Abstract
Sirtuin 3 (SIRT3) is a member of the sirtuin family of proteins that promote longevity in many organisms. Increased expression of SIRT3 has been linked to an extended life span in humans. Here, we have shown that Sirt3 protects the mouse heart by blocking the cardiac hypertrophic response. Although Sirt3-deficient mice appeared to have normal activity, they showed signs of cardiac hypertrophy and interstitial fibrosis at 8 weeks of age. Application of hypertrophic stimuli to these mice produced a severe cardiac hypertrophic response, whereas Sirt3-expressing Tg mice were protected from similar stimuli. In primary cultures of cardiomyocytes, Sirt3 blocked cardiac hypertrophy by activating the forkhead box O3a–dependent (Foxo3a-dependent), antioxidant–encoding genes manganese superoxide dismutase (MnSOD) and catalase (Cat), thereby decreasing cellular levels of ROS. Reduced ROS levels suppressed Ras activation and downstream signaling through the MAPK/ERK and PI3K/Akt pathways. This resulted in repressed activity of transcription factors, specifically GATA4 and NFAT, and translation factors, specifically eukaryotic initiation factor 4E (elf4E) and S6 ribosomal protein (S6P), which are involved in the development of cardiac hypertrophy. These results demonstrate that SIRT3 is an endogenous negative regulator of cardiac hypertrophy, which protects hearts by suppressing cellular levels of ROS.
Introduction
Cardiac hypertrophy is a common response of myocytes to a variety of physiologic and pathologic stimuli. Because mammalian cardiomyocytes lose their ability to divide soon after birth, the only way for them to deal with a sustained increase in workload is to undergo hypertrophy. During hypertrophy, myocytes not only grow in size but also add sarcomeres and induce the expression of a group of genes, which are usually expressed during fetal heart development. These changes may be compensatory initially to manage the increased workload on the heart; however, prolonged hypertrophy leads to congestive heart failure and sudden death due to arrhythmias (1).
At the molecular level, hypertrophy of cardiomyocytes is considered an outcome of imbalance between prohypertrophic and antihypertrophic factors and their downstream mechanisms controlling cell growth. In recent years, much progress has been made in our understanding of hypertrophic activators (1); however, relatively little is known about endogenous negative regulators of cardiac growth, which have potential to block the cardiac hypertrophic response (reviewed in ref. 2).
Recent studies have revealed a key role of histone acetyltransferases (HATs) and deacetylases (HDACs) in the control of cardiac hypertrophy. Whereas HATs, such as p300 and PCAF, are prohypertrophic, HDACs of different classes have been shown to have different effects on cardiac growth, resulting from stress stimuli (1, 3). SIRT3 belongs to class III of HDACs, commonly called sirtuins (SIRTs), which need NAD as a cofactor for their deacetylation reaction (4). SIRT3 was initially localized in mitochondria, but recent studies have indicated that it is expressed in the nucleus as well (5, 6). Both nuclear and mitochondrial substrates of SIRT3 have been identified, these include Histone 3 (H3), H4, and Ku70 of the nucleus and acetyl-CoA-synthetase 2 of the mitochondrion (5, 7, 8). SIRT3 expression has been shown to be activated by calorie restriction (CR), and the increased expression of the deacetylase in adipocytes was shown to induce the expression of genes involved in mitochondrial biogenesis (9). Expression of SIRT3 has also been implicated in the synthesis and maintenance of cellular ATP levels in many tissues, including heart, liver, and kidney (10).
Of the 7 SIRT analogues, SIRT3 is the only member whose increased expression has been linked to the longevity of humans. Polymorphism in the SIRT3 gene promoter, which leads to gene activation, has been found to be associated with an extended life span of man (11, 12). The molecular basis of SIRT3-dependent longevity is, however, not known. We have recently demonstrated that SIRT3 levels are elevated both in mitochondria and in the nucleus after stress of cardiomyocytes, and overexpression of SIRT3 protects myocytes from genotoxic and oxidative stress-mediated cell death (8).
Here, by using both Sirt3-deficient and Sirt3-overexpressing Tg mice as well as isolated cardiomyocytes, we show that Sirt3 is a negative regulator of cardiac hypertrophy. We demonstrate that SIRT3 is a stress-responsive deacetylase that blocks the cardiac hypertrophic response through activation of Foxo-dependent antioxidants, manganese superoxide dismutase (MnSOD) and catalase, as well as by suppressing ROS-mediated Ras activation and the downstream MAPK/ERK and PI3K/Akt signaling pathways.
Results
Sirt3 levels are elevated during hypertrophy of the heart.
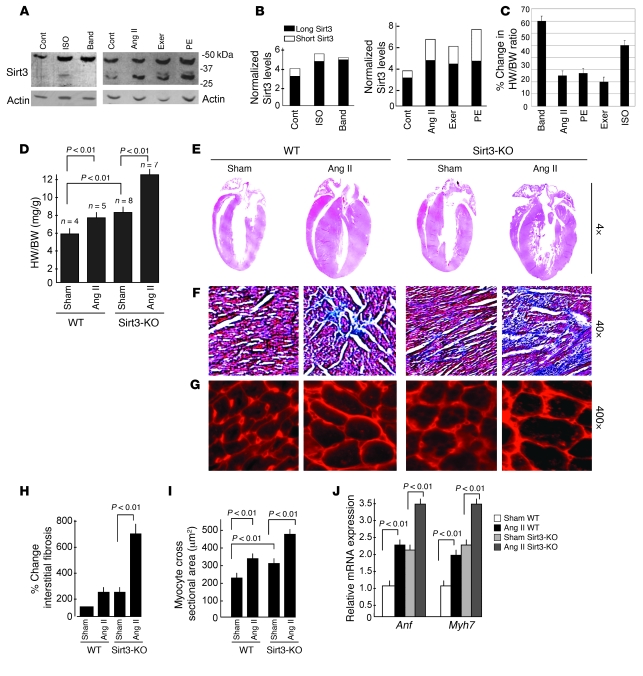
In mouse hearts, Sirt3 is expressed in 2 forms, a long form (~44 kDa) and a short form (~28 kDa) (8). To examine whether expression of these 2 forms of Sirt3 was changed after hypertrophy, we measured their expression levels in different models of cardiac hypertrophy. We found that both forms of Sirt3 were increased in hearts of mice subjected to chronic infusion of hypertrophy agonists, phenylephrine (PE) or Ang II, which produced nearly 25% cardiac hypertrophy (Figure 1, A and B). Similar increased expression of Sirt3 was also noticed in the hearts of mice subjected to a forced swimming exercise, which generated nearly 20% physiologic hypertrophy (Figure 1, A and B). In contrast, in hearts of mice that underwent (6 weeks) aortic banding, which produced nearly 60% cardiac hypertrophy, we found expression of only the long form of Sirt3, whereas the short form was either not changed or disappeared compared with sham controls (Figure 1, A and B). A similar change in the expression of Sirt3 isoforms was also noticed in hearts of mice subjected to chronic infusion of isoproterenol (ISO), which produced nearly 50% cardiac hypertrophy (Figure 1, A–C). These results indicated that, whereas both forms of Sirt3 are increased during mild hypertrophy, the short form of Sirt3 is downregulated during severe hypertrophy of the heart.
Figure 1. Sirt3 is required to block cardiac hypertrophic response.
(A) Expression levels of 2 forms of Sirt3 in different models of cardiac hypertrophy. Western blotting analysis of heart samples of mice subjected to chronic infusion of agonists, ISO, Ang II, or PE as well as of aortic banding (Band) or forced swimming exercise (exer). (B) Quantification of 2 forms of Sirt3 in different models of hypertrophy. (C) Percentage of cardiac hypertrophy in response to different stimuli. Values are mean ± SEM, n = 5–10. (D) HW/BW ratios of WT and Sirt3-KO mice infused with either vehicle (sham) or Ang II for 14 days. Values are mean ± SEM. (E) H&E-stained sections of hearts from WT and Sirt3-KO mice subjected to Ang II–mediated hypertrophy show gross changes of cardiac hypertrophy. (F) Sections of hearts stained with Masson’s trichrome to detect fibrosis (blue). (G) Heart sections stained with wheat germ agglutinin to demarcate cell boundaries. Original magnification, ×4 (E); ×40 (F); ×400 (G). (H and I) Quantification of fibrosis and myocyte cross-sectional area in control (sham) and Ang II–treated WT and Sirt3-KO mice hearts. (J) Anf and Myh7 mRNA levels in heart samples of control (sham) and Ang II–treated WT and Sirt3-KO mice. Mean ± SEM (n = 4–8). Cont, control.
