 in the absence of dox induction. A subset of female mice were bred for 3 sequential pregnancies and allowed to nurse for 3 weeks before weaning. Because no phenotype was detected in male animals, the remainder of this analysis reflects phenotypes arising exclusively in female animals.
Examination of transgene expression in multiparous animals using real-time quantitative PCR (qPCR) demonstrated that no HA-Six1 mRNA could be detected in the MTB dox-induced (+dox) cohort. Six1 transgene was, however, detectable in both the TOSix cohort that was not induced with dox (–dox) and TOSix +dox cohorts. Transgene levels were significantly higher in the induced cohort as compared with the uninduced cohort, and importantly, relative Six1 expression differences were much greater between the uninduced and induced TOSix animals than between the 4922 and 6239 transgenic lines (Figure 1C). Thus, we were able to examine dose-dependent effects of Six1 by comparing the control group (MTB +dox animals that did not express Six1, herein referred to as no Six1) with 2 different experimental groups: animals that express low levels of Six1 (herein called low Six1) (–dox animals from the 6239 and 4922 TOSix lines) and animals that express high levels of Six1 (herein called high Six1) (+dox animals from the 6239 and 4922 TOSix lines). Although Six1 mRNA expression could be detected by the sensitive method of qPCR in both young and old uninduced TOSix animals, nuclear Six1 protein expression could only be detected in older, uninduced TOSix animals (Figure 1D), suggesting an age-dependent increase in Six1 protein levels over time.
Long-term Six1 overexpression leads to precocious alveolar differentiation and hyperplasia.
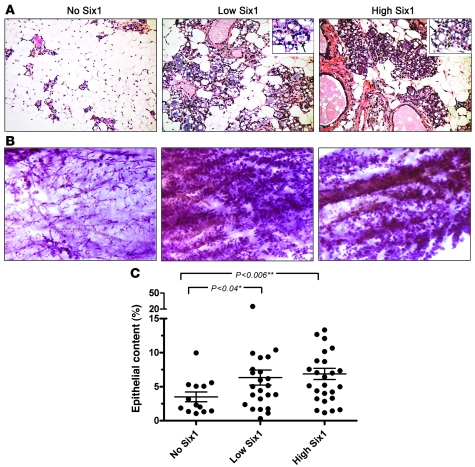
To determine the effect of Six1 overexpression on the mammary gland, mammary tissue from aged no Six1–, low Six1–, and high Six1–expressing animals was taken at least 4 weeks after completion of weaning from the third sequential pregnancy. H&E-stained mammary sections from animals expressing no Six1 showed normal alveolar histology, while alveoli from both low Six1– and high Six1–expressing animals appeared to be expanded and to contain lipid droplets characteristic of secretory differentiation (Figure 2A). Analysis of multiparous Six1-expressing and control animals showed normal postinvolution mammary gland histology after 6 months of dox treatment, suggesting that the alveolar phenotype is due to precocious differentiation, set on by long-term exposure to Six1, rather than a failure to properly involute (data not shown).
Figure 2. Six1 overexpression leads to hyperplasia and precocious alveolar development.
(A) H&E-stained mammary gland sections show lipid droplets and alveolar expansion in low Six1– and high Six1–expressing animals, as compared with no Six1 controls (original magnification, ×20). Higher power insets show lipid droplets in mammary alveoli (arrows) in both low Six1 and high Six1 mammary glands (original magnification, ×63). (B) Mammary whole-mount analysis confirms diffuse hyperplasia in low Six1– and high Six1–expressing animals as compared with no Six1 animals (original magnification, ×1.25). (C) Quantification of epithelial versus fat and stromal content taken from scanned H&E-stained mammary gland sections from no Six1, low Six1, and high Six1 animals. Each point represents quantification of 1 mammary gland. The middle horizontal lines represent the mean, and error bars represent mean ± SEM.
In addition to the precocious differentiation induced by Six1, presence of both low and high levels of Six1 resulted in marked mammary hyperplasia throughout the gland (Figure 2B). In the majority of cases, the hyperplastic phenotype appeared to affect the entire gland rather than discrete areas. To objectively quantify the degree of hyperplasia, H&E-stained sections were scanned and analyzed by a color-based algorithm in order to determine the epithelial versus fat contribution of each gland. Based on this analysis, mammary glands from both low Six1– and high Six1–expressing animals displayed a significantly higher (P < 0.04 and P < 0.006, respectively) epithelial content than age-matched control animals (>12 months of treatment) (Figure 2C).
Six-1 overexpression leads to aggressive mammary tumor formation in a dose-dependent manner.
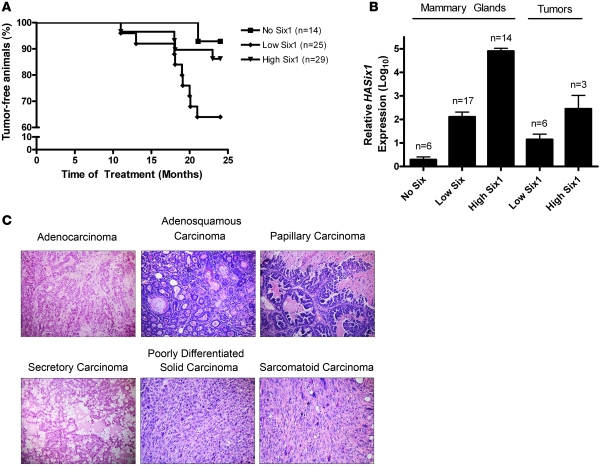
Overexpression of Six1 in the mammary gland of multiparous mice led to the development of overt mammary tumors in approximately 40% (9 of 25) of low Six1–expressing animals and approximately 15% (4 of 29) of high Six1–expressing animals. In contrast, only 7% (1 of 14) of control animals developed a mammary tumor, and this tumor did not occur until the mouse was over 2 years of age (Figure 3A). The average latency of tumor onset in the Six1-expressing animals was approximately 17.5 months after starting dox or vehicle treatment. Because aged, multiparous FVB animals are susceptible to mammary tumor formation due to prolactin-secreting pituitary abnormalities (37), we tested blood prolactin levels from a subset of low Six1– and high Six1–expressing animals that acquired mammary tumors to ensure that the tumors observed in aged animals were induced by Six1. All TOSix animals had blood prolactin levels within the normal range, providing evidence that the tumors arising in the Six1-overexpressing animals were not prolactin driven but were indeed induced by Six1 overexpression (data not shown). A small subset of nulliparous TOSix animals also developed mammary tumors with similar latency and histology, suggesting that mammary cancer initiation in TOSix animals is not dependent on parity. Although the sample size for the nulliparous study was insufficient to reach statistical significance, tumor histology from these nulliparous animals was similar to that from the multiparous study groups, and this group was therefore included in all subsequent histological analysis. It should be noted that although extensive analysis was not done on all tissues in the animals, no overt metastases were observed in tumor-bearing animals.
Figure 3. Mammary tumors arise in Six1-expressing animals in a dose-dependent manner and manifest histologically diverse phenotypes.
(A) Kaplan-Meier analysis of the percentage of tumor-free animals reveals that both low Six1– and high Six1–expressing animals develop tumors, and tumor frequency is higher in low Six1–expressing animals. (B) Transgene-specific qPCR analysis of mammary glands and tumors taken from TOSix and control animals reveal that HASix1 expression is lower in tumors than mammary glands (including glands contralateral to tumors) but higher than that observed in control MTB +dox (no Six1) mammary glands. Values were transformed using log10(value+1) equation and plotted using a linear axis. Error bars represent mean ± SEM. Analysis was performed on multiparous animals. (C) Representative images of H&E-stained tumor sections, demonstrating various histological patterns of tumors observed in TOSix animals (original magnification, ×20).
Because low Six1–expressing animals had a substantially higher tumor penetrance (40%) than high Six1–expressing animals (15%), we reasoned that there may be an optimal level of Six1 required to induce tumorigenesis. In order to further explore this finding, we used qPCR analysis to compare HA-Six1 transgene expression in all tumors to a subset of mammary glands from no Six1–, low Six1–, and high Six1–expressing animals. As expected, tumors from low Six1– and high Six1–expressing animals showed higher levels of transgene expression than the control (no Six1) mammary glands. However, the tumors expressed Six1 at lower levels than the low Six1– and high Six1–expressing mammary glands, including contralateral mammary glands taken from tumor-bearing animals (Figure 3B). This finding suggests that while overexpression of Six1 leads to tumor formation, an optimal range of Six1 expression may be necessary for tumor initiation.
