Abstract
Inappropriate activation of developmental pathways is a well-recognized tumor-promoting mechanism. Here we show that overexpression of the homeoprotein Six1, normally a developmentally restricted transcriptional regulator, increases TGF-β signaling in human breast cancer cells and induces an epithelial-mesenchymal transition (EMT) that is in part dependent on its ability to increase TGF-β signaling. TGF-β signaling and EMT have been implicated in metastatic dissemination of carcinoma. Accordingly, we used spontaneous and experimental metastasis mouse models to demonstrate that Six1 overexpression promotes breast cancer metastasis. In addition, we show that, like its induction of EMT, Six1-induced experimental metastasis is dependent on its ability to activate TGF-β signaling. Importantly, in human breast cancers Six1 correlated with nuclear Smad3 and thus increased TGF-β signaling. Further, breast cancer patients whose tumors overexpressed Six1 had a shortened time to relapse and metastasis and an overall decrease in survival. Finally, we show that the effects of Six1 on tumor progression likely extend beyond breast cancer, since its overexpression correlated with adverse outcomes in numerous other cancers including brain, cervical, prostate, colon, kidney, and liver. Our findings indicate that Six1, acting through TGF-β signaling and EMT, is a powerful and global promoter of cancer metastasis.
Introduction
Misexpression of embryonic transcription factors has been linked to cancer development and progression. Inappropriate expression of these transcription factors is thought to reinstitute developmental programs out of context, contributing to tumor formation and progression (1). Consistent with this hypothesis, the Six1 homeoprotein plays a critical role in the development of numerous organs (2, 3) and shows little to no expression in most non-neoplastic adult tissues (4), yet is overexpressed in a number of neoplasms where it increases cell proliferation (5, 6) and survival (7). In addition, Six1 overexpression correlates with advanced disease in breast (4) and ovarian cancer (7), hepatocellular carcinoma (8), and rhabdomyosarcoma (RMS) (6). Based on the observation that Six1 is overexpressed in 50% of primary breast cancers and 90% of metastatic lesions (9), we hypothesized that misexpression of Six1 in breast cancer activates cellular pathways that directly contribute to tumor progression and/or metastatic spread.
The TGF-β pathway has been implicated in breast cancer progression and metastasis (10). TGF-β is a ubiquitously expressed cytokine with a dual role in carcinogenesis. In early lesions, TGF-β is an important tumor suppressor preventing uncontrolled cell proliferation (11). However, many advanced tumors are resistant to the growth-inhibitory actions of TGF-β, and TGF-β can instead activate pro-metastatic pathways (12). Recent identification of a TGF-β response signature (TβRS) that correlates with breast cancer metastasis (13) has reinforced the role of TGF-β in promoting malignant breast cancer. One mechanism by which TGF-β contributes to cancer progression is the induction of an oncogenic epithelial-mesenchymal transition (EMT) (14). Epithelial cells undergoing EMT detach from neighboring cells, invade through the basement membrane, and move into surrounding stroma, which is characteristic of the developmental EMT that occurs during diverse morphogenic processes (15). The newly acquired phenotype of tumor cells that have undergone an EMT is thought to contribute to tumor cell spread and metastasis (16).
Metastasis is a multi-step process whereby tumor cells acquire properties that permit passage from the primary tumor into surrounding tissue, intravasate into local blood vessels, extravasate at a secondary site, and survive in a distant location (17). Early steps in the metastatic cascade, particularly migration away from the primary tumor and into the vasculature and lymphatics, have been likened to EMT, and multiple EMT-inducing proteins including the developmental transcription factors Goosecoid, Twist, and FoxC1 contribute to tumor progression (18). However, EMT-inducing proteins have also been implicated in the later stages of metastasis including extravasation of tumor cells from the vasculature (19–21). Interestingly, Six1 expression is important for developmental events in which epithelial plasticity is observed, including delamination of myogenic progenitor cells from the dermomyotome (22) and invasion of the ureteric bud into the metanephric mesenchyme during early renal morphogenesis (2). Together, these data suggest that Six1 overexpression in human breast cancer patients may contribute to cancer progression through induction of an oncogenic EMT.
In this study we identify increased TGF-β signaling in Six1-overexpressing mammary carcinoma cells. We then show that mammary epithelial cells overexpressing Six1 undergo EMT in a manner that is dependent on TGF-β signaling. Because both TGF-β signaling and EMT are associated with metastasis, we examine and show that Six1 overexpression promotes breast cancer–associated metastasis in both an orthotopic and an experimental mouse metastasis model. Furthermore, we demonstrate that the Six1-mediated increase in experimental metastasis, similar to the EMT, is also dependent on TGF-β signaling. In addition, the interaction between Six1 and TGF-β signaling is conserved in human breast cancer, in which we show that Six1 and nuclear Smad3 protein are co-expressed in human breast tumors. Importantly, we find that Six1 overexpression in breast cancer patients correlates with shortened time to metastasis and relapse and with shortened overall survival. Finally, we demonstrate that Six overexpression correlates with adverse outcomes in numerous additional types of cancer, including brain, cervical, prostate, colon, kidney, and liver. Together, these data strongly argue that Six1 is a critical mediator of breast cancer metastasis through its ability to induce a TGF-β–dependent EMT.
Results
Six1 expression activates TGF-β signaling in mammary epithelial cells.
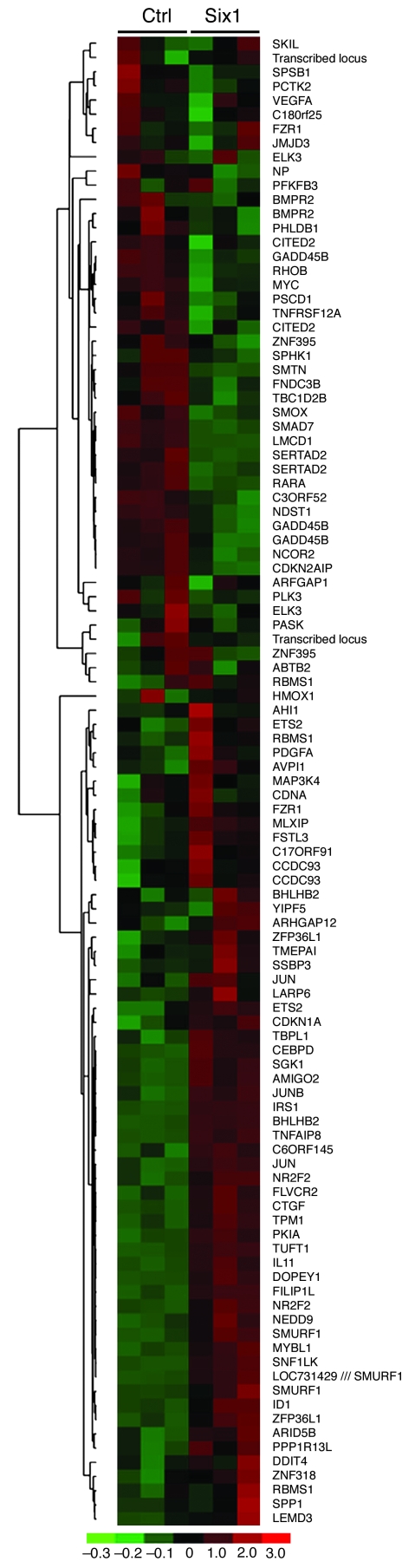
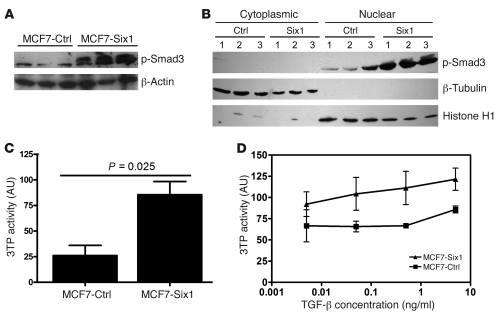
Because Six1 is expressed in a higher percentage of metastatic lesions compared with primary breast cancers, we attempted to identify pathways regulated by Six1 that might be involved in tumor progression. To this end, we performed microarray analysis of MCF7 cells overexpressing Six1 (MCF7-Six1 cells) or control transfected MCF7 cells (MCF7-Ctrl cells) (4). We chose MCF7 cells because they are a tumorigenic breast cancer cell line with low metastatic potential and because they express lower endogenous and, in our Six1-overexpressing cells, exogenous levels of Six1 compared with the endogenous levels of Six1 observed in other breast cancer cell lines (data not shown). Interestingly, we noted that a number of TGF-β target genes were upregulated in response to Six1 overexpression, including IL-11, CTGF, and p21. Recently, a TβRS has been described (13). We thus analyzed our microarray data for the genes described in the TβRS (Figure 1). Strikingly, most of the genes identified in the TβRS were differentially regulated between the control and Six1-expressing cells, which strongly suggests that TGF-β signaling is altered in response to Six1 expression. To confirm that TGF-β signaling was altered in MCF7-Six1 cells, we analyzed the levels of phosphorylation of Smad3, a downstream effector of the TGF-β pathway. Western blot analysis revealed increased levels of phosphorylated Smad3 (p-Smad3; Figure 2A) and increased nuclear accumulation of p-Smad3 (Figure 2B) consistent with activation of the TGF-β pathway. In addition, total levels of Smad3 were also slightly increased in MCF7-Six1 cells (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI37815DS1). To determine whether TGF-β–dependent transcription is activated in the MCF7-Six1 cells, we analyzed expression of a TGF-β–responsive 3TP-luciferase reporter in MCF7-Six1 versus MCF7-Ctrl cells. 3TP-luciferase activity was increased in MCF7-Six1 cells under both normal growth conditions (Figure 2C) and when treated with TGF-β under low-serum conditions (Figure 2D). Together, these data demonstrate that Six1 overexpression increases TGF-β signaling and Smad-mediated transcription.