To examine the role of Sirt3 in the development of cardiac hypertrophy, we subjected Sirt3-KO mice, along with their WT controls, to agonist-induced hypertrophy. Sirt3-KO mice did not exhibit any obvious cardiac phenotype; however, there was consistently an increased heart weight/body weight (HW/BW) ratio in Sirt3-KO mice compared with WT controls (Figure 1D). We also observed substantially higher levels of fibrosis and an increased cross-sectional area of cardiomyocytes in Sirt3-KO mice compared with WT controls, suggesting a propensity of these hearts to develop heart failure (Figure 1, F and G). Chronic Ang II infusion (3.0 mg/kg/day for 14 days) produced nearly 44% cardiac hypertrophy in Sirt3-KO mice, whereas WT mice produced only 22% cardiac hypertrophy, as assessed by the HW/BW ratio (Figure 1D). The expression levels of other hypertrophy markers such as mRNA levels of natriuretic peptide precursor type A (Anf) and myosin, heavy chain 7, cardiac muscle, β (Myh7) were also significantly higher in Sirt3-KO mice than in WT controls, and their levels increased further after stimulation with the agonist (Figure 1J). To confirm these findings, we examined the effect of 2 other hypertrophy agonists, ISO and PE and found results identical to those with Ang II treatment (data not shown). These results thus indicated that Sirt3 might be required for blocking of the cardiac hypertrophic response.
Sirt3 overexpression blocks the cardiac hypertrophic response in vitro and in vivo.
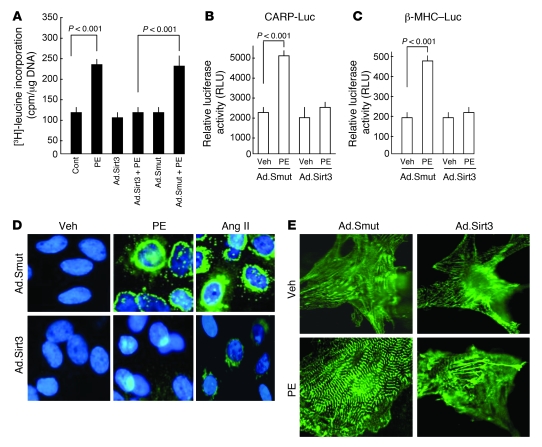
To test whether increased levels of Sirt3 during hypertrophy are protective, we overexpressed primary cultures of cardiomyocytes with Sirt3 WT (human SIRT3) or mutant, which lacks deacetylase activity, by adenoviral delivery (referred to herein as Ad.Sirt3 and Ad.Smut, respectively). At a dose of 10 MOI of virus for 16 hours of infection, nearly 4- to 8-fold induction of SIRT3 relative to endogenous levels was observed, which mimics the upregulation of SIRT3 during cardiac hypertrophy (data not shown). After infection with adenovirus vectors, cardiomyocytes were treated with PE (20 μM) or Ang II (2 μM) for 48 hours, and their hypertrophic response was measured by monitoring induction of protein synthesis, reorganization of sarcomeres, and activation of fetal gene expression. We found that Ad.Sirt3 overexpression significantly reduced the PE-induced protein accumulation, as measured by [3H]-leucine incorporation in total cellular protein (Figure 2A). Staining of these cells for α-actinin, a Z-disc–associated protein, indicated that Ad.Sirt3 overexpression, but not that of the mutant virus, was capable of blocking the PE-induced reorganization of sarcomeres (Figure 2E). The analysis of ANF release from nuclei demonstrated that, whereas it was highly expressed in the perinuclear region of PE- and Ang II–treated cardiomyocytes infected with the mutant virus, it was reduced to nearly undetectable levels in cells overexpressed with the Sirt3 WT virus (Figure 2D).
Figure 2. Sirt3 overexpression blocks cardiac hypertrophic response in vitro.
(A) Rat cardiomyocytes were overexpressed with Sirt3 WT (Ad.Sirt3) or mutant virus (Ad.Smut) and then treated with PE (20 μM) for 48 hours. Incorporation of [3H]-leucine into total cellular protein was determined and normalized to DNA content of the cells. Values are mean ± SEM (n = 5). (B and C) Cardiomyocytes expressing Ad.Sirt3 or Ad.Smut viruses were transfected with a CARP promoter/luciferase reporter vector (CARP-Luc) or β-MHC promoter/luciferase reporter vector (β-MHC–Luc). Cells were treated with vehicle (Veh) or PE (20 μM), and the luciferase activity was measured 48 hours after treatment. A β-gal/reporter plasmid was used as a reference control. Values are mean ± SEM (n = 3). (D) Cardiomyocytes were infected with the indicated adenoviruses and then stimulated with PE (20 μM), Ang II (2 μM), or vehicle for 48 hours. ANF release (green) was determined by staining cells with anti-ANF antibody. DAPI stain was used to mark the position of nuclei. (E) Reorganization of sarcomeres after PE treatment of cells. Cells were treated as in D and immunostained with α-actinin antibody for visualization of sarcomeres. Original magnification, ×630 (D); ×1,000 (E).
For further confirmation of the antihypertrophic potential of Sirt3, we examined its effect on the promoter activity of 2 hypertrophy signal-sensitive genes, CARP (cardiac ankyrin repeat protein) and Myh7. Results obtained from promoter/reporter gene analyses demonstrated that the promoter activity of both CARP and Myh7 genes was markedly induced after PE treatment of cells, as expected. Overexpression of Ad.Sirt3, but not of the mutant virus, prevented the PE-mediated activation of both promoters (Figure 2, B and C). These data demonstrated that Sirt3 is capable of blocking the agonist-mediated hypertrophic response of cardiomyocytes.
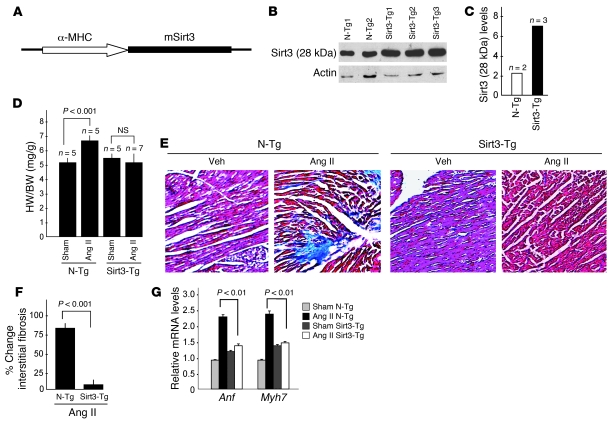
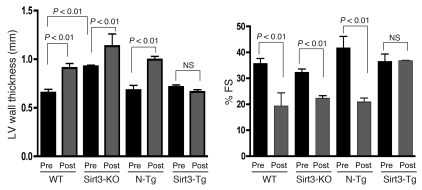
To demonstrate antihypertrophic effects of Sirt3 in vivo, we generated Tg mice overexpressing murine Sirt3 (mSirt3) (28 kDa short form of Sirt3) in the heart, under the control of α-MHC promoter. Three independent lines of Tg mice (Sirt3-Tg) were established. Total Sirt3 protein, reflecting endogenous and transgene expression, was increased 3- to 4-fold in all 3 Sirt3-Tg lines compared with non-Tg (N-Tg) controls of identical genetic background (Figure 3, A and B). Sirt3-Tg mice developed normally, without any apparent change in the HW/BW ratio compared with N-Tg controls, after up to 1 year of observation. To examine the effect of cardiac-specific overexpression of Ad.Sirt3 on the development of hypertrophy, we subjected mice to chronic infusion of hypertrophy agonists. Age- and gender-matched N-Tg mice were used as positive controls. Ang II infusion for 14 days resulted in the development of 25% of cardiac hypertrophy in N-Tg mice, whereas Sirt3-Tg mice showed no noticeable increase in the HW/BW ratio (Figure 3D). Similarly, Sirt3-Tg mice were resistant to Ang II–induced cardiac fibrosis, whereas it was robust in N-Tg controls (Figure 3, E and F). Ang II treatment also significantly induced the expression of fetal genes, Anf and Myh7, in the hearts of N-Tg animals; this also was not detected in Sirt3-Tg mice (Figure 3G). We obtained similar results by examining the effect of other hypertrophy agonists, e.g., ISO (Supplemental Figure 1; available online with this article; doi: 10.1172/JCI39162DS1). To monitor cardiac functions of these mice, we performed echocardiography before and after induction of hypertrophy. As shown in Figure 4, the LV wall thickness of Sirt3-KO mice was significantly higher than the WT controls, and it was increased further after agonist infusion. The LV wall thickness of Sirt3-Tg mice did not change significantly after ISO infusion, thus again suggesting that these mice were protected from hypertrophic stimuli. In these mice, quantification of LV fractional shortening was significantly reduced in controls and in Sirt3-KO mice, but it remained unchanged in Sirt3-Tg mice after treatment with hypertrophic stimuli, suggesting that LV functions of Sirt3-Tg mice were preserved (Figure 4). These results together indicated that the increased expression of Ad.Sirt3 was capable of blocking the pathologic cardiac hypertrophic response in vivo.
Figure 3. Sirt3-Tg mice are protected from agonist-mediated cardiac hypertrophy.
(A) A schematic of Tg construct used to generate mSirt3-Tg mouse lines. (B) Sirt3 (28 kDa) expression analysis in 2 N-Tg and 3 Sirt3-Tg mouse lines. (C) Quantitative analysis of expression of Sirt3 (28 kDa) in N-Tg and Sirt3-Tg mice lines. (D) N-Tg and mSirt3-Tg mice were treated with either vehicle (Sham) or Ang II (3.0 mg/kg per day for 14 days), and their HW/BW ratios were determined. (E) Representative heart sections stained with Masson’s trichrome for detecting fibrosis in N-Tg and Sirt3-Tg mice subjected to Ang II–mediated hypertrophy. Original magnification, ×40. (F) Quantification of fibrosis in N-Tg and Sirt3-Tg mouse hearts after Ang II treatment. (G) ANF and Myh7 mRNA levels in hearts of N-Tg and Sirt3-Tg mice treated with either vehicle (Sham) or Ang II. Values indicate relative expression levels to sham-operated group (mean ± SEM; n = 5 [D, F, and G]).
