Abstract
Vaccination has played a significant role in controlling and eliminating life-threatening infectious diseases throughout the world, and yet currently licensed vaccines represent only the tip of the iceberg in terms of controlling human pathogens. However, as we discuss in this Review, the arrival of the genome era has revolutionized vaccine development and catalyzed a shift from conventional culture-based approaches to genome-based vaccinology. The availability of complete bacterial genomes has led to the development and application of high-throughput analyses that enable rapid targeted identification of novel vaccine antigens. Furthermore, structural vaccinology is emerging as a powerful tool for the rational design or modification of vaccine antigens to improve their immunogenicity and safety.
Introduction: genomics, the foundation of new vaccine principles
Vaccination is considered to be one of the public health interventions that has had the greatest impact on world health, yet infectious disease remains the leading cause of death worldwide and the vaccines in use today represent only the tip of the iceberg when considering the number of diseases that need to be targeted (Figure 1A). However, the arrival of the genome era has radically changed the way identification of vaccine candidates is approached (Figure 1B). As a result of technological revolutions and shifting paradigms, we have entered a renaissance in vaccine development in which rapid targeted identification of novel vaccine antigens is possible through large-scale high-throughput genomic, transcriptomic, and proteomic analyses (Figure 2 and Table 1).
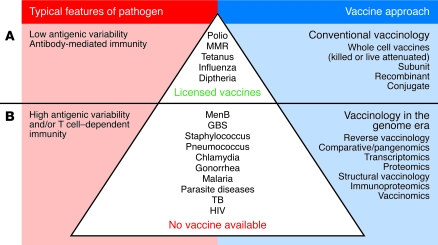
Figure 1. Schematic overview of conventional vaccinology versus vaccinology in the genome era.
(A) Most licensed vaccines target pathogens that have low antigenic variability and pathogens for which protection depends on antibody-mediated immunity. These vaccines have typically been developed using conventional vaccinology. (B) Several pathogens are shown for which no vaccine is available, due to either their high antigenic variability and/or the need to induce T cell–dependent immunity to elicit protection. New approaches are being applied to vaccine development for these pathogens in the genome era. Vaccines/diseases shown in the figure are selected examples of each category and are not a complete list. TB, M. tuberculosis.
Figure 2. Schematic overview of the way in which high-throughput analyses applied to various aspects of a pathogen and its interactions with the host immune system are used to identify vaccine candidates in the genome era.
From the point of view of the pathogen (starting at the lower left-hand corner and moving in a clockwise direction), vaccine candidates can be identified by analysis of the organism’s genome and/or pan-genome (the complete genetic content of the organism/species, which contains the complete repertoire of antigens that an organism/species is capable of expressing), transcriptome (the complete set of RNA transcripts expressed by an organism under a specified condition), proteome (the complete set of proteins expressed by an organism under a specified condition), surface proteome (the subset of proteins that are surface exposed), or structural genome (the 3D structure of the proteins of an organism, in particular the structural epitopes of immunogenic antigens). With respect to interactions between the pathogen and the host immune system, vaccine candidates can also be identified by analysis of the organism’s immunoproteome (the set of antigens that interact with the host immune system). The newer field of vaccinomics (the way in which individual host immune systems respond to a vaccine) will also aid in future vaccine development.
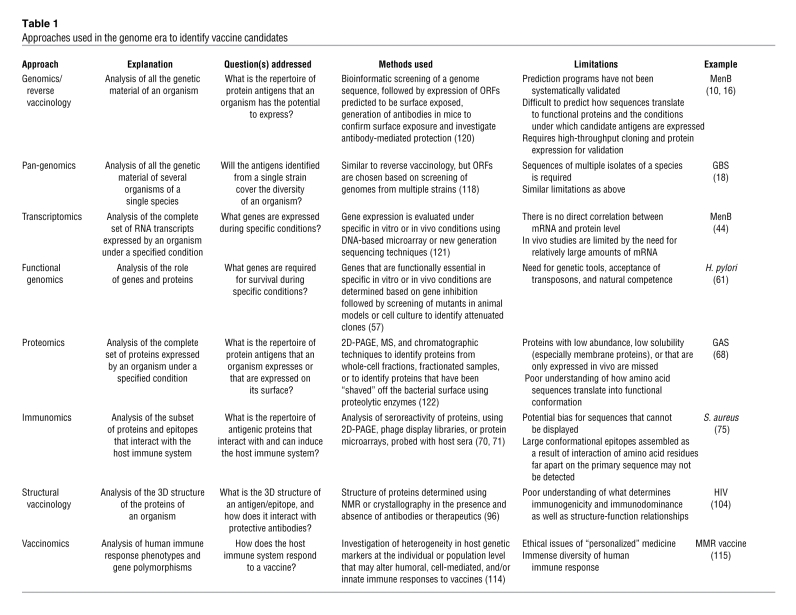
Table 1 .
Approaches used in the genome era to identify vaccine candidates
Since the origin of modern vaccination in 1796, with the discovery of the smallpox vaccine, there have been numerous technological advances and breakthroughs in the fight against infectious disease (1–3). However, most successful vaccines have been developed using conventional methods that follow the paradigm established by Pasteur over a century ago, namely to “isolate, inactivate, and inject” the disease-causing microorganism. Hence, most vaccines available for human use consist of either whole microorganisms (either killed or live attenuated) or purified subunits of a microorganism; only a small number are based on recombinantly produced antigens. Furthermore, available vaccines primarily target microorganisms that have little or no antigenic diversity or variability and for which vaccination induces antibody-mediated protective immunity (e.g., the microorganisms that cause polio and diphtheria) (Figure 1A) (4, 5). Conventional vaccinology has often proven to be inadequate in the development of vaccines for those pathogens that are antigenically diverse, those that cannot be cultivated in the laboratory, those that lack suitable animal models of infection, and/or those that are controlled by mucosal or T cell–dependent immune responses (Figure 1B) (4, 6).
The genome era, initiated with the completion of the first bacterial genome, that of Haemophilus influenzae in 1995 (7), catalyzed a long overdue revolution in vaccine development. Advances in sequencing technology and bioinformatics have resulted in an exponential growth of genome sequence information, and at least one genome sequence is now available for each major human pathogen. As of August 2009, more than 880 bacterial genomes have been completed and more than 2700 are ongoing (GOLD Genomes OnLine Database, http://www.genomesonline.org/gold.cgi; NCBI: Microbial Genomes, http://www.ncbi.nlm.nih.gov/genomes/MICROBES/microbial_taxtree.html; JCVI-CMR Comprehensive Microbial Resource, http://cmr.jcvi.org/tigr-scripts/CMR/shared/Genomes.cgi.; ref. 8). The application of genome analysis to vaccine development, a concept termed “reverse vaccinology,” initiated a positive feedback loop in terms of the development and application of novel approaches to the field of vaccinology. As a result, it is becoming possible to systematically examine almost every aspect of a pathogen and its interactions with the host immune system in the search for vaccine candidates (Figure 2; Table 1). Reverse vaccinology applied to the genome of a pathogen aims to identify the complete repertoire of antigens that an organism is capable of expressing on its surface. Transcriptomics and proteomics enable the investigation of the array of antigens actually expressed by a pathogen under specified conditions, by examining the mRNA and protein of the organism, respectively. Analysis can also focus on the subset of proteins that are surface exposed (surface proteome) or the subset of genes that are functionally important for infection (functional genomics). Newer fields of study are focused on elucidating the set of antigens that interact with the host immune system and the mechanisms involved in these interactions (immunomics), the structural epitopes of immunogenic antigens (structural vaccinology), and the way in which individual host immune systems respond to a vaccine (vaccinomics). While each of these approaches has limitations (Table 1), they have all emerged as powerful tools in vaccine development. Here, we outline these approaches and the way in which vaccinology in the genome era is bringing us closer to developing vaccines that were previously out of reach.
Reverse vaccinology
Classical reverse vaccinology: from one genome to a comprehensive serogroup B meningococcal vaccine.
