Abstract
Nicotinamide nucleotides (NAD and NADP) are important cofactors in many metabolic processes in living organisms. In this study, we analyzed transgenic Arabidopsis (Arabidopsis thaliana) plants that overexpress NAD kinase2 (NADK2), an enzyme that catalyzes the synthesis of NADP from NAD in chloroplasts, to investigate the impacts of altering NADP level on plant metabolism. Metabolite profiling revealed that NADP(H) concentrations were proportional to NADK activity in NADK2 overexpressors and in the nadk2 mutant. Several metabolites associated with the Calvin cycle were also higher in the overexpressors, accompanied by an increase in overall Rubisco activity. Furthermore, enhanced NADP(H) production due to NADK2 overexpression increased nitrogen assimilation. Glutamine and glutamate concentrations, as well as some other amino acids, were higher in the overexpressors. These results indicate that overexpression of NADK2 either directly or indirectly stimulates carbon and nitrogen assimilation in Arabidopsis under restricted conditions. Importantly, since neither up-regulation nor down-regulation of NADK2 activity affected the sum amount of NAD and NADP or the redox state, the absolute level of NADP and/or the NADP/NAD ratio likely plays a key role in regulating plant metabolism.
Carbon (C) and nitrogen (N) assimilation are closely linked fundamental processes for plant growth. The emerging details of C and N assimilation suggest that a regulatory system coordinates uptake and distribution of these nutrients in response to both metabolic and environmental cues (Coruzzi and Bush, 2001; Coruzzi and Zhou, 2001). Because the amounts of assimilated C and N largely influence plant growth and crop yields, there have been many attempts to engineer C and N assimilation by overexpression of a single enzyme. However, in only a few cases have significant improvements in C and N assimilation been achieved (Miyagawa et al., 2001; Sinclair et al., 2004), possibly because synchronous activation of a series of metabolic pathways might be necessary to influence assimilation. One successful example of metabolic engineering is in transgenic Arabidopsis (Arabidopsis thaliana) lines expressing the maize Dof1 (for DNA-binding with one finger 1) transcription factor, which have increased N contents and improved growth rates under N starvation conditions (Yanagisawa et al., 2004). An increase in the biosynthesis of cofactors could also be used to modify C and N assimilation, because high levels of a cofactor may stimulate multiple enzymatic reactions, resulting in synchronous metabolic pathway activation. However, the effectiveness of such a cofactor strategy, to our knowledge, has not been reported.
NAD and NADP are pyridine nucleotides that are essential for electron transport and that serve as cofactors in numerous metabolic processes in many organisms. We recently showed that modification of the NADP/NAD ratio impacted cell growth, implying that NAD and NADP play different roles in energy transduction (Takahashi et al., 2006b; Hashida et al., 2009). Although our previous data, obtained with an Arabidopsis mutant, suggested that the NADP/NAD ratio might affect metabolic processes as well (Takahashi et al., 2006b), this possibility remains to be evaluated.
NAD kinase (NADK; EC 2.7.1.23) catalyzes the de novo biosynthesis of NADP from NAD and ATP. Genes encoding NADK have been cloned from Homo sapiens (Lerner et al., 2001), Escherichia coli (Kawai et al., 2001a), Saccharomyces cerevisiae (Kawai et al., 2001b; Outten and Culotta, 2003), and other species (Kawai et al., 2000; Raffaelli et al., 2004; Sakuraba et al., 2005). Characterization of NADK isoforms indicates that the enzyme is involved in biotic and abiotic stress responses. For instance, cold stress induces calmodulin (CaM)-dependent NADK activity, but not CaM-independent NADK activity, in green bean (Phaseolus vulgaris) plants (Ruiz et al., 2002). Decreases in NADK activity in response to salinity and drought were observed in tomato (Solanum lycopersicum) and wheat (Triticum aestivum), respectively (Zagdańska, 1990; Delumeau et al., 2000). Furthermore, cultured cells of tobacco (Nicotiana tabacum) treated with elicitors, including cellulase, harpin, and incompatible bacteria, elevated CaM-dependent NADK activity and NADPH concentrations (Harding et al., 1997). Since NADPH is a substrate of NADPH oxidase, which regulates reactive oxygen species (ROS) production in defense signaling, these observations integrate NADK into the plant defense response system.