Figure 4. Sirt3-Tg mice subjected to agonist-mediated cardiac hypertrophy show preserved cardiac functions.
Echocardiography was performed on WT, Sirt3-KO, N-Tg controls, and Sirt3-Tg mice before (pre) and after (post) infusion of ISO (8.7 mg/kg/d) for 7 days. LV wall thickness and fractional shortening (FS) were measured as described in Methods. Values are mean ± SEM, n = 5.
SIRT3 suppresses both transcription and translation events involved in development of cardiac hypertrophy. Hypertrophy agonists are known to induce cardiomyocyte growth by activating several steps of transcription and translation controls of protein synthesis (1). To test whether Sirt3 has the ability to interfere with the transcription regulation of cardiac genes, we examined the effect of the deacetylase on 2 key transcription factors, GATA4 and NFAT, which are known to play major roles in the induction of cardiomyocyte hypertrophy (1, 3). The transcription activity of GATA4 and NFAT has been shown to be regulated by their nucleo-cytoplasmic shuttling. While the phosphorylation of GATA4 by ERK1/2 promotes its nuclear localization, the glycogen synthase kinase 3 β–mediated (GSK3β-mediated) phosphorylation causes export of GATA4 from the nucleus, and this blocks its transcription activity (13). The NFAT transcription factors are also exported out from the nucleus by phosphorylation, whereas dephosphorylation by calcineurin, a Ca-dependent phosphatase, promotes its nuclear localization and activates the transcription of genes harboring NFAT-binding sites (14, 15).
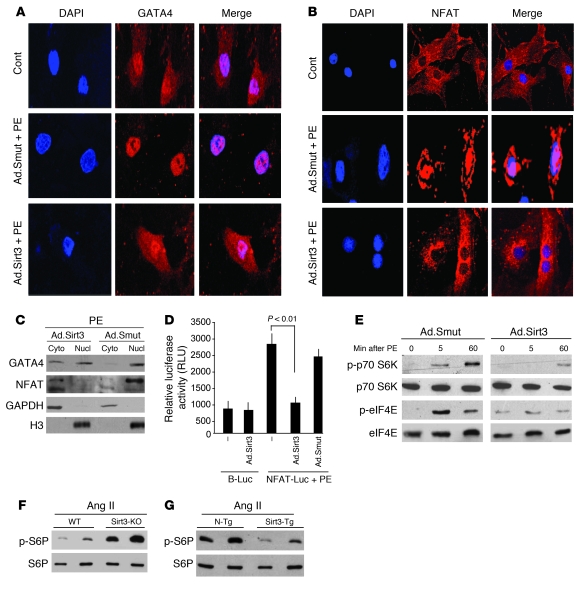
To test whether Sirt3 antagonizes the transcription activity of GATA4 and NFAT by promoting their export from the nucleus, we infected cardiomyocytes with Ad.Sirt3 or Ad.Smut, and we then stimulated them with PE for 2 hours. Immunostaining of cells for GATA4 and NFAT showed that both factors were localized in the nucleus as well as in the cytoplasm of control cells. PE stimulation resulted in localization of both factors exclusively in the nucleus, whereas overexpression of Ad.Sirt3, but not of the mutant virus, prevented the PE-mediated nuclear accumulation of both GATA4 and NFAT (Figure 5, A and B).
Figure 5. Sirt3 inhibits activation of transcription and translation regulators involved in development of hypertrophy.
(A and B) Rat cardiomyocytes were infected with Ad.Sirt3 or Ad.Smut viruses and then stimulated with either vehicle or PE (20 μM) for 2 hours. Cells were stained for GATA4 (A) or NFAT (B), and subcellular localization of factors was determined by confocal microscopy. Positions of nuclei were determined by DAPI stain (blue). Original magnification, ×630 (A and B). (C) Cardiomyocytes were infected with viruses and treated with PE as in A. Cytoplasmic (Cyto) and nuclear (Nucl) fractions of myocytes were generated and analyzed by Western blotting. (D) Cardiomyocytes expressing the indicated adenoviruses were transfected with a basic luciferase plasmid (B-Luc) or NFAT-responsive/luciferase (NFAT-Luc) reporter plasmid. On the second day after transfection, cells were treated with PE (20 μM), and the luciferase activity was determined 24 hours after transfection. Values are normalized with the protein content of the cell (mean ± SEM, n = 5). (E) Cardiomyocytes were infected with the indicated adenoviruses and induced with PE (20 μM). Cells were harvested at different time points after PE treatment, and the lysate was analyzed by Western blotting. (F and G) Heart extracts of mice subjected to Ang II–mediated hypertrophy were analyzed by Western blotting. Results are shown for 2 mice of the same group. WT and N-Tg mice are controls of the same genetic background for Sirt3-KO and Sirt3-Tg mice, respectively.
To obtain additional evidence for the effect of Sirt3 on the localization of GATA4, we analyzed cytoplasmic and nuclear fractions of cardiomyocytes by Western blotting. The results showed that PE treatment caused accumulation of GATA4 and NFAT mainly in the nuclear fraction. Overexpression of Ad.Sirt3 prevented PE-mediated nuclear accumulation of NFAT and made GATA4 be distributed equally between cytoplasmic and nuclear fractions, thus confirming the ability of Sirt3 to suppress the transcription activity of GATA4 and NFAT (Figure 5C).
We also confirmed the effect of Sirt3 on the transcription activity of NFAT by examining the expression of an NFAT-responsive promoter/reporter (NFAT-Luc) plasmid in a transient transfection assay. The results showed that the activity of the NFAT-Luc plasmid was dramatically reduced by overexpression of the Ad.Sirt3 WT, but not the mutant virus, thus again indicating that Sirt3 was capable of blocking the transcription activity of NFAT (Figure 5D). Together, these results demonstrated that Sirt3 has the ability to block the activity of cardiac gene transcription driven by GATA4 and NFAT, which contributes to development of myocyte hypertrophy.
We next asked whether Sirt3 also regulates the activity of translation factors and their related signaling molecules, which are known to be activated during hypertrophy of myocytes. The key steps in translational control of protein synthesis include activation of eukaryotic initiation factor 4E (eIF4E), p70.S6 kinase, and S6 ribosomal protein (S6P) (16, 17). We examined the effect of Sirt3 on the activity of these factors by measuring their phosphorylation status by Western blotting. We found that PE stimulation of cardiomyocytes enhanced the phosphorylation of p70.S6 kinase and eIF4E at different time points examined. Overexpression of Ad.Sirt3, but not of the mutant virus, notably reduced the PE-induced phosphorylation of p70.S6 kinase and eIF4E, indicating that Sirt3 was capable of blocking the PE-induced activation of translation events leading to upregulation of protein synthesis in cardiomyocytes (Figure 5E). To obtain evidence from an in vivo model of cardiac hypertrophy, we quantified the phosphorylation of S6P, a target of P70.S6 kinase, in heart lysates obtained from Sirt3-KO and WT mice subjected to chronic Ang II infusion. We found substantially high levels of S6P phosphorylation in Sirt3-KO hearts compared with WT hearts, indicating hyperactivation of translation events in Sirt3-KO mice (Figure 5F). A similar increased phosphorylation of S6P was noticed in heart samples of N-Tg mice but not in Sirt3-Tg mice subjected to Ang II treatment (Figure 5G). These results thus suggested a role of Sirt3 in blocking of the agonist-induced activation of the translation machinery involved in the development of cardiac hypertrophy.
SIRT3 inhibits the activation of MAPK/ERK and PI3K/Akt signaling pathways.
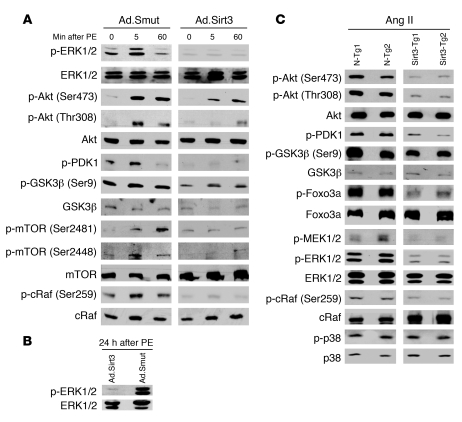
Knowing that both transcription and translation events inducing myocyte hypertrophy were blocked by SIRT3, we next searched for the signaling mechanism that contributed to this effect. In the case of activation of G protein–coupled receptors by agonists such as PE, ISO, and Ang II, sequential activation of MAPK, PI3K, Akt/PKB, and mTOR kinases has been documented (3). Among the MAPKs, ERK1/2 is considered to be a central regulator of agonist-mediated cardiac hypertrophy (3). We therefore examined the effect Sirt3 on the activation of ERK1/2 at different time points after PE stimulation of cells. The results showed that PE stimulation caused phosphorylation of ERK1/2 in cells expressed with the mutant virus; however, cells overexpressed with virus synthesizing Sirt3 WT were resistant to PE-mediated activation of ERK1/2 (Figure 6A). Some earlier reports have indicated that prolonged exposure of cells to PE is required to elicit alterations in gene expression and protein synthesis and that the initial peak of ERK1/2 activity is not sufficient to trigger the hypertrophic response (18). To address this issue, we studied phosphorylation of ERK1/2 in Sirt3 WT and mutant virus overexpressing cells after 24 hours of PE treatment. We again found that Sirt3 completely blocked the PE-mediated activation of ERK1/2, but that the mutant virus did not, suggesting that the deacetylase was capable of suppressing the activity of the MAPK/ERK1/2 signaling pathway (Figure 6B).