Six1-induced tumors are histologically diverse and include high-grade adenocarcinomas and sarcomatoid (spindle cell) tumors.
Histologic analysis of mammary tumors arising in both low Six1– and high Six1–expressing animals revealed neoplastic lesions of diverse histologies (Tables 1 and 2 and Figure 3C). The majority of tumors showed glandular (“adeno”) differentiation, as seen in human infiltrating ductal breast carcinoma. Two thirds of the tumors showed some degree of squamous differentiation. Additional histologic patterns, also with human correlates, included secretory and papillary differentiation. Two of fourteen tumors (14%) showed high-grade, poorly differentiated, solid areas, analogous to human poorly differentiated carcinoma. Most strikingly, 3 of 14 (21%) tumors exhibited highly aggressive spindle cell morphology, analogous to highly aggressive human sarcomatoid carcinoma (Table 1). While 43% of lesions exhibited features of intraepithelial neoplasia, 86% of tumors showed invasive features, characterized by loss of regular glandular architecture, stromal desmoplasia, and an infiltrative growth pattern. Interestingly, 5 of 14 (36%) individual tumors exhibited multiple histologic patterns (Table 2). Thus, overall, the neoplastic lesions appeared to have an in situ origin, show diverse differentiation, and exhibit progression to highly aggressive malignant neoplasms, as observed in human carcinoma of the breast.
Table 1 .
Summary of histological features of Six1-induced tumors
Table 2 .
Characteristics of tumors containing diverse histologies
The sarcomatoid histomorphology within some Six1-overexpressing tumors suggested that Six1 could induce an EMT in vivo, leading to a loss of polarized epithelial architecture and cell-cell contacts and a gain of mesenchymal-type cell characteristics. Thus, immunohistochemical analysis was performed on tumor sections displaying either sarcomatoid or epithelioid histomorphology (see H&E-stained sections in Figure 4A) for molecular markers of EMT. Strikingly, the sarcomatoid tumor regions displayed a complete loss of the epithelial marker E-cadherin (Figure 4B) and a concurrent gain of the mesenchymal markers SMA (Figure 4C) and nuclear Zeb1 (Supplemental Figure 2). The cells retained the expression of the epithelial marker cytokeratin 18 (CK18) (Figure 4D), demonstrating that the mesenchymal tumors arose from an epithelial origin, which is characteristic of human sarcomatoid carcinoma. Thus, Six1 misexpression is able to induce oncogenic EMT in vivo in a subset of tumors.
Figure 4. A subset of Six1 tumors show a complete EMT.
(A) H&E-stained sections of regions of tumor showing epithelial and sarcomatoid (spindle cell) morphology. (B) Immunohistochemistry with E-cadherin antibody shows strong cell-surface staining in the epithelial regions and complete absence of staining in the sarcomatoid regions. (C) Immunohistochemistry to detect the EMT marker SMA shows a gain of SMA expression in the sarcomatoid regions. (D) Immunohistochemistry performed using an antibody against the luminal epithelial marker CK18. Sarcomatoid tumors retain cytokeratin expression, supporting an epithelial origin. Original magnification, ×40 (A–D).
The majority of Six1-driven tumors exhibit a partial EMT and express downstream targets of activated Wnt signaling.
Although almost 80% of Six1-induced tumors did not display an overt sarcomatoid histomorphology, the majority did show focal loss of E-cadherin. In these tumors, E-cadherin loss correlated with the presence of nuclear β-catenin, a protein that colocalizes with E-cadherin in adherens junctions in noncancerous cells but moves to the nucleus in some tumor types (6). Nuclear β-catenin was detected in 67% (8 of 12) of tumors stained, including those arising in both nulliparous and multiparous animals, and in 80% (8 of 10) of tumors stained arising in the multiparous subset. As expected, regions exhibiting high nuclear β-catenin expression also expressed high levels of the β-catenin transcriptional target, cyclin D1 (Figure 5A). Together, these data suggested that partial loss of E-cadherin with concomitant activation of Wnt target genes is a predominant molecular phenotype in Six1-driven tumors. While this phenotype induced by Six1 is not an overt EMT as observed in the sarcomatoid tumors, it possesses many molecular features of an EMT. These data suggest that Six1 drives a partial EMT, concomitant with activated Wnt signaling in most tumors.
Figure 5. Wnt target genes are increased in the majority of Six1-driven tumors.
(A) Immunohistochemistry of serial sections from a TOSix mammary tumor shows area of concurrent and focal loss of E-cadherin, gain of nuclear and cytoplasmic β-catenin, and gain of the β-catenin transcriptional target, cyclin D1 (denoted by dotted line) (original magnification, ×63). (B) qPCR analysis of Wnt signaling transcriptional target expression, including Ccnd1, c-Myc, Axin2, and Tcf7 and (C) the cytoskeletal organizer, Ezr, in mammary glands taken from no Six1, low Six1–, and high Six1–expressing animals verses tumors arising in multiparous TOSix animals. Error bars represent mean ± SEM. ***P < 0.001, **P < 0.01, *P < 0.05; #, no significance.
In support of this notion, Six1-overexpressing tumors showed a statistically significant upregulation of several Wnt/β-catenin transcriptional targets, including Ccnd1, c-Myc, and Axin2 (38). The same trend is clear for another Wnt transcriptional target, Tcf7, where the data approaches statistical significance (P = 0.08) (38). Surprisingly, these β-catenin target genes were only upregulated in the tumors and not in the mammary glands overexpressing Six1 (Figure 5B), suggesting that Six1 does not directly activate Wnt signaling or Ccnd1 and c-Myc in the mammary gland, targets previously reported to be activated not only through Wnt signaling but directly by Six1 in other tumor types (34). Thus, activation of Wnt signaling may act as a preferred cooperating event for Six1-driven tumorigenesis, or alternatively, Six1 may only activate these targets in a tumorigenic context. In support of the hypothesis that altered Six1-mediated activation may occur in cancer, ezrin (Ezr), a reported direct transcriptional target of Six1 in rhabdomyosarcomas (34), is highly expressed in Six1-induced tumors when compared with control mammary glands and is only slightly, but not significantly, overexpressed in mammary glands overexpressing high levels of Six1 (Figure 5C). In contrast, a previously reported developmental target of Six1, c-Met (22), does not appear to be regulated by Six1 (Supplementary Figure 3).
Six1-overexpressing mammary glands are enriched for stem cells, and Six1-driven tumors exhibit progenitor cell characteristics.
Six1 is known to stimulate proliferation in progenitor populations during normal development (19–26), and recent evidence suggests that the closely related family member Six2 may contribute to the self-renewing potential of kidney stem cells (27). The majority of Six1-overexpressing tumors have activated the Wnt signaling pathway, a pathway that has been strongly implicated in mammary epithelial stem cell maintenance (39). Additionally, Six1-driven tumors display multiple histologies and the majority undergo an EMT, a process strongly associated with the gain of stem cell characteristics (14). Thus, we asked whether Six1 could promote a stem/progenitor cell phenotype in mammary epithelial cells and whether Six1-driven tumors would exhibit stem/progenitor cell characteristics.
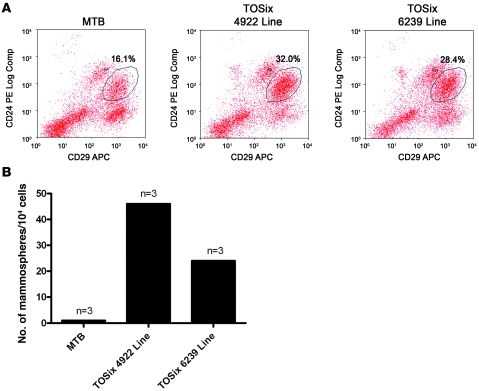
To determine whether Six1 overexpression alters the percentage of mammary epithelial stem cells, primary mammary epithelial cells were isolated from both 6239 and 4922 TOSix bitransgenic lines and MTB control females aged approximately 1.5 years. Epithelial cells were analyzed by flow cytometry using the stem cell markers CD24 and CD29 (39). TOSix animals from both transgenic lines displayed an increased CD29hiCD24+ mammary population compared with MTB controls (32.0% and 28.4% vs. 16.1%, respectively) (Figure 6A).