The genome sequence of a microorganism provides unprecedented access to the complete repertoire of its antigens, from which vaccine candidates can be selected through rapid and intelligent screening processes. Serogroup B Neisseria meningitidis (MenB), the most common cause of meningococcal disease in the developed world, is the prototypic example of an organism for which several decades of conventional vaccine development failed to produce a comprehensive vaccine (reviewed in ref. 9), yet the use of reverse vaccinology by Novartis Vaccines identified more vaccine candidates in 18 months than had been discovered during the previous 40 years (10), and this has driven vaccine development into clinical development (Figure 3).
Figure 3. Flow chart of MenB vaccine development.
Preclinical development was based on a reverse vaccinology approach, in which the genome sequence of the virulent MenB strain MC58 was used to identify ORFs predicted to encode proteins that were surface exposed (i.e., secreted [S] or located in the outer membrane [OM]), which were then expressed in E. coli, purified, and used to immunize mice. Antibodies generated in mice were then used to confirm surface exposure of the vaccine candidate by FACS and to identify proteins that induced bactericidal activity. This screening process resulted in identification of several novel vaccine candidates, including GNA1870 (which is fHBP), GNA1994 (which is NadA), GNA2132, GNA1030, and GNA2091. The formulation for the comprehensive MenB vaccine consists of four components: fHBP-GNA2091 and GNA2132-GNA1030 fusion proteins, NadA, and OMV from the New Zealand MeNZB vaccine strain. Clinical development using this formulation has shown in phase I and II trials that the vaccine is well tolerated and immunogenic. The vaccine induced bactericidal activity using human complement (hSBA) with titers greater than 1:4, which indicates the generation of antibodies able to kill the bacteria at a level that correlates with protection against the bacteria, in more than 90% of infants after the fourth dose. This vaccine entered phase III clinical trials in 2008. P, periplasm; IM, inner membrane; C, cytoplasm.
MenB vaccine candidates identified by conventional approaches, all of which have failed to produce a clinically useful vaccine, can be subdivided based on the reasons for the failure. First, although the capsule polysaccharides of N. meningitidis serogroups other than serogroup B (specifically serogroups A, C, Y, and W-135) have been successfully used to make conventional vaccines for these pathogens, the capsule polysaccharide of MenB failed to be a suitable vaccine candidate because it is identical to human polysialic acid and is therefore poorly immunogenic (11, 12). Second, a number of MenB vaccine candidates identified by conventional approaches proved to be hypervariable and/or poorly conserved between the diverse MenB strains that cause endemic disease, making it unlikely that they would provide broad protection against MenB (13).
In 2000, a new resource for meningococcal vaccine development became available with completion of the genome of the virulent MenB strain MC58, giving in silico access to 2158 predicted ORFs to screen for novel vaccine antigens (14). Assuming that surface-exposed antigens are the most suitable vaccine candidates, due to their potential to be recognized by the immune system, the draft MC58 genome was screened using bioinformatics tools, leading to the identification of 570 ORFs predicted to encode either surface-exposed or secreted proteins (10). Antigen selection then continued based on a number of criteria: the ability of candidates to be cloned and expressed in Escherichia coli as recombinant proteins (350 candidates); the confirmation of surface exposure by ELISA and FACS analysis; the ability of induced antibodies to elicit protective immunity, as measured by serum bactericidal assay and/or passive protection in infant rats (28 candidates); and screening to determine the conservation of antigens within a panel of diverse meningococcal strains, primarily containing disease-associated MenB strains.
Five antigens identified by reverse vaccinology, genome-derived Neisseria antigen 1870 (GNA1870; which is factor H–binding protein [fHBP]), GNA1994 (which is NadA), GNA2132 (GenBank accession number NP_275117), GNA1030 (GenBank accession number AAF41429), and GNA2091 (GenBank accession number NP_275079), and outer membrane vesicles (OMV) from the New Zealand MeNZB vaccine strain, which contains the immunogen PorA (15), have been combined to form the Novartis MenB vaccine that is now in phase III clinical trials. The vaccine formulation consists of a fHBP-GNA2091 fusion protein, a GNA2132-GNA1030 fusion protein, NadA, and OMV. The multivalent vaccine approach was taken due to the antigenic diversity of disease-causing MenB strains and should strengthen the protective activity of the vaccine, increase the breadth of MenB strains targeted by the vaccine, and prevent the selection of escape mutants (i.e., bacteria that have a mutation in a gene encoding an antigen that would allow them to escape killing or neutralization by vaccine-induced antibodies). When tested against a panel of 85 meningococcal isolates (predominantly MenB isolates) representative of the global population of disease-causing strains, the vaccine induced bactericidal antibodies in mice against 78% and 90% of strains when administered with the adjuvants aluminium hydroxide and MF59 (an oil-in-water emulsion), respectively (16). Initial phase II clinical results in adults and infants indicated that this vaccine was well tolerated and induced a protective immune response against three diverse MenB strains in 89%–96% of subjects following three vaccinations and 93%–100% after four vaccinations (17). This vaccine therefore has the potential to provide broad protection to infants against MenB infections, and phase III clinical testing began in the first quarter of 2008.
This initial success of reverse vaccinology in developing a vaccine for MenB served as a proof-of-concept for this approach and catalyzed a paradigm shift in vaccine development. This process is independent of several of the constraints of classical vaccinology, such as the need to culture the pathogen in vitro. Genome-based vaccine discovery projects have since been initiated for a range of pathogens, including Streptococcus agalactiae (18), Streptococcus pneumoniae (19), Porphyromonas gingivalis (20), Chlamydia pneumoniae (21), Bacillus anthracis (22, 23), and Brucella melitensis (24). These projects are based on stand-alone reverse vaccinology approaches, refined reverse vaccinology approaches (e.g., using pangenomics), and/or a combination of genomic, proteomic, and transcriptomic approaches.
Pan-genomic reverse vaccinology: a universal vaccine against group B Streptococcus.
Comparative analyses of the genomes of multiple isolates of a bacterial pathogen and of closely related pathogenic and nonpathogenic bacteria are of considerable scientific interest, not only for the information gained regarding genome size, gene content, and gene conservation or variability among different strains and their associated diseases, but also for the implications for effective vaccine and drug-discovery programs. Multi-genome comparisons can be performed using either genome sequence– or microarray-based methods.
S. agalactiae (also known as group B Streptococcus [GBS]) is a Gram-positive pathogen that causes life-threatening pneumonia, sepsis, and meningitis in newborn and young infants; there is currently no vaccine against GBS. In 2002, the complete genome sequences of two clinical isolates of GBS, the type III NEM316, responsible for a fatal case of septicemia, and the type V 2603V/R, an emerging capsular serotype, were determined (25, 26). The sequenced 2603V/R strain and 19 other GBS clinical isolates from several serotypes were analyzed by DNA microarray comparative genomic hybridization, which revealed substantial genetic heterogeneity, even among strains with the same serotype, and in particular between genes that are expected to play a role in disease, such as transcriptional regulators and surface proteins (26). Hence, the complete genome sequences of an additional six strains, representing the major serotypes causing invasive diseases, were generated (27). Comparative analysis of the eight complete GBS genomes represented a revolution in genomics and led to the introduction of the pan-genome concept to define the global gene repertoire of a species, with inevitable implications for pathogenesis, vaccine design, and definition of the species. Although the eight GBS strains revealed a similar number of predicted genes, the analysis revealed an unexpected degree of intraspecies diversity (27). Each strain contained a core genome that was present in all strains (an average of 1806 genes typically involved in housekeeping functions), a dispensable genome comprising a set of genes present in two or more strains (an average of 439 genes typically with hypothetical or unknown functions), and strain-specific genes found only in a single isolate. The most surprising finding that emerged from these analyses was that the GBS pan-genome is open and theoretically unlimited, and mathematical extrapolations enabled the estimation that for every new GBS genome sequenced, an average of 33 new strain-specific genes will be added to the GBS pan-genome (27, 28).
