Abstract
Research into the pathogenesis, prevention, and control of infectious and parasitic diseases remains a global priority, as these scourges continue to be a substantial cause of mortality and morbidity. The plethora of molecular tools that are now readily available has facilitated a genome-wide approach to studying the pathogenesis of such diseases, with direct implications for disease prevention and treatment. The articles in this Review Series describe how genome-wide approaches have provided insight into a range of human pathogens, leading to greater understanding of the human diseases that they cause, and highlight some of the challenges that must be overcome if we are to maximize what we learn from the wealth of genomic information now available.
Introduction
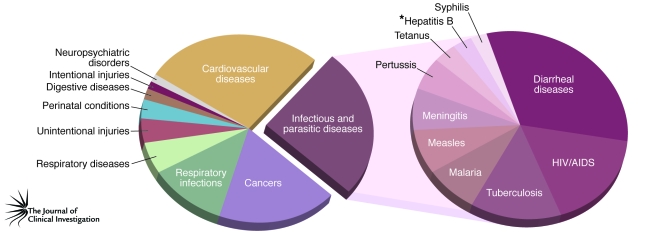
Infectious agents, including bacteria, viruses, fungi, and parasites, are a cause of substantial mortality and morbidity throughout the world. Indeed, the WHO has established that in 2004, the most recent year for which such information is currently available, infectious and parasitic diseases were the second leading cause of death worldwide (Figure 1) (1). They were also the leading cause of burden of disease, as determined by disability-adjusted life years, which are defined as the sum of the years of life lost due to premature mortality and the years of health and productivity lost due to disability (1). Although low-income countries carry the majority of the burden of infectious and parasitic diseases, research into the pathogenesis, prevention, and control of these diseases remains a global priority, with the immense economic benefits of controlling these diseases likely to be felt worldwide. The emergence of drug-resistant strains of bacteria, viruses, and other parasites means that diseases once believed to be under control have reemerged as global health concerns. Examples of this include the emergence of strains of Mycobacterium tuberculosis (the bacterium that causes tuberculosis) that are resistant to the drugs used as first-line treatment and strains of Staphylococcus aureus that are resistant to many commonly used antibiotics such as penicillin, methicillin, and vancomycin. Emerging diseases, in particular those caused by newly identified infectious agents or newly identified strains or forms of an infectious agent, also provide an ongoing global health concern. SARS and a potential influenza A H5N1 pandemic (avian flu) are usually the examples mentioned in relation to this point, but the ongoing influenza A H1N1 pandemic (swine flu) has highlighted how rapidly and unexpectedly such diseases can emerge.
Figure 1. The 2004 worldwide ten leading causes of death and ten leading causes of death from infectious and parasitic diseases.
The global burden of disease: 2004 update, published by the WHO in 2008 (1), provides estimates of mortality and burden of disease by cause for all regions of the world in 2004. The data in this publication, which were used to generate this figure, indicate that infectious and parasitic diseases were the second leading cause of death in the world in 2004, after cardiovascular diseases. Specifically, approximately 17 million and 9.5 million deaths were a result of cardiovascular diseases and infectious and parasitic diseases, respectively. Among those who died of infectious and parasitic diseases, diarrheal diseases were the leading cause of death, closely followed by HIV/AIDS. *These numbers exclude deaths from liver cancer and cirrhosis resulting from chronic HBV infection.
Recent technological advances mean that many new, high-throughput molecular tools are now available to those studying infectious and parasitic diseases at a reasonable price. Among these, genome sequencing and microarray technologies have enabled researchers to take a genome-wide approach to investigate pathogenesis and pathogen-host interactions. This approach has been termed by some “pathogenomics” (2).
The first complete bacterial genome sequence, that of Haemophilus influenzae, was reported in 1995 (3). Since then, the ability to rapidly sequence many millions of nucleotides has enabled researchers to generate a wealth of genomic data, with complete genomes of many eukaryotes and their pathogens (including each major human pathogen) now available. For example, at the time of writing (July 2009), the influenza genome sequencing project had made available through GenBank the complete sequences of 3,733 human and avian influenza isolates (National Institute of Allergy and Infectious Diseases; http://www3.niaid.nih.gov/LabsAndResources/resources/mscs/Influenza/) and more than 880 bacterial genomes had been completed (Genomes OnLine Database, version 2.0; http://www.genomesonline.org/gold.cgi). Although analysis of individual genomes, in particular the first complete genome for a given pathogen, can provide important new information about pathogenesis, many pathogenomic studies involve comparison of multiple strains and/or isolates of a single pathogen, as researchers seek to gain insight into specific disease phenotypes and genotype-phenotype relationships.