Figure 6. Six1-overexpressing mammary glands exhibit stem/progenitor cell characteristics.
(A) Flow cytometric analysis of mammary epithelial cells harvested from sucrose-treated, nulliparous TOSix animals (4922 and 6239 lines) and MTB animals aged approximately 1.5 years. Three animals were pooled for each group. Antibodies used to perform flow cytometry include CD24 and CD29, markers found on mammary epithelial stem cells. TOSix animals have substantially more stem cells than MTB controls. Percentages denote the CD29hiCD24+ population. APC, allophycocyanin. (B) Secondary mammosphere assays were performed using mammary epithelial cells isolated from the above groups. Mammosphere numbers are increased in TOSix animals compared with MTB controls. Experiments in A and B were repeated with mammary epithelial cells from either sucrose- or dox-treated animals, yielding similar results.
To determine whether Six1 overexpression led to an increased population of functional mammary epithelial stem cells, mammosphere assays were performed using epithelial cells isolated from TOSix (4922 and 6239 lines) and MTB animals. The secondary mammosphere assay, which measures capacity for self-renewal (40–42), revealed that Six1 bitransgenic mammary glands contain an increased number of functional mammary epithelial stem cells when compared with control mammary glands. Similar to the results from flow cytometry, mammary cells isolated from TOSix animals formed substantially more mammospheres than cells isolated from MTB controls (Figure 6B). Similar results were obtained for both flow cytometry and mammosphere assays, regardless of whether the animals were sucrose treated or induced with dox, such that bitransgenic Six1-expressing animals always displayed a higher percentage of stem cells than the MTB control counterparts (data not shown). Together, our results demonstrate that mammary glands overexpressing Six1 are enriched for stem cells, suggesting that Six1 may play a role in the establishment or maintenance of mammary epithelial stem cell populations.
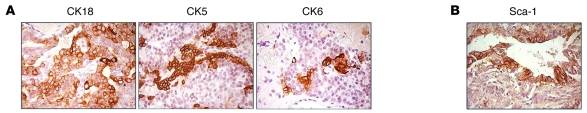
To determine whether Six1-driven tumors also manifest a stem/progenitor cell phenotype, we examined the TOSix tumors for expression of the luminal epithelial cell marker CK18, the myoepithelial cell marker CK5, and the mammary progenitor cell marker CK6 (43). Strikingly, 6 of 12 (50%) Six1-driven tumors exhibited areas of mixed cytokeratin expression, including CK18, CK5, and CK6 (Figure 7A). Additionally, 9 of 12 (75%) TOSix tumors expressed the stem cell marker Sca-1 (Figure 7B). These data, in conjunction with the observation that individual tumors driven by Six1 frequently show multiple histologic patterns and activation of Wnt signaling, suggest that Six1-induced tumors arise from a progenitor/stem cell–like cell population.
Figure 7. Six1-driven tumors display features of stem/progenitor cell origin.
(A) Immunohistochemistry of sections from a single TOSix mammary tumor with activated Wnt signaling displays mixed expression of cytokeratins marking different cell types, including CK18, which is expressed on luminal epithelial cells; CK5, which is expressed on myoepithelial cells; and CK6, which is expressed on the surface of mammary progenitor cells. (B) Immunohistochemistry of a section from a TOSix tumor that expresses the mammary progenitor cell marker, Sca-1. Original magnification, ×63 (A and B).
Six1 overexpression is observed in many human malignancies.
Inappropriate expression of Six1 has been reported in breast (19, 29), ovarian (26), cervical (30), and hepatocellular carcinomas (31) and in rhabdomyosarcoma (32–34) and Wilms tumor (35). The fact that Six1 is overexpressed in several, unrelated tumor types led us to examine its expression in an even more diverse set of tumor types. Using the Oncomine database (44), we evaluated Six1 expression in 23 microarray data sets that profile one or more tumor types against normal tissue. Strikingly, in 15 out of 20 tumor types that we were able to evaluate, Six1 was overexpressed compared with the normal tissue. In many cases this was confirmed in multiple datasets (Table 3). Interestingly, the levels of Six1 overexpression were relatively low, ranging from a 1.1- to 4-fold increase, but in each case the data were highly statistically significant (Table 3). This observation suggests that, similar to the results from our transgenic model, low levels of Six1 overexpression are more likely to promote tumorigenesis than high levels of Six1.
Table 3 .
Many human tumor types overexpress Six1
The malignancies in which Six1 was overexpressed included brain tumors (glioblastoma, oligodendroglioma, and astrocytoma), lung tumors (adenocarcinoma, squamous cell carcinoma, carcinoid, and small cell carcinoma), kidney tumors (clear cell carcinoma and Wilms tumor), and bladder, colon, tonsillar squamous cell, ovarian, pancreatic, and prostate carcinomas (Table 3). These data strongly support the notion that inappropriate expression of Six1 is an important tumor-initiating or -promoting factor in many human tumors.
Overexpression of Six1 and Ccnd1 correlates in human breast tumors and strongly predicts poor prognosis.
Because we observed an increase in cyclin D1 expression in regions undergoing EMT in Six1-induced mammary tumors, we sought to determine whether cyclin D1 and Six1 proteins are coexpressed in human breast cancer using tissue microarrays. Immunohistochemical staining using antibodies generated against Six1 and cyclin D1 was performed on arrays containing primary tumors (mixed invasive ductal carcinoma and invasive lobular carcinoma) from 160 patients, and staining was scored for intensity on a 0–4 scale. Representative examples of anti-Six1 and anti–cyclin D1 staining in human breast cancers are shown in Figure 8A. Importantly, intensity of cyclin D1 antibody staining increased with the intensity of Six1 antibody staining in a statistically significant manner (Figure 8B). These results demonstrate that Six1 and cyclin D1 immunoreactivity correlate in human breast cancer.
Figure 8. Anti-Six1 and anti-cyclin D1 immunoreactivity correlate in human breast cancers.
(A) Breast tumor tissue microarrays were subjected to immunohistochemistry using antibodies against Six1 and cyclin D1. They were then scored for staining intensity on a 0–4 scale. Representative examples are shown of tumor cores with low-intensity Six1 staining ([scored 0–1], core number AA6), medium-intensity Six1 staining ([scored 1.5–2.5], core number DB6), and high-intensity Six1 staining ([scored 3–4], core number DB8), with their corresponding intensity of cyclin D1 staining (original magnification, ×40). (B) The intensity of cyclin D1 staining increases with increasing intensity of Six1 staining, as scored on a 0–4 scale. Six1 staining intensity groups were established as described in A. Error bars represent mean ± SEM.
Our data show that Six1 is capable of initiating tumors that are highly invasive and aggressive, and clinical studies show that Six1 overexpression occurs in 90% of breast cancer metastatic lesions compared with 50% of primary breast tumors (19, 29). In addition, a recent examination of the van de Vijver dataset, profiling 295 early stage (I and II) breast carcinomas (45), and the Pawitan and Ivshina dataset, profiling 240 invasive breast carcinomas unselected for disease stage (46, 47), showed that Six1 is a predictor of poor prognosis in breast cancer (48). Together, these observations support a causal role for Six1 in human breast cancer progression.