In Arabidopsis, three genes encoding NADK have been identified (NADK1, NADK2, and NADK3; Berrin et al., 2005; Turner et al., 2005). NADK2 is localized in chloroplasts, whereas NADK1 and NADK3 are in the cytosol (Chai et al., 2005, 2006). Chloroplastic NADK2 transcripts are abundant in leaves and are not affected by abiotic stresses. The genes encoding the cytosolic NADKs, NADK1 and NADK3, are ubiquitously expressed and are induced by abiotic stresses such as oxidative stress (Berrin et al., 2005; Chai et al., 2006). Based on these observations, the physiological role of NADK2 is likely different from that of NADK1 and NADK3. Moreover, the activity of NADK2 appears to be responsible for a major part of NADP production in photosynthetic cells, because a knockout mutant of NADK2 (nadk2) produced far less NADK activity and NADP(H) (Chai et al., 2005; Takahashi et al., 2006b). NADP produced by NADK2 is reduced to NADPH in the photosynthetic electron transport chain in chloroplasts, and then NADPH is transported from chloroplasts to the cytosol and other organelles via the malate/oxaloacetate shuttle (Heineke et al., 1991; Scheibe, 2004). The reducing energy of NADPH is used for biological processes in chloroplasts, including chlorophyll synthesis and CO2 fixation in the Calvin cycle, and for various processes in the cytosol and other organelles. Thus, NADK2 likely plays a central role in the metabolism of photosynthetic cells.
The nadk2 mutant produces pale green rosette leaves and does not grow as well as wild-type Arabidopsis. This mutant also has lower leaf chlorophyll contents, reduced expression of the NADPH:protochlorophyllide oxidoreductase genes (Chai et al., 2005), and high accumulation of zeaxanthin under low-light conditions, probably through inhibition of zeaxanthin epoxidase activity (Takahashi et al., 2006b). Consequently, the nadk2 mutant has decreased photosynthetic activity, presumably due to both decreased chlorophyll contents and aberrant energy dissipation in the xanthophyll cycle (Takahashi et al., 2006b), suggesting a tight linkage between NADK2 activity and CO2 fixation. On the other hand, nadk1, nadk2, and nadk3 mutants all showed an increased sensitivity to ROS stress (Berrin et al., 2005; Chai et al., 2005, 2006), suggesting that the NADKs play a conserved role in defense response. Hence, unlike other NADKs, NADK2 might contribute to both sustainable growth and defense responses in Arabidopsis. These observations prompted us to evaluate the effects of overexpression of the chloroplastic NADK2 on the metabolic alteration in Arabidopsis plants.
In this work, we generated and characterized NADK2-overexpressing transgenic Arabidopsis plants to increase intracellular NADP(H) concentrations and then investigated the effects of this metabolic disturbance. We found that the concentrations of several Calvin cycle metabolites and some amino acids increased in these transgenic plants. Since the total amounts of NAD and NADP as well as the redox state were similar in the wild-type Arabidopsis, the nadk2 mutant, and the NADK2 overexpressors, either NADP and/or the NADP/NAD ratio could be the key factor regulating plant metabolism. Moreover, our results also indicated that elevation of NADP concentrations leads to coordinated activation of both C fixation and N assimilation in planta. Although observed metabolic enhancement would be pleiotropic, we propose here that genetic manipulation of cofactors is a viable strategy for engineering plant metabolism.
RESULTS
Generation of Transgenic Arabidopsis Plants Overexpressing NADK2
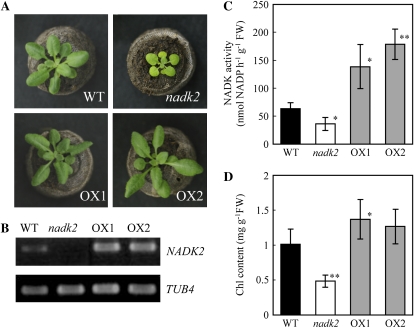
We initially generated NADK2-overexpressing transgenic Arabidopsis plants using full-length NADK2 cDNA (2,958 bp) under the control of the cauliflower mosaic virus 35S promoter to investigate the effect of NADK2-mediated NADP production on C and N assimilation. Two transgenic lines with significant expression of the transgene (NADK2-OX1 and -OX2; Fig. 1, A–C) were used for further analysis. The nadk2 mutant showed a severe morphological phenotype, with smaller rosette leaves and shorter and fewer siliques (Fig. 1A; Table I); however, there was almost no change in the soil-grown NADK2 overexpressor plants. Amplification of transgene-derived and endogenous NADK2 mRNA was much higher in leaves of both NADK2-OX1 and -OX2 than in wild-type plants, and there was no detectable PCR product from the T-DNA insertion nadk2 mutant (Fig. 1B; Chai et al., 2005). NADK activity in the nadk2 mutant was half the level of the wild type but 2.1- to 2.8-fold higher in the NADK2 overexpressors (Fig. 1C). Bolting and anthesis were delayed in the nadk2 mutant, but the timing of floral development was normal in the NADK2 overexpressors (Table I). Chlorophyll content was lower in rosette leaves of the nadk2 mutant, consistent with a previous observation (Chai et al., 2005), but tended to be higher in the NADK2 overexpressors (Fig. 1D).
