Abstract
The cellular prion protein (PrP) is a highly conserved, widely expressed, glycosylphosphatidylinositol-anchored (GPI-anchored) cell surface glycoprotein. Since its discovery, most studies on PrP have focused on its role in neurodegenerative prion diseases, whereas its function outside the nervous system remains unclear. Here, we report that human pancreatic ductal adenocarcinoma (PDAC) cell lines expressed PrP. However, the PrP was neither glycosylated nor GPI-anchored, existing as pro-PrP and retaining its GPI anchor peptide signal sequence (GPI-PSS). We also showed that the PrP GPI-PSS has a filamin A–binding (FLNa-binding) motif and interacted with FLNa, an actin-associated protein that integrates cell mechanics and signaling. Binding of pro-PrP to FLNa disrupted cytoskeletal organization. Inhibition of PrP expression by shRNA in the PDAC cell lines altered the cytoskeleton and expression of multiple signaling proteins; it also reduced cellular proliferation and invasiveness in vitro as well as tumor growth in vivo. A subgroup of human patients with pancreatic cancer was found to have tumors that expressed pro-PrP. Most importantly, PrP expression in tumors correlated with a marked decrease in patient survival. We propose that binding of pro-PrP to FLNa perturbs FLNa function, thus contributing to the aggressiveness of PDAC. Prevention of this interaction could provide an attractive target for therapeutic intervention in human PDAC.
Introduction
Although the normal prion protein (PrP) is a glycosylphosphatidylinositol-anchored (GPI-anchored) and ubiquitously expressed glycoprotein, and, when altered, is the causative agent of spongiform encephalopathy, its normal function remains mysterious (1, 2). The synthesis, processing, and transit of PrP to the cell surface are complex and not completely understood (3). Normally, PrP is present in lipid rafts and can function as a signaling molecule (4, 5). PrP has many binding partners, such as glycosaminoglycans, copper, laminin receptor, N-CAM, heat shock proteins, dystroglycan, stress-inducible protein, selectin, and glypican 1 (6–14). PrP also binds Grb2, an adapter protein, lipids, and nucleic acids (15–17). PrP plays a role in apoptosis in a cell context–dependent manner (18–22). PrP is involved in the proliferation of epithelial cells and in the distribution of junction associated proteins in human enterocytes in vitro and in intestine in vivo (23). On the other hand, since the PrP-deficient (Prnp–/–) mouse is viable and appears to be normal, the physiologic functions of PrP remain an enigma (24, 25).
Because PrP has been associated with cellular survival, proliferation, and differentiation, aberrant PrP function may also contribute to tumorigenesis. PrP is overexpressed in human gastric cancers (26). An expression microarray study found that PRNP is also overexpressed in human colorectal cancers (27) and is one of the 25 genes that is overexpressed in pancreatic cancer cell lines (28). However, the role PrP plays in tumorigenesis is not clear.
The most common human pancreatic cancer is pancreatic ductal adenocarcinoma (PDAC), the fourth leading cause of cancer deaths in the US. (29). The tumorigenesis of PDAC is complex and not completely understood (30, 31). Evolution of human PDAC correlates with histological changes, characterized by the progression from a flat, columnar epithelium to a papillary, mucinous epithelium with increasing loss of cellular architecture and with nuclear atypia (32, 33). These precursor lesions are commonly referred to as pancreatic intraepithelial neoplasia (PanIN-1, PanIN-2, and PanIN-3) (32, 33).
Substantial progress has been made in identifying molecular mechanisms underlying PDAC development (34–36). The most common genetic lesions found in human PDAC are mutations in KRAS, TP53, DPC4 (SMAD 4), and INK4A, suggesting that these genes are pivotal in the genesis of human PDAC. This interpretation is supported by studies in transgenic mouse models (37, 38). It was found that mutation in Kras in association with additional genetic lesions, such as deletion of Tp53, Ink4a, or Tgfbr2, is sufficient to drive PDAC formation (37, 38). However, other growth factor receptors, signaling molecules, and cell surface receptors have also been implicated in PDAC carcinogenesis (31, 36).
Since PRNP is overexpressed in PDAC cell lines (28), we investigated whether PrP is expressed in a group of 7 human PDAC cell lines, using a panel of well-characterized anti-PrP mAbs (39, 40). We found that all 7 PDAC cell lines expressed PrP. However, the PrP in the PDAC cell lines was neither glycosylated nor GPI anchored. Rather, the PrP exists as a pro-protein retaining its GPI anchor peptide signal sequence (GPI-PSS). Unexpectedly, the GPI-PSS of PrP contains a filamin A–binding (FLNa-binding) motif. FLNa is a cytolinker protein (41, 42). Binding of pro-PrP to FLNa disrupted the cytoskeleton and signaling events in the PDAC cell lines. Furthermore, in human pancreatic cancers, a subgroup of patient tumors expressed PrP, which correlated with markedly decreased survival. We hypothesize that binding of pro-PrP to FLNa confers pancreatic cancer with a growth advantage.
Results
PrP exists as pro-PrP in PDAC cell lines.
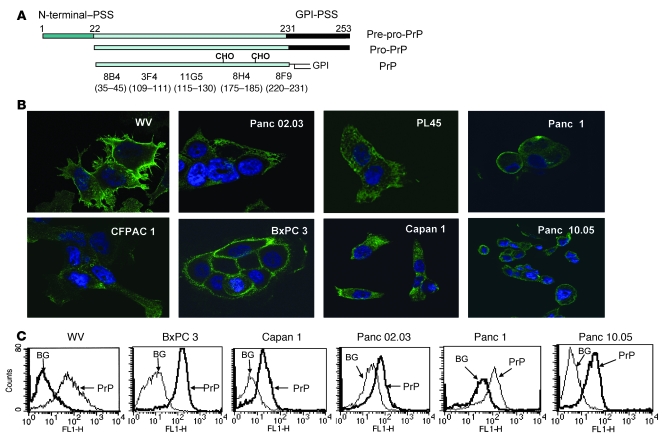
Human PrP is synthesized as a 253–amino acid long pre-pro-PrP (Figure 1A). The N terminus has a leader signal sequence. The C terminus has the GPI-PSS. These sequences are removed in the ER and thus are absent from mature PrP. The protein backbone of mature PrP has a MW of about 23 kDa. Addition of 2 N-linked glycans and a GPI anchor completes the maturation of GPI-anchored PrP.
Figure 1. Expression of PrP in the PDAC cell lines exists as pro-PrP.
(A) A diagram of the processing of GPI-anchored PrP and the epitopes of the mAbs (CHO, N-linked glycans). (B) Confocal microscopic images show that WV cells express PrP on the cell surface. All 7 PDAC cell lines express varying levels of PrP on the cell surface as well as in the cytoplasm. Original magnification, ×1,000. (C) Histograms show the presence of PrP on the cell surface of live PDAC cell lines. BG, background, cells stained with control irrelevant mAb D7C7.
When stained with a well-characterized anti-PrP mAb, 8H4 (39, 40), we found that PrP was expressed in a human neuroblastoma cell line, WV, as well as in a panel of 7 human PDAC cell lines which are as follows: BxPC 3, Panc 02.03, PL45, Capan 1, CFPAC 1, Panc 1, and Panc 10.05 (Figure 1B). While most of the PrP detected in WV cells was on the cell surface, in the human PDAC cell lines, PrP was detected on the cell surface and in the cytoplasm (Figure 1B). The level of PrP varies among PDAC cell lines; BxPC 3 cells appeared to have highest level of PrP on the cell surface. The results of staining of live PDAC cell lines with mAb 8H4 supports this interpretation (Figure 1C).
When immunoblotted with a N terminus–specific anti-PrP mAb 8B4 or a C terminus–specific anti-PrP mAb 8H4 (39, 40), PrP from WV cells migrated as a 33–34-kDa protein due to the addition of the N-linked glycans (Figure 2A). In contrast, PrP from the PDAC cell lines (n = 6) migrated as a 26-kDa protein (Figure 2A). Because PrP from all 6 PDAC cell lines has similar MW, in subsequent studies we concentrated our studies on 3 of the PDAC cell lines: BxPC 3, Panc 02.03, and Capan 1.
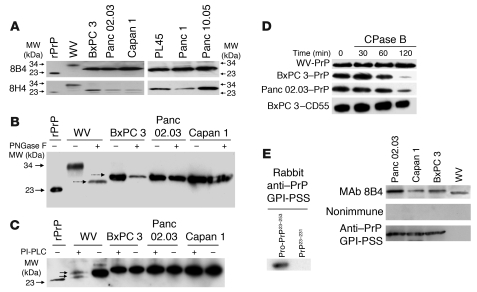
Figure 2. PrP in the PDAC cell lines exists as pro-PrP.