Figure 1. Six1 expression leads to differential regulation of genes comprising the TβRS.
The TβRS gene list was filtered for probesets with “present” calls in more than 50% of the microarrays, then clustered using hierarchical clustering. The color scale represents the expression level of a gene above (red), below (green), or at (black) the mean expression level of that gene across all samples. Ctrl, control.
Figure 2. Six1 induces increased TGF-β signaling.
(A and B) MCF7-Six1 cells displayed increased levels (A) and nuclear localized (B) p-Smad3. Western blot was performed on whole cell lysate or nuclear and cytoplasmic fractions using antibodies against p-Smad3, β-actin, β-tubulin (cytoplasmic marker), and histone H1 (nuclear marker). (C and D) MCF7-Six1 cells displayed increased TGF-β–responsive 3TP-luciferase reporter activity at baseline (C) and after treatment with the indicated concentrations of TGF-β for 24 hours under serum-free conditions (D). Values are normalized to renilla luciferase. Data are presented as the mean ± SD of 3 individual clones. P values represent statistical analysis using a paired t test.
Six1 expression in human mammary epithelial cells induces properties of EMT.
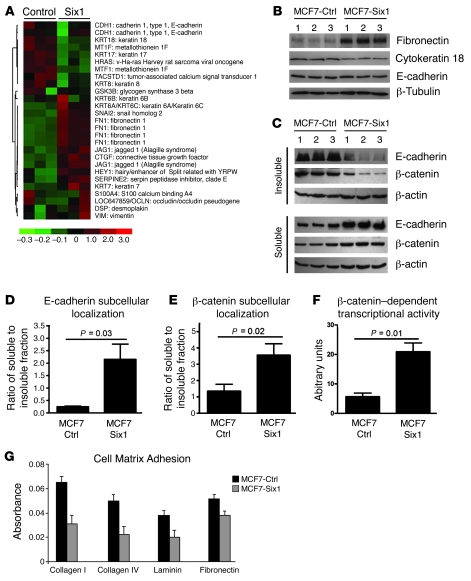
In epithelial cells, the TGF-β pathway is known to be a potent inducer of EMT (23). Because TGF-β signaling was activated in the MCF7-Six1 cells, we hypothesized that MCF7-Six1 cells would exhibit a more mesenchymal phenotype compared with the MCF7-Ctrl cells. To determine whether Six1 overexpression induces a mesenchymal phenotype, we analyzed our microarray data for molecular features of EMT including the downregulation of epithelial markers and the upregulation of mesenchymal markers. We generated a list of 56 genes, based on published evidence for their role in EMT (24–26). Twenty-six probesets passed our filtering criterion and were analyzed by hierarchical clustering based on their gene expression. Indeed, an EMT signature was observed in MCF7-Six1 cells when compared with control cells (Figure 3A). Epithelial-associated genes including keratin 8, 17, and 18 were decreased, while mesenchymal-associated genes including fibronectin, Serpine2, Slug, Hey1, Jag1, and CTGF were increased. These data strongly suggest that the MCF7-Six1 clones display molecular features of EMT.
Figure 3. Six1 induces features of EMT in MCF7 mammary carcinoma cells.
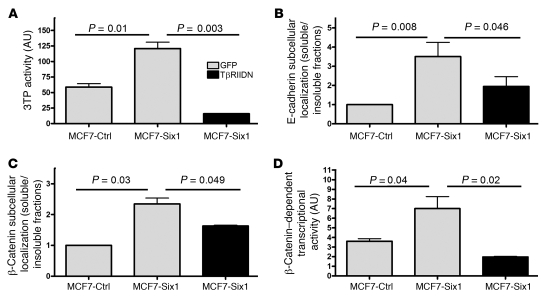
(A) MCF7-Six1 cells displayed differential regulation of EMT-associated genes. Gene expression from MCF7-Six1 and MCF7-Ctrl clones was analyzed by microarray analysis performed in duplicate for each clone. A gene list generated by a priori search of EMT-associated genes was filtered for probesets with a “present” call in at least 50% of the microarrays, then clustered using hierarchical clustering. The color scale represents the expression level of a gene above (red), below (green), or at (black) the mean expression level of that gene across all samples. (B) MCF7-Six1 clones displayed increased fibronectin, decreased cytokeratin 18, and no change in total E-cadherin, as determined by Western blot of whole cell lysates. (C) MCF7-Six1 cells showed a shift of E-cadherin and β-catenin from the insoluble (cytoskeleton-associated) fraction to the soluble (cytosolic) fraction. Fractions were analyzed by Western blot using antibodies against E-cadherin, β-catenin, and β-actin. (D and E) Quantification of Western blot of subcellular localization. (F) MCF7-Six1 cells had increased activity of the β-catenin–responsive luciferase reporter TOP-flash. Values were normalized to renilla luciferase activity. (G) Six1 expression decreased cell adhesion to matrix proteins in MCF7 cells. The relative number of adhering cells was quantified by crystal violet staining. For D–G, data are presented as mean ± SD of 3 individual clones. P values represent statistical analysis using a paired t test.
Using protein immunoblotting, we confirmed that MCF7-Six1 lines contained increased levels of the mesenchymal protein fibronectin and decreased levels of the epithelial proteins cytokeratin 18, claudin-1, and claudin-8 (Figure 3B and Supplemental Figure 2), consistent with a Six1-induced EMT. Similar analysis showed that total E-cadherin protein levels were not dramatically different between the MCF7-Six1 and MCF7-Ctrl lines (Figure 3B). Because an alteration in membranous E-cadherin is an important element of EMT, we further analyzed the subcellular localization of E-cadherin. Immunofluorescence staining of MCF7 cell lines showed a more diffuse staining of E-cadherin in the Six1-expressing cells compared with the highly membranous staining in the control lines (Supplemental Figure 3A). These data suggest that Six1 overexpression in MCF7 cells leads to a redistribution of E-cadherin from the membrane to the cytoplasm. Staining for β-catenin, which exists in adherens junctions along with E-cadherin, showed a similar redistribution from the membrane to the cytoplasm (Supplemental Figure 3B), representing a functional downregulation of E-cadherin and β-catenin in adherens junctions. Further, cellular fractionation demonstrated an approximately 9-fold and 3.5-fold increase in the ratio of soluble (cytoplasmic) to insoluble (cytoskeletal-associated) E-cadherin and β-catenin, respectively, in the MCF7-Six1 cells compared with the MCF7-Ctrl cells (Figure 3, C–E). Because cytoplasmic relocalization of β-catenin results in an increased pool of the protein that is able to move to the nucleus and stimulate transcription, we examined the transcriptional activation of the β-catenin reporter, TOP-flash, in Six1-overexpressing cells. As expected, a 4-fold increase in β-catenin–dependent transcriptional activity was observed in MCF7-Six1 cells compared with MCF7-Ctrl cells (Figure 3F). Six1 expression in MCF7 cells also reduced adhesion to several cell matrix proteins (Figure 3G). Together, these data demonstrate that Six1 can induce key cellular changes of EMT in cancerous mammary epithelial cells.
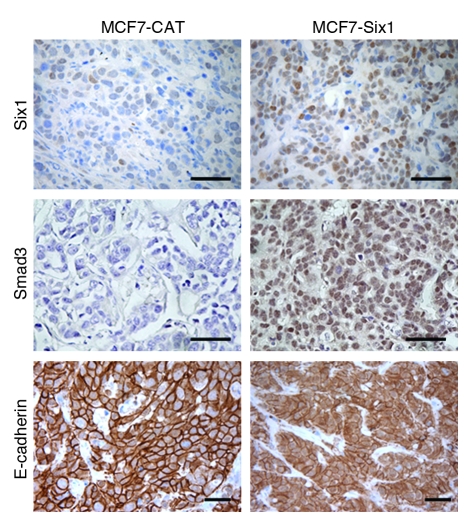
Consistent with our in vitro observations, when these cells were injected into the mammary fat pad of nude mice, the resulting tumors showed a dramatic increase in nuclear Smad3 in the Six1-expressing tumors (Figure 4). Interestingly, while the MCF7-Six1 cells in vitro showed a slight increase in total Smad3 levels, the MCF7-Six1 tumors had dramatically increased levels of total Smad3, which suggests that Six1 may regulate Smad3 levels, particularly in an in vivo context, and that this may, at least in part, contribute to Six1-induced upregulation of TGF-β signaling. Additionally, we observed more cytoplasmic (relative to membranous), staining of E-cadherin in the Six1-expressing tumors compared with the control tumors (Figure 4), indicating that increased TGF-β signaling and properties of EMT manifest in Six1-overexpressing cells in vivo and therefore may contribute to the malignant potential of these cells.