Figure 1.
Phenotype of Arabidopsis wild type (WT), the nadk2 mutant, and transgenic lines overexpressing NADK2 (OX1 and OX2) grown for 23 d under continuous light. A, Plants grown in soil. B, Levels of the NADK2 transcript in rosette leaves. RT-PCR was performed with 1 μg of total RNA. C, NADK activity in rosette leaves. D, Chlorophyll (Chl) contents of rosette leaves. Values are means ± sd calculated from three independent experiments. Significant differences from the wild type are evaluated by t test and shown by asterisks (* P < 0.05 or ** P < 0.01). FW, Fresh weight.
Table I.
Developmental characteristics of Arabidopsis wild type, the nadk2 mutant, and NADK2 overexpression (NADK2-OX1 and -OX2) lines under continuous light conditions
Size and weight of leaves were measured using the fifth rosette leaf of 23-d-old plants. All other parameters were measured on 90-d-old plants. The data are means ± sd of at least 30 plants.
| Parameter | Wild Type | nadk2 | NADK2-OX1 | NADK2-OX2 |
|---|---|---|---|---|
| Leaf length (mm) | 8.9 ± 1.4 | 6.5 ± 1.2 | 9.3 ± 0.7 | 9.6 ± 1.0 |
| Leaf width (mm) | 7.8 ± 1.3 | 6.5 ± 1.2 | 7.9 ± 0.8 | 8.1 ± 0.9 |
| Leaf fresh weight (mg) | 10.6 ± 2.0 | 5.5 ± 1.3 | 10.9 ± 1.5 | 12.0 ± 0.8 |
| Specific leaf fresh weight (mg cm−2) | 14.1 ± 1.1 | 11.1 ± 0.4 | 13.6 ± 1.2 | 13.9 ± 0.7 |
| Start of bolting (d) | 28.9 ± 1.3 | 31.3 ± 2.2 | 28.8 ± 1.8 | 28.3 ± 1.1 |
| Start of anthesis (d) | 32.5 ± 2.0 | 36.0 ± 2.6 | 33.5 ± 2.3 | 32.0 ± 1.6 |
| Number of siliques per plant | 32.2 ± 4.7 | 19.6 ± 6.0 | 35.6 ± 6.0 | 35.8 ± 5.8 |
| Silique length (mm) | 10.3 ± 1.4 | 9.3 ± 1.3 | 10.2 ± 1.6 | 10.3 ± 1.5 |
Overexpression of NADK2 Increases the NADP(H)/NAD(H) Ratio But Has No Effect on Redox State
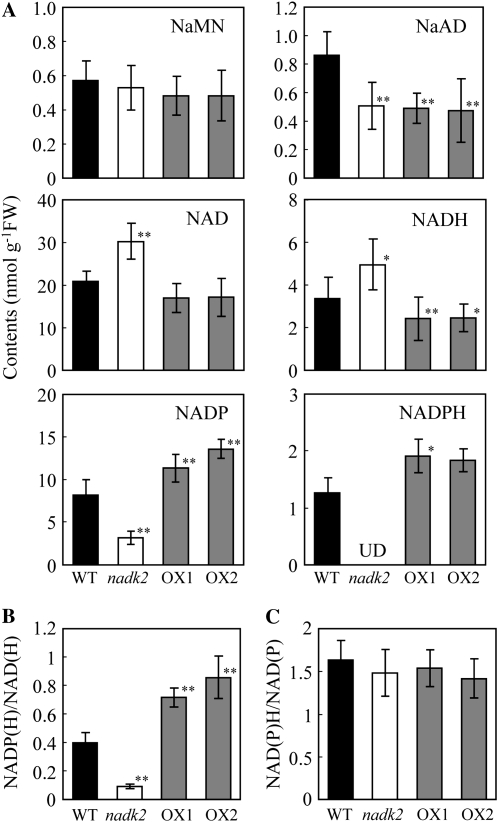
To measure the effect of NADK2 overexpression on pyridine nucleotide pools, NAD(P)(H) and their precursors, nicotinate mononucleotide (NaMN) and nicotinate adenine dinucleotide (NaAD), were quantified in leaf tissues by capillary electrophoresis mass spectrometry (CE-MS; Fig. 2A). NaMN concentrations were about the same in all plants analyzed, whereas NaAD was lower in both the nadk2 mutant and the NADK2 overexpressors. NAD and NADH concentrations were higher in the nadk2 mutant, but NADP and NADPH were significantly lower. This observation supports our previous suggestion that NADP synthesis in leaf tissue largely depends on NADK2 activity (Takahashi et al., 2006b). In the NADK2 overexpressors, the NAD content was unchanged, NADH concentrations were lower, and NADP and NADPH concentrations were higher, resulting in a 1.5- to 1.7-fold higher NADP(H)/NAD(H) ratio than in the wild type. The ratios of NADP(H)/NAD(H) in the wild type and nadk2 were 0.41 and 0.11, respectively (Fig. 2B). However, the sum of oxidative and reduced forms of NAD and NADP, and the (NADH+NADPH)/(NAD+NADP) ratio, which indicates the redox state, were not affected by modulation of NADK2 activity (Fig. 2C). These results indicate that modulation of NADK2 activity fundamentally influences only the NAD/NADP ratio, although the reason that NaAD was similarly reduced in both the nadk2 mutant and NADK2 overexpressors is unknown.