(A) Immunoblots show PrP from WV cells has a MW of 34 kDa, while PrP from the PDAC cell lines has a MW of 26 kDa. A recombinant PrP (rPrP) produced in E. coli is included as a control and MW marker. (B) Immunoblots show treatment of PrP from WV cells with endoglycosidase-F (PNGase F) reduces its MW from 34 kDa to 25.5 kDa. But identical treatment does not change the mobility of PrP from the PDAC cell lines. Deglycosylated PrP from WV cells migrated slightly faster than PrP from the PDAC cell lines (dashed arrows). (C) Immunoblots show PrP from WV cells is sensitive to PI-PLC, as shown by the appearance of a smaller PrP species (bottom arrow) in addition to the PNGase F–treated species (top arrow), but PrP from the PDAC cell lines is resistant to PI-PLC. (D) Immunoblots show that while PrP from the 2 PDAC cell lines is sensitive to CPase B, PrP from WV cells is resistant. CD55 from BxPC 3 cells is also resistant to CPase B. (E) Immunoblots show a rabbit antiserum specific for the PrP GPI-PSS reacts with recombinant pro-PrP (rPro-PrP23–253) but not with recombinant mature PrP (rPrP23–231). The anti–GPI-PSS antiserum also reacts with pro-PrP from the PDAC cell lines but does not react with the PrP from WV cells.
To determine whether PrP in the PDAC cell lines contains N-linked glycans, we treated the cell lysates with endoglycosidase-F (PNGase F) prior to immunoblotting. Deglycosylation reduced the MW of PrP from WV cells from 34 kDa to 25.5 kDa (Figure 2B). Identical treatment did not change the mobility of PrP from the PDAC cell lines. Hence, in the PDAC cell lines PrP is unglycosylated.
Deglycosylated PrP from WV cells migrated slightly faster than PrP from the PDAC cell lines (Figure 2B). We therefore determined whether PrP is GPI anchored in the PDAC cell lines. Affinity-purified, deglycosylated PrP was treated with phosphatidylinositol-specific PLC (PI-PLC) to remove the GPI anchor prior to immunoblotting. After treatment, PrP from WV cells separated into 2 species, 25.5 and 25 kDa (Figure 2C). In the 25-kDa PrP, the GPI anchor has been removed. This species represents 40%–60% of the total PrP in WV cells (n = 3). The 25.5-kDa species is the deglycosylated PrP that is not cleaved by PI-PLC. Some GPI anchors are resistant to PI-PLC, due to the acylation of an inositol hydroxyl group in the anchor (43). Identical treatment did not change the mobility of PrP from the PDAC cells. Thus, PrP in these PDAC cell lines is either not GPI anchored or its GPI anchor is resistant to PI-PLC. This conclusion is consistent with our finding that treatment of live BxPC 3 (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI39542DS1) and Panc 02.03 cells (data not shown) with PI-PLC did not reduce the level of cell surface PrP.
Carboxypeptidase (CPase) removes amino acids from the C termini of proteins (44). GPI-anchored proteins should be resistant to CPase, because their C termini are protected by the lipid anchors. If PrP from the PDAC cell lines lacks a GPI anchor, it should be susceptible to CPase. To test this hypothesis, affinity-purified, deglycosylated PrP from each cell line was treated for different periods of time with CPase B, prior to immunoblotting. As expected, PrP from WV cells was resistant to CPase B (Figure 2D). However, after incubating with CPase B for 2 hours, the levels of PrP from BxPC 3 and Panc 02.03 cells were reduced by 80% (n = 3). By contrast, CD55, another GPI-anchored protein in BxPC 3 cells, was resistant to CPase B. Furthermore, PrP from the PDAC cell lines but not PrP from WV cells was also sensitive to another CPase, CPase Y, which has distinct amino acid preference from CPase B (data not shown).
GPI-anchored proteins are present in lipid rafts (45, 46). Because PrP in the PDAC cell lines lacks a GPI anchor, PrP is no longer detected in lipid rafts in BxPC 3 cells, while flotillin 1, a lipid raft residential protein, still remains in lipid rafts (Supplemental Figure 1B).
Based on the SDS-PAGE mobility of PrP from the PDAC cell lines, we speculate that PrP in the PDAC cells may still have its GPI-PSS. To test this hypothesis, we generated a polyclonal antiserum in rabbits that is specific for the GPI-PSS of PrP. The antiserum reacted with a recombinant pro-PrP23–253, which contains the GPI-PSS, but not with a recombinant mature PrP23–231, which lacks the GPI-PSS (Figure 2E). The antiserum also reacted with affinity-purified PrP from all 3 PDAC cell lines but not with affinity-purified PrP from WV cells (Figure 2E). Pro-PrP is a precursor of mature PrP. The fact that no pro-PrP was detected in WV cells suggests that either the processing of PrP or the removal of the unprocessed pro-PrP is more rapid in WV cells. Collectively, these results provide conclusive evidence that in the PDAC cell lines PrP exists as pro-PrP.
Despite lacking a GPI anchor, some PrP was detected on the surface of PDAC cell lines (Figure 1C). In general, the GPI-PSS contains 15–25 small, hydrophobic amino acids, similar to a typical transmembrane domain. Some cell surface PrP may represent pro-PrP, with its GPI-PSS inserted into the membrane; the GPI-PSS is functioning as a surrogate transmembrane anchor domain, a scenario that has been suggested by others (47). This hypothesis is consistent with our findings that 4 different anti-PrP mAbs, which reacted with epitopes spread along the PrP, reacted with cell surface PrP (Supplemental Figure 2A). Furthermore, while the anti–PrP GPI-PSS antiserum reacted with fixed PDAC cells (Supplemental Figure 2B), it did not react with live PDAC cells (Supplemental Figure 2C). Therefore, on the cell surface the ectodomain of PrP is available to antibody binding but the GPI-PSS is not.
The PrP GPI-PSS binds to FLNa.
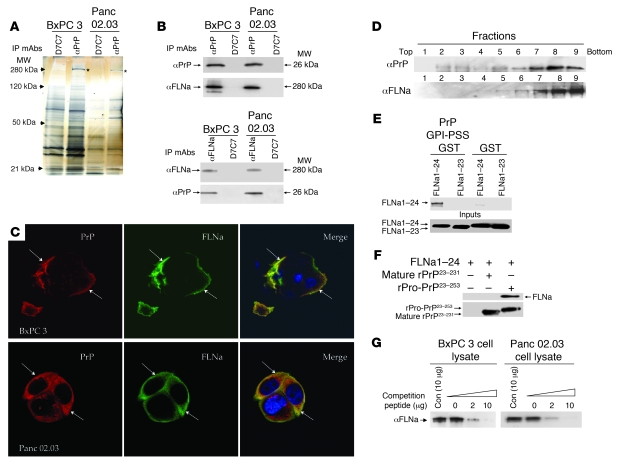
We next sought to identify cellular proteins that interact with PrP in the PDAC cell lines. Coimmunoprecipitation with anti-PrP mAb 8B4 but not an irrelevant mAb D7C7 identified a prominent band with a MW of 280 kDa in BxPC 3 and Panc 02.03 cell lysates (Figure 3A). The protein was sequenced by mass spectrometry and found to be FLNa, an actin-associated protein that integrates cell mechanics and signaling (41, 42).
Figure 3. FLNa binds to the GPI-PSS of pro-PrP.
(A) A silver-stained gel shows a band with MW of 280 kDa (*) is coimmunoprecipitated with anti-PrP mAb 8B4 but not with control mAb D7C7. (B) Immunoblots show the copurification of FLNa with PrP and vice versa. (C) Confocal microscopic images show colocalization of FLNa (green) and PrP (red) in PDAC cell lines. Arrows show area of colocalization. Original magnification, ×1,000. (D) Immunoblots show PrP and FLNa are present in similar fractions after centrifugation in sucrose gradient. (E) An in vitro pull-down experiment shows much stronger binding of full-length FLNa1–24 to a PrP GPI-PSS GST fusion protein than to a GST protein without the GPI-PSS. A FLNa1–23 monomer did not bind PrP GPI-PSS GST fusion protein. Immune complexes were pulled down with GST binding beads and immunoblotted with an anti-FLAG mAb to detect FLNa. (F) Immunoblots show binding of FLNa1–24 to recombinant pro-PrP23–253 but not mature recombinant PrP23–231. Anti-PrP mAb 8H4 was used to pull down the immune complexes. The immunoblot was done either with an anti-Flag mAb or anti-PrP mAb 8H4. (G) Immunoblots show competition of binding of FLNa to pro-PrP by a PrP-GPI-PSS synthetic peptide. Copurification of PrP and FLNa in the PDAC cell lysates was carried out in the presence of different concentrations of either a synthetic peptide corresponding to the GPI-PSS or a control synthetic peptide. Anti-PrP mAb 8B4 coimmunoprecipitated proteins were then immunoblotted with an anti-FLNa mAb. Con, control peptide.