Figure 4. Six1 activates TGF-β signaling and induces EMT in vivo.
Paraffin-embedded tissue sections from MCF7 tumors in nude mice were immunostained for Six1, E-cadherin, and Smad3. Six1 induced redistribution of E-cadherin from a membranous pattern in the control tumors to a more cytoplasmic pattern, and also increased Smad3 nuclear localization. Original magnification, ×400. Scale bar: 100 μm.
To confirm that Six1 overexpression can induce TGF-β signaling and EMT more generally in mammary epithelial cells, we analyzed the effect of Six1 expression in the immortalized but nontransformed mammary epithelial cell line MCF12A. Consistent with our observations in the MCF7-Six1 cells, the MCF12A-Six1 cells had increased p-Smad3, which indicated an increase in TGF-β signaling (Supplemental Figure 4A). Additionally, MCF12A-Six1 cells demonstrated an even more profound EMT than MCF7-Six1 cells, consistent with previously published results suggesting that EMT is difficult to induce in MCF7 cells (27). MCF12A-Six1 cells lost their epithelial morphology and took on a scattered, elongated appearance when compared with the MCF12A-Ctrl cells (Supplemental Figure 4B) and downregulated numerous epithelial markers while upregulating the mesenchymal marker N-cadherin (Supplemental Figure 4C). In addition, MCF12A-Six1 cells exhibited reduced cell-matrix adhesion and increased cellular invasion (Supplemental Figure 4, D and E). Together, these data demonstrate that Six1 overexpression stimulates both TGF-β signaling and EMT in nontransformed mammary epithelial cells as well as in mammary carcinoma cells.
To investigate whether Six1 is more generally associated with the mesenchymal phenotype in numerous breast cancer cell lines, we analyzed its expression in publicly available microarray gene expression datasets obtained from human breast cancer cell lines that were categorized into subgroups including basal A, basal B, or luminal (28). We found that the basal B subgroup of breast cancer cell lines, which is closely associated with cells that are highly invasive and display a mesenchymal phenotype, was significantly enriched for high levels of Six1 expression (P = 0.03), with 64% of the breast cancer lines displaying Six1 expression levels above the mean Six1 level across all cell lines. This is in contrast to the small percentage of basal A and luminal cancer cell lines (25% and 29%, respectively), that expressed Six1 levels above the mean. Together, these data demonstrate that Six1 is preferentially expressed in breast cancer cell lines displaying a more mesenchymal phenotype.
TGF-β signaling is required for properties of Six1-induced EMT.
To test whether TGF-β signaling is necessary for Six1-induced EMT, we expressed a dominant-negative TGF-β type II receptor (TβRIIDN) in MCF7-Six1 cells. TβRIIDN lacks its cytoplasmic tail and therefore cannot activate the TGF-β type I receptor and downstream signaling. As expected, TβRIIDN decreased TGF-β signaling as measured by 3TP-luciferase activity (Figure 5A). Importantly, inhibition of TGF-β signaling in MCF7-Six1 cells reversed the E-cadherin and β-catenin mislocalization (Figure 5, B and C). Additionally, expression of TβRIIDN reversed the Six1-induced increase in β-catenin–dependent transcriptional activity (Figure 5D). These data demonstrate that TGF-β signaling is necessary for elements of the Six1-induced EMT. Because TβRIIDN brought TGF-β signaling down to levels below those seen in control cells, we performed similar experiments using the TβRI kinase inhibitor SB-431542 as a second means to inhibit TGF-β signaling. Treatment of MCF7-Six1 cells with 3 μM SB-431542 decreased their 3TP-luciferase activity to a level comparable to the MCF7-Ctrl cells (Supplemental Figure 5A). Analysis of the β-catenin transcriptional activity (Supplemental Figure 5B) confirmed that increased TGF-β signaling is required for the increased β-catenin transcriptional activity associated with Six1-mediated EMT. Interestingly, neither TβRIIDN nor SB-431542 significantly reversed the ability of Six1 to decrease adhesion to cell matrix proteins, increase fibronectin, or decrease cytokeratin 18 (data not shown) suggesting either that, while TGF-β signaling is important for the effect of Six1 on cell-cell junctions, other pathways activated in response to Six1 are involved in cell matrix adhesion and regulation of epithelial and mesenchymal markers, or that these properties, once induced, are not reversible. Overall, activation of the TGF-β pathway is essential to maintaining some of the elements of epithelial plasticity induced by Six1.
Figure 5. Increased TGF-β signaling is necessary for elements of Six1-induced EMT.
(A) TβRIIDN inhibited TGF-β signaling, as assessed by the TGF-β–responsive promoter 3TP after normalizing to renilla luciferase. (B and C) Inhibition of TGF-β signaling with TβRIIDN reversed E-cadherin and β-catenin relocalization as assessed by fractionation. Graphs represent quantification of Western blots for the ratio of E-cadherin or β-catenin in the soluble versus the insoluble fractions. Data are presented as mean value ± SEM in at least 3 independent experiments. (D) Inhibition of TGF-β signaling with TβRIIDN reverses β-catenin–dependent transcription, as assessed by a β-catenin–responsive reporter (TOP-flash) after normalizing to renilla luciferase. Data are presented as mean value ± SD. P values represent statistical analysis using a paired t test.
Overexpression of Six1 promotes breast cancer associated metastasis.
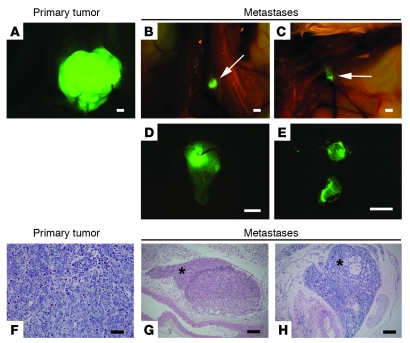
Based on the recognized role of both TGF-β signaling and EMT in tumor progression and on the previously reported correlation between Six1 overexpression and advanced disease in human tumors, we hypothesized that Six1 may also mediate metastasis, in part through its ability to induce an oncogenic EMT. Oncogenic EMT has been linked, in particular, to enabling early steps in the metastatic cascade, including dissemination from the primary tumor, invasion through the tumor stroma, and intravasation into the local vasculature (16). More recently however, oncogenic EMT has also been linked to later stages of metastasis, such as extravasation from the vasculature (20). To determine whether Six1 is causally involved in breast cancer metastasis, we used the tumorigenic but poorly aggressive human breast cancer cell line MCF7, in which Six1 induced TGF-β signaling and EMT. MCF7 cells provide an ideal model, as numerous studies have shown that, while tumorigenic, these cells metastasize infrequently in hormonally intact nude mice (29). A recent study by Horwitz and colleagues (30) showed that MCF7 cells metastasize when injected into ovariectomized mice supplemented with estrogens. Therefore, experiments were performed in hormonally intact mice under conditions in which MCF7 cells metastasize infrequently. The MCF7-Six1 and MCF7-Ctrl cells were fluorescently tagged to enhance our ability to visualize metastases, particularly micrometastases, in vivo. Two clonal isolates of each of the lines were injected into the mammary fat pad of the 4th mammary gland of NOD/Scid mice (Supplemental Figure 6). All mice were sacrificed once their tumors had reached a uniform size of approximately 2 cm3, in order to minimize differences in tumor burden that might affect metastasis (the mean time to reach 2 cm3 was 69 days for MCF7-Ctrl tumors and 81 days for MCF7-Six1 tumors). In contrast to previous tumorigenicity assays, in which MCF7-Six1 tumors grew faster than MCF7-Ctrl tumors after subcutaneous injection into the flank of nude mice (5), in the orthotopic model the MCF7-Six1 tumors grew slightly more slowly than the MCF7-Ctrl tumors, suggesting that the microenvironment of the mammary gland likely affects tumor growth. Nevertheless, in this model, 63% (15 of 24) of mice injected with the MCF7-Six1 cell lines developed metastases, while only 28% (7 of 25) of the MCF7-Ctrl cell lines metastasized, indicating that Six1 overexpression significantly increased metastatic spread (Figure 6, A–H; P = 0.04). Lymphatic metastases (Figure 6, G and H) were observed in multiple locations, but were most frequently found in the lumbar (Figure 6, B and D) and axillary lymph nodes (Figure 6, C and E). The increased metastatic capability of the MCF7-Six1 cell lines was confirmed in orthotopic xenograft injections in the less severely immunocompromised nude mice. Importantly, in this model, no metastases were observed in 19 nude mice injected orthotopically with MCF7-Ctrl clones. In contrast, metastases were observed in 40% (8 of 20) of nude mice injected orthotopically with MCF7-Six1 cells (Supplemental Figure 7, A–F; P = 0.003). In the nude mice, Six1-overexpressing tumor cells were also seen to invade lymphatics (Supplemental Figure 7B) and to metastasize to distant lymph nodes (Supplemental Figure 7, C and D) and on a rare occasion, to metastasize to the bone (Supplemental Figure 7E). Thus, these data provide direct experimental evidence that Six1 overexpression in breast cancer cells promotes metastasis and further demonstrate that Six1 overexpression leads to metastasis to sites relevant to human breast cancer.