Figure 6. Sirt3 blocks the agonist-induced signaling pathways involved in development of cardiac hypertrophy.
(A) Rat cardiomyocytes were infected with Ad.Sirt3 or Ad.Smut viruses and treated with PE (20 μM) for the indicated time intervals (0, 5, and 60 minutes). Cell lysate was analyzed by Western blotting with kinase-specific and phospho-kinase–specific antibodies. Numbers in parentheses indicate the position of the phospho–amino acid recognized by the antibody. (B) ERK1/2 phosphorylation was analyzed 24 hours after PE-treatment of cells. (C) Heart extract prepared from N-Tg and Sirt3-Tg mice subjected to Ang II–mediated hypertrophy was analyzed by Western-blotting with antibodies against different kinases as indicated. Results are shown for 2 mice of each group.
We then examined the role of Sirt3 in regulating the activity of the PI3K/Akt pathway. Akt is activated by various extracellular stimuli in a phosphatidylinositol-3 kinase–dependent manner by an upstream kinase, PDK1 (3). The downstream targets of Akt include S6P, Raf, GSK3β, FOXOs, and mTOR. During cardiac hypertrophy, GSK3β (Ser9) has been shown to be phosphorylated by Akt, leading to suppression of its kinase activity (19). By analyzing the phosphorylation status of proteins, we found that PE treatment induced phosphorylation of PDK1, Akt, GSK3β, and cRaf, which was blocked by overexpression of Sirt3 WT, but not by the mutant virus (Figure 6A). The activity of mTOR, a central kinase of Akt signaling pathway which controls protein synthesis, was also induced after PE treatment of cells. Ad.Sirt3 overexpression again suppressed the PE-mediated phosphorylation of mTOR, but the mutant virus did not (Figure 6A). These data indicated that Sirt3 was capable of blocking the activity of the Akt signaling pathway.
To confirm these results in an in vivo model of cardiac hypertrophy, we examined the phosphorylation status of these kinases in heart samples obtained from Sirt3-Tg and N-Tg mice subjected to Ang II–mediated hypertrophy. Again, we found that Sirt3-Tg reduced the phosphorylation of ERK1/2, Akt, cRaf, Foxo3a, and GSK3β compared with the effect in N-Tg mice challenged with the same agonist-treatment (Figure 6C). We, however, found no change in phosphorylation of P38 between N-Tg and Sirt3-Tg mice, which served as negative control. These data thus further confirmed the role of Sirt3 in blocking the activity of the ERK and Akt signaling pathways, which are known to be involved in the induction of cardiac hypertrophy.
Sirt3 blocks Ras activation and accumulation of mitochondrial free radicals.
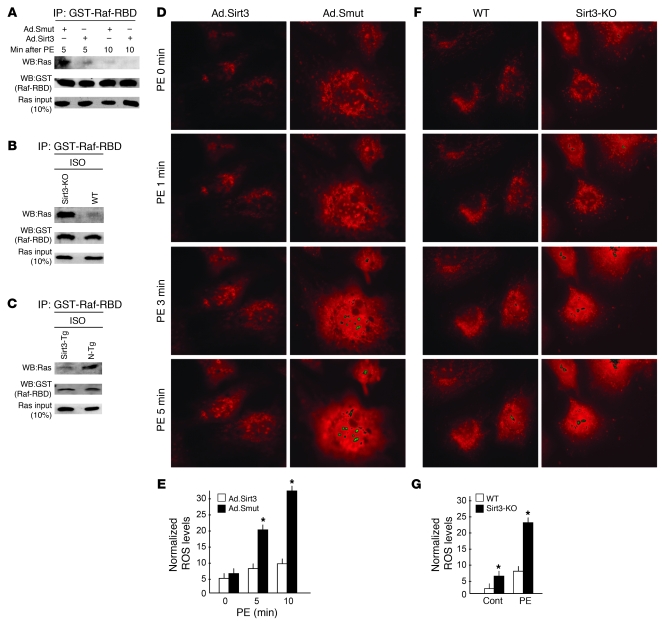
Because the activity of both the ERK1/2 and Akt signaling pathways was suppressed by Sirt3, we hypothesized that there might be a common upstream target that regulates the activity of both of these pathways and that is sensitive to Ad.Sirt3 overexpression. To this end, we focused on the Ras, a small (21 kDa) GTP-binding protein, which plays a pivotal role in the development of cardiac hypertrophy and which is capable of regulating the activity both the ERK1/2 and Akt pathways (20, 21). Ras is biologically active when bound to GTP and becomes inactive as a result of its innate GTPase activity, which hydrolyzes the bound GTP to GDP (22). Active Ras binds to Raf and activates it. To test whether Sirt3 regulates the activity of Ras, we examined the coprecipitation of active Ras with Ras-binding domain of Raf (Raf-RBD), which specifically binds to Ras-GTP (active Ras). As shown in Figure 7A, active Ras was readily pulled down by Raf-RBD from PE-treated cardiomyocytes infected with the mutant virus, but not from cells overexpressed with WT Ad.Sirt3 virus. To confirm these findings, we repeated this experiment with a cardiac extract prepared from Sirt3-KO and Sirt3-Tg mice subjected to chronic ISO infusion. We found that, in response to ISO stimulation, there was marked activation of Ras (as measured by coprecipitation with Raf-RBD) in Sirt3-KO hearts but not in hearts of Sirt3-Tg mice, when compared with their respective controls (Figure 7, B and C), thus suggesting that Sirt3 has the potential to block the agonist-mediated activation of Ras.
Figure 7. Sirt3 blocks Ras activation and mitochondrial ROS accumulation during hypertrophy.
(A) Rat cardiomyocytes were infected with the indicated adenoviruses and treated with PE (20 μM) for indicated time intervals. From the cell extract, Raf was immunoprecipitated with use of Raf-RBD beads, and the complex was analyzed for coprecipitation of active Ras by Western blotting (WB). Sirt3 overexpression suppressed the Ras-Raf binding both at 5 and 10 minutes after PE treatment, an indication of Ras deactivation. (B and C) Heart extracts of different groups of mice subjected to ISO-mediated hypertrophy were analyzed for coprecipitation of active Ras with Raf-RBD. Ras was highly activated in Sirt3-KO hearts, whereas it was suppressed in Sirt3-Tg hearts compared with controls. (D) Real-time measurement of mitochondrial ROS production in rat cardiomyocytes infected with Ad.Sirt3 or Ad.Smut virus. Cells were induced with 20 μM of PE in a thermoregulatory chamber of microscope, and the production of ROS (red fluorescence) from cells was determined at regular intervals with use of MitoSox dye by time-lapse confocal microscopy. (E) The increase of ROS was quantified by measurement of the intensity of fluorescence at different time intervals (mean ± SEM, n = 100 cells). *P < 0.01 compared with Ad.Sirt3-infected cells. (F) Cardiomyocytes cultured from neonatal hearts of Sirt3-KO and WT mice were stimulated with PE (20 μM) and ROS production measured as in D. (G) Quantification of ROS levels in 2 groups of cardiomyocytes shown in F (mean ± SEM, n = 50 cells). *P < 0.01 compared with WT cells. Original magnification, ×630 (D and F). GST, glutathione-S-transferase.
Recently, a number of reports have demonstrated that Ras is activated by ROS-mediated oxidative modification of thiol on Cys118 (23, 24). Because induction of ROS has been shown to be necessary for the activation of signaling pathways leading to cardiac hypertrophy (21, 24, 25) and we have recently shown that SIRT3 protects cardiomyocytes from oxidative stress (8), we reasoned that SIRT3 might regulate the activity of Ras by controlling the cellular ROS levels induced by hypertrophy stimuli. To test this hypothesis, we quantified the ROS accumulation in cardiomyocytes after different treatments, by using a mitochondrial ROS-sensitive dye and by performing time-lapse confocal microscopy. As expected, we found that PE treatment induced large quantities of ROS production in cardiomyocytes in a time-dependent manner. Interestingly, cells overexpressed with Ad.Sirt3 failed to generate significant amounts of ROS after PE treatment (Figure 7, D and E). We confirmed these results by measuring ROS levels in cardiomyocytes obtained from Sirt3-KO and WT mice. The results indicated that cardiomyocytes of Sirt3-KO mice were producing nearly 2-fold more ROS at the basal level, than did the myocytes of WT mice, suggesting that Sirt3 is required to regulate cellular ROS levels. We then measured ROS levels in mouse cardiomyocytes treated with hypertrophy agonists, and we found that in response to PE treatment, Sirt3-KO cardiomyocytes generated remarkably higher amounts of ROS compared with those cells prepared from WT mice (Figure 7, F and G). These results thus demonstrated that Sirt3 is capable of attenuating cellular ROS levels of cardiomyocytes during stress.