The identification of protein vaccine candidates suitable for a broadly protective vaccine against GBS infections represents the first important example of how reverse vaccinology has been refined from a classical approach based on one genome sequence to a pan-genome concept that is better able to describe a bacterial species (18). Comparing the complete genome sequences of the eight available GBS strains and using bioinformatics algorithms, Maione and coworkers at Novartis Vaccines selected a total of 589 genes encoding putative surface-exposed and secreted proteins from within the GBS pan-genome (18). Among the selected proteins, 396 belonged to the core genome and 193 were present in the dispensable genome. A total of 312 proteins were successfully expressed in E. coli as soluble recombinant proteins, and their capacity to elicit protection was evaluated in an active maternal immunization/neonatal challenge model. From this systematic screening, four antigens were selected for their capacity to elicit protection in infant mice. Only one of these antigens, the Sip protein, which had been previously described as a potential vaccine candidate (29), belonged to the core genome. The other three proteins were part of the dispensable portion of the GBS pan-genome. Nevertheless, the combination of these proteins conferred protection against a large panel of GBS strains representative of all circulating serotypes (18). This work showed the importance of screening more than one genome for the identification of a broadly protective vaccine against pathogens, such as GBS, with highly variable circulating strains.
The implications of open, closed, and finite pan-genomes in vaccine development.
With the exponential increase in recent years of the number of species for which multiple complete genome sequences are available, it has been possible to apply the GBS pan-genome model to many other pathogens (28). The open pan-genome model is valid for E. coli (30); however, the pan-genome of B. anthracis can be fully described by only four genomes and is therefore considered “closed” (31). This probably reflects the fact that B. anthracis is a highly clonal species that recently evolved from Bacillus cereus. Recently, a new “finite” pan-genome (named a supragenome) has been proposed based on comparison of 13 strains of H. influenzae (32) and 17 strains of S. pneumoniae (33). Using the assumption that dispensable genes are not accumulated in the population with equal probability, the authors predict that the overall number of genes within these species is limited and a finite number of new genomes would be sufficient to define the complete supragenome of these species. Irrespective of the pan-genome size, it is evident that a species cannot be characterized by a single genome sequence. Although the core genes, generally highly conserved and present in all strains, represent the most desirable source of potentially universal antigens, the group of dispensable genes, even though not present in all strains, might be an endless source of important virulence factors that exploited in appropriate combinations might elicit a broad immune response.
Discovery of new structures and functions: pili in pathogenic streptococci as promising vaccine candidates.
The recent identification of pili (long filamentous appendages that extend from the bacterial cell surface; ref. 34) in the main pathogenic strains of streptococci, facilitated by the identification of a characteristic gene organization in pilus islands, represents an example of the impact of genomics in accelerating the discovery of promising vaccine candidates. Three protective antigens of GBS identified at Novartis Vaccines by pan-genomic reverse vaccinology assemble into high–molecular weight polymers visible by EM as pilus-like structures (35). Furthermore, three pilus islands have been discovered in GBS that encode structurally distinct pilus types (36). Each pilus contains two antigens capable of eliciting protective immunity in mice (37). Typical pilus regions have also been identified in the complete available genomes of group A Streptococcus (GAS), and a combination of recombinant pilus proteins was shown to confer protection in mice against mucosal challenge with virulent GAS isolates (38). The availability of multiple complete genome sequences for S. pneumoniae has also allowed the discovery of two pilus islands that contribute to adherence to lung epithelial cells as well as to colonization in a murine model of infection, where they elicit host inflammatory responses (39, 40). In addition, the S. pneumoniae pilus subunits are immunogenic in mice and are able to confer protection in passive and active immunization models (41).
Although the components of pili have long been regarded as potential targets of vaccine development due to their essential role in colonizing host tissue, the pilus-based vaccines tested so far have unfortunately failed due to the high variability of their protein antigens and their inability to induce protection against heterologous strains (34). Nevertheless, the presence of pili that contain protective antigens in all three principal streptococcal pathogens provides support for the hypothesis that these structures play an important role in virulence, a notion that has proven controversial. Of clear interest is the recent demonstration that, at least for GBS, the variability of pili is limited and that a combination of only three pilin subunits induces broad protective immunity against GBS strains in mice: information that was extrapolated to calculate that a vaccine containing pilus components from all three islands could confer protection against 94% of GBS strains (37). Further, it has been estimated, using sequence and epidemiologic data, that a vaccine comprising a combination of 12 pilin variants could theoretically protect against more than 90% of currently circulating GAS strains (42).
Transcriptomics: expression analysis reveals vaccine candidates
The availability of complete microbial genome sequences has facilitated the design of comprehensive DNA-based microarray chips, which have been exploited in various ways for vaccine development. The transcriptome of a pathogen (the complete set of RNA transcripts expressed under a specified condition) can be analyzed to identify and characterize the expression of potential antigens important for pathogenesis and/or survival in the host (43).
Once again, MenB serves as a prototypic example of the application of microarray-based transcriptional profiling to identify novel vaccine candidates (44). Analysis of the MenB transcriptome during adhesion to host epithelial cells led to the identification of 189 genes with increased expression under conditions that mimicked in vivo host-pathogen interactions (44). Twelve of these genes were confirmed by FACS analysis to express surface proteins accessible to the immune system (with four of these being detected only after adhesion to epithelial cells), five of which induced protective antibodies in mice. Transcriptional profiling of MenB, performed during exposure to human serum and endothelial cells (45), iron limitation (46), and oxygen starvation (47), has identified several additional genes predicted to encode proteins involved in pathogenesis and has implied the potential function of several uncharacterized genes based on their expression during specific experimental conditions.
Several interesting variations of microarray-based approaches have since been used and are highlighted here. Genes expressed by the cholera-causing bacterium Vibrio cholerae during human infection were identified using bacteria directly isolated from diseased individuals (48). Comparison of the transcriptome of V. cholerae isolated from patient stool samples with that of bacteria grown in vitro greatly increased the understanding of the hyperinfectious state that is seen after passage of bacteria through the human gastrointestinal tract (48). Furthermore, transcriptomic comparison of the Staphylococcus aureus clinical isolate UAMS-1 with the prototype laboratory strain RN6390 revealed substantial differences in expression of genes encoding surface proteins (high in UAMS-1) and those encoding proteins involved in exotoxin production (low in UAMS-1) (49), providing insights into pathogenesis and highlighting the importance of studying clinically relevant strains for vaccine development. The transcriptome of S. pneumoniae during interaction with human macrophages (50) and human lung epithelial cells (51) has been determined, and additional analyses have been undertaken to compare the transcriptome of an encapsulated pathogenic strain with that of an unencapsulated avirulent strain during association with lung cells to help understand differences between pathogenic and commensal strains of the bacterium. Further, the transcription profile of Mycobacterium tuberculosis during the course of early tuberculosis in immunocompetent BALB/c and SCID mice revealed a set of 67 genes activated exclusively in the lungs of immunocompetent BALB/c mice, providing an insight into the bacterial response to the host immune system (52).
The application of transcriptome analysis to vaccine development is expected to greatly advance with improving technologies for differentially extracting microbial RNA from tissues during in vivo experiments (53, 54). The increasing availability of microarrays (e.g., arrays for 39 different pathogens are available free of cost from the J. Craig Venter Institute: Pathogen Functional Genomics Resource Center; http://pfgrc.jcvi.org/index.php/microarray/available_microarrays.html) and the new wave of microarray-independent gene-expression analyses based on advances in ultra-high–throughput pyrosequencing technology, which have enabled the rapid sequencing of cDNA and quantification of sequence reads (55, 56), should enable further transcriptome-based advances in vaccine development.
Functional genomics: essential genes as vaccine candidates
High-throughput functional screens can now be performed on a genomic scale by combining whole-genome microarrays and comprehensive ordered libraries of mutants, enabling identification of specific vaccine candidates based on genes essential for microorganism survival and/or pathogenesis. These screens have also proven highly valuable in assigning functions to the numerous uncharacterized ORFs identified in genome sequences and in identifying mutants that could serve as live vaccines or delivery systems for heterologous antigens (57).