The authors of the articles in this Review Series on genomic approaches to infectious disease seek to highlight the advances made in understanding the pathogenesis of a select number of important human pathogens using genomic technologies and indicate how such techniques can lead to greater understanding of human diseases. We hope that these Reviews will highlight the utility and current limitations of the pathogenomics approach.
Using pathogenomics and more
As indicated above, more than 880 bacterial genomes have been completely sequenced (Genomes OnLine Database, version 2.0; http://www.genomesonline.org/gold.cgi). Among these are full genome sequences for 13 strains of group A Streptococcus (GAS), a Gram-positive bacterium responsible for several diseases in humans, ranging from mild conditions, such as pharyngitis, tonsillitis, and impetigo, to the more nefarious, such as toxic shock–like syndrome and necrotizing fasciitis (often known as flesh-eating disease). In the first article in this Review Series (4), James Musser and Samuel Shelburne III discuss how these genome sequences, together with microarray technology, high-throughput proteomics, and enhanced bioinformatics, have been used to provide molecular insight into GAS virulence, clone emergence, and disease specificity.
There are also at least 14 complete genome sequences for S. aureus, another Gram-positive bacterium that is a leading cause of bacterial infections of the bloodstream, lower respiratory tract, and skin and soft tissue in the United States. These infections can give rise to diseases that range from mild conditions, such as impetigo and cellulitis, to those that are life threatening, such as pneumonia, meningitis, toxic shock syndrome, and septicemia. Methicillin-resistant S. aureus (MRSA) is a growing threat worldwide, and an increasing number of cases of MRSA infection occur outside healthcare facilities, in otherwise healthy people, and are known as community-associated MRSA (CA-MRSA) infections. In their Review (5), Frank DeLeo and Henry Chambers focus on the growing threat of CA-MRSA in the United States and highlight how genome-wide approaches are beginning to provide insight into the emergence and virulence of this pathogen. However, they note that more work needs to be done, and their hope is that the complete sequencing of many more S. aureus genomes might help firmly establish how new, more virulent strains emerge.
When the first complete genome sequence of Helicobacter pylori was published in 1997 (6), it was the seventh completely sequenced bacterial genome. There are currently at least 7 complete genome sequences for this Gram-negative bacterium that colonizes the human stomach, causing peptic ulceration, gastric lymphoma, and gastric adenocarcinoma. In the third article in this Review Series (7), John Atherton and Martin Blaser discuss, from a genetic and molecular perspective, how H. pylori has adapted to humans (a species they have colonized for over 50,000 years) and how H. pylori–human interactions shape disease pathogenesis. As a corollary to this, they suggest that the increasing absence of H. pylori from the stomach throughout the life of many individuals might have led to human physiological changes and contributed to recent increases in esophageal adenocarcinoma.
As with bacterial pathogens, complete genomic sequence data has been generated in large amounts for many RNA viruses, probably because their genomes are quite small (approximately 10,000 nucleotides), making the process relatively easy and cheap. In his Review (8), Edward Holmes uses three very different RNA viruses that infect humans — influenza virus, HIV, and dengue virus — as examples to put forward the case that while the abundance of genomic data has taught us much about the evolution and epidemiology of these viruses, it has yet to provide insight into disease pathogenesis, prevention, and control. He argues that, at least in the case of RNA viruses, the potential of genomics has yet to be fully harnessed, because much sequence data is often collected and stored out of context of other key data, including epidemiological, clinical, and immunological data. He proffers the hope that future integration of these variables and increasing use of metagenomics (analysis of all the DNA of all the microbes recovered in an environmental sample) will help pathogenomic studies provide crucial insight into viral disease pathogenesis, prevention, and control.
While complete genome sequences for bacteria and RNA viruses that infect humans have been generated in abundance, genomic approaches to studying parasitic diseases have lagged behind. For example, the complete sequence of the genome of Plasmodium falciparum, the parasite that causes the most deadly form of malaria, was not published until 2002 (9). That same year, a full genome sequence for Anopheles gambiae, a particularly important mosquito vector for the Plasmodium species, was also published (10). As Thomas Wellems, Karen Hayton, and Rick Fairhurst note in the fifth article in this Review Series (11), it is hoped that these genomic advances will provide insight into the molecular processes underlying P. falciparum transmission and infection and new avenues to explore to overcome the difficult challenges of malaria control. However, they also devote substantial discussion to human genetic polymorphisms, such as that responsible for sickle-cell hemoglobin, that have been selected for by the life-threatening complications of infection with P. falciparum.