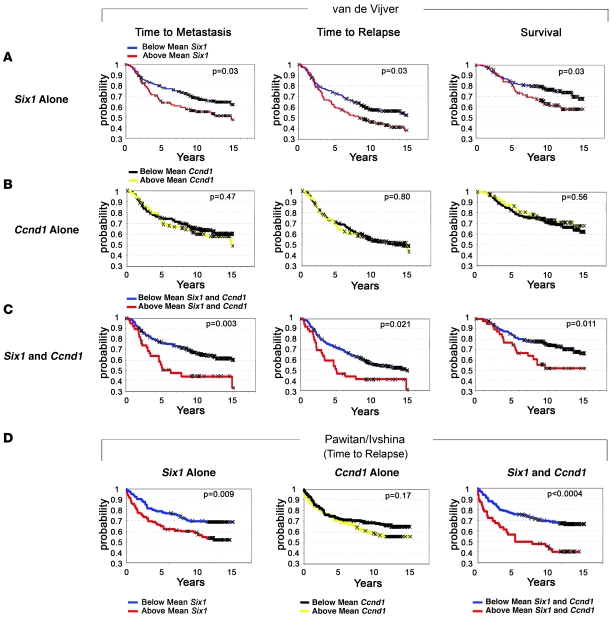
Because results from our mouse model and from human breast cancer samples suggest that Six1 and cyclin D1 are often coexpressed in mammary cancer, we used the aforementioned datasets to test the hypothesis that cyclin D1 may cooperate with Six1 in human breast cancer. In the van de Vijver dataset, patients whose tumors expressed above mean levels of Ccnd1 exhibited no significant trends in any of the prognostic indicators, including time to metastasis, time to relapse, and overall survival due to breast cancer–related death (Figure 9B). However, in patients whose tumors had above mean expression of both Ccnd1 and Six1, adverse outcomes were exacerbated and concomitant overexpression of both Six1 and Ccnd1 increased the prognostic value when compared with Six1 overexpression alone, with respect to the following parameters: shortened time to metastasis (P = 0.003 for Six1 and Ccnd1 vs. P = 0.03 for Six1 alone), shortened time to relapse (P = 0.021 for Six1 and Ccnd1 vs. P = 0.03 for Six1 alone), and shortened survival due to breast cancer–related death (P = 0.011 for Six1 and Ccnd1 vs. P = 0.03 for Six1 alone) (Figure 9, A vs. C). In the Pawitan and Ivshina dataset, while Ccnd1 overexpression alone did not predict shortened time to relapse, above mean levels of Six1 and Ccnd1 together cooperatively worsened prognosis and were more strongly associated with shortened time to relapse than Six1 alone (P < 0.0004 for Six1 and Ccnd1 vs. P = 0.009 for Six1 alone) (Figure 9D). These results, obtained with 2 independent datasets comprising a total of 535 patients, show that the combination of Six1 and Ccnd1 are powerful predictors of poor prognosis.
Figure 9. Six1 overexpression significantly correlates with poor clinical outcome in breast cancers, and this significance is increased by coexpression of Ccnd1.
(A) In a study of 295 women with early-stage invasive breast carcinoma (45), high Six1 expression is associated with shortened time to metastasis, shortened time to relapse, and shortened breast cancer–specific survival (survival) (48). (B) In the same dataset, Ccnd1 overexpression alone does not correlate with shortened time to metastasis or relapse, or with shortened survival. (C) When Six1 and Ccnd1 are both overexpressed, their correlation to clinical parameters is exacerbated and the significance for poor prognosis is increased. (D) In a study of 240 patients diagnosed with invasive breast cancer of any stage (Pawitan and Ivshina) (46, 47), high Six1 expression is strongly associated with shortened time to relapse (48). Ccnd1 overexpression alone has no correlation with shortened time to relapse. When Six1 and Ccnd1 are both overexpressed, their correlation to shortened time to relapse is more significant. In both of these datasets, the mean value for Six1 and Ccnd1 expression in human breast cancers was used to divide the patients into high (above mean) and low (below mean) Six1– and Ccnd1–expressing animals.
Discussion
Six1 is aberrantly expressed in a number of cancers, including breast (19, 28, 29), ovarian (26), cervical (30), and hepatocellular carcinomas (31) as well as in rhabdomyosarcomas (32–34) and Wilms tumors (35). In these cancers, reexpression of Six1 is believed to reinstate a developmental program out of context, increasing proliferation, migration, invasion, and survival to stimulate cancer initiation as well as progression. To determine whether Six1 is in fact able to initiate breast cancer, we established a mammary-specific transgenic mouse model that allowed us to study the effects of both low and high levels of Six1 overexpression. In both low Six1– and high Six1–expressing animals, the transgene induced precocious alveolar differentiation and mammary epithelial hyperplasia. In addition, chronic Six1 overexpression, particularly in the low Six1–expressing animals, resulted in the formation of frank tumors, most of which had activated Wnt signaling and exhibited EMT and stem cell features.
qPCR results revealed that Six1 transgene expression, while increased compared with control MTB +dox (no Six1) mammary glands, is consistently overexpressed at lower levels in the TOSix tumors as compared with TOSix mammary glands. Transgene silencing has been reported to occur in MMTV-LTR–driven mouse models in cases in which the transgene facilitates an EMT (49). In these cases, as cells undergo an EMT, epithelial-specific promoters, such as MMTV-LTR, become silenced. Because Six1 is capable of initiating an EMT or partial EMT in the majority of the mammary tumors, this silencing mechanism may result in reduced levels of the Six1 transgene expression within the tumors as compared with that in the contralateral mammary glands. Alternatively, selective pressure may contribute to the outgrowth of tumor cells expressing the optimal level of Six1 required to promote tumorigenesis. Regardless of mechanism, the consistent and striking correlation between low levels, as opposed to high levels, of Six1 overexpression and tumor frequency suggests that there is an optimal level of Six1 for inducing tumorigenesis.
Interestingly, recent evidence suggests that Six1 can confer dose-dependent effects in cranial placodes during development (50). Small-to-moderate increases in the levels of homeobox gene expression are known to have substantial effects on target gene transcription with significant phenotypic consequences. For example, a 1.5-fold increase of Oct3/4 expression is sufficient to activate and repress multiple target genes (51), and both Oct3/4 and another homeoprotein, Nanog, display dose-dependent effects during development (51–53). Additionally, Oct3/4 influences the genesis of germ cell tumors in a dose-dependent manner (54). We hypothesize that the effects of Six1 are dose dependent, at least in part, due to its required interactions with available cofactors within the cell. Six1 has no intrinsic activation or repression domains and thus is dependent on the Eya and Dach transcriptional cofactors to activate and/or repress transcription (21). When Six1 exists in a complex with Dach, it represses transcription. However, if Eya is added to the complex, it can switch Six1 from a repressor to an activator (21). Like Six1, Eya1 also acts in a dose-dependent manner during development (55). Thus, it is likely that the stoichiometry of Six1 with its cofactors, as well as the availability of cofactors in the cell, modulates the ability of Six1 to activate and/or repress transcription and to induce tumorigenesis. Future experiments are required to determine how the ratio of Six1 to its cofactors may differentially affect tumorigenesis.
Tumors arising in TOSix animals have diverse histologies, ranging from relatively well-differentiated tumors (squamous, secretory, and papillary) to poorly differentiated solid and sarcomatoid tumors. Interestingly, the sarcomatoid tumors undergo a complete EMT, as determined by loss of E-cadherin, gain of mesenchymal markers Zeb1 and SMA, and retention of the epithelial marker CK18. Although this is the first evidence to our knowledge that Six1 is directly capable of driving an EMT, a recent Six1/Six4–double-knockout study is suggestive of a role for Six1 in EMT during normal development. While Six4-knockout mice display no phenotypic abnormalities, myogenic precursor cells in Six1/Six4–double-knockout mice fail to delaminate from the dermomyotome (an epithelial structure) and migrate to the limb bud (22). This defect is suggestive of a failure to undergo a developmental EMT. Furthermore, the Six2 protein, which is 96% conserved to Six1 across the Six domain and homeodomain at the amino acid level, is critical for maintaining the mesenchymal progenitor cell population in the developing kidney, and loss of Six2 results in premature and ectopic differentiation of mesenchymal cells into kidney epithelial cells (56). While these developmental studies suggest a role for Six1 in regulating EMT, our model provides what we believe to be unique evidence that overexpression of Six1 can induce an EMT in vivo, particularly in a tumorigenic context.