The identity of FLNa was confirmed by immunoblotting of proteins copurified with PrP with a FLNa-specific mAb (Figure 3B). Conversely, immunoblotting of proteins copurified with FLNa with an anti-PrP mAb also revealed the presence of PrP (Figure 3B). Furthermore, PrP and FLNa also partially colocalized in BxPC 3 and Panc 02.03 cells (Figure 3C) and were present in similar fractions in a sucrose gradient (Figure 3D). In WV cells, PrP did not copurify with FLNa, because WV cells do not express FLNa (data not shown).
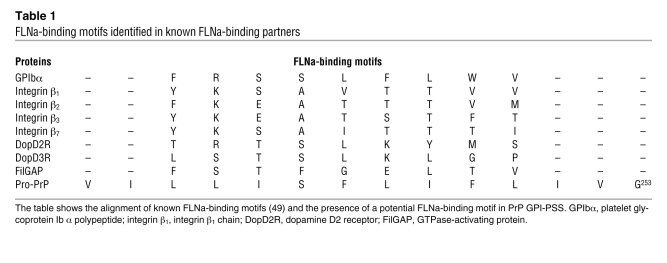
Native FLNa is a homodimer; each subunit contains a spectrin-related F-actin–binding domain, followed by 24 Ig-like domains (41, 42). Each Ig-like domain has about 96 amino acids and has 7 β-sheet strands (A to G). The faces of strands C and D are common binding sites for all FLNa-binding partners for which atomic structures have been resolved (48–50). These FLNa binding partners share a conserved, hydrophobic amino acid motif (49). Interestingly, ClustalW alignment suggests that the GPI-PSS of pro-PrP contains the FLNa-binding motif (Table 1). We thus investigated whether FLNa indeed binds the GPI-PSS of PrP.
Table 1 .
FLNa-binding motifs identified in known FLNa-binding partners
First, in an in vitro pull-down experiment, we found that a full-length FLNa1–24 dimer binds much more PrP GPI-PSS glutathione-S-transferase (GST) fusion protein than control GST protein without the PrP GPI-PSS. On the other hand, a FLNa1–23 monomer, which lacks the last Ig-like dimerization domain, did not bind the PrP GPI-PSS GST fusion protein (Figure 3E). Second, this observation was confirmed by using full-length pro-PrP23–253 and mature PrP23–231, a full length FLNa dimer binds pro-PrP23–253 but not mature PrP23–231 (Figure 3F). Third, these findings were further confirmed in BxPC 3 and Panc 02.03 cells. The levels of FLNa copurified with pro-PrP in these cell lines could be competed with a PrP GPI-PSS synthetic peptide, but not with a control peptide (Figure 3G). Similar results were obtained with Capan 1 cells (data not shown). Together, these experiments provide strong evidence that FLNa binds to the GPI-PSS on pro-PrP.
PrP, but not FLNa, is readily detected in the membrane fraction when PDAC cell lysate was fractionated with a membrane protein extraction reagent kit (data not shown). Thus, PrP but not FLNa is embedded in the membrane. The high concentration of salts and detergent in the extraction buffer has probably prevented the co-fractionation of FLNa and PrP. We next determined whether FLNa, which is present near the inner membrane leaflet (41), interacts with membrane PrP as proposed in Supplemental Figure 3A. We labeled the cell surface of PDAC cell lines with biotin and then immunoprecipitated the biotinylated proteins with avidin-conjugated beads, using the coimmunoprecipitation buffer. Bound proteins were then eluted and immunoblotted with mAbs specific for PrP, FLNa, or Hsp-90. Hsp-90 is a cytosolic protein and is used as a control to determine whether contaminating cytosolic proteins are present in the cell surface protein preparation. It is clear that proteins bound to avidin beads contain PrP and FLNa but not Hsp-90. On the other hand, all 3 proteins were present in the flow-through fraction, which includes cytosolic proteins (Supplemental Figure 3B). In another series of experiments, we showed that PrP but not FLNa was readily biotinylated on the cell surface (data not shown). Collectively, these results suggest that FLNa interacts with cell surface PrP.
Downregulation of PrP alters the distribution of FLNa in PDAC cell lines.
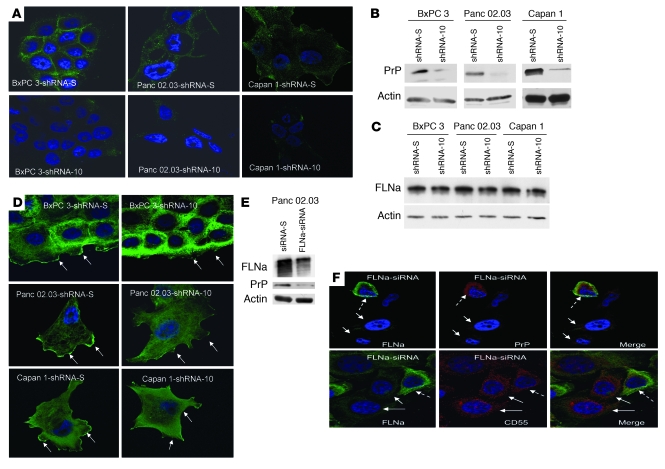
To study the possible consequences of the binding of pro-PrP to FLNa, we used shRNA to reduce PrP expression in the 3 PDAC cell lines. We identified 3 potential PrP-specific shRNA target sequences, and each shRNA was then introduced into BxPC 3, Panc 02.03, and Capan 1 cells to establish stable cell lines (51). As controls, stable cell lines expressing a scrambled shRNA-S were also established. One of the PrP-specific sequences, shRNA-10, inhibited the expression of PrP by more than 90%, as judged by immunofluorescent staining (Figure 4A), immunoblotting (Figure 4B), flow cytometry (Supplemental Figure 4A) as well as by the amount of soluble PrP released by the tumor cells into the culture medium (Supplemental Figure 4B). Two other PrP-specific shRNA sequences, shRNA-2 and shRNA-4, inhibited the expression of PrP in BxPC 3 cells by 50% and 20%, respectively (Supplemental Figure 4, A and B).
Figure 4. Downregulation of PrP or FLNa expression in the PDAC cell lines.
(A) Immunofluorescence staining and confocal microscopic images show the PDAC cell lines with shRNA-10 have reduced levels of PrP. Original magnification, ×1,000. (B) Immunoblots show the PrP-downregulated shRNA-10 cells have reduced levels of PrP. (C) Immunoblots show the level of FLNa does not change in PrP-downregulated cells. (D) Immunofluorescence staining and confocal microscopic images show that knocking down PrP alters the spatial distribution of FLNa. Arrows show staining of membrane ruffles and leading edges. Original magnification, ×1,000. (E) Immunoblots show that when expression of FLNa is inhibited, the expression of PrP is also reduced in Panc 02.03 cells. (F) Immunofluorescence staining and confocal microscopic images show the expression of FLNa modulates PrP but not CD55 expression. In the top left panel, the dashed arrow identifies a cell with FLNa (green); solid arrows identify 2 cells lacking FLNa. In the top center panel, 2 solid arrows identify 2 cells lacking PrP; the dashed arrow identifies 1 cell with PrP (red). The top right panel is the merge of the left and center panels; 2 arrows identify 2 cells lacking both PrP and FLNa, and a dashed arrow identifies 1 cell with both PrP and FLNa. In the bottom panels, 2 FLNa-negative cells (left panel, solid arrows) still express high levels of CD55 (red; center panel, solid arrows), although some cells have both FLNa and CD55 stain (left and center panels, dashed arrows). Original magnification, ×1,000.
Reducing PrP does not alter the expression levels of FLNa.
The levels of FLNa in control cells and cells in which PrP has been downregulated (referred to herein as PrP-downregulated cells) are comparable (Figure 4C). However, reducing PrP expression does alter the spatial distribution of FLNa. In control cells, FLNa is concentrated in areas lining the plasma membrane and in membrane ruffles as well as diffusely in the cytosol (Figure 4D, arrows indicate membrane ruffles). In the 3 PrP-downregulated cell lines, FLNa is greatly reduced in the membrane ruffles and is more concentrated in the cytosol. These results were confirmed in multiple independently established PrP-downregulated cell lines (n > 3) and cell lines with scrambled shRNA-S (n > 3).