Figure 6. Six1 overexpression promotes metastasis in an orthotopic xenograft model.
(A–H) NOD/Scid mice injected orthotopically with MCF7-Six1 cells exhibited distant metastases to lymph nodes, as detected by whole body imaging of ZsGreen fluorescence (A–E) and confirmed in a subset of mice by histology (H&E stain; F–H). (A) Representative ZsGreen fluorescent image of primary tumor. (B and C) Fluorescent image of tumor cells within lumbar (B, arrow) and axillary (C, arrow) lymph nodes. (D and E) Fluorescent image of lumbar (D) and axillary (E) lymph nodes upon dissection (original magnification, ×5). (F) H&E staining of primary tumor showing a poorly differentiated carcinoma (original magnification, ×400). (G) Tumor deposits within distant lymph node (asterisk indicates tumor cells in subcapsular sinus; original magnification, ×100). (H) Large axillary metastasis (asterisk), consistent with lymph node replaced by tumor (original magnification, ×100). Scale bars: 2 mm (A–C), 1 mm (D and E), 100 μm (F), 200 μm (G and H).
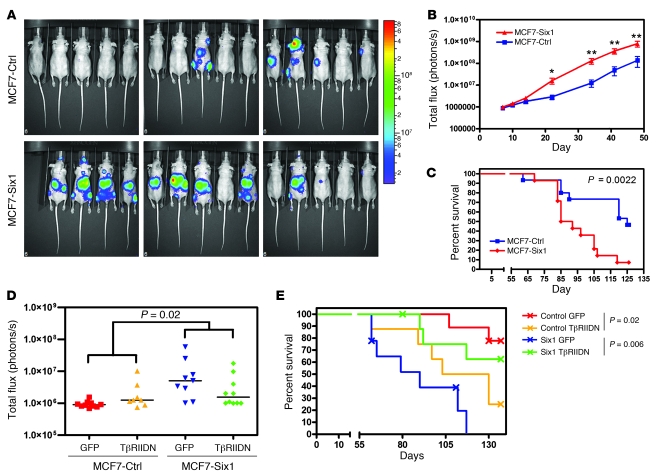
Recently numerous EMT-inducing genes including Ras (31), Zeb1 (20), Ilei (32), and Tgfb (13, 33) have been implicated in the later stages of metastasis including extravasation, suggesting that EMT may not only be required for the early steps of metastasis. Indeed, recent data strongly suggest that EMT is in fact necessary for vascular extravasation (20). Because Six1 induces an EMT that is dependent on TGF-β signaling, and because TGF-β signaling can regulate metastasis via its ability to induce extravasation of tumor cells from the vasculature (13, 21), we hypothesized that Six1 may also contribute to the later stages of metastasis. However, our orthotopic xenograft model produced primarily lymphatic metastases and almost no distant metastasis, thus making it difficult to assess whether Six1 can affect metastatic dissemination via its ability to influence exit from the bloodstream. We therefore used an experimental metastasis model to test whether Six1 is specifically critical to later stages of metastasis in which TGF-β signaling and EMT may be involved. The MCF7-Six1 and MCF7-Ctrl cell lines were tagged with firefly luciferase to permit tracking of these cells in vivo and then injected directly into the left ventricle and the arterial bloodstream of nude mice. Importantly, mice injected with MCF7-Six1 cells had a significantly increased metastatic burden beginning 22 days after injection, as measured by total body bioluminescence, exhibiting an average bioluminescent signal at 34 days after injection that was more than 10-fold above that of mice injected with MCF7-Ctrl cells (Figure 7, A and B). In addition, the mice injected with MCF7-Six1 cells had an overall shortened survival compared with mice injected with MCF7-Ctrl cells (Figure 7C), demonstrating that Six1 not only induces lymphatic metastases from the orthotopic site but also contributes to the later stages of metastasis once the tumor cells have entered the bloodstream.
Figure 7. Six1 increases metastatic burden in an experimental metastasis model dependent on TGF-β signaling.
(A) Bioluminescent imaging of nude mice 48 days after intracardiac injection of either MCF7-Ctrl or MCF7-Six1 cells into the left ventricle. The luminescence signal is represented by the overlaid false-color image, with intensity of the signal indicated by the scale. The anesthetized mice were imaged 10 minutes after intraperitoneal injection of D-luciferin using the IVIS200 (Caliper LS). (B) Quantification of the total body luminescent signal (in photons/second) for each group at the indicated days after injection, presented as the mean ± SEM. *P < 0.05; **P < 0.01. (C) Kaplan-Meier curve representing the overall survival of the injected mice. Statistical analysis was performed using the log-rank test. (D) Quantification of luminescent signal (in photons/second) on day 48 after injection, with the cell lines indicated. Data points represent the luminescent signal of individual animals. Horizontal bars represent the median values. Statistical analysis of luminescence data from days 4–62 was performed using linear mixed models with group-by-time interaction and an unstructured variance-covariance matrix. Interaction contrasts across the 4 animal groups were obtained for each of the 5 days after injection. (E) Kaplan-Meier curve representing the overall survival of the injected mice. Statistical analysis was performed using the log-rank test.
TGF-β signaling is necessary for the increase in experimental metastasis induced by Six1.
To test whether TGF-β signaling is necessary for Six1-induced metastasis, we performed intracardiac injections of luciferase-tagged MCF7-Six1 and MCF7-Ctrl cells expressing either a GFP control or TβRIIDN to inhibit TGF-β signaling. We chose to examine the role of TGF-β signaling in Six1-induced metastasis in the experimental metastasis model, as opposed to the orthotopic metastasis model, to avoid any potential confounding effects of inhibiting TGF-β signaling on primary tumor growth, and because TGF-β signaling was previously shown to play a role in tumor cell extravasation (13, 21). To assess the interaction specifically between Six1 and TGF-β signaling in metastasis, we compared the combined effect on metastatic burden of injection with TβRIIDN-expressing MCF7-Ctrl cells and injection of TβRIIDN-expressing MCF7-Six1 cells. Notably, whereas injection of TβRIIDN-expressing MCF7-Ctrl cells slightly increased the metastatic burden compared with the control, TβRIIDN expression in the context of Six1 expression significantly decreased the metastatic burden, demonstrating that TGF-β signaling is necessary for the increased metastasis observed with Six1 overexpression (P = 0.02; Figure 7D). Additionally, inhibition of TGF-β signaling in the MCF7-Six1 cells significantly increased the overall survival of mice compared with the MCF7-Six1 cells expressing the GFP control (Figure 7E). Interestingly, intracardiac injection of TβRIIDN-expressing MCF7-Ctrl cells decreased the survival of these mice compared with the GFP-expressing MCF7-Ctrl cells (P = 0.02), suggesting that TGF-β signaling acts to suppress metastasis in a context where Six1 is not overexpressed, perhaps by inhibiting cell proliferation, a well-known function of TGF-β (11). However in the mice injected with MCF7-Six1 cells, inhibition of TGF-β signaling significantly improved survival compared with the mice injected with the GFP-expressing MCF7-Six1 cells (P = 0.006). Therefore, Six1 overexpression appears to enhance the pro-metastatic functions of TGF-β signaling while overcoming its normal tumor suppressive role. Together these results indicate that TGF-β signaling is necessary for the Six1-induced increase in metastasis in breast cancer cells and further suggest that Six1 may be involved in mediating the switch of TGF-β from tumor suppressive to tumor promotional in late-stage tumors.
Six1 significantly correlates with activated TGF-β signaling in human breast cancer.
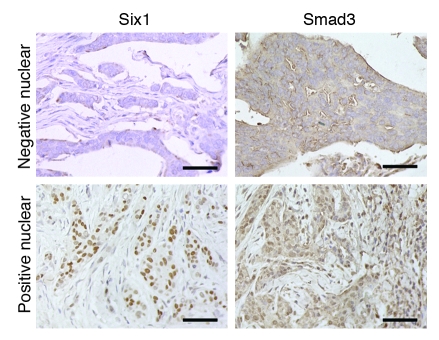
Based on our results suggesting that Six1 can activate TGF-β signaling in mammary carcinoma cell lines, we sought to determine whether these results could be extended to human breast cancer. Using antibodies generated against Six1 and Smad3, we performed immunohistochemical analysis on 58 cases of invasive ductal carcinoma. Indeed, we found a significant correlation between positive nuclear staining with the Six1 antibody and nuclear localized Smad3, an indicator of activated TGF-β signaling (P = 0.005; Figure 8). These results suggest that Six1 may also regulate TGF-β signaling in human breast cancer and reinforce the hypothesis that TGF-β signaling underlies the pro-metastatic activity of Six1 in breast cancer.
Figure 8. Six1 antibody staining correlates with activated TGF-β signaling in human breast cancer.