Although a subset of Six1-driven tumors undergo an oncogenic EMT accompanied by morphologic changes, the majority of tumors maintain an epithelial histology, with discrete areas of E-cadherin loss and concomitant gain of nuclear β-catenin, suggesting that a partial EMT is occurring. Increases in transcriptional targets of β-catenin, including Ccnd1, Axin2, c-Myc, and Tcf7 are only detected in tumors arising in TOSix animals and are not detected in contralateral Six1-overexpressing mammary glands. Although Ccnd1 and c-Myc as well as the cytoskeletal regulator Ezr are reported to be direct transcriptional targets of Six1 in rhabdomyosarcoma, the absence of their expression in Six1-overexpressing mammary glands suggests that Six1 does not directly regulate these targets in the normal mammary gland. Thus, regulation of these targets by Six1 may be tissue specific, or alternatively, Six1 may regulate the aforementioned targets primarily in a tumorigenic, but not normal, context (34). Likewise, because the majority of Six1-driven tumors display characteristics of Wnt signaling, activation of this signaling pathway is most likely either a preferred cooperating oncogenic pathway for Six1 or the result of Six1 activation that only occurs in the tumorigenic state.
Wnt and its downstream effector β-catenin are known to induce tumorigenesis when overexpressed in the mouse mammary gland. Although some characteristics of Wnt or activated β-catenin transgenic mice phenocopy TOSix mammary phenotypes, such as precocious lobuloalveolar development, Six1-driven tumors display greater histologic diversity than the tumors arising from animal models of activated Wnt signaling (57–60). Additionally, while Wnt signaling is known to induce EMT in development and in cell culture models (7–11), oncogenic EMT is not induced by activated Wnt signaling in the mammary gland, as animals overexpressing Wnt in this context maintain E-cadherin on the surface of the mammary tumor cells (57). These data suggest that it is Six1, or Six1 in concert with Wnt, that is the primary driving force for the complete and partial EMTs observed.
Because recent studies demonstrate that cells undergoing EMT take on stem cell characteristics (14), and because Six1 drives the formation of tumors with multiple histomorphologies, we sought to determine whether Six1 is involved in the regulation of mammary epithelial stem cells. Recent evidence demonstrates that Six2 regulates a stem cell population in the kidney, suggesting that the Six family may play important roles in stem cell regulation (27). Interestingly, our findings indicate that Six1 may play a role in mammary epithelial stem cells, both in the normal gland as well as in tumorigenesis. Mammary epithelial cells from Six1-overexpressing mice are enriched for stem cells and have increased mammosphere-forming capability. Additionally, Six1-driven tumors contain cells of multiple mammary lineages and express Sca-1. Together, these data strongly suggest that Six1 promotes a stem cell phenotype in the normal mammary gland and that Six1-driven tumors arise from a stem/progenitor cell population.
Finally, since Six1 is not normally expressed in most adult tissues, and since its misexpression can initiate mammary tumorigenesis, we investigated whether inappropriate expression of Six1 may be involved in malignancies in addition to breast cancer. We found that a large number of cancers overexpress Six1 compared with the expression of the normal tissue from which they arise. Although the increase in Six1 transcript levels in these tumors is moderate (1.1- to 4-fold increase), at least as measured on microarray platforms, it is highly significant and is observed in multiple tumor types. These data strongly support a critical role for Six1 in human tumorigenesis and further support the model whereby low levels of Six1 overexpression drive tumorigenesis.
Given the increased expression of cyclin D1 in tumors arising from animals overexpressing Six1, we hypothesized that Six1 and cyclin D1 may be coexpressed in human breast cancers. Indeed, immunohistochemical analysis of breast cancer tissue microarrays revealed that tumors that stained intensely with an anti-Six1 antibody also stained intensely with an anti–cyclin D1 antibody. It should be noted, however, that the Six1 antibody (Atlas Antibodies) used for this analysis is made against a region of Six1 that may allow cross-reactivity with other Six family members. Repeated attempts to generate antibodies uniquely specific to Six1 have resulted in antibodies that do not work for immunohistochemical analysis. Thus, we can only confidently say that Six family expression correlates with cyclin D1 in human tumors. However, since Six1 and Ccnd1 mRNA together are highly predictive of poor prognosis (see Figure 9), we conclude that Six1 and cyclin D1 are likely coexpressed in human breast tumors.
In work described elsewhere (48), we demonstrate that Six1 is an indicator of poor prognosis in human breast cancers. When Six1 overexpression is combined with overexpression of Ccnd1, poor prognosis in breast cancer patients, as assessed by shortened time to metastasis, shortened time to relapse, and decreased survival, is exacerbated, even though Ccnd1 alone is not predictive of poor prognosis. These results strongly suggest that the cooperation occurring between Six1 and cyclin D1 in our mouse model is recapitulated in human breast cancers. In our analysis, we also attempted to identify a correlation with other Wnt signaling targets, including Axin, Tcf7, and c-Myc. However, overexpression of these molecules did not seem to cooperate with Six1 as prognostic indicators. These data suggest that, unlike what is observed in the mouse model, Six1 may not cooperate with canonical Wnt signaling in human breast cancer, but rather, only the association with cyclin D1 is conserved. However, more association studies must be performed to fully rule out a cooperative interaction between Six1 and Wnt signaling in human breast cancer.
In closing, our data are the first to our knowledge to demonstrate that Six1 overexpression is sufficient to induce tumorigenesis when expressed out of context in a normal adult cell, leading to highly aggressive and invasive mammary tumors with EMT and stem cell features. We conclude that Six1 is promoting tumorigenesis by reemploying its developmental program to drive not only proliferation and survival but also migration and invasion through EMT. Additionally, our data are the first to our knowledge to suggest that Six1 acts to promote a stem cell phenotype. Because Six1 is overexpressed in a large number of different cancers, we propose that this homeoprotein may act to initiate numerous types of cancer in addition to breast cancer, acting as a more global regulator of tumorigenesis than originally anticipated.
Methods
Generation of mice.
The full-length human HA-Six1 cDNA was inserted into pTetSplice vector using EcoRI adapters to generate the transgenic plasmid. Microinjections were performed by the Cancer Center Transgenic Mouse Core Facility at University of Colorado Denver (UCD) Anschutz Medical Campus. Transgenic progeny were identified by Southern blot analysis using a full-length Six1 cDNA probe and standard protocols (29). Four lines transmitted the transgene through their germline (4910, 4922, 6239, and 6245). Although all lines expressed Six1 after dox induction, the 2 lines with highest expression levels were chosen for further analysis (4922 and 6239). PCR genotyping was performed at 3 weeks of age using genomic DNA isolated from a tail biopsy and the REDExtract-N-Amp Tissue PCR Kit (Sigma-Aldrich). Primers used to detect the TetSix transgene were 5′-AGCTCCAAGACTCTCTGCTC-3′ (forward) and 5′-TCCGGCGAATTTCTGCCATT-3′ (reverse).
The mice were housed at the Center for Comparative Medicine at the UCD Anschutz Medical Campus and treated in accordance with the Guide for the humane care and use of laboratory animals (NIH publication no. 86-23. Revised 1985). All animal protocols were approved by the UCD-IACUC. Hemizygous MMTV-rtTA (MTB) mice, originally generated in the laboratory of L.A. Chodosh (36), were intercrossed with hemizygous TetSix animals to obtain the following genotypes: wild type, MTB, TetSix, and TOSix. Littermates (including females and males) from the MTB and TOSix genotypes started the following treatment regimen at 12 weeks of age: all MTB and a subset of TOSix animals were treated with water containing 2 mg/ml dox (Sigma-Aldrich) and 5% sucrose. A subset of TOSix animals were treated with water containing 2% sucrose as a vehicle control (5% sucrose caused increased intake of control water). Water was provided to animals in a light-protected bottle and changed weekly. A cohort of female animals from the MTB and TOSix genotypes were bred for 3 sequential pregnancies and were allowed to nurse for 3 weeks before weaning. Animals on long-term tumor studies (treated for 11–22 months) were palpated weekly. TOSix (4922 and 6239 lines) and MTB nulliparous animals that began 2% sucrose treatment at 12 weeks of age were used to perform flow cytometry and mammosphere assays.
Whole-mount staining analysis of mammary gland.