Figure 2.
Pyridine nucleotide pools in leaves of the wild type (WT), the nadk2 mutant, and NADK2 overexpressors (OX1 and OX2). A, Quantification of the nucleotide contents. B, NADP(H)/NAD(H) ratio. C, (NADPH+NADH)/(NADP+NAD) ratios. Analyses were performed on leaves harvested from 23-d-old plants grown in soil, and nucleotide levels were quantified by CE-MS. Each value represents the mean ± se of three independent extracts. Significant differences from the wild type are shown (* P < 0.05, ** P < 0.01; t test). FW, Fresh weight; UD, under the detection limit.
NADK2 Overexpression Affects Rubisco Activity and Calvin Cycle Intermediates
The effective quantum yield of electron transport through PSII was lower in the nadk2 mutant than in the wild type (Takahashi et al., 2006b) but was the same as in the wild type in NADK2 overexpressors (data not shown). However, CO2 uptake under constant light conditions indicated a tendency to higher rates in the NADK2 overexpressors (data not shown). The activities of enzymes in the Calvin cycle were thus measured. Both the initial and total activities of Rubisco (EC 4.1.1.39) in the nadk2 mutant were approximately 1.1- and 1.2-fold lower than in the wild type (Table II). In contrast, Rubisco activities in the NADK2 overexpressors were 1.1- or 1.2-fold higher than in the wild type. The activities of the other Calvin cycle enzymes, including phosphoglycerate kinase (PGK; EC 2.7.2.3), NADP-dependent glyceraldehyde-3-phosphate dehydrogenase (NADP-GAPDH; EC 1.2.1.13), aldolase (EC 4.1.2.13), stromal fructose-1,6-bisphosphatase (FBPase; EC 3.1.3.11), and phosphoribulokinase (PRK; EC 2.7.1.19), were not affected.
Table II.
Activity of Calvin cycle enzymes in rosette leaves of the wild type, the nadk2 mutant, and NADK2 overexpression (NADK2-OX1 and -OX2) lines
Activities were determined in 4-week-old leaves. Data presented are means ± se of measurements from five independent plants per genotype. Statistical significances relative to the wild type by t test are as follows: * P < 0.01, ** P < 0.05, *** P < 0.1.