Immunohistochemical staining of human breast cancer tissue arrays. Representative images show a tumor with little Six1 expression, as measured with the Atlas Six1 antibody, or nuclear Smad3, as measured using the Zymed Smad3 antibody (top row) and a tumor with both Six1 expression and nuclear Smad3 (bottom row). Original magnification, ×400. Scale bars: 100 μm.
Six1 expression in human breast cancer correlates with decreased time to metastasis and overall decreased survival.
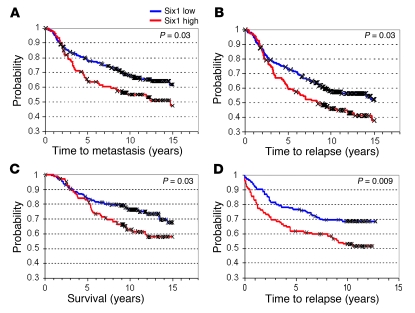
If Six1 is an important contributor to metastatic spread, then we expected that Six1 expression would correlate with advanced disease and poor prognosis in human breast cancer patients. Unfortunately, associated clinical prognostic data were not available for the breast cancer samples that were analyzed using the Six1 and Smad3 antibodies. Thus, to address whether Six1 is associated with adverse outcomes in human breast cancer, we utilized publicly available breast tumor gene expression datasets that were categorized based on clinical outcome. Two datasets were examined: The van de Vijver (34) dataset profiling 295 early-stage (stages I and II) breast carcinomas and the Pawitan/Ivshina (35, 36) dataset profiling 240 invasive breast carcinomas unselected for disease stage. In the van de Vijver dataset, patients whose tumors had high Six1 expression exhibited a significantly shorter time to metastasis (Figure 9A; P = 0.03) and to relapse (Figure 9B; P = 0.03) and had significantly shortened survival due to breast cancer–related deaths (Figure 9C; P = 0.03). We also observed a positive correlation between Six1 expression and the 70-gene “poor prognosis” signature in this dataset (MammaPrint; P < 0.0019). While information with respect to metastasis or to disease-specific survival was not available for the Pawitan/Ivshina dataset, high Six1 expression was more strongly associated with shortened time to relapse in this set of patients (P = 0.009) (Figure 9D), suggesting that the predictive value of Six1 may increase in late-stage disease.
Figure 9. Six1 overexpression correlates with a shortened time to metastasis and relapse and decreased survival.
(A–C) In a study of 295 women with early-stage invasive breast carcinoma (34), high Six1 expression was associated with (A) shortened time to metastasis, (B) shortened time to relapse, and (C) shortened breast cancer–specific survival (survival). (D) In a study of 240 patients diagnosed with invasive breast cancer unselected for disease stage (35, 36), high Six1 expression was strongly associated with shortened time to relapse. In both of these datasets, the median value for Six1 expression was used to divide the samples into high (above median) and low (below median) Six1 expressors. P values were calculated by log-rank analysis.
Further examination of the Pawitan/Ivshina dataset showed that Six1 expression was strongly associated with lymph node positivity (P = 0.001) and larger tumor size (P = 0.002) but was not significantly associated with grade, estrogen receptor status, or age. As a single predictor, Six1 expression was significantly associated with accelerated disease relapse (P = 0.007). However, in a multiple-predictor Cox proportional hazards (Cox PH) model, Six1 status did not improve prediction when combined with lymph node status, grade, tumor size, and age. When paired with any of these parameters individually (i.e., 2 predictor Cox PH models), Six1 was a significant predictor of shortened time to relapse (P = 0.05). Thus, Six1 significantly correlates with adverse outcomes in human breast cancer.
Six1 overexpression in multiple human malignancies correlates with disease progression.
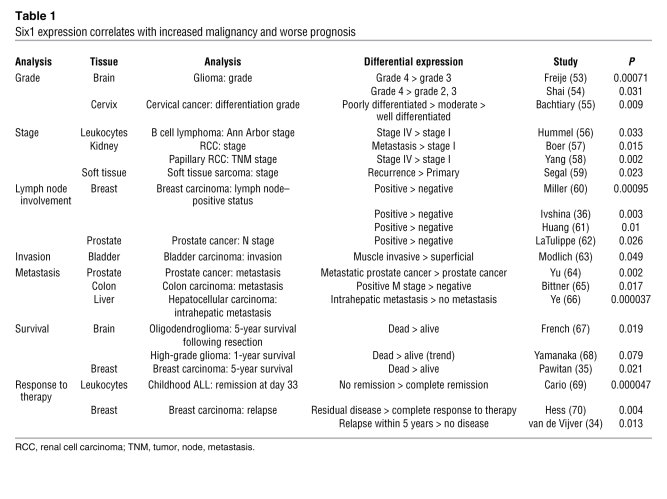
Six1 is expressed in tissues of numerous origins in both development and neoplastic disease. In addition, previous studies have demonstrated that Six1 expression correlates with disease progression in several types of cancer: in ovarian carcinoma and alveolar RMS with increased disease stage, in hepatocellular and ovarian carcinoma with poor survival (6–8), and in breast cancer, where Six1 overexpression occurs in 50% of primary tumors and 90% of metastatic lesions (9). These data in conjunction with our data, which demonstrate a causal role for Six1 in TGF-β signaling, EMT, and breast cancer metastasis, led us to evaluate Six1 expression levels in published microarray datasets that examined parameters of tumor aggressiveness. Six1 expression was significantly associated with increased aggressiveness in 21 of the studies we examined. It was associated with higher-grade malignancy in cervical cancer and glioma, more advanced stage in renal cell carcinoma and B cell lymphoma, lymph node involvement in breast and prostate cancers, invasion in bladder carcinoma, and metastasis in prostate, colon, and hepatocellular carcinomas (Table 1). Higher Six1 expression correlated significantly with poor survival in glioma, oligodendroglioma, and breast carcinoma and with relapse in childhood acute lymphoblastic leukemia (ALL) and breast carcinoma (Table 1). These results clearly demonstrate that increased Six1 expression is associated with aggressive disease in many cancers, implicating the gene as a global regulator of disease progression and metastasis.
Table 1 .
Six1 expression correlates with increased malignancy and worse prognosis
Discussion
Mutation or misexpression of homeobox genes is common in neoplastic disease (1), but a causal role for these genes in tumorigenesis has been a subject of debate. Recently, a role for homeobox genes in tumorigenesis has been strongly supported by the demonstration that several homeobox genes play an active role in breast and other cancers (37). Because homeobox genes regulate developmental programs that coordinate many different cell behaviors important in embryogenesis, it is not surprising that their misexpression in differentiated tissues can result in the acquisition of numerous tumor-promoting properties such as proliferation, migration, invasion, and survival.
The Six1 homeobox gene plays an important role in the development of multiple different tissues by regulating proliferation and survival (2, 3). We have previously shown that Six1 is overexpressed in breast cancer (4), where it stimulates proliferation, causes genomic instability, and transforms immortalized human mammary epithelial cells (38). In addition, we have previously reported that Six1-mediated upregulation of cyclin A1 is critical for the ability of Six1 to increase proliferation of breast cancer cells (5). In the present study, we identified Six1-mediated alterations in the expression of a number of genes included in the TβRS. We further showed that Six1 overexpression in MCF7 cells led to activation of the TGF-β pathway, a previously unreported target pathway of Six1. Together, these data demonstrate that Six1, in addition to upregulating cyclin A1, also increases TGF-β signaling in mammary carcinoma cells.
The TGF-β pathway plays a dual role in tumor progression. Evidence from numerous experimental systems has defined a tumor suppressive role for TGF-β based upon the growth-inhibitory activity of TGF-β in normal cells and early neoplastic lesions (11). However, in later stages of tumorigenesis, resistance to the growth-inhibitory effects permits the unmasking of pro-metastatic functions. In breast cancer patients, the TβRS has been correlated with metastasis (13), suggesting an association between increased TGF-β signaling and advanced disease. Based on its established pro-proliferative activity in mammary carcinoma cells coupled with its activation of TGF-β signaling, Six1 expression has the potential to selectively promote the pro-metastatic activity of TGF-β while antagonizing its growth-inhibitory function. Therefore, in the context of Six1 expression, TGF-β signaling may be a potent contributor to tumor progression.
TGF-β signaling is believed to play a role in breast cancer progression by stimulating metastatic spread, in part through induction of EMT (12). Treatment of numerous cell lines with TGF-β induces EMT, and the effects of TGF-β include the dissolution of adherens junctions and tight junctions, increased migration and invasion, regulation of epithelial and mesenchymal markers, and alteration of cell matrix adhesion (23). In our studies, in addition to increased TGF-β signaling we observed numerous features of EMT in both MCF7 mammary carcinoma cells and in the nontransformed mammary epithelial cell line MCF12A, indicating a general role for Six1 in induction of an EMT in mammary epithelial cell lines. Consistent with these results, analysis of MCF7-Six1–derived tumors displayed both increased cytoplasmic E-cadherin and nuclear Smad3 compared with MCF7-Ctrl tumors, confirming our in vitro observations in vivo. While our study is the first to describe a role for Six1 in oncogenic EMT, a recent Six1/Six4 double-knockout study in mice does suggest a possible role for the Six family proteins in regulating developmental EMT during the delamination of migratory muscle precursor cells from the dermomyotome and subsequent migration to the limb bud (22). In addition, the highly related family member Six2 is critical for maintaining the mesenchymal progenitor cell population in the developing kidney, and loss of Six2 results in premature and ectopic differentiation of mesenchymal cells into epithelia (39). Together, these data implicate the Six family of homeoproteins as critical regulators of EMT in both developmental and oncogenic contexts.