Inguinal mammary fad pads were excised from euthanized mice and stretched on a histologic glass slide. The glands were fixed for at least 4 hours in Carnoy’s fixative and then transferred to a cassette and placed in 70% ethanol for 15 minutes. Glands were rinsed with water and carmine-stained overnight. Tissue was dehydrated in graded alcohols and xylene and then coverslipped using Permount mounting media (Fisher).
Histology and immunohistochemistry.
Tumors and mammary glands were fixed in 4% paraformaldehyde, processed on a standard histology processor, embedded in paraffin, and cut into 5 micron sections. For histologic analysis, sections were H&E stained. For immunohistochemistry, paraffin sections were dewaxed in xylene and rehydrated in decreasing concentrations of alcohol. Zeb-1 immunostaining was performed as previously described (61). For Sca-1 immunostaining, TSA biotin system (Perkin Elmer) was used according to the recommended protocol and an anti-rat biotinylated secondary antibody was obtained from Vector Laboratories (used at 1:250). For remaining targets, sections were exposed to citrate buffer and heat antigen retrieval and then blocked and incubated with appropriate primary antibody, using the recommended protocol for ImmPRESS kit or rabbit primary antibodies and M.O.M. kit for mouse primary antibodies (Vector Laboratories). Primary antibodies used included those against Six1 (1:100; Atlas Antibodies), β-catenin (1:100; BD Biosciences — Transduction Laboratories), E-cadherin (1:500; BD Biosciences — Transduction Laboratories), cyclin D1 (1:25; Thermo Scientific), CK18 (1:50; Epitomics), CK5 (1:500; Abcam), CK6 (1:500; Covance), Zeb-1 (1:1,500; gift from Doug Darling, University of Louisville, Louisville, Kentucky, USA), Sca-1 (Ly-6A/E, clone D7) (1:100; BD Biosciences — Pharmingen), and SMA (1:500; Sigma-Aldrich). DAB substrate (Dako) was used to develop slides. Slides were counterstained with hematoxylin (Dako) and coverslipped using Permount mounting media (Fisher).
Mammary epithelial versus fat content algorithm analysis.
H&E-stained mammary gland sections were scanned using an Aperio T3 Scanscope at an effective resolution of 0.46 microns/pixel square. Resulting images were sampled down to a resolution of 2.43 microns/pixel square to facilitate subsequent analysis. Image analysis was performed using ImageJ version 1.37 (http://rsbweb.nih.gov/ij/), running on Java version 1.5. All image analysis steps were executed using custom written ImageJ plugins. Preliminary analysis steps isolated mammary gland biopsy tissue from white field background. Additional steps identified and isolated breast epithelial tissue based on specified signal color values. The composition of mammary gland epithelial tissue in the biopsy tissue was expressed as a percentage of the whole-tissue sample.
RNA isolation.
Tissues were excised and placed in 4 ml of TRIzol Reagent (Invitrogen) and homogenized using a power homogenizer, and total RNA isolation was performed per manufacturer’s recommendation. All RNA samples were treated with DNase I in order to eliminate genomic DNA contamination using RNeasy Micro Kit (Qiagen). Two micrograms of total RNA per sample were used to generate cDNA using random hexamer primers and SuperScript III Reverse Transcriptase (Invitrogen).
qPCR and Northern blot analysis.
qPCR was performed using a model 7500 Real-Time PCR instrument (Applied Biosystems). Amplicons were detected using TaqMan fluorescence probes per manufacturer’s protocol (Applied Biosystems). The primers and probes used for this study were as follows: HASix1 5′-GCTTGCCACCATGGGATATC-3′ (sense), 5′-ACGCACGCCACTTGCTC-3′ (antisense), and 5′-CCTGATTATGCTATGTCGATGCTGCCGTCGT-3′ (probe); Axin2 5′-ACGCACTGACCGACGATTC-3′ (sense), 5′-CCATGCGGTAAGGAGGGAC-3′ (antisense), and 5′-TGTCCATGACGGACAGTAGCGTAGATGG-3′ (probe); and Tcf7 5′-AAGGTCATTGCTGAGTGCACAC-3′ (sense), 5′-TGCATGCCACCTGCGAC-3′ (antisense), and 5′-AGAGCGCTGCCATCAACCAGATCCT-3′ (probe). TaqMan Gene Expression Assays were purchased for Ccnd1 (Mm00432359_m1), c-Myc (Mm00487804_m1), Ezr (Mm01307018_m1), and c-Met (Mm01156980_m1) (Applied Biosystems). Target genes were analyzed using standard curves to determine relative levels of gene expression, and individual cDNA samples were normalized according to the levels of cyclophilin B using following primers and probe: 5′-GCAAAGTTCTAGAGGGCATGGA-3′, 5′-CCCGGCTGTCTGTCTTGGT-3′, and 5′-TGGTACGGAAGGTGGAG-3′ (probe). Analysis was performed using Sequence Detection Software (SDS) version 1.3.1 (Applied Biosystems). Northern blot analysis was performed as previously described (19).
Preparation of primary mouse mammary epithelial cells.
Primary mouse mammary epithelial cells were isolated as previously described (62), except that cells were digested in collagenase overnight, rather than for 1 hour, to enrich for stem cells, as has been performed by other groups when examining stem cell populations (63, 64). Mammary cells were isolated from TOSix (4922 and 6239) and MTB females, aged 1.5 years (sucrose treated) (3 animals per group). Cells were used to perform flow cytometry analysis and mammosphere assays (described below).
Flow cytometry.
Isolated cells were cultured overnight in DMEM/F12 medium with l-glutamine (Hyclone), containing 5% FBS (Hyclone), penicillin-streptomycin (Hyclone), 10 mM HEPES (Cellgro), and 50 μg/ml gentamicin (Cellgro). Mammary epithelial cells were washed in PBS containing 0.5% FBS. For 30 minutes 106 cells were stained in 20 μl of antibody solution (1:100 dilution) on ice. Cells were washed in 1 ml of 0.5% FBS-PBS and resuspended in 400 μl of 1 μg/ml DAPI/0.5% FBS-PBS. The following antigens were used: FITC-linked anti-Ter119 (catalog no. 557915; BD Biosciences — Pharmingen), anti-CD45 (catalog no. 553079; BD Biosciences — Pharmingen), anti-CD31 (catalog no. 558738; BD Biosciences — Pharmingen), biotin-linked anti-CD24 (catalog no. 553260; BD Biosciences — Pharmingen), PE-linked streptavidin (catalog no. 554061; BD Biosciences — Pharmingen), and allophycocyanin-linked anti-CD29 (catalog no. 102215; BioLegend). Fluorescence was detected with CyAn (Beckman Coulter).
Mammosphere assays.
Primary mammary epithelial cells were plated immediately after isolation in 6-well, ultra-low attachment plates (2 ml per well) at a concentration of 10,000–30,000 cells/ml in serum-free DMEM/F12 media (Hyclone), supplemented with 20 ng/ml bFGF (BD Biosciences), 20 ng/ml EGF (BD Biosciences), 4 μg/ml heparin (Sigma), penicillin-streptomycin (Hyclone), and B27 (Gibco) as described previously (40–42). The cells were fed every 3–4 days. After 7 days, following trypsin digestion using 0.05% trypsin and 0.53 mM EDTA-4Na (Invitrogen), single cells from the primary mammospheres were plated at 1,000 cells per well to perform secondary mammosphere assays. The mammospheres (colonies >50 nm) were counted 7–10 days after passage 2.
Tissue microarray immunohistochemistry.
High-density breast cancer tissue microarrays were obtained from Biomax (BR1921), and immunohistochemistry was performed using the protocols detailed above for Six1 (Atlas Antibodies) and cyclin D1 (Thermo Scientific). Staining intensity was blindly scored by a collaborating pathologist on a 0–4 scale, and tumors (160 samples) were divided into groups according to the following scale: 0–1, low Six1 or cyclin D1 staining intensity; 1.5–2.5, medium Six1 or cyclin D1 staining intensity; and 3–4, high Six1 or cyclin D1 staining intensity.
Microarray data analysis.