How human genetics affects the outcome of infection with pathogenic agents is the focus of the Review by Alexandre Alcaïs, Laurent Abel, and Jean-Laurent Casanova (12). As Casanova and colleagues point out, although infectious diseases are thought by many to be solely environmental diseases, variability in susceptibility to and the clinical manifestations of disease among individuals in a population who are infected with the same infectious agent indicates other factors are probably at play. Substantial evidence now indicates that one of these factors is human genetics and that there are numerous forms of genetic susceptibility to infectious disease, from those inherited in a monogenic manner to those inherited in a multigenic fashion. The authors even speculate that “infectious diseases are largely genetically determined, probably more so than most other human diseases” (12).
In the final article in this Review Series (13), Rino Rappuoli, Kate Seib, and their colleagues discuss how genomic approaches can be harnessed for vaccine development. Most vaccines currently in use in humans were developed using conventional culture-based methods. However, the authors argue that the use of large-scale high-throughput genomic analyses to generate vaccines, an approach termed reverse vaccinology, will open up the possibility of developing vaccines for infectious agents that could not be targeted using conventional vaccinology approaches (13). This approach has been used to develop a vaccine that is currently in phase III clinical trials against serogroup B Neisseria meningitidis (MenB), the most common cause of meningococcal disease in the developed world. The use of other technologies, such as transcriptomics, proteomics, and structural vaccinology, to complement the genomic approaches to vaccine development is also highlighted.
Future directions
Despite the brief amount of time since the sequencing of the complete genome of H. influenzae (3), it is already becoming difficult to imagine approaching issues related to infectious diseases without considering the genomic data now available in abundance, and the articles in this Review Series highlight some of the questions that have been answered by such data. However, this is an ongoing process, and the wealth of pathogenomic data has also raised an immense number of new questions, many of which researchers would not even have been able to formulate before the ready availability of high-throughput genomic and microarray technologies. Further technological advancement, such as the recent use of massively parallel sequencing in picoliter-size reaction vessels to sequence the complete diploid genome of a single individual, James D. Watson, (14) is likely to produce even more genomic information in the future, facilitating yet more questions.
How do we move forward? In his Review (8), Holmes suggests that genomic data must be integrated with other relevant variables to provide clues to disease pathogenesis, prevention, and control. Assimilating all relevant information for an individual infectious agent and disease will require enormous cooperation, and it is hoped that readers working in any discipline will be stimulated to contribute.
Footnotes
Conflict of interest: The author has declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 119:2452–2454 (2009). doi:10.1172/JCI40662
References
- 1.WHO. 2008. The global burden of disease: 2004 update. http://www.who.int/healthinfo/global_burden_disease/2004_report_update/en/index.html .
- 2.Guinane C.M., et al. Pathogenomic analysis of the common bovine Staphylococcus aureus clone (ET3): emergence of a virulent subtype with potential risk to public health. J. Infect. Dis. 2008;197:205–213. doi: 10.1086/524689. [DOI] [PubMed] [Google Scholar]
- 3.Fleischmann R.D., et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 4.Musser J.M., Shelburne S.A., III A decade of molecular pathogenomic analysis of group A Streptococcus. . J. Clin. Invest. 2009;119:2455–2463. doi: 10.1172/JCI38095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeLeo F.R., Chambers H.F. Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. . J. Clin. Invest. 2009;119:2464–2474. doi: 10.1172/JCI38226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomb J.F., et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 7.Atherton J.C., Blaser M.J. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. . J. Clin. Invest. 2009;119:2475–2487. doi: 10.1172/JCI38605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holmes E.C. RNA virus genomics: a world of possibilities. J. Clin. Invest. 2009;119:2488–2495. doi: 10.1172/JCI38050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner M.J., et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holt R.A., et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 11.Wellems T.E., Hayton K., Fairhurst R.M. The impact of malaria parasitism: from corpuscles to communities. J. Clin. Invest. 2009;119:2496–2505. doi: 10.1172/JCI38307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alcaïs A., Abel L., Casanova J.-L. Human genetics of infectious diseases: between proof of principle and paradigm. J. Clin. Invest. 2009;119:2506–2514. doi: 10.1172/JCI38111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rinaudo C.D., Telford J.L., Rappuoli R., Seib K.L. Vaccinology in the genome era. J. Clin. Invest. 2009;119:2515–2525. doi: 10.1172/JCI38330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wheeler D.A., et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]