| Enzyme | Activity
|
|||
|---|---|---|---|---|
| Wild Type | nadk2 | NADK2-OX1 | NADK2-OX2 | |
| μmol min−1 g−1 fresh weight | ||||
| Rubisco | ||||
| Initial | 9.5 ± 0.2 | 9.0 ± 0.1*** | 10.5 ± 0.3** | 11.2 ± 0.3* |
| Total | 14.4 ± 0.4 | 11.6 ± 0.4* | 15.7 ± 0.3*** | 17.5 ± 0.4* |
| PGK | 47.6 ± 4.8 | 52.4 ± 4.5 | 51.0 ± 4.2 | 46.4 ± 2.1 |
| NADP-GAPDH | 13.7 ± 2.5 | 13.6 ± 2.7 | 13.1 ± 1.0 | 11.2 ± 0.1 |
| Aldolase | 2.0 ± 0.1 | 1.7 ± 0.4 | 2.0 ± 0.2 | 2.0 ± 0.2 |
| Stromal FBPase | 2.5 ± 0.4 | 2.3 ± 0.8 | 2.1 ± 0.5 | 2.1 ± 0.5 |
| PRK | 14.9 ± 2.1 | 16.6 ± 2.2 | 15.3 ± 2.0 | 12.8 ± 1.1 |
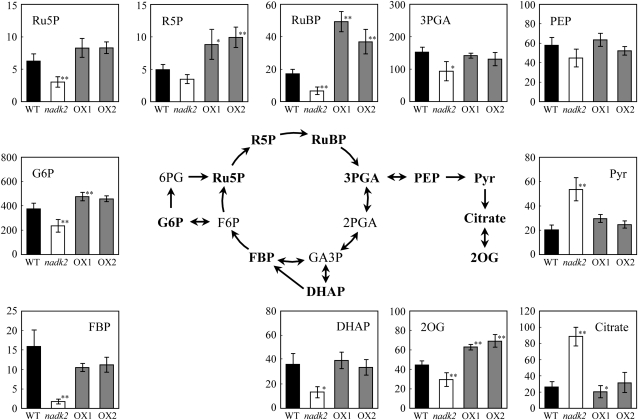
Rubisco catalyzes the carboxylation of ribulose-1,5-bisphosphate (RuBP) and produces 3-phosphoglycerate (3PGA), which is reduced to triosephosphates, which are then converted to intermediates in glycolysis, the TCA cycle, and other pathways. Therefore, we measured the concentrations of intermediates of these central metabolic pathways (Fig. 3). The nadk2 mutant accumulated about half the wild-type concentrations of most intermediates, except for pyruvate and citrate. However, Glc-6-P, Rib-5-P, ribulose-5-phosphate, RuBP, pyruvate, and 2-oxoglutarate were all higher in the NADK2 overexpressors. The exceptions were Fru-1,6-bisphosphate, dihydroxyacetone phosphate, 3PGA, and phosphoenolpyruvate, which were unchanged. Despite an increase in total Rubisco activity, RuBP levels were much higher in the NADK2 overexpressors than in the wild type. On the other hand, no significant difference was observed in the metabolite contents of the other pathways, including the shikimate, glutathione, and polyamine pathways (data not shown).
Figure 3.
Glycolysis, TCA cycle, and Calvin cycle intermediates in leaves of the wild type (WT), the nadk2 mutant, and NADK2 overexpression (OX1 and OX2) lines. Analyses were performed with rosette leaves harvested from 23-d-old plants grown in soil. Each value represents the mean ± se of three independent experiments (nmol g−1 fresh weight). Significant differences from the wild type are shown (* P < 0.05, ** P < 0.01; t test). DHAP, Dihydroxyacetone phosphate; FBP, Fru-1,6-bisphosphate; GA3P, glyceraldehyde-3-phosphate; G6P, Glc-6-P; 2OG, 2-oxoglutarate; PEP, phosphoenolpyruvate; Pyr, pyruvate; Ru5P, ribulose-5-phosphate; R5P, Rib-5-phosphate.
Increased Accumulation of Glu Family Amino Acids by NADK2 Overexpression
In plants, amino acids are synthesized from intermediates of the central metabolic pathway. Therefore, there is a tight relationship between amino acid biosynthesis and Calvin cycle activity (Stitt and Schulze, 1994; Coruzzi and Bush, 2001; Coruzzi and Zhou, 2001). Amino acids can be categorized into the Asp, Glu, Ser, pyruvate, and His families and the aromatic amino acids. Amino acids were measured in the nadk2 mutant and in the NADK2 overexpressors, and the largest differences were found in the Glu family amino acids, especially Gln and Glu (Table III). Glu, and to a larger extent Gln, concentrations were higher in both NADK2 overexpression lines in proportion to NADK activity. Because Gln and Glu are good molecular markers for N utilization efficiency (Foyer et al., 2003), this result indicates that NADK2 overexpression also affects N assimilation. Total amino acid contents were also higher in the NADK2 overexpressors and lower in the nadk2 mutant, possibly reflecting the overall influence of N in the metabolic system. Furthermore, NADK2 activity levels affected the levels of other amino acid families. Thr, Cys, Gly, and Ser concentrations were higher in the NADK2 overexpressors. Ala concentrations were higher in the nadk2 mutant.
Table III.