Several functional genomics approaches are based on the inhibition of genes using transposon mutagenesis approaches (i.e., strategies that utilize the ability of a transposon to insert into a gene and inactivate it), such as signature-tagged mutagenesis (STM), genome analysis and mapping by in vitro transposition (GAMBIT), and transposon site hybridization (TraSH), followed by the screening of mutants in animal models or cell culture to identify attenuated clones (reviewed in refs. 58, 59). STM has been applied to more that 30 pathogens, including MenB, where 65 novel genes required for infection that causes septicemia in infant rats were identified (60), and Helicobacter pylori, where 47 genes essential for colonization of the gerbil stomach were identified (61). Also of interest have been studies using TraSH to identify the mechanisms by which M. tuberculosis persists in the host (62) and those using GAMBIT to determine the complete set of genes required by H. influenzae for growth and viability in vitro (63). For pathogens that do not readily accept transposons, such as S. aureus (64) and Staphylococcus epidermidis (65), antisense RNA–mediated transcriptional attenuation has been used to identify genes that are essential for infection and/or pathogenesis.
In vivo expression technology (IVET) and recombinase-based in vivo expression technology (RIVET) are alternative gene-expression techniques that have greatly benefited from the availability of genome sequences and advances in screening methods. These approaches enable identification of microbial promoters, from a library of transcriptional fusions of genomic DNA to a reporter gene, that are specifically induced during infection (66). For example, application of RIVET to an attenuated V. cholerae strain (CVD110) identified 217 genes induced in human volunteers; many of these were specifically induced following human infection and not in mouse models of infection (67).
Proteomics: a complement to genomics for vaccine development
In the genomic era, proteomics-based approaches have rapidly developed and are now widely considered to be effective technologies that are complementary to classical genomic-based approaches for discovering surface-associated, immunogenic proteins that could be potential vaccine candidates. The availability of a constantly increasing number of complete genome sequences has enabled the proteomic community to rapidly and specifically identify proteins of interest from discrete cell compartments. 2D-PAGE coupled to mass spectrometry (MS), chromatographic techniques, and protein arrays are the principal proteomic methods used for analyzing, usually in high-throughput mode, the complete protein profile of a microorganism, including protein localization, protein-protein interactions, posttranslation modifications, and differential expression in specified conditions.
For most bacterial pathogens, proteins able to elicit a protective immune response are either secreted proteins or surface-exposed proteins, and these represent the most promising vaccine candidates. In silico analysis of the complete sequence of several bacterial genomes predicts that surface-associated proteins constitute 30%–40% of all bacterial proteins. Using this information, a novel proteomic-based approach was developed by Rodriguez-Ortega et al. (68) to specifically isolate bacterial surface proteins. The method uses proteolytic enzymes to “shave” the bacterial surface under conditions that preserve cell viability, and the peptides released are analyzed by MS. The peptide sequences are then matched with predicted gene sequences from published genomes, permitting a fast and selective identification of all proteins partially or entirely exposed on the bacterial surface. The effectiveness of this approach is supported by the fact that 95% of the 72 proteins identified in this way from the completely sequenced M1-SF370 strain of GAS (69) were predicted to be cell wall proteins, lipoproteins, transmembrane-spanning proteins, and secreted proteins and included most of the protective antigens described to date plus a novel protective antigen. Surface exposure of most of these proteins was confirmed by FACS analysis (68).
Immunomics: identification of immunogenic proteins
Circulating human antibodies induced by exposure of the host immune system to a pathogen represent a molecular imprint of the specific set of proteins able to elicit a humoral immune response during the course of infection. Therefore, a combination of genomic/proteomic-based approaches and serological analysis can identify potential vaccine candidates and provide effective validation of these candidates. The set of proteins identified by this approach, which are able to elicit a humoral immune response during the course of infection, represents the “immunoproteome” or “antigenome” of that pathogen (70, 71). A number of methods have been developed to enable high-throughput display of the proteome of a pathogen to the host immune system.
Immunomics (also known as serological proteome analysis [SERPA]) combines proteomic-based approaches with serological analysis and has been widely applied for antigen discovery and vaccine development (72–74). Applied to S. aureus, this approach led to the identification of 15 highly immunogenic proteins, including known and novel vaccine candidates (75). Surface proteins derived from S. aureus grown in vitro under various conditions were first resolved by 2D-PAGE, transferred onto a membrane, and then blotted with sera from either healthy individuals or S. aureus–infected patients preselected for the presence of antibodies specific for staphylococcal proteins. Highly reactive protein spots were then identified by MS analysis (75). Another example of SERPA combined with in silico analysis, applied to B. anthracis, has permitted the identification of a set of 84 immunogenic proteins that are expressed in vivo, providing a pool of potential candidates for vaccine development and diagnostic and therapeutic purposes (76). This approach has also been applied to select immunoreactive proteins from Klebsiella pneumoniae (77), Streptococcus pyogenes (78), S. pneumoniae (79), Streptococcus suis (80), and Clostridium difficile (81).
A limitation of the immunomic approaches is that differentially expressed proteins, such as those dependent on pathogen-host interaction, will not be detected. The use of phage display methods and protein microarray technologies can overcome this limitation and lead to the identification of the complete immunoproteome, where immunogenic polypeptides and proteins can be identified independently of their level of expression in vitro. Bacterial and phage-based expression libraries of either large polypeptides or small peptides encoded by random synthetic oligonucleotides of identical length have frequently been used (82). A novel genome-based approach that combines serological antigen identification and the use of a comprehensive genomic peptide library, potentially expressing all genome-encoded amino acid sequences, has been recently developed and termed antigenomics (83). For library construction, the pathogen genome was randomly sheared into 30- to 300-bp DNA fragments by mechanical and/or enzymatic treatments. The fragments were displayed as diversely sized peptides on the surface of E. coli via fusion to one of two outer membrane proteins (LamB and FhuA) and probed with convalescent sera selected for high antibody titers to identify immunogenic antigens. Using this approach, it is theoretically possible to define the global repertoire of all immunogenic proteins, including their antibody-binding sites, that are expressed by a pathogen in vivo, targeted by the human immune system, and therefore likely to induce antibodies in humans if used as vaccine antigens. The limitation of this approach is that conformational epitopes that are assembled as a result of interactions of amino acid residues far apart on the primary sequence may not be detected. Applied for the first time to S. aureus (83), this technology has since been extended to other human pathogens, S. epidermidis, S. agalactiae, S. pyogenes, and S. pneumoniae (71, 84). In addition to identifying most known protective proteins, it has also allowed the identification of novel classes of highly conserved antigens that are potential vaccine candidates. Based on the analysis of the antigenomes of five staphylococcal and streptococcal species, it has been estimated that the antigenome of a particular pathogen typically consists of approximately 100–200 antigens, most of them surface-exposed or secreted proteins (71).
Using protein microarray technologies with comprehensive high-throughput cloning and expression approaches may also rapidly lead to generation of the potential global immunoproteome of an infectious microorganism. All individual proteins are spotted onto microarray chips that are probed with sera to obtain immunodominant antigen profiles (85). Examples of protein microarray application are represented by determination of humoral immune response to Plasmodium falciparum infection (86), Francisella tularensis (87, 88), and V. cholerae (89). Another important application of protein microarrays permits the high-throughput examination of protein function by assessing their biochemical activities and interactions with other molecules (90).
The increasing knowledge regarding highly immunoreactive antigens offers useful information for further investigation of potential associations between specific immune responses and diseases (91–94). In addition, the comparison of multiple immunoproteomes may allow the discovery of immunogenic structural features shared and conserved between different pathogens, which could form the basis of broadly protective multispecies vaccines.