Sirt3 enhances antioxidant mechanisms of cardiomyocytes.
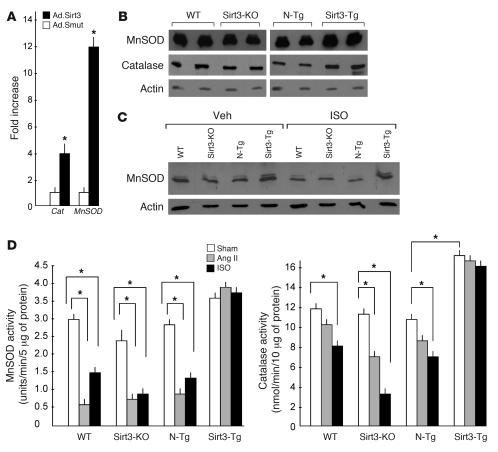
Factors capable of antagonizing cellular ROS production (antioxidants) have been found to block the cardiac hypertrophic response (24). There are mainly 2 kinds of antioxidant mechanisms in a cell that counteract ROS. The first mechanism includes superoxide dismutases, catalase, and peroxidases, which convert superoxide to water. The other mechanism includes glutathione and thioredoxin, which reduce the thiol group of oxidized proteins (26). To determine whether Sirt3 was capable of suppressing cellular ROS levels by modulating the activity of an antioxidant, we conducted a real-time PCR analysis of almost 80 transcripts from cardiomyocytes infected with Ad.Sirt3 WT or mutant virus. We found elevated mRNA transcripts of a series of antioxidant genes. Among them, the levels of MnSOD and Cat were highly elevated following Ad.Sirt3 overexpression (Figure 8A). To confirm these results, we quantified MnSOD and catalase levels in hearts of Sirt3-Tg, N-Tg, Sirt3-KO, and WT mice, and we found that the expression of these antioxidants was notably higher in hearts of Sirt3-Tg mice compared with N-Tg controls (Figure 8, B and C). We also analyzed the consequence of hypertrophy-agonist stimulation on the expression levels of MnSOD and catalase. We found that, whereas the levels of expression of these antioxidants were reduced in Sirt3-KO mice, they were generally protected in the hearts of Sirt3-Tg mice after agonist treatment compared with their sham controls, thus suggesting that Sirt3 regulates the expression levels of MnSOD and catalase in the heart (Figure 8C).
Figure 8. Sirt3 enhances the synthesis of the antioxidants, MnSOD and catalase.
(A) Level of induction of mRNA of MnSOD and Cat in rat cardiomyocytes infected with Ad.Sirt3 or Ad.Smut viruses. Values are mean ± SEM of triplicates (*P < 0.01) (B) Western analysis of MnSOD and catalase levels in cardiac extracts of different group of mice (2 hearts in each group). (C) Western analysis of MnSOD levels from different groups of mice subjected to ISO-mediated cardiac hypertrophy. (D) Enzymatic activities of MnSOD and catalase in hearts of different groups of mice, subjected to chronic infusion of vehicle, Ang II, or ISO (mean ± SEM, n = 6; *P < 0.01). Sirt3 overexpression prevented the agonist-mediated decline of cardiac levels of MnSOD and catalase.
To obtain additional evidence for these findings, we measured enzymatic activity of MnSOD and catalase in different groups of hearts. We found that, like that for protein levels, the activity of MnSOD and catalase was significantly reduced in heart samples of WT, N-Tg, and Sirt3-KO mice challenged with hypertrophy agonists, whereas it was consistently maintained in the hearts of Sirt3-Tg mice, thus confirming a role of Sirt3 in the regulation of activity of MnSOD and catalase (Figure 8D).
Sirt3 deacetylates and activates the Foxo3a transcription factor.
Factors belonging to the Foxo subfamily have been shown to inhibit ROS generation by enhancing the activities of MnSOD and catalase (27, 28). Because SIRT3 and SIRT1 have been found to have redundant effects of protecting cells during stress, we postulated that, like SIRT1, SIRT3 might have the ability to control the activity of Foxo factors (8, 29). To test this hypothesis, we first examined the binding ability of Sirt3 to Foxo3a, a major form of Foxo analogue present in the heart. We found that Sirt3 and Foxo3a were able to bind to each other in vivo (Figure 9, A and B). We then examined the deacetylation of Foxo3a by Sirt3. For this purpose, cells were overexpressed with Flag.Foxo3a and then treated with H2O2 to induce acetylation of Foxo3a. Subsequently, Foxo3a was immunoprecipitated from cell lysates, and it was tested for deacetylation by Sirt3 and by Sirt1. We found that both Sirt3 and Sirt1 substantially deacetylated Foxo3a in an NAD-dependent manner, suggesting that, like Sirt1, Sirt3 is capable of deacetylating Foxo3a (Figure 9C).
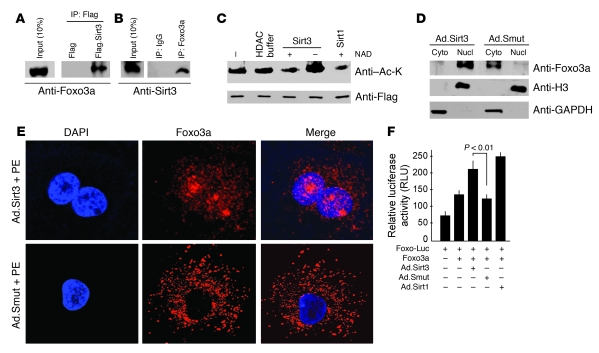
Figure 9. Sirt3 binds, deacetylates, and activates Foxo3a.
(A) Lysate of cells overexpressed with Flag or Flag.Sirt3 was subjected to immunoprecipitation with Flag-M2 agarose beads. The resulting complex was analyzed by Western blotting with anti-Foxo3a antibody. (B) Lysate of cells infected with Ad.Sirt3 virus was subjected to immunoprecipitation with anti-Foxo3a antibody, and the resulting complex was analyzed by Western blotting with anti-Sirt3 antibody. (C) Sirt3 deacetylates Foxo3a. Cells were overexpressed with Flag.Foxo3a and then treated with H2O2 to induce protein acetylation. Flag.Foxo3a was immunoprecipitated and subjected to deacetylation with Sirt3 or Sirt1 in the presence or absence of NAD in the buffer. Protein acetylation was determined by Western blotting with anti–Ac-K and anti-Flag antibodies. (D) Cytoplasmic and nuclear fractions of cardiomyocytes infected with Ad.Sirt3 or Ad.Smut viruses were prepared and analyzed by Western blotting with Foxo3a antibody. Histone 3 and GAPDH were utilized as nuclear and cytoplasmic markers, respectively. (E) Confocal analysis of Foxo3a localization (red) in cardiomyocytes infected with Ad.Sirt3 or Ad.Smut and treated with PE. Positions of nuclei were determined by DAPI stain (blue). Original magnification, ×1,000. (F) Overexpression of Sirt3 activates Foxo3a-dependent promoters. Neonatal rat cardiomyocytes were infected with viruses synthesizing Sirt1, Sirt3, or a mutant protein. The next morning cells were transfected with a Foxo3a expression plasmid and a Foxo3a responsive/luciferase reporter plasmid in different combinations as indicated. Luciferase activity was determined 48 hours after transfection. Sirt3 overexpression significantly activated Foxo3a-dependent promoter. Mean ± SEM, n = 4.
The transcription activity of Foxo factors has been shown to be controlled by their nucleo-cytoplasmic shuttling. Akt-mediated phosphorylation of Foxos allows their binding to the chaperone protein 14-3-3, which promotes their export from the nucleus into the cytoplasm (30). However, deacetylation of Foxo factors by Sirt1 overrides the phosphorylation-dependent nuclear export and renders Foxos immobile within the nucleus, thereby promoting the transcription of Foxo-dependent genes (29). Because our earlier experiments showed that the phosphorylation of Foxo3a was notably reduced in Sirt3-Tg compared with N-Tg hearts (Figure 6C), we posited that Sirt3 might be able to block the phophorylation-dependent nuclear export of the protein. To test this hypothesis, we generated nuclear and cytoplasmic fractions of cardiomyocytes overexpressed with Sirt3 WT or the mutant virus. Analysis of these fractions revealed that Ad.Sirt3 overexpression increased the nuclear accumulation of Foxo3a but the mutant virus did not (Figure 9D). To confirm these findings, we performed confocal microscopy of cardiomyocytes infected with Ad.Sirt3 or mutant viruses. As shown in Figure 9E, Ad.Sirt3-overexpressing cells had Foxo3a localized preferentially in the nucleus, whereas it was completely localized in the cytoplasm of control cells infected with the mutant virus. To obtain further proof that Sirt3 controls the transcription activity of Foxo3a, we performed a luciferase-based promoter/reporter assay. We found that the activity of the Foxo3a-dependent reporter gene was substantially increased by overexpression of Ad.Sirt3 as well as by Sirt1 but not by the mutant virus (Figure 9F). Collectively, these data demonstrated that Sirt3 was capable of promoting the nuclear localization of Foxo3a and enhancing the transcription of Foxo3a-dependent genes.