The role of TGF-β signaling in EMT is well established. Our study shows that inhibition of the TGF-β signaling reversed elements of Six1-induced EMT. These data demonstrate that Six1-induced EMT is, at least in part, dependent on TGF-β signaling. Interestingly, inhibition of TGF-β signaling was unable to completely reverse Six1-induced EMT, indicating that Six1 activates additional pathways responsible for inducing elements of EMT, or alternatively that the cells exhibit a more permanent EMT that is no longer dependent on increased TGF-β signaling (31). In support of the involvement of additional EMT-promoting pathways, Six1 has been implicated in the Sonic hedgehog pathway (40), the Notch pathway (41), and the Wnt/β-catenin pathway during normal development (42), all of which can contribute to EMT (19).
Recent evidence has convincingly established a role for EMT in metastatic dissemination and tumor progression (16). The loss of epithelial properties and simultaneous acquisition of mesenchymal properties permits detachment from the neighboring cells, invasion through the underlying basement membrane, movement into the surrounding stroma, and eventual intravasation into local blood or lymphatic vessels, setting the stage for metastatic spread (15). We demonstrate, for what we believe is the first time, in an animal model that Six1 overexpression not only activates TGF-β signaling and induces EMT, but that it also promotes lymph node metastasis and, on rare occasion, bone metastasis, of breast cancer cells from an orthotopic site. This is remarkable, as one of the commonly cited drawbacks of xenograft and other mouse models of cancer is that metastatic cells preferentially colonize the lungs and usually fail to colonize other sites common for human breast cancer, including the lymph nodes, liver, bone, and brain (43). Indeed, considerable effort has been directed toward developing xenograft models that can be used to study bone metastasis, as most human-derived cells lines do not metastasize to the bone when grown orthotopically in mice (44). In addition, many orthotopic xenograft models used to study metastasis debulk the primary tumors to allow more time for distant metastases to develop. The fact that Six1 is sufficient to induce metastasis to lymph nodes and bone within the time span of primary tumor growth suggests that it is a powerful regulator of the metastatic process. However, while the orthotopic model implicates Six1 in the early stages of metastasis, it does not adequately assess whether Six1 contributes to the later stages of metastasis, including extravasation and distant site colonization, stages in which regulators of EMT and TGF-β signaling are involved (13, 19). Therefore, we also tested the role of Six1 in inducing experimental metastasis using an intracardiac injection model. We demonstrate that intracardiac injection of MCF7-Six1 cells significantly increased metastatic burden and decreased overall survival when compared with injection of MCF7-Ctrl cells. Together these results demonstrate that Six1 is not only a major regulator of lymphatic metastasis, but that it also contributes to later stages of metastasis, potentially by increasing survival in the bloodstream, extravasation, or proliferation at a distant site.
Importantly, we demonstrate that TGF-β signaling is a critical mediator of Six1-induced experimental metastasis. Interestingly, in the intracardiac injection model, Six1 regulates the TGF-β response in a manner that is consistent with the notion that Six1 expression antagonizes the tumor-suppressive activity of TGF-β and mediates a switch to its pro-metastatic activity. In mice injected with the MCF7-Ctrl cells, inhibition of TGF-β signaling significantly decreased survival while increasing the metastatic burden, as would be expected if this signaling pathway was acting in a tumor-suppressive manner. In contrast, in the mice injected with Six1-expressing cells, inhibition of TGF-β signaling decreased their metastatic burden and significantly increased their survival, suggesting that in vivo Six1 may be important for promoting the pro-metastatic effects of TGF-β while inhibiting its tumor-suppressive effects. Consistent with this hypothesis, Six1 has been previously shown to increase proliferation of breast cancer cells (5), which would antagonize the well-established growth-inhibitory and tumor-suppressive activity of TGF-β (11). In combination, the ability of Six1 to activate TGF-β signaling while attenuating its growth-inhibiting effects would preferentially activate the pro-metastatic activity of TGF-β, including EMT.
As is the case with many cancers, breast cancer metastasis represents the most deadly stage of tumor progression and is currently the least treatable stage. An understanding of the underlying mechanisms of the metastatic process will provide the opportunity for targeted therapeutics and also for better screening of breast cancer patients most at risk for spread. Our findings that overexpression of Six1 mRNA in human breast cancers is significantly associated with shortened time to metastasis and to relapse, and with decreased survival, reinforces the role of Six1 in human breast cancer progression. These clinical findings, coupled with our experimental and clinical findings that Six1 activates TGF-β signaling, that Six1 and nuclear Smad3 immunoreactivity correlate in human breast tumors, and that Six1 induces EMT and promotes metastatic spread in immunocompromised mice, establish Six1 as a viable candidate for future studies as a potential drug target in breast cancer. Additionally, our results show that the pro-metastatic properties of Six1 may not be confined specifically to breast cancer. On the contrary, in a surprisingly large number of cancers, Six1 expression is significantly associated with disease progression or poor outcome. Thus, we propose that Six1 is a global regulator of tumor metastasis, at least in part due to its ability to increase TGF-β signaling and induce EMT.
Methods
Cell culture.
MCF7-Six1 and MCF7-Ctrl stable transfectants were generated as previously described (4) to overexpress Six1 (MCF-Six1) or the irrelevant protein chloramphenicol transferase (CAT), respectively. In addition to the CAT transfectants, a clone transfected with Six1 that did not express Six1 mRNA or protein was used as a third control line. Each clonal isolate was transduced with the pLNCX2-ZsGreen retrovirus (30) and then selected for stable expression of ZsGreen by FACS. MCF7-Six1 and MCF7-Ctrl clones were transduced with MSCV-GFP or MSCV-TβRIIDN-IRES-GFP retrovirus, selected for stable expression of GFP by FACS, then transduced with MSCV-luciferase-puromycin retrovirus and selected with puromycin for stable luciferase expression. MCF12A-Ctrl and MCF12A-Six1 stable transfectants had been previously generated (38).
In vivo metastasis assays.
For the orthotopic model, 1 × 106 cells in 100 μl of growth factor–reduced Matrigel (BD Biosciences) were injected underneath the nipple of the number 4 mammary fat pad of 6-week-old female nude or NOD/Scid mice. At the same time, these mice were implanted with pellets containing 2 mg 17β-estradiol (Sigma-Aldrich) and 8 mg α-cellulose (Sigma-Aldrich). Mice were sacrificed when the primary tumor reached a volume of 2 cm3 (volume = 0.5 × width2 × length). The Fisher’s exact test was performed to determine statistical significance in the number of mice with metastases from MCF7-Ctrl versus MCF7-Six1 tumors. For the intracardiac model, 1 × 105 cells in 100 μl of PBS were injected into the left ventricle of 5-week-old female nude mice. During the injection, entry into the left ventricle was monitored based on a flash of bright red blood in the hub of the needle and a pulsatile force on the syringe. These mice were also implanted with estrogen pellets as described above. Mice were monitored using the IVIS200 (Caliper LS) imaging systems. The mice were injected with D-luciferin, anesthetized with isoflurane, and imaged 10 minutes after luciferin injection. Quantification was performed using LivingImage version 2.6 software. The mice were monitored over the course of the experiment and euthanized when moribund. The bioluminescence values at days 4, 20, 33, 48, and 62 were compared across the 4 groups of mice (control GFP, control TβRIIDN, Six1 GFP, and Six1 TβRIIDN) using a linear mixed model with group by time interaction and an unstructured variance-covariance matrix. Interaction contrasts of means were obtained for each time point. Analyses were performed using SAS version 9.2 (45). All animal studies were performed according to protocols reviewed and approved by the Institutional Animal Care and Use Committee at the University of Colorado Denver.
MCF7 microarray analysis.
Total RNA was isolated using TRIzol reagent and analyzed using Agilent Bioanalyzer to ensure quality. Microarray analysis was performed using the Affymetrix GeneChip HT-U133A, with the assistance of the Broad Institute at the Massachusetts Institute of Technology. Intensity values were scaled such that the overall fluorescence intensity of each microarray was equivalent. Present calls were determined by MAS5 software analysis. Gene lists were filtered for probesets with present calls in more than 50% of the analyzed microarrays (46). The red, green, and black color scale in Figures 1 and 3 represents the expression level of a gene above, below, or equal to, respectively, the mean expression level for that gene across all samples. Analysis of gene list expression and hierarchical clustering was performed using D-chip software (47).
Histology and immunohistochemistry.