Next, we used the approach of cell surface biotinylation, as described in Supplemental Figure 3B, to determine whether the amount of FLNa copurified with biotinylated cell surface protein is reduced in PrP-downregulated cells. It is clear that compared with control cells, the level of FLNa copurified with cell surface protein is markedly reduced in PrP-downregulated cells (Supplemental Figure 3C). Collectively, these results suggest that without PrP, much less FLNa is able to reach the inner membrane leaflet area.
Reducing FLNa expression diminishes the expression of PrP.
We were unable to establish stable FLNa knockdown PDAC cell lines. Therefore, we used siRNA to transiently reduce FLNa expression in Panc 02.03 cells. We achieved 60%–80% (n = 3) reduction in FLNa expression (Figure 4E); in these cells the level of PrP was also reduced (about 50%), as shown by immunoblotting (Figure 4E). As shown by confocal microscopy, cells lacking FLNa also lacked PrP (Figure 4F). On the other hand, cells that expressed FLNa also expressed PrP (Figure 4F). This effect is specific for PrP, because cells lacking FLNa still have detectable CD55 (Figure 4F).
Downregulation of PrP alters the organization of actin filaments and signaling events in PDAC cell lines.
FLNa regulates actin polymerization and signaling (41, 42). Therefore, we next stained control and PrP-downregulated cells for F-actin as an indicator of cytoskeletal organization. We also stained cells with an antibody specific for phosphorylated tyrosine, p-Tyr, as a generic indicator of signaling events.
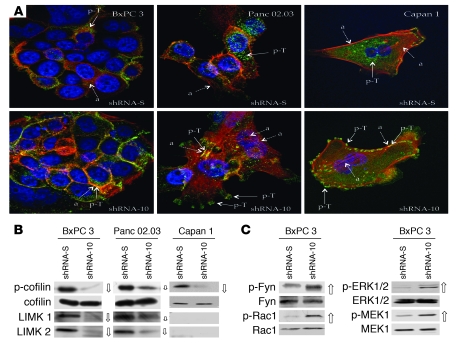
Downregulation of PrP drastically alters the staining patterns of both actin (red) and p-Tyr (green) in all 3 PDAC cell lines (Figure 5A; for individual actin and p-Tyr staining see Supplemental Figures 5 and 6). In control BxPC 3 cells, actin and p-Tyr were mainly in the cytosol and tended not to colocalize. By contrast, in PrP-downregulated BxPC 3 cells, most of the actin and p-Tyr were colocalized in cell-cell contact areas. In control Panc 02.03 cells, actin was detected both in the cell membrane and in the cytosol, whereas p-Tyr was mainly in the cytosol. In PrP-downregulated Panc 02.03 cells, a more complex actin network was seen in the cytosol and in filopodia-like structures. In these cells, much of the p-Tyr was in the plasma membrane and colocalized with actin. Similarly, in control Capan 1 cells, most of the p-Tyr was in the cytosol. In contrast, in PrP-downregulated Capan 1 cells, most of the p-Tyr was in the plasma membrane, in a punctate pattern colocalized with actin. Thus, a reduction in PrP causes actin reorganization and alters signal transduction in all 3 PDAC cell lines, each with distinct phenotypes.
Figure 5. Binding of pro-PrP to FLNa alters actin organization and signaling events.
(A) Immunofluorescence staining and confocal microscopic images show that knocking down PrP modifies the spatial distribution of actin filaments (red) and p-Tyr (green) in 3 PDAC cell lines. (a, actin, shown with dashed arrows; p-T, p-Tyr, shown with solid arrows). Original magnification, ×1,000. (B) Immunoblots of PrP-downregulated BxPC 3 and Panc 02.03 cells show that the levels of p-cofilin, LIMK1, and LIMK2 are markedly reduced (open arrows, the size of the arrow indicates the degree of change) compared with control cells. P-cofilin is also reduced in PrP-downregulated Capan 1 cells. (C) Immunoblots show upregulation of p-Fyn, p-Rac1, p-ERK1/2, and p-MEK1 (open arrows) in PrP-downregulated BxPC 3 cells.
Cofilin regulates actin organization by controlling its polymerization (52). Two kinases, LIMK1 and LIMK2, phosphorylate and inactivate cofilin (53). This kinase activity is counteracted by a family of phosphatases, such as slingshot and chronophin, which dephosphorylate cofilin (54). We next determined whether changes in PrP levels modulate the levels of cofilin and p-cofilin in the PDAC cell lines. We observed that the levels of p-cofilin but not cofilin were reduced by 90%, 50%, and 90% in PrP-downregulated BxPC 3, Panc 02.03, and Capan 1 cells, respectively (Figure 5B). The levels of LIMK1 and LIMK2 were also similarly reduced in PrP-downregulated BxPC 3 and Panc 02.03 cells. However, neither LIMK1 nor LIMK2 was detectable in Capan 1 cells. The levels of slingshot and chronophin were either unchanged or undetectable in these PDAC cell lines (data not shown). Hence, while the decrease in p-cofilin levels in BxPC 3 and Panc 02.03 cells can be explained by a reduction in LIMK1 and LIMK2, the upstream event that regulates p-cofilin in Capan 1 cells is not known.
In addition to cofilin, a large family of Rho-GTPases and kinases is involved in regulating cytoskeletal organization (55, 56). We therefore investigated whether PrP influences the expression of some of the upstream signaling molecules in BxPC 3 cells. We observed that p-Rac1, a Rho-GTPase; p-ERK1/2 and p-MEK1, 2 serine/threonine kinases in the MAPK pathway; and p-Fyn, a Src family tyrosine kinase, are markedly increased in PrP-downregulated cells (Figure 5C). Thus, PrP downregulation affects multiple signaling pathways in BxPC 3 cells.
PrP modulates the proliferation, invasiveness, and growth of PDAC cell lines.
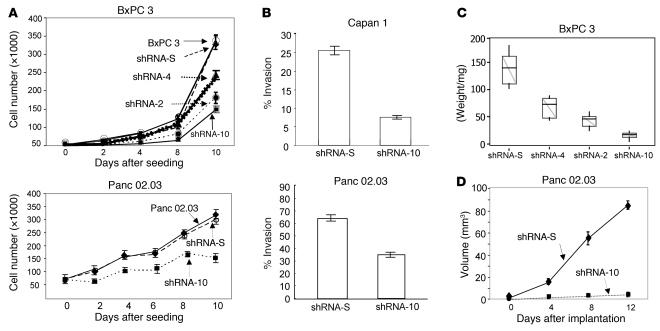
We next investigated the effects of knocking down PrP on PDAC cell behavior. PrP-downregulated BxPC 3–shRNA-10 and Panc 02.03–shRNA-10 cells proliferated more slowly than control cells with scrambled shRNA-S or control cells without any shRNA (Figure 6A). The reduction in cellular proliferation correlates with the levels of PrP expression; BxPC 3–shRNA-10 cells, which expressed the lowest level of PrP, had the slowest proliferation rate, followed by BxPC 3–shRNA-2 cells, and then BxPC 3–shRNA-4 cells. PrP-downregulated Capan 1–shRNA-10 and Panc 02.03–shRNA-10 cells were also less invasive in vitro than control cells (Figure 6B). We then inoculated nude mice with different PrP-downregulated BxPC 3–shRNA cell lines. Similar to that found for in vitro proliferation, BxPC 3–shRNA-10 cells also had the slowest growth rate, followed by BxPC 3–shRNA-2 cells, and then BxPC 3–shRNA-4 cells (Figure 6C). When inoculated into nude mice, the growth of Panc 02.03–shRNA-10 cells was also retarded (Figure 6D).
Figure 6. Downregulation of PrP influences the in vitro and in vivo behavior of the PDAC cell lines.
(A) Proliferation of PrP-downregulated cells is reduced compared with control cells with scrambled shRNA-S or cells without any shRNA. Parental BxPC 3 cells (open circles); BxPC 3–shRNA-S cells (filled diamonds); BxPC 3–shRNA-4 cells (filled triangles); BxPC 3–shRNA-2 cells (filled circles); BxPC 3–shRNA-10 cells (filled squares); parental Panc 02.03 cells (open circles); Panc 02.03–shRNA-S cells (filled diamonds); Panc 02.03–shRNA-10 cells (filled squares). The results presented are the mean of triplicate wells ± SD. (B) In vitro invasiveness of PrP-downregulated Capan 1–shRNA-10 cells and Panc 02.03–shRNA-10 cells in Matrigel is reduced. The results presented are the mean of triplicate wells ± SD. (C) In vivo growth of PrP-downregulated BxPC 3 cells in nude mice depends on the levels of PrP expression (n = 10/group, composite of 2 experiments; n = 5/experiment). The upper whisker extends to the highest value within the upper limit. The lower whisker extends to the lowest value within the lower limit. The top of the box is the third quartile. The bottom of the box is the first quartile. The median of the data is also indicated by the horizontal line. (D) The growth of PrP-downregulated Panc 02.03–shRNA-10 cells in nude mice is inhibited. Panc 02.03–shRNA-S cells (filled diamonds); Panc 02.03–shRNA-10 cells (filled squares). The results presented are the mean of 10 mice/group ± SD (composite of 2 experiments, n = 5/experiment).