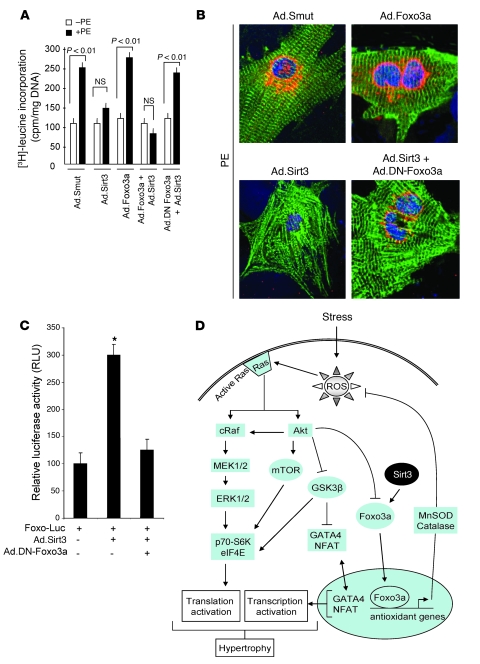
Finally, to test whether Sirt3 requires Foxo3a to block the cardiac hypertrophic response, we examined the effect Sirt3 in cells in which Foxo3a was inactivated by using a dominant-negative mutant of Foxo3a (DN-Foxo3a) (31). For this purpose, Sirt3-deficient cardiomyocytes were infected with different adenovirus vectors and then treated with PE for 48 hours. The hypertrophic response of cells was measured by [3H]-leucine incorporation into total protein (Figure 10A) as well as by examination of the sarcomere organization and of ANF release from nuclei (Figure 10B). We found that, whereas Ad.Sirt3 overexpression blocked the hypertrophic response of Sirt3–/– cells; Foxo3a failed to do so, suggesting that Foxo3a alone (in absence of Sirt3) was not sufficient to antagonize hypertrophy (Figure 10A). We then examined the effect of Sirt3 in combination with a DN-Foxo3a. The results showed that overexpression of the DN-Foxo3a eliminated the antihypertrophic effects of Sirt3, thus suggesting that the endogenous Foxo3a is needed for the protective effect of Sirt3 in cardiomyocytes (Figure 10, A and B). To confirm that the DN-Foxo3a was indeed capable of blocking the effect of endogenous protein, we analyzed its effect on the expression of a Foxo3a-dependent promoter/reporter plasmid. The results indicated that, whereas Sirt3 alone increased the activity of the Foxo3a-dependent promoter, in combination with the DN-Foxo3a, it was incapable of doing so (Figure 10C), thus indicating that the DN-Foxo3a used in our experiments was competent for inactivating the endogenous Foxo3a, consistent with other reports (31). These results together demonstrated that the negative hypertrophic effect of Sirt3 is, in part, dependent upon activation of Foxo3a.
Figure 10. A DN-Foxo3a eliminates the antihypertrophic effect of Sirt3.
Cardiomyocytes were prepared from neonatal Sirt3-KO mice. They were infected with different combinations of adenoviruses as indicated and then stimulated with PE (20 μM) for 48 hours. (A) Measurement of [3H]-leucine incorporation into total cellular protein (mean ± SEM, n = 5; P < 0.01). (B) Immunostaining of cells with anti-ANF (red) and anti–α-actinin (green) antibodies. Positions of nuclei were determined by DAPI staining (blue). Original magnification, ×1,000. (C) The DN-Foxo3a inhibits the activity of endogenous Foxo3a. Cardiomyocytes were overexpressed with a Foxo3a responsive/luciferase reporter plasmid and viruses synthesizing Sirt3 or DN-Foxo3a in different combinations, as indicated. The luciferase activity was determined 48 hours after transfection. All values are normalized to protein content of the cell (mean ± SEM, n = 4; *P < 0.01). The DN-Foxo3a was capable of blocking Sirt3-dependent activation of Foxo-promoter activity. (D) Scheme illustrating signaling pathways modified by Sirt3 to block the cardiac hypertrophic response. Sirt3 levels are elevated during stress of cardiomyocytes, which deacetylates Foxo3a and traps it inside the nucleus to enhance the transcription of Foxo-dependent antioxidant genes, MnSOD and Cat. Increased expression of MnSOD and catalase suppresses ROS levels generated by stress stimuli. Because ROS is the second messenger of hypertrophic signaling pathways, suppression of ROS levels shuts down major signaling pathways involved in activation of transcription and translation events contributing to the cardiac hypertrophic response.
Discussion
This study was designed to investigate the role of SIRT3 in the development of cardiac hypertrophy. By using different in vivo and in vitro models as well as Tg animals, we showed that Sirt3 is a negative regulator of cardiac hypertrophy. Both forms of Sirt3 were elevated during mild hypertrophy, whereas the short form was downregulated during severe hypertrophy and heart failure. The overexpression of Sirt3 prevented a hypertrophic response by shutting down the major signaling pathways that induce cardiac hypertrophy. Experiments carried out for identifying the major upstream target of Sirt3 led us to deduce that Sirt3 blocks cardiac hypertrophy by enhancing the activity of antioxidants MnSOD and catalase, thereby reducing the cellular ROS levels. These studies revealed that Sirt3 protects cardiomyocytes by promoting the antioxidant defense mechanism of the cell.
Increased Sirt3 activity is necessary for preventing cardiac hypertrophy.
SIRT3 is highly expressed in metabolically active tissues, such as the heart, liver, kidney, and adipose tissue (9). In agreement with our results, its levels have been shown to be increased during mild stress conditions, such as CR (9). The mechanism of increased expression of SIRT3 during stress has not been examined; however, based on studies with other sirtuins, it can be presumed that a change in the cellular NAD/NADH ratio following stress may, in part, contribute to increased levels of SIRT3. It has been reported that decreased NADH levels following CR, which increased the NAD/NADH ratio, elevated levels of yeast Sir2 (32). Another report has shown that increased utilization of nicotinamide, a noncompetitive inhibitor of SIRT1, by the enzyme nicotinamide phosphoribosyl transferase (Nampt), which recycles nicotinamide in the NAD de novo cycle, regulates the SIRT1 levels (33). The levels of Nampt have been shown to be upregulated during mild stress of cardiomyocytes, thus supporting the possibility of involvement of the NAD salvage pathway in the regulation of SIRT1 levels during mild stress situations (34). This type of regulation is likely to participate in the induction of SIRT3 levels during physiologic and mild hypertrophy, in which NAD/NADH levels are likely to be elevated (35), but not during severe pathologic hypertrophy and heart failure, in which NAD levels decline due to increased consumption of NAD by overactivation of poly(ADP-ribose) polymerase-1 (36). There are other mechanisms that can also regulate SIRT1 levels at the level of gene transcription as well as at the posttranscription level by stabilization of mRNA (37). Some evidence has also suggested that increased SIRT levels during mild stress are related to stabilization of the protein by diminished degradation. Kanfi et al. have reported that inhibition of proteosomal degradation by MG-132 increases SIRT6 levels to an extent similar to that observed after nutrient deprivation (38). It is therefore likely that in different types and magnitudes of hypertrophy SIRT3 levels are regulated by different mechanisms. Future studies directed toward understanding the mechanism of regulation of SIRT3 levels during stress should be able to shed more light on this subject.
Whatever the mechanism of its increased levels may be, our data presented here demonstrated that Sirt3 is required to prevent cardiac hypertrophy. Sirt3-deficient mice looked normal, with no apparent signs of any disorder. Their body weight and activity level were also within the normal range. However, we found that they expressed cardiac stress markers and had significantly higher HW/BW ratios and interstitial fibrosis compared with their WT cohorts, suggesting that they had a basal level of cardiac hypertrophy at 8 weeks of age. Cardiomyocytes cultured from Sirt3-deficient hearts produced higher levels of ROS than did myocytes cultured from WT hearts. Moreover, Sirt3-deficient mice exhibited a pronounced cardiac hypertrophic response when subjected to agonist treatments, suggesting that Sirt3 is needed to prevent stress-induced cardiac hypertrophy. A recent study conducted with Sirt3-deficient mice has indicated that Sirt3 is an important regulator of basal ATP levels in the heart. Ahn et al. have shown that hearts of Sirt-KO mice have nearly 50% less ATP at the resting stage compared with WT controls (10). Because the ATP concentration in the heart is in the millimolar range, even at a 50% reduction in ATP concentration, its levels would be substantially above the Km of most of the enzymes to sustain heart function at the resting stage. However, during stress situations, when the ATP demand of the heart is expected to rise, decreased ATP levels alone could be sufficient to cause heart dysfunction, with the consequence of developing cardiac hypertrophy.
Regulation of cardiac signaling pathways by Sirt3.