The Oncomine (www.oncomine.org) database tool was used to analyze mRNA expression microarray data from several cancer studies (44). Standardized normalization techniques and statistical calculations are provided on the Oncomine website. Standard analyses were applied to raw microarray data using Robust Multichip Average for Affymetrix data and Loess for cDNA arrays. If not already performed by the author of the original study, reference data was incorporated at this step. Oncomine then applies a z-score normalization to scale data and allows comparison of multiple independent studies. Normalization includes log2 transformation and setting the array median to 0 and standard deviation to 1. Fold change was calculated by dividing the average of the normalized tumor values by the average of the normalized normal tissue values.
Analyses of clinical outcome data.
Clinical follow-up and gene expression data were obtained from 2 independent, publicly available data sets (45–47). Clinical outcome information from the Pawitan study (47) was obtained from data published in the Ivshina study (46). Expression data for Six1 and Ccnd1 were extracted for each tumor and were mean centered across all samples in the data set. Samples were then segregated into 2 groups for each analysis. The first group comprised samples in which Six1 and/or Ccnd1 expression was above the mean, and the second group comprised the rest of the samples; each data set was analyzed separately.
Statistics.
Statistical analysis of all qPCR data was performed using a paired 2-tailed Student’s t test. Statistics for mammary epithelial content analysis were calculated using an unpaired, 2-tailed t test with Welch’s correction. For Six1 and cyclin D1 tissue array correlations, the P value was determined using 1-way analysis of variance. All of the above statistical analysis was performed using Prism 4 (GraphPad), and P values of less than 0.05 were considered statistically significant.
Six1 was assessed for differential expression in the Oncomine database with t-statistics using Total Access Statistics 2002 (FMS Inc.). The t tests were conducted as 2 sided for differential expression analysis.
Kaplan-Meier survival curves representative of clinical outcome data were generated using the software package WINSTAT FOR EXCEL (R. Fitch Software), and P values were calculated by log-rank analysis. Cox proportional hazards regression was used to assess the contribution of Six1 and Ccnd1 in multiple predictor statistical models of disease-free and disease-specific survival, and predictors were judged to be statistically significant when P values were less than 0.05.
Supplementary Material
Acknowledgments
This work was funded by grants to H.L. Ford from the National Cancer Institute (2RO1-CA095277), the American Cancer Society (RSG-07-183-01-DDC), and the Susan G. Komen Foundation (BCTR0504466). E.L. McCoy was funded by a predoctoral fellowship from the Department of Defense Breast Cancer Research Program (W81XWH-06-1-0409). A.L. Welm was supported by an Era of Hope Scholar Award from the Department of Defense Breast Cancer Research Program (#BC075015). We thank Raedun Clark and other members of the Ford laboratory for technical help and advice, Storey Wilson and Scott Lucia for performing the imaging analysis, and John Tentler for performing assays to detect blood prolactin levels in the animals. We also thank Kim Christensen for her assistance in the preparation of this manuscript as well as Peggy Neville for her thoughtful comments related to the manuscript, and the laboratories of Lori Sussel, Lee Niswander, and Jennifer Richer for technical assistance.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 119:2663–2677 (2009). doi:10.1172/JCI37691.
Karen A. Heichman’s present address is: ARUP Laboratories, Salt Lake City, Utah, USA.
See the related Commentary beginning on page 2528.
References
- 1.Samuel S., Naora H. Homeobox gene expression in cancer: insights from developmental regulation and deregulation. Eur. J. Cancer. 2005;41:2428–2437. doi: 10.1016/j.ejca.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 2.Pearson J.C., Lemons D., McGinnis W. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 3.Fidler I.J. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 4.Brabletz T., Jung A., Spaderna S., Hlubek F., Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat. Rev. Cancer. 2005;5:744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 5.Guarino M., Rubino B., Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39:305–318. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- 6.Thiery J.P. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 7.Huber M.A., Kraut N., Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr. Opin. Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Kemler R., et al. Stabilization of beta-catenin in the mouse zygote leads to premature epithelial-mesenchymal transition in the epiblast. Development. 2004;131:5817–5824. doi: 10.1242/dev.01458. [DOI] [PubMed] [Google Scholar]
- 9.Mohamed O.A., Clarke H.J., Dufort D. Beta-catenin signaling marks the prospective site of primitive streak formation in the mouse embryo. Dev. Dyn. 2004;231:416–424. doi: 10.1002/dvdy.20135. [DOI] [PubMed] [Google Scholar]
- 10.Yang L., Lin C., Liu Z.R. P68 RNA helicase mediates PDGF-induced epithelial mesenchymal transition by displacing Axin from beta-catenin. Cell. 2006;127:139–155. doi: 10.1016/j.cell.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 11.Yook J.I., et al. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat. Cell Biol. 2006;8:1398–1406. doi: 10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
- 12.Grego-Bessa J., Diez J., Timmerman L., de la Pompa J.L. Notch and epithelial-mesenchyme transition in development and tumor progression: another turn of the screw. Cell Cycle. 2004;3:718–721. [PubMed] [Google Scholar]
- 13.Zavadil J., Bottinger E.P. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 14.Mani S.A., et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamada S., et al. Bone morphogenetic protein 4 induces epithelial-mesenchymal transition through MSX2 induction on pancreatic cancer cell line. J. Cell. Physiol. 2007;213:768–774. doi: 10.1002/jcp.21148. [DOI] [PubMed] [Google Scholar]
- 16.Peinado H., Portillo F., Cano A. Transcriptional regulation of cadherins during development and carcinogenesis. Int. J. Dev. Biol. 2004;48:365–375. doi: 10.1387/ijdb.041794hp. [DOI] [PubMed] [Google Scholar]
- 17.Hallaq H., et al. A null mutation of Hhex results in abnormal cardiac development, defective vasculogenesis and elevated Vegfa levels. Development. 2004;131:5197–5209. doi: 10.1242/dev.01393. [DOI] [PubMed] [Google Scholar]
- 18.Pan G., Thomson J.A. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17:42–49. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- 19.Ford H.L., Kabingu E.N., Bump E.A., Mutter G.L., Pardee A.B. Abrogation of the G2 cell cycle checkpoint associated with overexpression of HSIX1: a possible mechanism of breast carcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12608–12613. doi: 10.1073/pnas.95.21.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ozaki H., et al. Six1 controls patterning of the mouse otic vesicle. Development. 2004;131:551–562. doi: 10.1242/dev.00943. [DOI] [PubMed] [Google Scholar]
- 21.Li X., et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- 22.Grifone R., et al. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development. 2005;132:2235–2249. doi: 10.1242/dev.01773. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda K., et al. Six1 is essential for early neurogenesis in the development of olfactory epithelium. Dev. Biol. 2007;311:53–68. doi: 10.1016/j.ydbio.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 24.Zheng W., et al. The role of Six1 in mammalian auditory system development. Development. 2003;130:3989–4000. doi: 10.1242/dev.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coletta R.D., et al. Six1 overexpression in mammary cells induces genomic instability and is sufficient for malignant transformation. Cancer Res. 2008;68:2204–2213. doi: 10.1158/0008-5472.CAN-07-3141. [DOI] [PubMed] [Google Scholar]
- 26.Behbakht K., et al. Six1 overexpression in ovarian carcinoma causes resistance to TRAIL-mediated apoptosis and is associated with poor survival. Cancer Res. 2007;67:3036–3042. doi: 10.1158/0008-5472.CAN-06-3755. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi A., et al. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3:169–181. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coletta R.D., et al. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6478–6483. doi: 10.1073/pnas.0401139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reichenberger K.J., Coletta R.D., Schulte A.P., Varella-Garcia M., Ford H.L. Gene amplification is a mechanism of Six1 overexpression in breast cancer. Cancer Res. 2005;65:2668–2675. doi: 10.1158/0008-5472.CAN-04-4286. [DOI] [PubMed] [Google Scholar]