Structural vaccinology: the next generation of epitope mapping and vaccine design
Structural biology is increasingly being applied to vaccine development (structural vaccinology), focusing on determining and understanding the structural basis of immunodominant and immunosilent antigens, to enable the rational design of peptide mimetics of bactericidal epitopes (95, 96). The explosion of genome and proteome data, as well as improved protein expression, purification, and structural determination technologies, has led to the rapid development of the field of structural vaccinology (97). Approximately 30,000 high-resolution protein structures are available in public databases (predominantly for soluble proteins), and several initiatives have been established to pursue high-throughput characterization of protein structures on a genome-wide scale; hence, this approach is often referred to as “structural genomics” (98). The structure-based design of antiviral therapeutics is a well-established approach that has led to the development of drugs directed toward the active sites of the HIV-1 protease (99) and influenza neuraminidase (100).
Structural vaccinology is now being used to increase understanding of previously identified vaccine candidates; to identify new vaccine candidates; and to optimize the immunogenicity, stability, safety, and ease of production of promising vaccine antigens for many pathogens (reviewed in ref. 96). For example, structural and antigenic characterization of the HIV envelope has led to an increased understanding of the way in which the viral spike (which is composed of the viral proteins gp120 and gp41) evades the host antibody response (101, 102). For gp120, conformational antibody epitopes were found to be CD4 induced (103). To base vaccines on epitopes that exist only transiently during certain stages of the infection cycle, such as during contact with CD4, it is necessary to stabilize these antigen conformations by adding either disulfide bonds or other cross-links. However, high-resolution mapping of gp120-antibody complexes has revealed that a neutralizing antibody, b12, targets a conformationally invariant region of gp120 that is involved in binding to CD4 (104). Furthermore, another broadly neutralizing antibody, 2G12, which recognizes a carbohydrate epitope on gp120, has recently been mutated based on structural information, leading to an increased ratio of 2G12 dimer to monomer formation, thereby increasing its neutralization potency (105).
Even though these broadly protective antibodies have been identified, the problem remains that infection with HIV typically leads to an immune response against highly variable immunodominant epitopes (e.g., the V1, V2, and V3 loops of the envelope glycoprotein gp160) that does not provide protection against diverse strains. However, immune refocusing technology (i.e., engineering of an antigen to direct the immune system to new epitopes) has been used to remove or dampen the immunodominant gp160 epitopes in order to allow the host to respond to epitopes that were previously subdominant (ref. 106; reviewed in ref. 107). Engineered gp160, in which amino acids in V1 and V3 have been glycosylated, deleted, and/or substituted, induces comparable levels of gp160-specific antibodies as the wild-type unmodified antigen. The antibodies induced by both the modified and unmodified gp160 have similar neutralizing activity against homologous strains; however, those induced by the modified protein have improved activity against heterologous strains due to the fact that a portion of the immune response is redirected to epitopes that are conserved between strains (106). This approach is also being used to engineer more broadly protective vaccine candidates for influenza, rhinovirus, and foot-and-mouth disease virus (107).
The structure of fHBP, one of the MenB antigens identified by reverse vaccinology to elicit protective human immune responses, has enabled the identification of epitopes recognized by bactericidal fHBP-specific antibodies raised against fHBP variants, and these epitopes have been used as the basis for engineering a chimeric fHBP vaccine candidate (108–110). The application of available structural information, without the need for a complete antigen-antibody structure, has also proven valuable in mapping protective epitopes through analysis of phage display libraries (111) and mutants that escape immune neutralization (112). An alternative approach involving the high-throughput modification of proteins and their screening for immunogenicity is predicted to evolve with advancing techniques (96).
Vaccinomics: human responses to vaccination
On the other side of the coin, human genomics may have a role in vaccine design. Vaccinomics refers to the investigation of heterogeneity in host genetic markers at the individual or population level that may result in variations in humoral, cell-mediated, and/or innate immune responses to vaccines, with the hope of predicting and optimizing vaccine outcomes (i.e., maximizing the immune response and minimizing vaccine failure and adverse events) (113, 114). This field has emerged in light of the availability of the human genome sequence, the International HapMap Project (which is investigating genetic similarities and differences between humans), and new tools that allow high-throughput detection of gene variations, such as SNP and linkage-disequilibrium maps. For example, HLA genotyping has revealed associations between HLA class and MMR (measles, mumps, and rubella) vaccine outcomes (115). The ability to predict vaccine responses may eventually allow physicians to determine whether to give a vaccine (i.e., is a patient genetically susceptible to a disease or the possible adverse effects of the vaccine?) and if so, what dose and schedule to use.
Conclusions and future directions
The arrival of the genome era and the inception of reverse vaccinology have shifted the paradigm of vaccine development from conventional culture-based methods to high-throughput genome-based approaches (Figure 1 and Table 1). Several years into the genome-based vaccine revolution, we have a previously unimaginable insight into a pathogen’s genome, transcriptome, proteome, surface proteome, and immunoproteome, all of which can be analyzed to discover novel vaccine antigens (Figure 2). Furthermore, the approaches used for vaccine development are continually being refined based on improved understanding of microbial physiology, epidemiology, evolution, virulence, and host-pathogen interactions as well as increased understanding of the complexity of microbial communities and the scale of microbial intra- and interspecies diversity (116–119). One, or a combination, of these approaches now typically drives vaccine discovery projects, with the approach used being heavily dependent on the characteristics of the target pathogen. For example, a classical reverse vaccinology screening approach may be sufficient for a species that cannot be cultured and has low antigenic diversity, whereas a refined reverse vaccinology approach using pan-genomic analysis may be required for antigenically variable and diverse species. On the other hand, a transcriptomic, functional genomic, or proteomic approach may provide a faster route to antigen selection in some cases by narrowing the pool of candidates to be assessed, but these approaches may be limited by species diversity and may also be dependent on the availability of a valid animal model to ensure that the antigens expressed under the conditions studied are relevant to human infection. Finally, the ability to screen antigens for their immunogenicity and to rationally design antigens for increased immunogenicity will aid vaccine development as the fields of immunomics and structural vaccinology continue to advance. The progress made in the genomic and postgenomic era has finally put the realization of vaccines for many pathogens within reach. Indeed, in the coming years, vaccines are set to have an even greater impact on world health than they currently do.