Amino acid accumulation in rosette leaves of the wild type, the nadk2 mutant, and NADK2 overexpression (NADK2-OX1 and -OX2) lines
Values indicate means obtained from three independent experiments. se values are in all cases less than 35% of mean values. Statistical significances relative to the wild type by t test are as follows: * P < 0.01, ** P < 0.05, *** P < 0.1
| Metabolite | Concentration
|
|||
|---|---|---|---|---|
| Wild Type | nadk2 | NADK2-OX1 | NADK2-OX2 | |
| nmol g−1 fresh weight | ||||
| Asp family | ||||
| Asn | 1,618.4 ± 54.7 | 671.7 ± 54.9** | 2,074.2 ± 234.0 | 1,582.9 ± 350.5 |
| Asp | 1,238.4 ± 180.1 | 2,944.0 ± 477.6*** | 1,628.5 ± 168.9 | 1,719.6 ± 170.8 |
| Lys | 106.5 ± 13.9 | 50.2 ± 5.2** | 90.9 ± 6.4 | 81.0 ± 23.4 |
| Met | 14.9 ± 3.0 | 7.4 ± 0.2*** | 19.0 ± 1.3 | 16.6 ± 0.4 |
| Thr | 635.5 ± 43.1 | 374.6 ± 22.5** | 922.6 ± 53.6** | 951.1 ± 104.6*** |
| Glu family | ||||
| Arg | 4,519.7 ± 581.6 | 443.4 ± 75.5** | 5,890.5 ± 382.0** | 6,375.7 ± 1,258.9 |
| Gln | 4,236.9 ± 537.0 | 1,071.5 ± 55.6** | 6,490.2 ± 546.7* | 6,555.0 ± 813.8*** |
| Glu | 2,786.6 ± 265.3 | 2,460.3 ± 519.2 | 3,917.8 ± 190.5** | 4,388.1 ± 142.8** |
| Pro | 984.1 ± 99.7 | 137.6 ± 9.1* | 2,070.1 ± 641.2 | 1,075.5 ± 251.4 |
| Ser family | ||||
| Cys | 4.8 ± 0.6 | 13.1 ± 1.1** | 7.6 ± 0.2** | 9.1 ± 1.9 |
| Gly | 259.6 ± 72.1 | 113.2 ± 14.1*** | 485.1 ± 56.7** | 282.9 ± 80.4 |
| Ser | 3,961.0 ± 272.0 | 2,971.5 ± 172.4** | 5,069.3 ± 40.9** | 4,300.2 ± 211.1 |
| Pyruvate family | ||||
| Ala | 271.5 ± 54.7 | 822.2 ± 129.8** | 368.5 ± 49.9 | 385.6 ± 29.2** |
| Leu | 75.2 ± 14.7 | 69.3 ± 5.9 | 66.9 ± 6.7 | 58.0 ± 2.5 |
| Val | 97.1 ± 12.1 | 116.7 ± 12.6 | 114.1 ± 12.6 | 102.3 ± 5.5 |
| Aromatic amino acids | ||||
| Phe | 36.3 ± 6.0 | 36.3 ± 3.9 | 45.7 ± 2.1 | 35.2 ± 2.5 |
| Trp | 4.6 ± 1.2 | 8.6 ± 0.7*** | 4.8 ± 0.9 | 6.3 ± 1.0 |
| Tyr | 15.1 ± 1.8 | 12.6 ± 0.5 | 16.2 ± 1.6 | 12.7 ± 0.8 |
| His family | ||||
| His | 116.7 ± 9.4 | 78.8 ± 9.7*** | 130.5 ± 12.4 | 108.6 ± 17.7 |
| Total | 21,027.8 ± 1,867.6 | 12,425.6 ± 1,014.1** | 29,502.4 ± 482.6** | 28,124.8 ± 1,927.1*** |
NADK2 Overexpression Increases Light-Dependent Accumulation of Gln and Glu
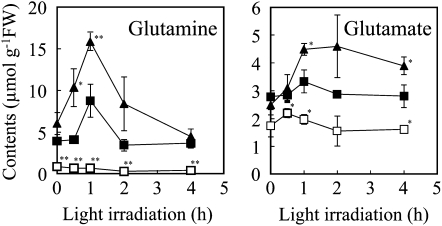
To further characterize amino acid biosynthesis in the NADK2 overexpressors, we examined time-dependent changes in leaf concentrations of Gln and Glu with exposure to light irradiation (60 μmol m−2 s−1; Fig. 4). Because N assimilation is a light energy-dependent process, an expanded capacity for N assimilation should lead to more rapid production of larger amounts of Gln and Glu in the overexpressor line with light irradiation. Initial Gln concentrations were higher in NADK2-OX2 and by 30 min were already increasing. Wild-type Gln concentrations began to increase starting at the 1-h time point. Initial Gln concentrations were lower in the nadk2 mutant than in the wild type. Concentrations of Gln peaked in all three plant lines at 1 h, although the nadk2 mutant produced very little.