Factors controlling gene transcription and translation have been shown to be end-point targets of the signaling mechanism that causes cardiac hypertrophy (1). Recent studies have shown that ROS production is necessary and is a common mechanism for a variety of stimuli inducing cardiac hypertrophy (24, 25). Our data showed that Sirt3 overexpression shuts down most of the signaling pathways leading to hypertrophy by controlling ROS accumulation in the heart. The well-characterized ROS-sensitive signaling pathways are the MAPK/ERK and PI3K/AkT pathways, and both of these pathways have been shown to play a leading role in the development of cardiac hypertrophy by different stimuli, including pressure overload, mechanical stretch, and GPCR agonists (22, 24, 25, 28). It has also been shown that Ras is an upstream target of both the MAPK/ERK and PI3K/Akt signaling pathways and activation of Ras is necessary for development of cardiac hypertrophy (21, 25). In this study, we observed simultaneous attenuation of the MAPK/ERK and PI3K/Akt pathways and Ras activation in Sirt3-Tg hearts and Sirt3-overexpressing cultures of cardiomyocytes. Furthermore, downregulation of these signaling pathways correlated well with reduced ROS levels achieved by Sirt3 overexpression, thus suggesting that ROS lies upstream of MAPK/ERK and PI3K/Akt pathways and plays a central role in the ability of Sirt3 to block the cardiac hypertrophic response.
A role of SIRT3 in regulation of cellular ROS levels has been documented before. Constitutive expression of SIRT3 was shown to reduce ROS levels in adipocytes (9). Our data presented here show that Sirt3 reduces ROS levels by enhancing the expression of antioxidative genes, MnSOD and Cat. The expression levels of both MnSOD and catalase have been shown to be controlled by the Foxo group of transcription factors (39, 40). Consistent with a previous report in which SIRT1 was shown to activate Foxo1, we found that Sirt3 is also capable of activating Foxo3a, by promoting its nuclear localization (29). A scheme is presented in Figure 10D, showing how Sirt3 activation blocks cardiac hypertrophic response by activation of Foxo3a-dependent antioxidants, MnSOD and catalase. These data point to a role of SIRT3 inside the nucleus and support recent studies in which SIRT3 was found to be localized in the nucleus as well as mitochondria (5, 8). Our study cannot and does not rule out other mechanisms such as reduced synthesis of ROS from mitochondria or reduced activity of intermediates of signaling pathways, which might be directly targeted by SIRT3. However, the fact that the expression levels of antioxidant genes were elevated by overexpression of Sirt3 in cardiomyocytes, and this data was supported by the results obtained from Sirt3-Tg mice, provides strong evidence that enhanced antioxidant mechanisms of cells play an important role in the cardioprotective effects of Sirt3.
SIRT3 is a longevity factor, and it is the only member of the sirtuin family whose increased expression has been linked to an extended life span of humans. Our data presented here show that Sirt3 is capable of blocking cardiac hypertrophy by reducing cellular ROS levels. These findings have profound implications not only for the management of heart failure, but also for many other diseases which are associated with increased production of ROS.
Methods
Tg mice generation.
All animal protocols were reviewed and approved by the University of Chicago Institutional Animal Care and Use Committee. Mice with cardiac-restricted expression of Sirt3 were generated by use of a cDNA encoding mSirt3 (encoding 28 kDa short form). Mouse Sirt3 was amplified from mRNA isolated from the adult heart using the following primers: 5′-GGCCGTCGACCGTTTGGCGAGGACTAGTGT-3′ and 5′-GGCCAAGCTTGCATGAAGTCTTGCTGGACA-3′, which contain restriction sites for SalI and HindIII, respectively. The amplified PCR fragment was cloned directly into SalI and HindIII restriction sites of expression plasmid, containing the pBluescript II KS plasmid (Stratagene), containing α-MHC promoter and human growth hormone poly(A) signal (a gift from J. Robbins, Cincinnati Children’s Hospital, Cincinnati, Ohio, USA) (41). The orientation of Tg construct was confirmed by sequencing. Constructs were linearized and the PBS backbone was digested out of the plasmid with the use of the enzyme NotI. The purified DNA construct was injected in the pronuclear stage zygotes of the CD1 mouse strain, according to the standard Tg procedure of the University of Chicago Tg facility. At 2–3 weeks of age, tail DNA was analyzed to confirm which mice were Tg. The following primers were used for genotyping: forward 5′-CTTCCAGCCCTCTCTTTCTC-3′; reverse 5′-TAGTGCCCAGGGTACAGCTC-3′; forward 5′-TTCACATTGCATGTGTGG-3′; reverse 5′-TAGCCTGCGTAGTGTTGGTG-3′. The expected band sizes for endogenous and Tg mice are 423 and 620 bps, respectively. Sirt3-KO mice were provided by the laboratory of F.W. Alt (Harvard Medical School, Boston, Massachusetts, USA) (42).
Induction of hypertrophy in mice.
The adult mice were subjected to forced swimming exercise for 12 weeks. The constriction of thoracic aorta (band) was done for 6 weeks as described earlier (36). The development of hypertrophy was judged noninvasively by use of echocardiography. Ang II (American Peptide Company Inc.), PE, and ISO (Sigma-Aldrich) were dissolved in 150 mM NaCl and 1 mM acetic acid, and they were delivered (Ang II, 3.0 mg/kg/d for 14 days; PE, 75 mg/kg/d for 14 days; ISO, 8.7 mg/kg/d for 7 days) by implanting of Osmotic Minipumps (model 2002, 2001; ALZET) into the abdomens of adult mice. Control mice underwent the same procedure, except that the respective pumps were filled only with vehicle (150 mM NaCl and 1 mM acetic acid).
Echocardiography of mice.
Transthoracic echocardiography in mice was performed under inhaled isoflurane (~1%) for anesthesia, delivered via nose cone. Chest hairs were removed with a topical depilatory agent. Limb leads were attached for electrocardiogram gating, and the animals were imaged in the left lateral decubitus position with a VisualSonics Vevo 770 machine, using a 30 MHz high-frequency transducer. Body temperature was maintained using a heated imaging platform and warming lamps. Two-dimensional images were recorded in parasternal long- and short-axis projections, with guided M-mode recordings at the midventricular level in both views. LV cavity size and wall thickness were measured in at least 3 beats from each projection and averaged. LV wall thickness (interventricular septum [IVS] and posterior wall [PW] thickness) and internal dimensions at diastole and systole (LVIDd and LVIDs, respectively) were measured. LV fractional shortening ([LVIDd – LVIDs]/LVIDd) and relative wall thickness ([IVS thickness + PW thickness]/LVIDd) were calculated from the M-mode measurements.
Primary cultures of cardiomyocytes, transfection/infection, and ROS measurement.
Neonatal rat or mouse cardiomyocytes were cultured and infected with adenoviral vectors as described earlier (8). NFAT-luciferase and Foxo-luciferase plasmids contain multiple binding sites of NFAT and Foxo3a, respectively. Cells were transfected with use of Tfx-20 Reagent (Promega). The luciferase assay was performed with use of a Luciferase Activity Kit from Promega. MitoSox (Invitrogen) was used for detecting ROS generation in cardiomyocytes as per the manufacturer’s protocol.
Subcellular fractionation and enzyme activity assays.
Nuclear and cytoplasmic fractions of cardiomyocytes were generated as described elsewhere (8). The enzymatic activity of MnSOD and catalase was measured by use of a kit (Cayman Inc.) according to the manufacturer’s protocol.
Histology and immunohistochemistry.
Heart sections were stained with wheat germ agglutinin coupled to tetramethylrhodamine isothiocyanate (Sigma-Aldrich) for detection of cell size. The cell size of myocytes was measured by using NIH ImageJ software (http://rsbweb.nih.gov/ij/). Fibrosis was detected with Masson’s Trichrome Stain (Sigma-Aldrich) by standard techniques. Sarcomere reorganization and ANF release were measured by staining of cardiomyocytes with antibodies specific for α-actinin and ANF, respectively. For the analysis of cellular distribution of NFAT, GATA4 and FoxO3a cells were immunostained with protein-specific antibodies, and the protein was localized by use of confocal microscopy (8).
Antibodies.
Sirt3 antibodies were bought from Abgent and Orbigen Inc. Antibodies against GAPDH, Histone 3, GATA4, NFAT, glutathione-S-transferase, Actin, and Tubulin were from Santa Cruz Biotechnology Inc. Antibodies against α-actinin, Catalase, and ANF were purchased from Sigma-Aldrich. Anti-Ras and anti-MnSOD antibodies were from Upstate, and antibodies against FoxO3a were purchased from Abcam Inc. Antibodies against signaling molecules were purchased from Cell Signaling Technology Inc.
Real-time PCR analysis of mRNA levels.
Anf, Myh7, Cat, and MnSOD mRNA levels were quantified by SYBR Green Real-Time PCR (Invitrogen) per the protocol described earlier (43). Briefly, total cellular RNA was isolated from cardiomyocytes using the TRIzol Reagent (Invitrogen). Residual genomic DNA was digested by incubating the RNA preparation with 0.5 units of RNase-free DNase-I per μg of total RNA in 1x reaction buffer for 15 minutes at room temperature, followed by heat inactivation at 90°C for 5 minutes. The quality of DNase-I–treated RNA was tested by formaldehyde-agarose gel electrophoresis. Two micrograms of DNase-I–treated RNA was reverse transcribed using the SuperScript III Kit (Invitrogen). The resultant cDNA was diluted 10-fold prior to PCR amplification. A reverse transcriptase minus reaction served as a negative control.