Acknowledgments
K.L. Seib is the recipient of an Australian National Health and Medical Research Council C.J. Martin Fellowship. We would like to thank Giorgio Corsi for artwork.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 119:2515–2525 (2009). doi:10.1172/JCI38330
References
- 1. Levine, M.M., and Lagos, R. 2004. Vaccines and vaccination in historical perspective. InNew generation vaccines . M.M. Levine, J.B. Kaper, R. Rappouli, M.A. Liu, and M.F. Good, editors. Marcel Dekker Inc. New York, New York, USA. 1–10. [Google Scholar]
- 2.Plotkin S.A. Six revolutions in vaccinology. Pediatr. Infect. Dis. J. 2005;24:1–9. doi: 10.1097/01.inf.0000148933.08301.02. [DOI] [PubMed] [Google Scholar]
- 3.Rappuoli R., Dougan G., Delany I.2009Developing vaccines in the era of reverse vaccinology. In New generation vaccines. 4th edition. A. Levine, editor. [Google Scholar]
- 4.Lipsitch M., O’Hagan J.J. Patterns of antigenic diversity and the mechanisms that maintain them. J. R. Soc. Interface. 2007;4:787–802. doi: 10.1098/rsif.2007.0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rappuoli R. Bridging the knowledge gaps in vaccine design. Nat. Biotechnol. 2007;25:1361–1366. doi: 10.1038/nbt1207-1361. [DOI] [PubMed] [Google Scholar]
- 6.Telford J.L. Bacterial genome variability and its impact on vaccine design. Cell Host Microbe. 2008;3:408–416. doi: 10.1016/j.chom.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Fleischmann R.D., et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 8.Liolios K., Mavromatis K., Tavernarakis N., Kyrpides N.C. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2008;36:D475–D479. doi: 10.1093/nar/gkm884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cartwright, K. 1995. Introduction and historical perspectives. InMeningococcal disease . K. Cartwight, editor. Chichester: Wiley. 1–9. [Google Scholar]
- 10.Pizza M., et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 11.Finne J., Bitter-Suermann D., Goridis C., Finne U. An IgG monoclonal antibody to group B meningococci cross-reacts with developmentally regulated polysialic acid units of glycoproteins in neural and extraneural tissues. J. Immunol. 1987;138:4402–4407. [PubMed] [Google Scholar]
- 12.Nedelec J., Boucraut J., Garnier J.M., Bernard D., Rougon G. Evidence for autoimmune antibodies directed against embryonic neural cell adhesion molecules (N-CAM) in patients with group B meningitis. J. Neuroimmunol. 1990;29:49–56. doi: 10.1016/0165-5728(90)90146-E. [DOI] [PubMed] [Google Scholar]
- 13.Seib K.L., Rappuoli R.2009Difficulties in developing neisserial vaccines. In Neisseria: molecular mechanisms of pathogenesis. C.A. Genco, and L.M. Wetzler, editors. Horizon Scientific Press. Norwich, United Kingdom. [Google Scholar]
- 14.Tettelin H., et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 15.Martin D.R., Ruijne N., McCallum L., O’Hallahan J., Oster P. The VR2 epitope on the PorA P1.7-2,4 protein is the major target for the immune response elicited by the strain-specific group B meningococcal vaccine MeNZB. Clin. Vaccine Immunol. 2006;13:486–491. doi: 10.1128/CVI.13.4.486-491.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giuliani M.M., et al. A universal vaccine for serogroup B meningococcus. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10834–10839. doi: 10.1073/pnas.0603940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rappuoli, R. 2008. The application of reverse vaccinology, Novartis MenB vaccine developed by design [abstract]. Presented at the 16th International Pathogenic Neisseria Conference. Rotterdam, The Netherlands. http://neisseria.org/ipnc/2008/ . [Google Scholar]
- 18.Maione D., et al. Identification of a universal Group B streptococcus vaccine by multiple genome screen. Science. 2005;309:148–150. doi: 10.1126/science.1109869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wizemann T.M., et al. Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect. Immun. 2001;69:1593–1598. doi: 10.1128/IAI.69.3.1593-1598.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross B.C., et al. Identification of vaccine candidate antigens from a genomic analysis of Porphyromonas gingivalis. Vaccine. 2001;19:4135–4142. doi: 10.1016/S0264-410X(01)00173-6. [DOI] [PubMed] [Google Scholar]
- 21.Montigiani S., et al. Genomic approach for analysis of surface proteins in Chlamydia pneumoniae. Infect. Immun. 2002;70:368–379. doi: 10.1128/IAI.70.1.368-379.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ariel N., et al. Search for potential vaccine candidate open reading frames in the Bacillus anthracis virulence plasmid pXO1: in silico and in vitro screening. Infect. Immun. 2002;70:6817–6827. doi: 10.1128/IAI.70.12.6817-6827.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ariel N., et al. Genome-based bioinformatic selection of chromosomal Bacillus anthracis putative vaccine candidates coupled with proteomic identification of surface-associated antigens. Infect. Immun. 2003;71:4563–4579. doi: 10.1128/IAI.71.8.4563-4579.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X., Hudson M., Walters N., Bargatze R.F., Pascual D.W. Selection of protective epitopes for Brucella melitensis by DNA vaccination. Infect. Immun. 2005;73:7297–7303. doi: 10.1128/IAI.73.11.7297-7303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glaser P., et al. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 2002;45:1499–1513. doi: 10.1046/j.1365-2958.2002.03126.x. [DOI] [PubMed] [Google Scholar]
- 26.Tettelin H., et al. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12391–12396. doi: 10.1073/pnas.182380799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tettelin H., et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tettelin H., Riley D., Cattuto C., Medini D. Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol. 2008;11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 29.Brodeur B.R., et al. Identification of group B streptococcal Sip protein, which elicits cross-protective immunity. Infect. Immun. 2000;68:5610–5618. doi: 10.1128/IAI.68.10.5610-5618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasko D.A., et al. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. . J. Bacteriol. 2008;190:6881–6893. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Medini D., Donati C., Tettelin H., Masignani V., Rappuoli R. The microbial pan-genome. Curr. Opin. Genet. Dev. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 32.Hogg J.S., et al. Characterization and modeling of the Haemophilus influenzae core and supragenomes based on the complete genomic sequences of Rd and 12 clinical nontypeable strains. Genome Biol. 2007;8:R103. doi: 10.1186/gb-2007-8-6-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiller N.L., et al. Comparative genomic analyses of seventeen Streptococcus pneumoniae strains: insights into the pneumococcal supragenome. J. Bacteriol. 2007;189:8186–8195. doi: 10.1128/JB.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Telford J.L., Barocchi M.A., Margarit I., Rappuoli R., Grandi G. Pili in gram-positive pathogens. Nat. Rev. Microbiol. 2006;4:509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- 35.Lauer P., et al. Genome analysis reveals pili in Group B Streptococcus. Science. 2005;309:105. doi: 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- 36.Rosini R., et al. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol. Microbiol. 2006;61:126–141. doi: 10.1111/j.1365-2958.2006.05225.x. [DOI] [PubMed] [Google Scholar]
- 37.Margarit I., et al. Preventing bacterial infections with pilus-based vaccines: the group B streptococcus paradigm. J. Infect. Dis. 2009;199:108–115. doi: 10.1086/595564. [DOI] [PubMed] [Google Scholar]
- 38.Mora M., et al. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15641–15646. doi: 10.1073/pnas.0507808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barocchi M.A., et al. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2857–2862. doi: 10.1073/pnas.0511017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bagnoli F., et al. A second pilus type in Streptococcus pneumoniae is prevalent in emerging serotypes and mediates adhesion to host cells. J. Bacteriol. 2008;190:5480–5492. doi: 10.1128/JB.00384-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gianfaldoni C., et al. Streptococcus pneumoniae pilus subunits protect mice against lethal challenge. Infect. Immun. 2007;75:1059–1062. doi: 10.1128/IAI.01400-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falugi F., et al. Sequence variation in group A streptococcus pili and association of pilus backbone types with lancefield T serotypes. J. Infect. Dis. 2008;198:1834–1841. doi: 10.1086/593176. [DOI] [PubMed] [Google Scholar]