Tumors and multiple organs were fixed in 4% paraformaldehyde, embedded in paraffin, and cut into 5-μm sections. For histologic analysis, sections were stained with H&E or immunohistochemical staining was performed as previously described (30), with primary antibodies against Six1 (1:100; Atlas antibodies), E-cadherin (1:2000; BD Biosciences, Transduction Laboratories), and Smad3 (5 μg/ml; Zymed). Tumor arrays (US Biomax) were stained using Six1 (1:100; Atlas antibodies) and Smad3 (5 μg/ml; Zymed) as previously described (30).
Western blot analysis.
Western blot was performed on whole cell lysates prepared with RIPA buffer as previously described (48) or with nuclear and cytoplasmic extracts (Pierce Biotechnology Inc.). Primary antibodies against E-cadherin, β-catenin, α-catenin, γ-catenin, fibronectin, and N-cadherin were from BD Biosciences Transduction Laboratories; β-actin and β-tubulin were from Sigma-Aldrich; p-Smad3/p-Smad1 were from Cell Signaling Technology; cytokeratin 18 was from Epitomics; and the Six1 antibody was made as previously described (48). For cell fractionation experiments, cell extracts were prepared as previously described (49).
Immunofluorescence.
Cells were grown on glass chamber slides fixed with 4% paraformaldehyde/PBS++ (PBS supplemented with 0.5 mM MgCl2 and 0.9 mM CaCl2), permeabilized with 0.2% Triton X-100/PBS++, and blocked with 10% goat serum in PBS++. Cells were incubated with primary antibodies against E-cadherin (1:200) and β-catenin (1:200) (BD Biosciences, Transduction Laboratories), then incubated with a FITC-conjugated anti-mouse IgG antibody (Sigma-Aldrich) and stained with DAPI.
3TP and TOP-flash luciferase assays.
MCF7 clones were cotransfected with pTOP-flash (50) or 3TP and renilla luciferase construct containing a cryptic promoter using FuGENE 6 (Roche) in triplicate. Clones were treated with 3 μM SB-431542 (Sigma-Aldrich) for 24 hours or various concentrations of TGF-β1 (R&D Systems) for 24 hours. After 48 hours, lysates were prepared in cell culture lysis reagent (Promega) and analyzed with the Dual Luciferase Kit (Promega) on a Monolight 3010 Luminometer (BD Biosciences). Paired t tests were performed to determine statistical significance between MCF7-Ctrl and MCF7-Six1 cells.
Cell adhesion and invasion assays.
We blocked 96-well plates coated with laminin, fibronectin, collagen I, or collagen IV (BD Biosciences, Biocoat) with 1% BSA for 1 hour. Cells were incubated for 1 hour at 37°C. The wells were washed 3 times with PBS, fixed for 10 minutes with ice-cold methanol, and stained with 0.05% crystal violet. The plates were washed and the dye solubilized with 10% glacial acetic acid. Absorbance was determined at 570 nm on a microplate reader (Molecular Devices). Transwell invasion assays were performed using the CytoSelect Cell Invasion Assay (8 μm, fluorometric format) (Cell Biolabs). Then, 104 cells per well in DMEM plus 5% BSA were incubated in the 96-well invasion plates in at 37°C for 48 hours with complete media as a chemoattractant. Cells that invaded through the filter were stained with CyQuant Green according to the manufacturer’s recommendations.
Analysis of clinical outcome data.
Gene expression and clinical outcome data were obtained from 2 independent, publicly available data sets (34–36). Gene expression information for Six1 was obtained for each tumor, and all samples in the data set were mean centered. Samples were then segregated into 2 groups for each analysis: samples in which Six1 expression was above the mean (Six1 “high”), and the remaining samples (Six1 “low”). Each data set was analyzed separately. Clinical outcome information from the Pawitan study (35) was obtained from data published in the Ivshina study (36). Kaplan-Meier survival curves were generated using WinStat for Excel (R. Fitch Software). P values were calculated by log-rank analysis. Cox PH regression was used to assess the contribution of Six1 in multiple predictor statistical models of disease-free and disease-specific survival. Predictors were judged significant at P ≤ 0.05. Analysis was completed with R statistical software (51).
Examination of public microarray datasets.
Microarray data sets of breast cancer cell line gene expression studies (28) were accessed from the NCBI GEO web site. Six1 expression values were obtained and the average Six1 expression over all cell lines was calculated. The number of cell lines exhibiting Six1 expression above the mean Six1 expression value was determined for each subset of breast cancer cell lines. The P value was calculated using Fisher’s exact test, comparing the basal B subtype with all others. mRNA expression microarray data from several cancer studies were analyzed using Oncomine (52). Details of standardized normalization techniques and statistical calculations can be found on the Oncomine web site (https://www.oncomine.com/). First, standard analyses were applied to raw microarray data using either Robust Multichip Average for Affymetrix data or Loess for cDNA arrays. If a reference was used, it was applied at this step (if it was not already performed by the author of the original study). To scale the data and allow comparison of multiple independent studies, Oncomine then applied z score normalization. This included a log2 transformation, setting the array median to 0 and standard deviation to 1. To determine whether Six1 was differentially expressed, 2-sided t tests were conducted using Total Access Statistics 2002 (FMS Inc.).
Supplementary Material
Acknowledgments
This work was funded by grants from the National Cancer Institute (2RO1-CA095277), The American Cancer Society (RSG-07-183-01-DDC), The Susan G. Komen Foundation (BCTR0707562), and the Cancer League of Colorado, to H.L. Ford. D.S. Micalizzi was funded by a predoctoral fellowship from the Department of Defense Breast Cancer Research Program (W81XWH-06-1-0757). K.L. Christensen was funded by a predoctoral fellowship from the National Science Foundation. A.L. Welm was funded by an Era of Hope Scholar Award from the Department of Defense Breast Cancer Research Program (BC075015). We would also like to thank Andrew Thorburn for critical reading of the manuscript.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 119:2678–2690 (2009). doi:10.1172/JCI37815.
Ricardo D. Coletta’s present address is: Department of Oral Diagnosis, School of Dentistry, State University of Campinas, Piracicaba, São Paulo, Brazil.
Karen A. Heichman’s present address is: ARUP Laboratories, Salt Lake City, Utah, USA.
See the related Commentary beginning on page 2528.
References
- 1.Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat. Rev. Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- 2.Xu P.X., et al. Six1 is required for the early organogenesis of mammalian kidney. Development. 2003;130:3085–3094. doi: 10.1242/dev.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laclef C., et al. Altered myogenesis in Six1-deficient mice. Development. 2003;130:2239–2252. doi: 10.1242/dev.00440. [DOI] [PubMed] [Google Scholar]
- 4.Ford H.L., Kabingu E.N., Bump E.A., Mutter G.L., Pardee A.B. Abrogation of the G2 cell cycle checkpoint associated with overexpression of HSIX1: a possible mechanism of breast carcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12608–12613. doi: 10.1073/pnas.95.21.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coletta R.D., et al. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6478–6483. doi: 10.1073/pnas.0401139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu Y., et al. Expression profiling identifies the cytoskeletal organizer ezrin and the developmental homeoprotein Six-1 as key metastatic regulators. Nat. Med. 2004;10:175–181. doi: 10.1038/nm966. [DOI] [PubMed] [Google Scholar]
- 7.Behbakht K., et al. Six1 overexpression in ovarian carcinoma causes resistance to TRAIL-mediated apoptosis and is associated with poor survival. Cancer Res. 2007;67:3036–3042. doi: 10.1158/0008-5472.CAN-06-3755. [DOI] [PubMed] [Google Scholar]
- 8.Ng K.T., et al. Clinicopathological significance of homeoprotein Six1 in hepatocellular carcinoma. Br. J. Cancer. 2006;95:1050–1055. doi: 10.1038/sj.bjc.6603399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reichenberger K.J., Coletta R.D., Schulte A.P., Varella-Garcia M., Ford H.L. Gene amplification is a mechanism of Six1 overexpression in breast cancer. Cancer Res. 2005;65:2668–2675. doi: 10.1158/0008-5472.CAN-04-4286. [DOI] [PubMed] [Google Scholar]
- 10.Serra R., Crowley M.R. TGF-beta in mammary gland development and breast cancer. Breast Dis. 2003;18:61–73. doi: 10.3233/bd-2003-18107. [DOI] [PubMed] [Google Scholar]
- 11.Siegel P.M., Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat. Rev. Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 12.Jakowlew S.B. Transforming growth factor-beta in cancer and metastasis. Cancer Metastasis Rev. 2006;25:435–457. doi: 10.1007/s10555-006-9006-2. [DOI] [PubMed] [Google Scholar]
- 13.Padua D., et al. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133:66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galliher A.J., Neil J.R., Schiemann W.P. Role of transforming growth factor-beta in cancer progression. Future Oncol. 2006;2:743–763. doi: 10.2217/14796694.2.6.743. [DOI] [PubMed] [Google Scholar]
- 15.Larue L., Bellacosa A. Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3ι kinase/AKT pathways. Oncogene. 2005;24:7443–7454. doi: 10.1038/sj.onc.1209091. [DOI] [PubMed] [Google Scholar]