The nucleotide sequences of the PCR primers used were as follows: ANF, (position 602F–739R), forward 5′-TAAGCCCTTGTGGTGTGTCA-3′ and reverse 5′-GCAAGACCCCACTAGACCAC-3′; β-MHC, (position 5929F–6005R), forward 5′-AAGGGCCTGAATGAGGAGTA-3′ and reverse 5′-AAAGGCTCCAGGTCTGAGG-3′; MnSOD, (position 193F–263R), forward 5′-TTAACGCGCAGATCATGCA-3′ and reverse 5′-GGTGGCGTTGAGATTGTTCA-3′; Catalase, (position 1593F–1655R), forward 5′-TGAGAAGCCTAAGAACGCAATTC-3′ and reverse 5′-CCCTTCGCAGCCATGTG-3′; β-actin, forward 5′-CAAGATCATTGCTCCTCCTG-3′ and reverse 5′-TCATCGTACTCCTGCTTGCT-3′. The expression of genes involved in antioxidant defense was quantified by a RT2 Profiler PCR Array (PARN-065; SABiosciences Inc.) per the manufacture’s instructions.
Statistics.
Paired 2-tailed Student’s t test was applied to determine statistical significance between 2 groups. P values of less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank F.W. Alt for providing Sirt3-KO mice. We also thank C. Labno and S. Bond for their technical assistance in microscopic analyses. This study was partially supported by NIH grants RO1 HL-68083, HL-77788, and HL-83423 (to M.P. Gupta).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 119:2758–2771 (2009). doi:10.1172/JCI39162.
References
- 1.Frey N., Katus H.A., Olson E.N., Hill J.A. Hypertrophy of the heart: a new therapeutic target? Circulation. 2004;109:1580–1589. doi: 10.1161/01.CIR.0000120390.68287.BB. [DOI] [PubMed] [Google Scholar]
- 2.Fiedler B., Wollert K.C. Interference of antihypertrophic molecules and signaling pathways with the Ca2+-calcineurin-NFAT cascade in cardiac myocytes. Cardiovasc. Res. 2004;63:450–457. doi: 10.1016/j.cardiores.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Heineke J., Molkentin J.D. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 4.Blander G., Guarente L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 5.Scher M.B., Vaquero A., Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21:920–928. doi: 10.1101/gad.1527307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwer B., North B.J., Frye R.A., Ott M., Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hallows W.C., Lee S., Denu J.M. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sundaresan N.R., Samant S.A., Pillai V.B., Rajamohan S.B., Gupta M.P. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol. Cell. Biol. 2008;28:6384–6401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi T., Wang F., Stieren E., Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J. Biol. Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 10.Ahn B.H., et al. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellizzi D., et al. A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics. 2005;85:258–263. doi: 10.1016/j.ygeno.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Rose G., et al. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp. Gerontol. 2003;38:1065–1070. doi: 10.1016/S0531-5565(03)00209-2. [DOI] [PubMed] [Google Scholar]
- 13.Akazawa H., Komuro I. Roles of cardiac transcription factors in cardiac hypertrophy. Circ. Res. 2003;92:1079–1088. doi: 10.1161/01.RES.0000072977.86706.23. [DOI] [PubMed] [Google Scholar]
- 14.Ichida M., Finkel T. Ras regulates NFAT3 activity in cardiac myocytes. J. Biol. Chem. 2001;276:3524–3530. doi: 10.1074/jbc.M004275200. [DOI] [PubMed] [Google Scholar]
- 15.Rao A., Luo C., Hogan P.G. Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 16.Gingras A.C., Raught B., Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 17.Pyronnet S. Phosphorylation of the cap-binding protein eIF4E by the MAPK-activated protein kinase Mnk1. Biochem. Pharmacol. 2000;60:1237–1243. doi: 10.1016/S0006-2952(00)00429-9. [DOI] [PubMed] [Google Scholar]
- 18.Barron A.J., Finn S.G., Fuller S.J. Chronic activation of extracellular-signal-regulated protein kinases by phenylephrine is required to elicit a hypertrophic response in cardiac myocytes. Biochem. J. 2003;371:71–79. doi: 10.1042/BJ20021395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugden P.H., Fuller S.J., Weiss S.C., Clerk A. Glycogen synthase kinase 3 (GSK3) in the heart: a point of integration in hypertrophic signalling and a therapeutic target? A critical analysis. Br. J. Pharmacol. 2008;153(Suppl. 1):S137–S153. doi: 10.1038/sj.bjp.0707659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Proud C.G. Ras, PI3-kinase and mTOR signaling in cardiac hypertrophy. Cardiovasc. Res. 2004;63:403–413. doi: 10.1016/j.cardiores.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Sugden P.H. Ras, Akt, and mechanotransduction in the cardiac myocyte. Circ. Res. 2003;93:1179–1192. doi: 10.1161/01.RES.0000106132.04301.F5. [DOI] [PubMed] [Google Scholar]
- 22.Sugden P.H., Clerk A. Activation of the small GTP-binding protein Ras in the heart by hypertrophic agonists. Trends Cardiovasc. Med. 2000;10:1–8. doi: 10.1016/S1050-1738(00)00038-4. [DOI] [PubMed] [Google Scholar]
- 23.Kuster G.M., et al. Alpha-adrenergic receptor-stimulated hypertrophy in adult rat ventricular myocytes is mediated via thioredoxin-1-sensitive oxidative modification of thiols on Ras. Circulation. 2005;111:1192–1198. doi: 10.1161/01.CIR.0000157148.59308.F5. [DOI] [PubMed] [Google Scholar]
- 24.Sawyer D.B., et al. Role of oxidative stress in myocardial hypertrophy and failure. J. Mol. Cell. Cardiol. 2002;34:379–388. doi: 10.1006/jmcc.2002.1526. [DOI] [PubMed] [Google Scholar]
- 25.Sugden P.H., Clerk A. Oxidative stress and growth-regulating intracellular signaling pathways in cardiac myocytes. Antioxid. Redox. Signal. 2006;8:2111–2124. doi: 10.1089/ars.2006.8.2111. [DOI] [PubMed] [Google Scholar]
- 26.Berndt C., Lillig C.H., Holmgren A. Thiol-based mechanisms of the thioredoxin and glutaredoxin systems: implications for diseases in the cardiovascular system. Am. J. Physiol. Heart Circ. Physiol. 2007;292:H1227–H1236. doi: 10.1152/ajpheart.01162.2006. [DOI] [PubMed] [Google Scholar]
- 27.Daitoku H., et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan W.Q., Wang K., Lv D.Y., Li P.F. Foxo3a inhibits cardiomyocyte hypertrophy through transactivating catalase. J. Biol. Chem. 2008;283:29730–29739. doi: 10.1074/jbc.M805514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frescas D., Valenti L., Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J. Biol. Chem. 2005;280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 30.Huang H., Tindall D.J. Dynamic FoxO transcription factors. J. Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 31.Cui M., Huang Y., Zhao Y., Zheng J. Transcription factor FOXO3a mediates apoptosis in HIV-1-infected macrophages. J. Immunol. 2008;180:898–906. doi: 10.4049/jimmunol.180.2.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin S.J., Ford E., Haigis M., Liszt G., Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Revollo J.R., Grimm A.A., Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 34.Yang H., et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Opie L.H., Owen P. Effects of increased mechanical work by isolated perfused rat heart during production or uptake of ketone bodies. Assessment of mitochondrial oxidized to reduced free nicotinamide-adenine dinucleotide ratios and oxaloacetate concentrations. Biochem. J. 1975;148:403–415. doi: 10.1042/bj1480403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pillai J.B., Isbatan A., Imai S., Gupta M.P. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. J. Biol. Chem. 2005;280:43121–43130. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- 37.Kwon H.S., Ott M. The ups and downs of SIRT1. Trends Biochem. Sci. 2008;33:517–525. doi: 10.1016/j.tibs.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 38.Kanfi Y., et al. Regulation of SIRT1 protein levels by nutrient availability. FEBS Lett. 2008;582:2417–2423. doi: 10.1016/j.febslet.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 39.Kops G.J., et al. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 40.Nemoto S., Finkel T. Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science. 2002;295:2450–2452. doi: 10.1126/science.1069004. [DOI] [PubMed] [Google Scholar]
- 41.Gulick J., Subramaniam A., Neumann J., Robbins J. Isolation and characterization of the mouse cardiac myosin heavy chain genes. J. Biol. Chem. 1991;266:9180–9185. [PubMed] [Google Scholar]
- 42.Lombard D.B., et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong J., Sulik K.K., Chen S.Y. Nrf2-mediated transcriptional induction of antioxidant response in mouse embryos exposed to ethanol in vivo: implications for the prevention of fetal alcohol spectrum disorders. Antioxid. Redox. Signal. 2008;10:2023–2033. doi: 10.1089/ars.2007.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.