- 43.Dhiman N., Bonilla R., O’Kane D.J., Poland G.A. Gene expression microarrays: a 21st century tool for directed vaccine design. Vaccine. 2001;20:22–30. doi: 10.1016/S0264-410X(01)00319-X. [DOI] [PubMed] [Google Scholar]
- 44.Grifantini R., et al. Previously unrecognized vaccine candidates against group B meningococcus identified by DNA microarrays. Nat. Biotechnol. 2002;20:914–921. doi: 10.1038/nbt728. [DOI] [PubMed] [Google Scholar]
- 45.Kurz S., et al. Transcriptome-based antigen identification for Neisseria meningitidis. Vaccine. 2003;21:768–775. doi: 10.1016/S0264-410X(02)00596-0. [DOI] [PubMed] [Google Scholar]
- 46.Grifantini R., et al. Characterization of a novel Neisseria meningitidis Fur and iron-regulated operon required for protection from oxidative stress: utility of DNA microarray in the assignment of the biological role of hypothetical genes. Mol. Microbiol. 2004;54:962–979. doi: 10.1111/j.1365-2958.2004.04315.x. [DOI] [PubMed] [Google Scholar]
- 47.Bartolini E., et al. Role of FNR and FNR-regulated, sugar fermentation genes in Neisseria meningitidis infection. Mol. Microbiol. 2006;60:963–972. doi: 10.1111/j.1365-2958.2006.05163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merrell D.S., et al. Host-induced epidemic spread of the cholera bacterium. Nature. 2002;417:642–645. doi: 10.1038/nature00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cassat J., et al. Transcriptional profiling of a Staphylococcus aureus clinical isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiology. 2006;152:3075–3090. doi: 10.1099/mic.0.29033-0. [DOI] [PubMed] [Google Scholar]
- 50.Song X.M., Connor W., Hokamp K., Babiuk L.A., Potter A.A. Transcriptome studies on Streptococcus pneumoniae, illustration of early response genes to THP-1 human macrophages. Genomics. 2008;93:72–82. doi: 10.1016/j.ygeno.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 51.Song X.M., Connor W., Jalal S., Hokamp K., Potter A.A. Microarray analysis of Streptococcus pneumoniae gene expression changes to human lung epithelial cells. Can. J. Microbiol. 2008;54:189–200. doi: 10.1139/W07-133. [DOI] [PubMed] [Google Scholar]
- 52.Talaat A.M., Lyons R., Howard S.T., Johnston S.A. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4602–4607. doi: 10.1073/pnas.0306023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hinton J.C., Hautefort I., Eriksson S., Thompson A., Rhen M. Benefits and pitfalls of using microarrays to monitor bacterial gene expression during infection. Curr. Opin. Microbiol. 2004;7:277–282. doi: 10.1016/j.mib.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 54.de Wet S.C., et al. An improved method for RNA extraction from carcass samples for detection of viable Escherichia coli O157:H7 by reverse-transcriptase polymerase chain reaction. Lett. Appl. Microbiol. 2008;47:399–404. doi: 10.1111/j.1472-765X.2008.02462.x. [DOI] [PubMed] [Google Scholar]
- 55.Morozova O., Marra M.A. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 56.Nielsen K.L. DeepSAGE: higher sensitivity and multiplexing of samples using a simpler experimental protocol. Methods Mol. Biol. 2008;387:81–94. doi: 10.1007/978-1-59745-454-4_6. [DOI] [PubMed] [Google Scholar]
- 57.Mazurkiewicz P., Tang C.M., Boone C., Holden D.W. Signature-tagged mutagenesis: barcoding mutants for genome-wide screens. Nat. Rev. Genet. 2006;7:929–939. doi: 10.1038/nrg1984. [DOI] [PubMed] [Google Scholar]
- 58.Saenz H.L., Dehio C. Signature-tagged mutagenesis: technical advances in a negative selection method for virulence gene identification. Curr. Opin. Microbiol. 2005;8:612–619. doi: 10.1016/j.mib.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 59.Scarselli M., Giuliani M.M., Adu-Bobie J., Pizza M., Rappuoli R. The impact of genomics on vaccine design. Trends Biotechnol. 2005;23:84–91. doi: 10.1016/j.tibtech.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 60.Sun Y.H., Bakshi S., Chalmers R., Tang C.M. Functional genomics of Neisseria meningitidis pathogenesis. Nat. Med. 2000;6:1269–1273. doi: 10.1038/81380. [DOI] [PubMed] [Google Scholar]
- 61.Kavermann H., et al. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J. Exp. Med. 2003;197:813–822. doi: 10.1084/jem.20021531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sassetti C.M., Boyd D.H., Rubin E.J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 63.Akerley B.J., et al. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. U. S. A. 2002;99:966–971. doi: 10.1073/pnas.012602299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Forsyth R.A., et al. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 2002;43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- 65.Zhang Y.Q., et al. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 2003;49:1577–1593. doi: 10.1046/j.1365-2958.2003.03671.x. [DOI] [PubMed] [Google Scholar]
- 66.Rediers H., Rainey P.B., Vanderleyden J., De Mot R. Unraveling the secret lives of bacteria: use of in vivo expression technology and differential fluorescence induction promoter traps as tools for exploring niche-specific gene expression. Microbiol. Mol. Biol. Rev. 2005;69:217–261. doi: 10.1128/MMBR.69.2.217-261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lombardo M.J., et al. An in vivo expression technology screen for Vibrio cholerae genes expressed in human volunteers. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18229–18234. doi: 10.1073/pnas.0705636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodriguez-Ortega M.J., et al. Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat. Biotechnol. 2006;24:191–197. doi: 10.1038/nbt1179. [DOI] [PubMed] [Google Scholar]
- 69.Ferretti J.J., et al. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U. S. A. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.De Groot A.S. Immunome-derived vaccines. Expert Opin. Biol. Ther. 2004;4:767–772. doi: 10.1517/14712598.4.6.767. [DOI] [PubMed] [Google Scholar]
- 71.Meinke A., Henics T., Hanner M., Minh D.B., Nagy E. Antigenome technology: a novel approach for the selection of bacterial vaccine candidate antigens. Vaccine. 2005;23:2035–2041. doi: 10.1016/j.vaccine.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 72.Sinha S., et al. Immunogenic membrane-associated proteins of Mycobacterium tuberculosis revealed by proteomics. Microbiology. 2005;151:2411–2419. doi: 10.1099/mic.0.27799-0. [DOI] [PubMed] [Google Scholar]
- 73.Prokhorova T.A., et al. Novel surface polypeptides of Campylobacter jejuni as traveller’s diarrhoea vaccine candidates discovered by proteomics. Vaccine. 2006;24:6446–6455. doi: 10.1016/j.vaccine.2006.05.085. [DOI] [PubMed] [Google Scholar]
- 74.Hsu C.A., et al. Immunoproteomic identification of the hypothetical protein NMB1468 as a novel lipoprotein ubiquitous in Neisseria meningitidis with vaccine potential. Proteomics. 2008;8:2115–2125. doi: 10.1002/pmic.200700574. [DOI] [PubMed] [Google Scholar]
- 75.Vytvytska O., et al. Identification of vaccine candidate antigens of Staphylococcus aureus by serological proteome analysis. Proteomics. 2002;2:580–590. doi: 10.1002/1615-9861(200205)2:5<580::AID-PROT580>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 76.Chitlaru T., et al. Identification of in vivo-expressed immunogenic proteins by serological proteome analysis of the Bacillus anthracis secretome. Infect. Immun. 2007;75:2841–2852. doi: 10.1128/IAI.02029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kurupati P., Teh B.K., Kumarasinghe G., Poh C.L. Identification of vaccine candidate antigens of an ESBL producing Klebsiella pneumoniae clinical strain by immunoproteome analysis. Proteomics. 2006;6:836–844. doi: 10.1002/pmic.200500214. [DOI] [PubMed] [Google Scholar]
- 78.Cole J.N., et al. Surface analyses and immune reactivities of major cell wall-associated proteins of group A Streptococcus. Infect. Immun. 2005;73:3137–3146. doi: 10.1128/IAI.73.5.3137-3146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ling E., et al. Glycolytic enzymes associated with the cell surface of Streptococcus pneumoniae are antigenic in humans and elicit protective immune responses in the mouse. Clin. Exp. Immunol. 2004;138:290–298. doi: 10.1111/j.1365-2249.2004.02628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang A., Xie C., Chen H., Jin M. Identification of immunogenic cell wall-associated proteins of Streptococcus suis serotype 2. Proteomics. 2008;8:3506–3515. doi: 10.1002/pmic.200800007. [DOI] [PubMed] [Google Scholar]
- 81.Wright A., Drudy D., Kyne L., Brown K., Fairweather N.F. Immunoreactive cell wall proteins of Clostridium difficile identified by human sera. J. Med. Microbiol. 2008;57:750–756. doi: 10.1099/jmm.0.47532-0. [DOI] [PubMed] [Google Scholar]