Figure 4.
Gln and Glu synthesis stimulated by light irradiation. Rosette leaves of 25-d-old plants were harvested at each time point indicated after light irradiation (60 μmol m−2 s−1), and Gln and Glu contents of the leaves were quantified. Black squares, the wild type; white squares, the nadk2 mutant; black triangles, the NADK2-OX2 line. Each value represents the mean ± se of three independent experiments. Significant differences from the wild type are shown (* P < 0.05, ** P < 0.01; t test). FW, Fresh weight.
Glu concentrations were somewhat higher in NADK2-OX2 than in wild-type Arabidopsis at the 1-h time point. The rates of Glu synthesis were 551 nmol g−1 fresh weight h−1 in the wild type and 1,992 nmol g−1 fresh weight h−1 in NADK2-OX2. The Glu concentration in the nadk2 mutant was only slightly increased at the 30-min time point.
Effects of NADK2 Overexpression on Gene Expression Associated with the GS/GOGAT Pathway
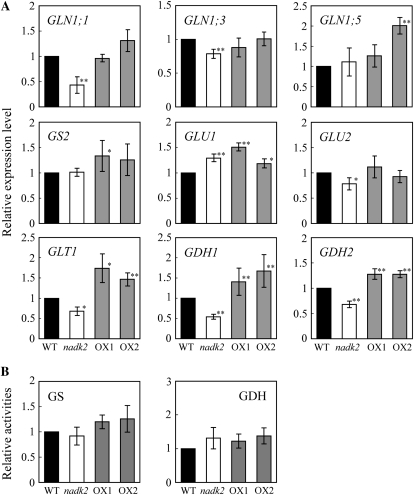
In plants, N assimilation proceeds through the Gln synthetase (GS)/Glu synthase (GOGAT) pathway (Lam et al., 1996). In order to determine whether the increased amino acid biosynthesis of the NADK2 overexpressors was caused, at least in part, by affecting the expression levels of GS/GOGAT pathway genes, the expression levels of several related genes were analyzed by real-time PCR. Levels of transcripts from the Glu dehydrogenases (GDH1 and GDH2; EC 1.4.1.3) were also compared. The expression of NADH-dependent GOGAT (GLT1; EC 1.4.1.13), GDH1, and GDH2 consistently mirrored the amino acid contents of the wild type, the nadk2 mutant, and the NADK2 overexpressor lines (Fig. 5A). Expression levels decreased among the isoenzymes of cytosolic GS (GLN1; EC 6.3.1.2), GLN1;1 (high-affinity form) and GLN1;3 (low-affinity form), in the nadk2 mutant, whereas no significant difference was observed in overexpressors. The expression levels of GLN1;5, chloroplastic GS (GS2; EC 6.3.1.2), and ferredoxin-dependent GOGAT (GLU1 and GLU2; EC 1.4.7.1) differed some from the wild type in both the nadk2 mutant and overexpressors, but not with any readily discernible pattern. Furthermore, we investigated GS and GDH activities in leaves (Fig. 5B). However, both GS and GDH activities were similar in the wild type, the nadk2 mutant, and NADK2 overexpressors.
Figure 5.
Effects of NADK2 overexpression on GS/GOGAT pathway-related gene expression. A, Levels of GLN1;1, GLN1;3, GLN1;5, GS2, GLU1, GLT1, GDH1, and GDH2 transcripts in rosette leaves of 23-d-old wild-type plants (WT), the nadk2 mutant, and NADK2 overexpressors (OX1 and OX2) were compared. Values were obtained from real-time PCR and expressed as relative to the wild type. B, GS and GDH activities in leaves are expressed relative to the wild type. GS and GDH activities in the wild type are 692.9 nmol Gln min−1 g−1 fresh weight and 36.3 nmol Glu min−1 g−1 fresh weight, respectively. Each value represents the mean ± sd calculated from three replicates. Significant differences from the wild type are shown (* P < 0.05, ** P < 0.01; t test).