- 16.Thiery J.P. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 17.Steeg P.S. Tumor metastasis: mechanistic insights and clinical challenges. Nat. Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 18.Lee J.M., Dedhar S., Kalluri R., Thompson E.W. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J. Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huber M.A., Kraut N., Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr. Opin. Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Drake J.M., Strohbehn G., Bair T.B., Moreland J.G., Henry M.D. ZEB1 Enhances Transendothelial Migration and Represses the Epithelial Phenotype of Prostate Cancer Cells. Mol. Biol. Cell. 2009;20:2207–2217. doi: 10.1091/mbc.E08-10-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siegel P.M., Shu W., Cardiff R.D., Muller W.J., Massague J. Transforming growth factor beta signaling impairs Neu-induced mammary tumorigenesis while promoting pulmonary metastasis. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8430–8435. doi: 10.1073/pnas.0932636100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grifone R., et al. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development. 2005;132:2235–2249. doi: 10.1242/dev.01773. [DOI] [PubMed] [Google Scholar]
- 23.Zavadil J., Bottinger E.P. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 24.Jechlinger M., et al. Expression profiling of epithelial plasticity in tumor progression. Oncogene. 2003;22:7155–7169. doi: 10.1038/sj.onc.1206887. [DOI] [PubMed] [Google Scholar]
- 25.Huang Y., et al. Epithelial to mesenchymal transition in human breast epithelial cells transformed by 17beta-estradiol. Cancer Res. 2007;67:11147–11157. doi: 10.1158/0008-5472.CAN-07-1371. [DOI] [PubMed] [Google Scholar]
- 26.Mani S.A., et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown K.A., et al. Induction by transforming growth factor-beta1 of epithelial to mesenchymal transition is a rare event in vitro. Breast Cancer Res. 2004;6:R215–R231. doi: 10.1186/bcr778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neve R.M., et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurebayashi J., et al. Quantitative demonstration of spontaneous metastasis by MCF-7 human breast cancer cells cotransfected with fibroblast growth factor 4 and LacZ. Cancer Res. 1993;53:2178–2187. [PubMed] [Google Scholar]
- 30.Harrell J.C., et al. Estrogen receptor positive breast cancer metastasis: altered hormonal sensitivity and tumor aggressiveness in lymphatic vessels and lymph nodes. Cancer Res. 2006;66:9308–9315. doi: 10.1158/0008-5472.CAN-06-1769. [DOI] [PubMed] [Google Scholar]
- 31.Janda E., et al. Ras and TGF[beta] cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J. Cell Biol. 2002;156:299–313. doi: 10.1083/jcb.200109037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waerner T., et al. ILEI: a cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell. 2006;10:227–239. doi: 10.1016/j.ccr.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 33.Oft M., Heider K.H., Beug H. TGFbeta signaling is necessary for carcinoma cell invasiveness and metastasis. Curr. Biol. 1998;8:1243–1252. doi: 10.1016/S0960-9822(07)00533-7. [DOI] [PubMed] [Google Scholar]
- 34.van de Vijver M.J., et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 35.Pawitan Y., et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivshina A.V., et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 37.Samuel S., Naora H. Homeobox gene expression in cancer: insights from developmental regulation and deregulation. Eur. J. Cancer. 2005;41:2428–2437. doi: 10.1016/j.ejca.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 38.Coletta R.D., et al. Six1 overexpression in mammary cells induces genomic instability and is sufficient for malignant transformation. Cancer Res. 2008;68:2204–2213. doi: 10.1158/0008-5472.CAN-07-3141. [DOI] [PubMed] [Google Scholar]
- 39.Self M., et al. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J. 2006;25:5214–5228. doi: 10.1038/sj.emboj.7601381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonnin M.A., et al. Six1 is not involved in limb tendon development, but is expressed in limb connective tissue under Shh regulation. Mech. Dev. 2005;122:573–585. doi: 10.1016/j.mod.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez S., et al. Notch2 is required for maintaining sustentacular cell function in the adult mouse main olfactory epithelium. Dev. Biol. 2008;314:40–58. doi: 10.1016/j.ydbio.2007.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petropoulos H., Skerjanc I.S. Beta-catenin is essential and sufficient for skeletal myogenesis in P19 cells. J. Biol. Chem. 2002;277:15393–15399. doi: 10.1074/jbc.M112141200. [DOI] [PubMed] [Google Scholar]
- 43.Vargo-Gogola T., Rosen J.M. Modelling breast cancer: one size does not fit all. Nat. Rev. Cancer. 2007;7:659–672. doi: 10.1038/nrc2193. [DOI] [PubMed] [Google Scholar]
- 44.Rose A.A., Siegel P.M. Breast cancer-derived factors facilitate osteolytic bone metastasis. Bull. Cancer. 2006;93:931–943. [PubMed] [Google Scholar]
- 45.SAS Institute. 2008. SAS/STAT® 9.2 User’s Guide. SAS Institute Inc. Cary, North Carolina, USA. http://support.sas.com/documentation/onlinedoc/stat/index.html. [Google Scholar]
- 46.McClintick J.N., Edenberg H.J. Effects of filtering by Present call on analysis of microarray experiments. BMC Bioinformatics. 2006;7:49. doi: 10.1186/1471-2105-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li C., Wong W.H. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. U. S. A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ford H.L., et al. Cell cycle-regulated phosphorylation of the human SIX1 homeodomain protein. J. Biol. Chem. 2000;275:22245–22254. doi: 10.1074/jbc.M002446200. [DOI] [PubMed] [Google Scholar]
- 49.Shtutman M., Levina E., Ohouo P., Baig M., Roninson I.B. Cell adhesion molecule L1 disrupts E-cadherin-containing adherens junctions and increases scattering and motility of MCF7 breast carcinoma cells. Cancer Res. 2006;66:11370–11380. doi: 10.1158/0008-5472.CAN-06-2106. [DOI] [PubMed] [Google Scholar]
- 50.Korinek V., et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC–/– colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 51.RDC Team. 2008. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria. http://cran.r-project.org/doc/manuals/refman.pdf. [Google Scholar]
- 52.Rhodes D.R., et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Freije W.A., et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 54.Shai R., et al. Gene expression profiling identifies molecular subtypes of gliomas. Oncogene. 2003;22:4918–4923. doi: 10.1038/sj.onc.1206753. [DOI] [PubMed] [Google Scholar]
- 55.Bachtiary B., et al. Gene expression profiling in cervical cancer: an exploration of intratumor heterogeneity. Clin. Cancer Res. 2006;12:5632–5640. doi: 10.1158/1078-0432.CCR-06-0357. [DOI] [PubMed] [Google Scholar]
- 56.Hummel M., et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N. Engl. J. Med. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- 57.Boer J.M., et al. Identification and classification of differentially expressed genes in renal cell carcinoma by expression profiling on a global human 31,500-element cDNA array. Genome Res. 2001;11:1861–1870. doi: 10.1101/gr.184501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang X.J., et al. A molecular classification of papillary renal cell carcinoma. Cancer Res. 2005;65:5628–5637. doi: 10.1158/0008-5472.CAN-05-0533. [DOI] [PubMed] [Google Scholar]
- 59.Segal N.H., et al. Classification and subtype prediction of adult soft tissue sarcoma by functional genomics. Am. J. Pathol. 2003;163:691–700. doi: 10.1016/S0002-9440(10)63696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller L.D., et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang E., et al. Gene expression predictors of breast cancer outcomes. Lancet. 2003;361:1590–1596. doi: 10.1016/S0140-6736(03)13308-9. [DOI] [PubMed] [Google Scholar]
- 62.LaTulippe E., et al. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- 63.Modlich O., et al. Identifying superficial, muscle-invasive, and metastasizing transitional cell carcinoma of the bladder: use of cDNA array analysis of gene expression profiles. Clin. Cancer Res. 2004;10:3410–3421. doi: 10.1158/1078-0432.CCR-03-0134. [DOI] [PubMed] [Google Scholar]
- 64.Yu Y.P., et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J. Clin. Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 65.Bittner M. A window on the dynamics of biological switches. Nat. Biotechnol. 2005;23:183–184. doi: 10.1038/nbt0205-183. [DOI] [PubMed] [Google Scholar]
- 66.Ye Q.H., et al. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat. Med. 2003;9:416–423. doi: 10.1038/nm843. [DOI] [PubMed] [Google Scholar]
- 67.French P.J., et al. Gene expression profiles associated with treatment response in oligodendrogliomas. Cancer Res. 2005;65:11335–11344. doi: 10.1158/0008-5472.CAN-05-1886. [DOI] [PubMed] [Google Scholar]
- 68.Yamanaka R., et al. Identification of expressed genes characterizing long-term survival in malignant glioma patients. Oncogene. 2006;25:5994–6002. doi: 10.1038/sj.onc.1209585. [DOI] [PubMed] [Google Scholar]
- 69.Cario G., et al. Distinct gene expression profiles determine molecular treatment response in childhood acute lymphoblastic leukemia. Blood. 2005;105:821–826. doi: 10.1182/blood-2004-04-1552. [DOI] [PubMed] [Google Scholar]
- 70.Hess K.R., et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J. Clin. Oncol. 2006;24:4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.