Pro-PrP is detected in a subgroup of resectable human PDAC cases and expression is associated with poorer prognosis.
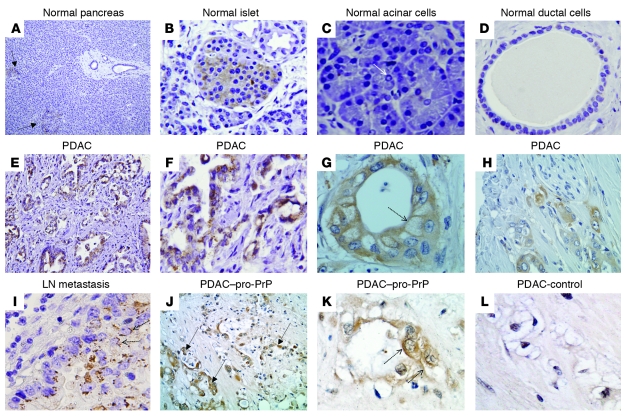
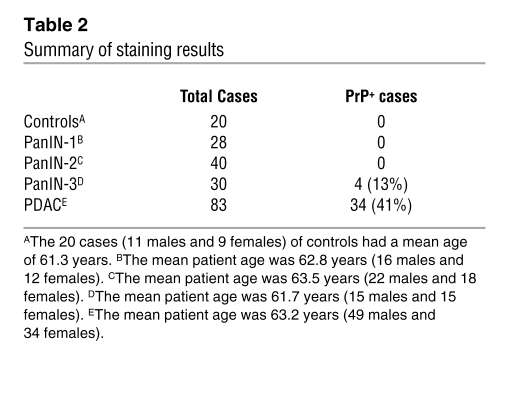
To determine whether our findings in cell models have clinical relevance, we carried out a retrospective study on the expression of PrP in human PDAC biopsies by immunohistochemistry. Tissues from patients with chronic pancreatitis or PanIN lesions served as controls. In normal human pancreas (Figure 7, A–D), only islet cells (Figure 7B) showed moderate PrP staining; neither acinar (Figure 7C) nor ductal epithelial cells (Figure 7D) stained for PrP. PrP was also undetectable in the duct cells in chronic pancreatitis (n = 20), PanIN-1 (n = 28) and PanIN-2 (n = 40) (data not shown). Four of thirty (13.3%) PanIN-3 specimens showed weak staining for PrP (data not shown). Among the 83 resectable PDAC cases, 34 (41%) showed strong staining for PrP (Figure 7) (summarized in Table 2). PrP immunoreactivity was also detected in the corresponding LN metastases (Figure 7I). All PDAC tumor cells reacted strongly with the anti–GPI-PSS antiserum, but the stromal cells surrounding the tumor cells showed only background staining (Figure 7, J and K). The anti–GPI-PSS antiserum also failed to react with normal ducts in the same tissue biopsies (Supplemental Figure 7). Staining of the PDAC with the control antiserum was also negative (Figure 7L). Thus, as in the PDAC cell lines, PrP exists as pro-PrP in human PDAC lesions.
Figure 7. PrP is present in PDAC lesions but not in normal ductal cells.
Immunohistochemical staining shows that in normal pancreas (A–D) only islet cells express PrP. (A) Two arrows identify 2 islets (original magnification, ×100). (B) A PrP-positive islet (original magnification, ×400). (C) Neither acinar cells, an arrow shows a centroacinar cell (original magnification, ×400), nor (D) ductal cells (original magnification, ×400) express PrP. (E–H) In PDAC, malignant ductal cells express PrP (original magnification, ×200 [E]; ×400 [H]). F and G are from 2 additional PDAC patients (original magnification, ×400). (G) The dashed arrow shows immunoreactivity on the cell surface. (I) PDAC lymph node metastases express PrP (original magnification, ×400). (J) PrP in PDAC reacted with the anti–PrP-GPI-PSS antibody; 3 arrows identify tumors (original magnification, ×200). (K) Dashed arrows in K indicate PDAC cell surface immunoreactivity (original magnification, ×400). (L) The control antiserum only has background immunoreactivity (original magnification, ×400).
Table 2 .
Summary of staining results
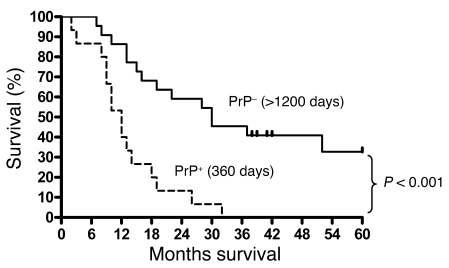
We next investigated whether PrP expression correlates with the clinical outcome in the group of 37 patients who had surgery done between 2001 and 2003. We observed that the expression of PrP is associated with shorter survival (Figure 8). Patients (n = 16) whose tumor showed strong PrP immunoreactivity had a shorter median survival time of 360 days, whereas patients (n = 21) whose tumor did not show PrP immunoreactivity had a mean survival time of more than 1,200 days (P < 0.001). Furthermore, we did not find any other factors, such as age, gender, tumor size, or differentiation, that are clearly associated with prognosis.
Figure 8. Expression of PrP is associated with poorer prognosis.
The 37 patients had surgery done from 2001 to 2003. Patients (n = 16) whose tumors expressed PrP (PrP+) had a median survival time of 360 days. On the other hand, of the 21 patients whose tumors lacked PrP (PrP–), 6 of these patients are still alive as of October of 2008. Four of these patients have already passed 5 years after surgery; 2 others will have passed 5 years in late November of 2008 (2 of the spikes). The other 2 spikes indicate a patient who died 41 months after surgery and a patient who died 52 months after surgery, respectively. This cohort of patients has mean survival time of more than 1,200 days (P < 0.001).
Discussion
Instead of being a GPI-anchored glycoprotein, PrP in human PDAC exists as pro-PrP, which binds to FLNa. This fatal attraction disrupts the normal functions of FLNa and confers a growth advantage to PDAC.
The GPI anchor modification process is complex-, protein-, and cell type–dependent (43, 45). The reason why the GPI-PSS of PrP is not removed in the PDAC cell lines and tumor biopsies is not known. A mutation in the PRNP gene is not the cause; sequencing of PRNP in 6 PDAC cell lines revealed no mutation (data not shown). Furthermore, since CD55 is GPI anchored in the PDAC cell lines, the failure to remove the GPI-PSS is not characteristic of all GPI-anchored proteins.
Interestingly, it has been reported that a normally GPI-anchored protein, carbonic anhydrase IV (CA-IV), exists as pro–CA-IV, retaining its GPI-PSS in normal human pancreatic ductal cells and in Capan 1 cell (57, 58), a cell line we also studied here. Another normally GPI-anchored protein, alkaline phosphatase, remains GPI anchored. Therefore, it appears that in Capan 1 cells a subset of normally GPI-anchored proteins, such as CD55 (see Figures 2 and 4) and alkaline phosphatase (57) are GPI anchored, while the other subset, including CA-IV (57) and PrP (see Figures 2 and 4), are not. It should be noted that in contrast to CA-IV, PrP is not detected in normal human pancreatic ductal cells. Even if some of the GPI-PSS were not removed from other GPI-anchored proteins, such as CA-IV, they would not be expected to disrupt the functions of FLNa. We examined 14 GPI-PSSs from other GPI-anchored proteins, including CA-IV, and found that only the GPI-PSS of PrP has the FLNa-binding motif.
Using an in vitro GPI anchor attachment assay, the GPI-PSS of PrP was found to be intrinsically inefficient (14%) in accepting the GPI anchor compared with 9 GPI-PSSs from other GPI-anchored proteins (ranging from 24% to 60%) (59). Thus, a slight defect in the GPI anchor attachment machinery in PDAC cell lines may have a more dramatic effect on PrP than on other GPI-anchored proteins, which have a more efficiently processed GPI-PSS, such as CD55 (50%) or alkaline phosphatase (30%).