- 30.Wan F., et al. Gene expression changes during HPV-mediated carcinogenesis: a comparison between an in vitro cell model and cervical cancer. Int. J. Cancer. 2008;123:32–40. doi: 10.1002/ijc.23463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng K.T., et al. Clinicopathological significance of homeoprotein Six1 in hepatocellular carcinoma. Br. J. Cancer. 2006;95:1050–1055. doi: 10.1038/sj.bjc.6603399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan J., et al. cDNA microarrays detect activation of a myogenic transcription program by the PAX3-FKHR fusion oncogene. Proc. Natl. Acad. Sci. U. S. A. 1999;96:13264–13269. doi: 10.1073/pnas.96.23.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Y., et al. Expression profiling identifies the cytoskeletal organizer ezrin and the developmental homeoprotein Six-1 as key metastatic regulators. Nat. Med. 2004;10:175–181. doi: 10.1038/nm966. [DOI] [PubMed] [Google Scholar]
- 34.Yu Y., Davicioni E., Triche T.J., Merlino G. The Homeoprotein Six1 transcriptionally activates multiple protumorigenic genes but requires ezrin to promote metastasis. Cancer Res. 2006;66:1982–1989. doi: 10.1158/0008-5472.CAN-05-2360. [DOI] [PubMed] [Google Scholar]
- 35.Li C.M., et al. Gene expression in Wilms’ tumor mimics the earliest committed stage in the metanephric mesenchymal-epithelial transition. Am. J. Pathol. 2002;160:2181–2190. doi: 10.1016/S0002-9440(10)61166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gunther E.J., et al. A novel doxycycline-inducible system for the transgenic analysis of mammary gland biology. FASEB J. 2002;16:283–292. doi: 10.1096/fj.01-0551com. [DOI] [PubMed] [Google Scholar]
- 37.Wakefield L.M., et al. Spontaneous pituitary abnormalities and mammary hyperplasia in FVB/NCr mice: implications for mouse modeling. Comp. Med. 2003;53:424–432. [PubMed] [Google Scholar]
- 38.Klaus A., Birchmeier W. Wnt signalling and its impact on development and cancer. Nat. Rev. Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 39.Shackleton M., et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 40.Chen M.S., et al. Wnt/beta-catenin mediates radiation resistance of Sca1+ progenitors in an immortalized mammary gland cell line. J. Cell Sci. 2007;120:468–477. doi: 10.1242/jcs.03348. [DOI] [PubMed] [Google Scholar]
- 41.Dontu G., et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Youn B.S., et al. Large-scale expansion of mammary epithelial stem cell aggregates in suspension bioreactors. Biotechnol Prog. 2005;21:984–993. doi: 10.1021/bp050059f. [DOI] [PubMed] [Google Scholar]
- 43.Li Y., et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15853–15858. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhodes D.R., et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van de Vijver M.J., et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 46.Ivshina A.V., et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 47.Pawitan Y., et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Micalizzi D.S., et al. The Six1 homeoprotein induces human mammary carcinoma cells to undergo epithelial to mesenchymal transition and metastasis in mice through increasing TGF-β signaling. J. Clin. Invest. 2009;119:2678–2690. doi: 10.1172/JCI37815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White D.E., Cardiff R.D., Dedhar S., Muller W.J. Mammary epithelial-specific expression of the integrin-linked kinase (ILK) results in the induction of mammary gland hyperplasias and tumors in transgenic mice. Oncogene. 2001;20:7064–7072. doi: 10.1038/sj.onc.1204910. [DOI] [PubMed] [Google Scholar]
- 50.Schlosser G., et al. Eya1 and Six1 promote neurogenesis in the cranial placodes in a SoxB1-dependent fashion. Dev. Biol. 2008;320:199–214. doi: 10.1016/j.ydbio.2008.05.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niwa H., Miyazaki J., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 52.Chambers I., et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/S0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 53.Mitsui K., et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/S0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 54.Gidekel S., Pizov G., Bergman Y., Pikarsky E. Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer Cell. 2003;4:361–370. doi: 10.1016/S1535-6108(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 55.Zou D., et al. Eya1 gene dosage critically affects the development of sensory epithelia in the mammalian inner ear. Hum. Mol. Genet. 2008;17:3340–3356. doi: 10.1093/hmg/ddn229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Self M., et al. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J. 2006;25:5214–5228. doi: 10.1038/sj.emboj.7601381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miyoshi K., et al. Activation of different Wnt/beta-catenin signaling components in mammary epithelium induces transdifferentiation and the formation of pilar tumors. Oncogene. 2002;21:5548–5556. doi: 10.1038/sj.onc.1205686. [DOI] [PubMed] [Google Scholar]
- 58.Varmus H. Regulation of HIV and HTLV gene expression. Genes Dev. 1988;2:1055–1062. doi: 10.1101/gad.2.9.1055. [DOI] [PubMed] [Google Scholar]
- 59.Lane T.F., Leder P. Wnt-10b directs hypermorphic development and transformation in mammary glands of male and female mice. Oncogene. 1997;15:2133–2144. doi: 10.1038/sj.onc.1201593. [DOI] [PubMed] [Google Scholar]
- 60.Imbert A., Eelkema R., Jordan S., Feiner H., Cowin P. Delta N89 beta-catenin induces precocious development, differentiation, and neoplasia in mammary gland. J. Cell Biol. 2001;153:555–568. doi: 10.1083/jcb.153.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spoelstra N.S., et al. The transcription factor ZEB1 is aberrantly expressed in aggressive uterine cancers. Cancer Res. 2006;66:3893–3902. doi: 10.1158/0008-5472.CAN-05-2881. [DOI] [PubMed] [Google Scholar]
- 62.Welm A.L., Kim S., Welm B.E., Bishop J.M. MET and MYC cooperate in mammary tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4324–4329. doi: 10.1073/pnas.0500470102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moraes R.C., et al. Constitutive activation of smoothened (SMO) in mammary glands of transgenic mice leads to increased proliferation, altered differentiation and ductal dysplasia. Development. 2007;134:1231–1242. doi: 10.1242/dev.02797. [DOI] [PubMed] [Google Scholar]
- 64.Stingl J., et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 65.Sanchez-Carbayo M., Socci N.D., Lozano J., Saint F., Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J. Clin. Oncol. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 66.Sun L., et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 67.French P.J., et al. Gene expression profiles associated with treatment response in oligodendrogliomas. Cancer Res. 2005;65:11335–11344. doi: 10.1158/0008-5472.CAN-05-1886. [DOI] [PubMed] [Google Scholar]
- 68.Graudens E., et al. Deciphering cellular states of innate tumor drug responses. Genome Biol. 2006;7:R19. doi: 10.1186/gb-2006-7-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chung C.H., et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5:489–500. doi: 10.1016/S1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- 70.Stearman R.S., et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am. J. Pathol. 2005;167:1763–1775. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beer D.G., et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 72.Bhattacharjee A., et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Powell C.A., et al. Gene expression in lung adenocarcinomas of smokers and nonsmokers. Am. J. Respir. Cell. Mol. Biol. 2003;29:157–162. doi: 10.1165/rcmb.2002-0183RC. [DOI] [PubMed] [Google Scholar]
- 74.Wachi S., Yoneda K., Wu R. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics. 2005;21:4205–4208. doi: 10.1093/bioinformatics/bti688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adib T.R., et al. Predicting biomarkers for ovarian cancer using gene-expression microarrays. Br. J. Cancer. 2004;90:686–692. doi: 10.1038/sj.bjc.6601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ishikawa M., et al. Experimental trial for diagnosis of pancreatic ductal carcinoma based on gene expression profiles of pancreatic ductal cells. Cancer Sci. 2005;96:387–393. doi: 10.1111/j.1349-7006.2005.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vanaja D.K., Cheville J.C., Iturria S.J., Young C.Y. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res. 2003;63:3877–3882. [PubMed] [Google Scholar]
- 78.Yu Y.P., et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J. Clin. Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 79.Cutcliffe C., et al. Clear cell sarcoma of the kidney: up-regulation of neural markers with activation of the sonic hedgehog and Akt pathways. Clin. Cancer Res. 2005;11:7986–7994. doi: 10.1158/1078-0432.CCR-05-1354. [DOI] [PubMed] [Google Scholar]
- 80.Boer J.M., et al. Identification and classification of differentially expressed genes in renal cell carcinoma by expression profiling on a global human 31,500-element cDNA array. Genome Res. 2001;11:1861–1870. doi: 10.1101/gr.184501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lenburg M.E., et al. Previously unidentified changes in renal cell carcinoma gene expression identified by parametric analysis of microarray data. BMC Cancer. 2003;3:31. doi: 10.1186/1471-2407-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.