- 82.Pereboeva L.A., Pereboev A.V., Wang L.F., Morris G.E. Hepatitis C epitopes from phage-displayed cDNA libraries and improved diagnosis with a chimeric antigen. J. Med. Virol. 2000;60:144–151. doi: 10.1002/(SICI)1096-9071(200002)60:2<144::AID-JMV7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 83.Etz H., et al. Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 2002;99:6573–6578. doi: 10.1073/pnas.092569199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Giefing C., et al. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J. Exp. Med. 2008;205:117–131. doi: 10.1084/jem.20071168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Davies D.H., et al. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc. Natl. Acad. Sci. U. S. A. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Doolan D.L., et al. Profiling humoral immune responses to P. falciparum infection with protein microarrays. Proteomics. 2008;8:4680–4694. doi: 10.1002/pmic.200800194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Eyles J.E., et al. Immunodominant Francisella tularensis antigens identified using proteome microarray. Proteomics. 2007;7:2172–2183. doi: 10.1002/pmic.200600985. [DOI] [PubMed] [Google Scholar]
- 88.Sundaresh S., et al. From protein microarrays to diagnostic antigen discovery: a study of the pathogen Francisella tularensis. Bioinformatics. 2007;23:i508–i518. doi: 10.1093/bioinformatics/btm207. [DOI] [PubMed] [Google Scholar]
- 89.Rolfs A., et al. Production and sequence validation of a complete full length ORF collection for the pathogenic bacterium Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 2008;105:4364–4369. doi: 10.1073/pnas.0712049105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.LaBaer J., Ramachandran N. Protein microarrays as tools for functional proteomics. Curr. Opin. Chem. Biol. 2005;9:14–19. doi: 10.1016/j.cbpa.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 91.Haas G., et al. Immunoproteomics of Helicobacter pylori infection and relation to gastric disease. Proteomics. 2002;2:313–324. doi: 10.1002/1615-9861(200203)2:3<313::AID-PROT313>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 92.Lin Y.F., et al. Comparative immunoproteomics of identification and characterization of virulence factors from Helicobacter pylori related to gastric cancer. Mol. Cell. Proteomics. 2006;5:1484–1496. doi: 10.1074/mcp.M600111-MCP200. [DOI] [PubMed] [Google Scholar]
- 93.Bernardini G., Braconi D., Lusini P., Santucci A. Helicobacter pylori: immunoproteomics related to different pathologies. Expert Rev. Proteomics. 2007;4:679–689. doi: 10.1586/14789450.4.5.679. [DOI] [PubMed] [Google Scholar]
- 94.Bernardini G., Braconi D., Santucci A. The analysis of Neisseria meningitidis proteomes: Reference maps and their applications. Proteomics. 2007;7:2933–2946. doi: 10.1002/pmic.200700094. [DOI] [PubMed] [Google Scholar]
- 95.Serruto D., Rappuoli R. Post-genomic vaccine development. FEBS Lett. 2006;580:2985–2992. doi: 10.1016/j.febslet.2006.04.084. [DOI] [PubMed] [Google Scholar]
- 96.Dormitzer P.R., Ulmer J.B., Rappuoli R. Structure-based antigen design: a strategy for next generation vaccines. Trends Biotechnol. 2008;26:659–667. doi: 10.1016/j.tibtech.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lundstrom K. Structural genomics and drug discovery. J. Cell Mol. Med. 2007;11:224–238. doi: 10.1111/j.1582-4934.2007.00028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Todd A.E., Marsden R.L., Thornton J.M., Orengo C.A. Progress of structural genomics initiatives: an analysis of solved target structures. . J. Mol. Biol. 2005;348:1235–1260. doi: 10.1016/j.jmb.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 99.Kim C.U., et al. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: design, synthesis, and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. . J. Am. Chem. Soc. 1997;119:681–690. doi: 10.1021/ja963036t. [DOI] [PubMed] [Google Scholar]
- 100.Kaldor S.W., et al. Viracept (nelfinavir mesylate, AG1343): a potent, orally bioavailable inhibitor of HIV-1 protease. J. Med. Chem. 1997;40:3979–3985. doi: 10.1021/jm9704098. [DOI] [PubMed] [Google Scholar]
- 101.Douek D.C., Kwong P.D., Nabel G.J. The rational design of an AIDS vaccine. Cell. 2006;124:677–681. doi: 10.1016/j.cell.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 102. Prabakaran, P., Dimitrov, A.S., Fouts, T.R., Dimitrov, D.S., and KuanTeh, J. 2007. Structure and function of the HIV envelope glycoprotein as entry mediator, vaccine immunogen, and target for inhibitors. InAdvances in pharmacology. Academic Press. San Diego, California, USA/London, United Kingdom. 33–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kwong P.D., et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhou T., et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.West A.P., Jr., et al. Design and expression of a dimeric form of the human immunodeficiency virus type 1 antibody 2G12 with increased neutralization potency. J. Virol. 2008;83:98–104. doi: 10.1128/JVI.01564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Garrity R.R., et al. Refocusing neutralizing antibody response by targeted dampening of an immunodominant epitope. J. Immunol. 1997;159:279–289. [PubMed] [Google Scholar]
- 107.Tobin G.J., et al. Deceptive imprinting and immune refocusing in vaccine design. Vaccine. 2008;26:6189–6199. doi: 10.1016/j.vaccine.2008.09.080. [DOI] [PubMed] [Google Scholar]
- 108.Cantini F., et al. Solution structure of the immunodominant domain of protective antigen GNA1870 of Neisseria meningitidis. J. Biol. Chem. 2006;281:7220–7227. doi: 10.1074/jbc.M508595200. [DOI] [PubMed] [Google Scholar]
- 109.Beernink P.T., Granoff D.M. Bactericidal antibody responses induced by meningococcal recombinant chimeric factor H-binding protein vaccines. Infect. Immun. 2008;76:2568–2575. doi: 10.1128/IAI.00033-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cantini F., et al. Solution structure of the factor H binding protein, a survival factor and protective antigen of Neisseria meningitidis. J. Biol. Chem. 2009;284:9022–9026. doi: 10.1074/jbc.C800214200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Smothers J.F., Henikoff S., Carter P. Tech.Sight. Phage display. Affinity selection from biological libraries. Science. 2002;298:621–622. doi: 10.1126/science.298.5593.621. [DOI] [PubMed] [Google Scholar]
- 112.Gulati U., et al. Antibody epitopes on the neuraminidase of a recent H3N2 influenza virus (A/Memphis/31/98). J. Virol. 2002;76:12274–12280. doi: 10.1128/JVI.76.23.12274-12280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Poland G.A., Ovsyannikova I.G., Jacobson R.M., Smith D.I. Heterogeneity in vaccine immune response: the role of immunogenetics and the emerging field of vaccinomics. Clin. Pharmacol. Ther. 2007;82:653–664. doi: 10.1038/sj.clpt.6100415. [DOI] [PubMed] [Google Scholar]
- 114.Poland G.A., Ovsyannikova I.G., Jacobson R.M. Personalized vaccines: the emerging field of vaccinomics. Expert Opin. Biol. Ther. 2008;8:1659–1667. doi: 10.1517/14712598.8.11.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ovsyannikova I.G., Jacobson R.M., Vierkant R.A., Pankratz V.S., Poland G.A. HLA supertypes and immune responses to measles-mumps-rubella viral vaccine: findings and implications for vaccine design. Vaccine. 2007;25:3090–3100. doi: 10.1016/j.vaccine.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 116.Gill S.R., et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Raskin D.M., Seshadri R., Pukatzki S.U., Mekalanos J.J. Bacterial genomics and pathogen evolution. Cell. 2006;124:703–714. doi: 10.1016/j.cell.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 118.Muzzi A., Masignani V., Rappuoli R. The pan-genome: towards a knowledge-based discovery of novel targets for vaccines and antibacterials. Drug Discov. Today. 2007;12:429–439. doi: 10.1016/j.drudis.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 119.Medini D., et al. Microbiology in the post-genomic era. Nat. Rev. Microbiol. 2008;6:419–430. doi: 10.1038/nrmicro1901. [DOI] [PubMed] [Google Scholar]
- 120.Masignani V., Rappuoli R., Pizza M. Reverse vaccinology: a genome-based approach for vaccine development. Expert Opin. Biol. Ther. 2002;2:895–905. doi: 10.1517/14712598.2.8.895. [DOI] [PubMed] [Google Scholar]
- 121.Conway T., Schoolnik G.K. Microarray expression profiling: capturing a genome-wide portrait of the transcriptome. Mol. Microbiol. 2003;47:879–889. doi: 10.1046/j.1365-2958.2003.03338.x. [DOI] [PubMed] [Google Scholar]
- 122.Zhu H., Bilgin M., Snyder M. Proteomics. Annu. Rev. Biochem. 2003;72:783–812. doi: 10.1146/annurev.biochem.72.121801.161511. [DOI] [PubMed] [Google Scholar]