The processing of pre–pro-PrP is tightly regulated by a quality control system (3, 60). Potentially, a disturbance in the protein quality control system in the ER or the proteasome system could also contribute to the accumulation of pro-PrP.
We propose that pro-PrP is present on the cell surface using the GPI-PSS as a surrogate transmembrane domain (Supplemental Figure 3A). There is a precedent for this hypothesis. Qa2 is normally a GPI-anchored cell surface protein; substitution of a single amino acid in the GPI-PSS of Qa2 prevents its GPI anchor modification. Nevertheless, Qa2 is still expressed on the cell surface as an integral membrane protein, using the GPI-PSS as a transmembrane domain (47). The central region of PrP (amino acid 113–128) can also function as a transmembrane domain (61). However, if this pro-PrP isoform does exist on the cell surface, it represents only a small fraction of the total cell surface PrP, because mAbs 3F4 (recognizing amino acid 109–112) and 11G5 (recognizing amino acid 115–130) react with cell surface PrP (Supplemental Figure 2A).
We speculate that binding of pro-PrP to FLNa occurs close to the plasma membrane (Supplemental Figure 3A), because PrP and FLNa colocalize in membrane ruffles, and when PrP is downregulated, FLNa appears to retract from membrane ruffles. Nevertheless, since pro-PrP is also present in the cytosol, pro-PrP and FLNa could also interact in other cellular compartments.
In vitro pull-down experiments show that pro-PrP binds FLNa directly. However, a direct link between pro-PrP and FLNa in the cell has not been established. Potentially, an intermediate molecule may facilitate binding of PrP to FLNa. Caveolin 1 interacts with normal PrP and FLNa (4, 42). However, while both BxPC 3 and Panc 02.03 cells express caveolin 1, pro-PrP and caveolin 1 did not copurify in these cell lines (data not shown).
Binding to pro-PrP is likely to interfere with the ability of FLNa to interact with some of its normal ligands, such as integrins or Smad (41, 42), which are important in cell proliferation, signaling, and migration. Pro-PrP could either directly compete for the binding site on FLNa or relocate FLNa from its normal surroundings, rendering it unavailable for binding its normal ligands. While downregulation of pro-PrP does not alter the expression of FLNa, downregulation of FLNa does reduce the levels of PrP, at least in Panc 02.03 cells. When pro-PrP is bound to FLNa, it may protect pro-PrP from proteolytic degradation.
The actin filament is regulated by a large family of molecules, one of which is cofilin (52–56). Accordingly, the levels of p-cofilin were reduced in PrP-downregulated PDAC cell lines. However, the upstream events that regulate p-cofilin in these cell lines remain unresolved. Because, while the level of p-cofilin is regulated by LIMK1 and LIMK2 in PrP-downregulated BxPC 3 and Panc 02.03 cells, neither LIMK1 nor LIMK2 is detected in Capan 1 cells. Therefore, there is an alternative pathway for regulating cofilin in Capan 1 cells. Downregulation of PrP in BxPC 3 cells also affects other signaling pathways, such as the MAPK pathway, the Rho GTPase pathways, and the GPCR pathway. However, a much more detailed biochemical study will be needed to delineate the pathways that are affected and the mechanisms by which these pathways affect the PDAC cell lines.
Palladin is another cytolinker that regulates actin filament and cell motility (62). Mutation in palladin has been linked to familial pancreatic cancer (63). However, this mutation was not found in other families with familial pancreatic cancer (64, 65). Therefore, the role palladin plays in PDAC remain unresolved.
Four of the PanIN-3 lesions (14%) showed weak PrP immunoreactivity. It is known that some of the PanIN-3 lesions already have carcinomas (33). The incident increases to 41% in resectable PDAC lesions with much stronger immunoreactivity. PDAC lesions also reacted strongly with anti–PrP GPI-PSS antiserum. Therefore, similar to that in the PDAC cell lines, the PrP present in PDAC lesions is pro-PrP.
PrP is not essential for PDAC initiation because only 41% of the PDAC cases have detectable PrP. However, the presence of PrP is associated with poorer clinical outcome, suggesting that PDAC cells with PrP have a growth advantage. This interpretation is consistent with our findings that reducing PrP expression diminishes the proliferation and invasiveness of the PDAC cell lines in vitro and their growth in vivo.
Little is known about the regulation of PrP expression either at the gene or protein level. PRNP has been reported to be upregulated in BxPC 3, Capan 1, and 5 other PDAC cell lines (28). However, other gene profiling studies have not identified PRNP as a contributing factor in human PDAC (66, 67). The reasons for these discrepancies are not known.
It is unlikely that a mutation in KRAS or inactivation of INK4A by itself is responsible for the expression of PrP in PDAC, because more than 90% of PDAC cases have a KRAS mutation and inactivation of INK4A (31, 35); yet only 41% of the resectable PDAC cases have detectable pro-PrP. Other genetic anomalies, such as mutation in TP53 and/or DPC4, which are found in about 50% of PDAC patients (31, 35), must have occurred to facilitate the expression of PrP in PDAC.
We hypothesize that binding of pro-PrP to FLNa is the reason that PrP-positive PDAC cells have a growth advantage. Currently, there is no marker for early diagnosis of human PDAC; detection of pro-PrP could offer such a marker. Furthermore, prevention of the interaction between pro-PrP and FLNa could provide a novel target for therapeutic intervention in PDAC. Finally, it appears that the existence of pro-PrP in pancreatic cancer is not an isolated incident; pro-PrP is also detected in human colon carcinoma cell lines (n = 3) (data not shown) and in hepatocarcinoma cell lines (n = 5) (data not shown).
Methods
Cell lines, mAbs, and reagents.
All 7 PDAC cell lines, BxPC 3, Panc 02.03, Capan 1, PL 45, CFPAC 1, Panc 1, and Panc 10.05, were obtained from ATCC. WV is a human neuroblastoma cell line that was originally generated in the laboratory of R. Petersen of Case Western Reserve University (68). Anti-PrP mAbs 8H4, 11G5, and 8B4 were generated in our laboratory (39, 40). The rabbit anti–PrP GPI-PSS antiserum was generated by immunizing rabbits repeatedly with a synthetic peptide corresponding to the GPI-PSS of pro-PrP (GSSMVLFSSPPVILLISFIFLVG) in CFA. The antiserum was affinity purified. All other mAbs and reagents were purchased from commercial sources and used according to the recommendations of the vendors. Mature PrP, pro-PrP, and PrP GPI-PSS GST fusion proteins were prepared using conventional techniques.
Immunofluorescence staining for confocal microscopy.
Tumor cell lines were cultured in poly-d-lysine–coated glass bottom Petri dishes (MatTek Corporation) overnight. Cells were then rinsed 3 times with ice-cold PBS and fixed in 4% paraformaldehyde for 15 minutes at 20°C. PrP or FLNa was detected with anti-PrP mAb 8H4 or anti-FLNa mAb PM6/317 (0.01 μg/μl). Bound Ab was detected with an Alexa Fluor 488 nm–conjugated (Invitrogen) goat anti-mouse Ig-specific antibody. Nuclei were stained with DAPI. To detect FLNa in PrP-downregulated cells, cells were fixed and then permeabilized with 0.3% Triton X-100 in PBS for 10 minutes at 20°C. The subsequent steps were carried out as described in above. F-actin was detected with Texas Red–conjugated (Invitrogen) phalloidin. Samples were analyzed on a LSM 510 META confocal microscope (Zeiss) at Case Comprehensive Cancer Center, Image Core Facility. All experiments have been repeated twice with comparable results.
Experimental details.
For details regarding immunoprecipitation, immunoblotting, identification of proteins copurified with PrP by mass spectrometry, in vitro pull-down assays, in vivo competition assay, fractionation of lipid raft–associated proteins, establishment of PrP-downregulated and control PDAC cell lines, and siRNA downregulation of FLNa in PDAC cell lines, see the Supplemental Methods.
In vitro proliferation.
An identical number (1 × 104) of cells were cultured in vitro in 24-well plates in triplicate. At different days after culture, the numbers of cells in each well were counted. The results presented were the mean of the triplicate wells ± SD at each time point. These results were confirmed with 3 independently generated control and PrP-downregulated cell lines.
In vitro invasion assay.
In vitro invasion assays were performed in the BioCoat Growth Factor Reduced Matrigel Invasion Chamber (BD Bioscience), using protocols provided by the supplier. The results presented were the mean of the triplicate wells ± SD. These results were confirmed with 3 independently generated control and PrP-downregulated cell lines.
Growth of tumor cells in nude mice.
Tumor cells were grown in vitro to 90% confluence, washed twice in cold PBS buffer, harvested, washed with cold PBS 3 times, counted, and kept on ice prior to injection. Then, 1 × 107 cells in 0.1 ml of PBS were injected subcutaneously into the back of nude mice. In the BxPC 3 experiment, at 21 days after implantation, the tumor mass from each individual mouse was surgically removed and weighed. In the Panc 02.03 experiment, at various times after tumor cell implantation (5 mice/group/tumor cell line), the length and width of the tumors were measured using a digital caliper. The results presented were the mean of the weights of the tumors or the length × width2/2 of the tumor ± SD. These results were confirmed with 3 independently generated control and PrP-downregulated cell lines.
Tissue samples and immunohistochemical staining.
Paraffin-embedded blocks of 83 surgically resected, primary infiltrating PDACs, resected between 2001 and 2006, were collected from the Surgical Pathology Files of University Hospital of Cleveland. Clinical and pathological data were obtained from detailed chart review, which included age, gender, race, tumor size, tumor location, lymph node metastasis status, and histological subtype of the invasive carcinoma. The H&E-stained slides from each case were visually inspected by light microscopy, and representative sections were selected for immunostaining.
Immunohistochemical staining of 5-μm sections was carried out using conventional methods. An isotype control, irrelevant mAb D7C7, and a nonimmune polyclonal rabbit antiserum were included as negative controls. Each slide was coded and evaluated independently by 2 pathologists (W. Xei and A.A. Petrolla.). The cytoplasmic and membrane staining intensity and distribution of each sample were graded as positive (>50% neoplastic cells stained strongly positive), weakly positive (5%–50% neoplastic cells stained weakly), or negative (<5% neoplastic cells stained). The identity of the case was revealed only after a score had been given. Similar results were obtained using 2 different anti-PrP mAbs 8B4 and 8H4. All studies have been approved by the Institutional Review Board for Human Investigation (UH IRB No. 08-05-29) of the University Hospital Case Medical Center, Cleveland, Ohio, USA.
Statistics.
The frequencies of PrP immunostaining among normal pancreas, pancreatic precursor lesions, and cancer samples were analyzed by the χ2 test or Fisher’s exact test to account for frequency values of less than 5. For purposes of statistical analysis, all PrP-positive carcinomas were combined for comparison to PrP-negative specimens. The Kaplan-Meier method was used to determine overall survival with respect to PrP expression. All 37 patients analyzed had surgery done in years from 2001 to 2003. None of these patients had presurgical chemotherapy or radiation therapy. P values of less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported in part by a grant from the NIH (R21CA133559-01 to M.-S. Sy), by Pathology Association of University Hospital of Cleveland Research grant, and startup funds from Department of Pathology of Case Western Reserve University (to W. Xin). We would like to thank Michael Lamm, Thomas P. Stossel, and Robert B. Petersen for suggestions and discussion. M.-S. Sy would like to thank P. Ling for support and editorial help.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 119:2725–2736 (2009). doi:10.1172/JCI39542.
References
- 1.Prusiner S.B. Prions. Proc. Natl. Acad. Sci. U. S. A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brockes J.P. Topics in prion cell biology. Curr. Opin. Neurobiol. 1999;9:571–577. doi: 10.1016/S0959-4388(99)00016-1. [DOI] [PubMed] [Google Scholar]
- 3.Hegde R.S., Rane N.S. Prion protein trafficking and the development of neurodegeneration. Trends Neurosci. . 2003;26:337–339. doi: 10.1016/S0166-2236(03)00143-7. [DOI] [PubMed] [Google Scholar]
- 4.Mouillet-Richard S., et al. Signal transduction through prion protein. Science. 2000;289:1925–1928. doi: 10.1126/science.289.5486.1925. [DOI] [PubMed] [Google Scholar]
- 5.Taylor D.R., Hooper N.M. The prion protein and lipid rafts. Mol. Membr. Biol. 2006;23:89–99. doi: 10.1080/09687860500449994. [DOI] [PubMed] [Google Scholar]
- 6.Caughey B., Brown K., Raymond G.J., Katzenstein G.E., Thresher W. Binding of the protease-sensitive form of PrP (prion protein) to sulfated glycosaminoglycan and congo red (corrected). J. Virol. 1994;68:2135–2141. doi: 10.1128/jvi.68.4.2135-2141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown D.R., et al. The cellular prion protein binds copper in vivo. Nature. . 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 8.Rieger R., Edenhofer F., Lasmezas C.I., Weiss S. The human 37-kDa laminin receptor precursor interacts with the prion protein in eukaryotic cells. Nat. Med. 1997;3:1383–1388. doi: 10.1038/nm1297-1383. [DOI] [PubMed] [Google Scholar]
- 9.Schmitt-Ulms G., et al. Binding of neural cell adhesion molecules (N-CAMs) to the cellular prion protein. J. Mol. Biol. 2001;314:1209–1225. doi: 10.1006/jmbi.2000.5183. [DOI] [PubMed] [Google Scholar]
- 10.Edenhofer F., et al. Prion protein PrPc interacts with molecular chaperones of the Hsp60 family. J. Virol. 1996;70:4724–4728. doi: 10.1128/jvi.70.7.4724-4728.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keshet G.I., Bar-Peled O., Yaffe D., Nudel U., Gabizon R. The cellular prion protein colocalizes with the dystroglycan complex in the brain. J. Neurochem. . 2000;75:1889–1897. doi: 10.1046/j.1471-4159.2000.0751889.x. [DOI] [PubMed] [Google Scholar]
- 12.Zanata S.M., et al. Stress-inducible protein 1 is a cell surface ligand for cellular prion that triggers neuroprotection. EMBO J. 2002;21:3307–3316. doi: 10.1093/emboj/cdf325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C., et al. Normal cellular prion protein is a ligand of selectins: binding requires Le x but is inhibited by sLe x. Biochem. J. 2007;406:333–341. doi: 10.1042/BJ20061857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mani K., et al. Prion, amyloid beta-derived Cu(II) ions, or free Zn(II) ions support S-nitroso-dependent autocleavage of glypican-1 heparan sulfate. J. Biol. Chem. 2003;278:38956–38965. doi: 10.1074/jbc.M300394200. [DOI] [PubMed] [Google Scholar]
- 15.Lysek D.A., Wuthrich K. Prion protein interaction with the C-terminal SH3 domain of Grb2 studied using NMR and optical spectroscopy. Biochemistry. 2004;43:10393–10399. doi: 10.1021/bi0494828. [DOI] [PubMed] [Google Scholar]
- 16.Mahfoud R., et al. Identification of a common sphingolipid-binding domain in Alzheimer, prion, and HIV-1 proteins. J. Biol. Chem. 2002;277:11292–11296. doi: 10.1074/jbc.M111679200. [DOI] [PubMed] [Google Scholar]
- 17.Gabus C., et al. The prion protein has DNA strand transfer properties similar to retroviral nucleocapsid protein. J. Mol. Biol. 2001;307:1011–1021. doi: 10.1006/jmbi.2001.4544. [DOI] [PubMed] [Google Scholar]
- 18.Chiarini L.B., et al. Cellular prion protein transduces neuroprotective signals. EMBO J. 2002;21:3317–3326. doi: 10.1093/emboj/cdf324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paitel E., Fahraeus R., Checler F. Cellular prion protein sensitizes neurons to apoptotic stimuli through Mdm2-regulated and p53-dependent caspase 3-like activation. J. Biol. Chem. 2003;278:10061–10066. doi: 10.1074/jbc.M211580200. [DOI] [PubMed] [Google Scholar]
- 20.Kuwahara C., et al. Prions prevent neuronal cell-line death. Nature. 1999;400:225–226. doi: 10.1038/22241. [DOI] [PubMed] [Google Scholar]
- 21.Bounhar Y., Zhang Y., Goodyer C.G., LeBlanc A. Prion protein protects human neurons against Bax-mediated apoptosis. J. Biol. Chem. 2001;276:39145–39149. doi: 10.1074/jbc.C100443200. [DOI] [PubMed] [Google Scholar]
- 22.Diarra-Mehrpour M., et al. Prion protein prevents human breast carcinoma cell line from tumor necrosis factor alpha-induced cell death. Cancer Res. 2004;64:719–727. doi: 10.1158/0008-5472.CAN-03-1735. [DOI] [PubMed] [Google Scholar]
- 23.Morel E., et al. The cellular prion protein PrP is involved in the proliferation of epithelial cells and in the distribution of junction-associated proteins. PLoS ONE. 2008;3:e3000. doi: 10.1371/journal.pone.0003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bueler H., et al. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature. 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 25.Westergard L., Christensen H.M., Harris D.A. The cellular prion protein (PrP(C)): its physiological function and role in disease. Biochim. Biophys. Acta. 2007;1772:629–644. doi: 10.1016/j.bbadis.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang J., et al. Over-expression of PrPC and its antiapoptosis function in gastric cancer. Tumour Biol. 2006;27:84–91. doi: 10.1159/000092488. [DOI] [PubMed] [Google Scholar]
- 27.Antonacopoulou A.G., et al. POLR2F, ATP6V0A1 and PRNP expression in colorectal cancer: new molecules with prognostic significance? Anticancer Res. 2008;28:1221–1227. [PubMed] [Google Scholar]
- 28.Han H., et al. Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray. Cancer Res. 2002;62:2890–2896. [PubMed] [Google Scholar]
- 29.Jemal A., et al. Cancer statistics, 2003. CA Cancer J. Clin. 2003;53:5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- 30.Li D., Xie K., Wolff R., Abbruzzese J.L. Pancreatic cancer. Lancet. 2004;363:1049–1057. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- 31.Hezel A.F., Kimmelman A.C., Stanger B.Z., Bardeesy N., Depinho R.A. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. . 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 32.Hruban R.H., et al. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am. J. Surg. Pathol. 2001;25:579–586. doi: 10.1097/00000478-200105000-00003. [DOI] [PubMed] [Google Scholar]
- 33.Hruban R.H., Wilentz R.E., Maitra A. Identification and analysis of precursors to invasive pancreatic cancer. Methods Mol. Med. 2005;103:1–13. doi: 10.1385/1-59259-780-7:001. [DOI] [PubMed] [Google Scholar]
- 34.Deramaudt T., Rustgi A.K. Mutant K-RAS in the initiation of pancreatic cancer. Biochim. Biophys. Acta. . 2005;1756:97–101. doi: 10.1016/j.bbcan.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Welsch T., Kleeff J., Friess H. Molecular pathogenesis of pancreatic cancer: advances and challenges. Curr. Mol. Med. 2007;7:504–521. doi: 10.2174/156652407781387082. [DOI] [PubMed] [Google Scholar]
- 36.Maitra A., Hruban R.H. Pancreatic cancer. Annu. Rev. Pathol. . 2008;3:157–188. doi: 10.1146/annurev.pathmechdis.3.121806.154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hingorani S.R., et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. . 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 38.Ijichi H., et al. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev. . 2006;20:3147–3160. doi: 10.1101/gad.1475506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zanusso G., et al. Prion protein expression in different species: analysis with a panel of new mAbs. Proc. Natl. Acad. Sci. U. S. A. 1998;95:8812–8816. doi: 10.1073/pnas.95.15.8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li R., et al. Identification of an epitope in the C terminus of normal prion protein whose expression is modulated by binding events in the N terminus. J. Mol. Biol. 2000;301:567–573. doi: 10.1006/jmbi.2000.3986. [DOI] [PubMed] [Google Scholar]
- 41.Stossel T.P., et al. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. . 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 42.Feng Y., Walsh C.A. The many faces of filamin: a versatile molecular scaffold for cell motility and signaling. Nat. Cell Biol. . 2004;6:1034–1038. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- 43.Maeda Y., Ashida H., Kinoshita T. CHO glycosylation mutants: GPI anchor. Methods Enzymol. . 2006;416:182–205. doi: 10.1016/S0076-6879(06)16012-7. [DOI] [PubMed] [Google Scholar]
- 44.Ambler R.P. Enzymatic hydrolysis with carboxypeptidases. Methods Enzymol. 1967;11:155–166. doi: 10.1016/S0076-6879(67)11016-1. [DOI] [PubMed] [Google Scholar]
- 45.Ikezawa H. Glycosylphosphatidylinositol (GPI)-anchored proteins. Biol. Pharm. Bull. 2002;25:409–417. doi: 10.1248/bpb.25.409. [DOI] [PubMed] [Google Scholar]
- 46.Rajendran L., Simons K. Lipid rafts and membrane dynamics. J. Cell Sci. 2005;118:1099–1102. doi: 10.1242/jcs.01681. [DOI] [PubMed] [Google Scholar]
- 47.Waneck G.L., Stein M.E., Flavell R.A. Conversion of a PI-anchored protein to an integral membrane protein by a single amino acid mutation. Science. 1988;241:697–699. doi: 10.1126/science.3399901. [DOI] [PubMed] [Google Scholar]
- 48.Kiema T., et al. The molecular basis of filamin binding to integrins and competition with talin. Mol. Cell. 2006;21:337–347. doi: 10.1016/j.molcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 49.Nakamura F., et al. The structure of the GPIb-filamin A complex. Blood. . 2006;107:1925–1932. doi: 10.1182/blood-2005-10-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakamura F., Osborn T.M., Hartemink C.A., Hartwig J.H., Stossel T.P. Structural basis of filamin A functions. J. Cell Biol. 2007;179:1011–1025. doi: 10.1083/jcb.200707073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim D.H., Rossi J.J. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 52.Bamburg J.R., Wiggan O.P. ADF/cofilin and actin dynamics in disease. Trends Cell Biol. 2002;12:598–605. doi: 10.1016/S0962-8924(02)02404-2. [DOI] [PubMed] [Google Scholar]
- 53.Scott R.W., Olson M.F. LIM kinases: function, regulation and association with human disease. J. Mol. Med. 2007;85:555–568. doi: 10.1007/s00109-007-0165-6. [DOI] [PubMed] [Google Scholar]
- 54.Huang T.Y., DerMardirossian C., Bokoch G.M. Cofilin phosphatases and regulation of actin dynamics. Curr. Opin. Cell Biol. . 2006;18:26–31. doi: 10.1016/j.ceb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 55.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 56.Pollard T.D., Borisy G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/S0092-8674(03)00120-X. [DOI] [PubMed] [Google Scholar]
- 57.Fanjul M., Alvarez L., Hollande E. Expression and subcellular localization of a 35-kDa carbonic anhydrase IV in a human pancreatic ductal cell line (Capan-1). J. Histochem. Cytochem. 2007;55:783–794. doi: 10.1369/jhc.6A7112.2007. [DOI] [PubMed] [Google Scholar]
- 58.Fanjul M., et al. Evidence for a membrane carbonic anhydrase IV anchored by its C-terminal peptide in normal human pancreatic ductal cells. Histochem. Cell Biol. 2004;121:91–99. doi: 10.1007/s00418-003-0616-2. [DOI] [PubMed] [Google Scholar]
- 59.Chen R., Knez J.J., Merrick W.C., Medof M.E. Comparative efficiencies of C-terminal signals of native glycophosphatidylinositol (GPI)-anchored proproteins in conferring GPI-anchoring. J. Cell Biochem. 2001;84:68–83. doi: 10.1002/jcb.1267. [DOI] [PubMed] [Google Scholar]
- 60.Rane N.S., Yonkovich J.L., Hegde R.S. Protection from cytosolic prion protein toxicity by modulation of protein translocation. EMBO J. . 2004;23:4550–4559. doi: 10.1038/sj.emboj.7600462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hegde R.S., et al. A transmembrane form of the prion protein in neurodegenerative disease. Science. 1998;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- 62.Goicoechea S.M., Arneman D., Otey C.A. The role of palladin in actin organization and cell motility. Eur. J. Cell Biol. 2008;87:517–525. doi: 10.1016/j.ejcb.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pogue-Geile K.L., et al. Palladin mutation causes familial pancreatic cancer and suggests a new cancer mechanism. PLoS Med. . 2006;3:e516. doi: 10.1371/journal.pmed.0030516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Slater E., et al. Palladin mutation causes familial pancreatic cancer: absence in European families. PLoS Med. 2007;4:e164. doi: 10.1371/journal.pmed.0040164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zogopoulos G., et al. The P239S palladin variant does not account for a significant fraction of hereditary or early onset pancreas cancer. Hum. Genet. . 2007;121:635–637. doi: 10.1007/s00439-007-0361-z. [DOI] [PubMed] [Google Scholar]
- 66.Iacobuzio-Donahue C.A., et al. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am. J. Pathol. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Logsdon C.D., et al. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. . 2003;63:2649–2657. [PubMed] [Google Scholar]
- 68.Petersen R.B., Parchi P., Richardson S.L., Urig C.B., Gambetti P. Effect of the D178N mutation and the codon 129 polymorphism on the metabolism of the prion protein. J. Biol. Chem. 1996;271:12661–12668. doi: 10.1074/jbc.271.21.12661. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.