NADK2 Overexpression Affects Both Gene Expression Associated with Nitrate Reduction and Nitrate Accumulation
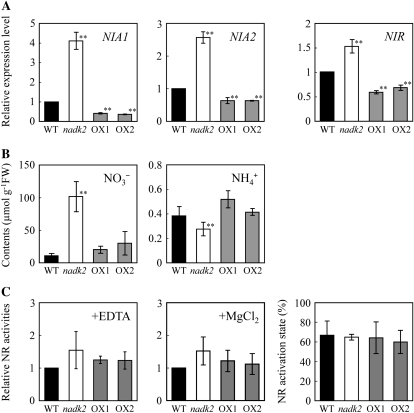
Nitrate absorbed from soils is reduced to ammonium in sequential reactions catalyzed by nitrate reductase (NR; EC 1.7.1.1) and nitrite reductase (EC 1.7.7.1). This nitrate reduction process is essential to N assimilation. For example, overexpression of nitrite reductase affects N assimilation in tobacco plants (Takahashi et al., 2001). Although the overexpression of NR scarcely influences N assimilation (Ferrario-Méry et al., 1998), it has recently been reported that overexpression of the constitutively active form of NR leads to high N assimilation, implying a critical role for posttranslational regulation of NR in N assimilation (Lea et al., 2006). To investigate the possibility that NADK2 overexpression increases N assimilation via nitrate reduction, we examined the expression of genes related to nitrate reduction and the contents of inorganic N in leaves. The transcript levels of NIA1 and NIA2, which encode the two isoforms of the NR, were 4- and 2.5-fold higher in the nadk2 mutant, respectively, but decreased in the NADK2 overexpressors (Fig. 6A). Nitrite reductase gene (NIR) expression was also higher in the nadk2 mutant but lower in the NADK2 overexpressors. This result ruled out the possibility that up-regulation of NIA and NIR caused the increase in N assimilation by the NADK2 overexpressors but suggested that the expression levels of NIA and NIR are responsive to the modified metabolic balances in nadk2 and the NADK2 overexpressors. NO3− induces NIA and NIR expression (Pouteau et al., 1989; Lin et al., 1994; Scheible et al., 1997). In fact, NO3− concentrations were approximately 10-fold higher in the nadk2 mutant (Fig. 6B). Although NO3− was also 2- to 3-fold higher in the NADK2 overexpressors, concentrations of Gln, a known repressor of NIA (Vincentz et al., 1993), were also higher in these lines. NH4+ was about the same in the wild type and NADK2 overexpressors but slightly lower in the nadk2 mutant. However, NR activity of the unphosphorylated form, total NR activity, and the NR activation state were unchanged in the nadk2 mutant and in the NADK2 overexpressors (Fig. 6C).
Figure 6.
Effects of NADK2 overexpression on nitrate reduction. Transcript and metabolite levels associated with nitrate reduction in rosette leaves of 23-d-old wild-type plants (WT), the nadk2 mutant, and NADK2 overexpression (OX1 and OX2) lines were analyzed. A, Transcript levels of nitrate reductase (NIA1 and NIA2) and nitrite reductase (NIR) genes estimated by real-time PCR are expressed relative to the wild type. Each value represents the mean ± sd calculated from three replicates. B, Accumulation of NO3− and NH4+ in leaves determined by ion chromatography. C, NR activity and the ratio in the activation state. Values are expressed relative to the wild type in the presence of EDTA (668.0 nmol nitrite h−1 g−1 fresh weight [FW]) and in the presence of MgCl2 (454.8 nmol nitrite h−1 g−1 fresh weight), respectively. Each value represents the mean ± se of three independent experiments. Significant differences from the wild type are shown ** P < 0.01; t test).
Requirement for Abundant N in NADK2 Overexpression-Mediated Activation of N Assimilation
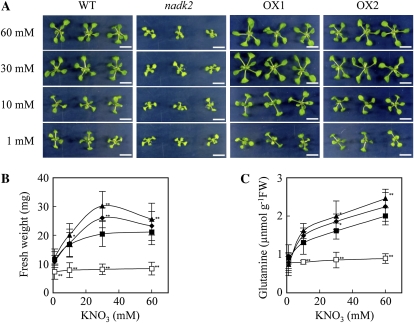
Transgenic Arabidopsis lines that are able to assimilate N at higher rates than the wild type grew better under low-N conditions (Yanagisawa et al., 2004). Therefore, we examined the growth of Arabidopsis plants with different NADK activities under different N conditions. Arabidopsis plants were grown on plates containing 1, 10, 30, or 60 mm KNO3 (Fig. 7A). Both NADK2 overexpressors grew better than the wild type with 30 mm KNO3. The improved growth of the NADK2-OX1 line was observed even under the 10 and 60 mm KNO3 conditions (Fig. 7B). The growth of NADK2 overexpressors with 60 mm KNO3 was not as good as with half the KNO3, implying that 60 mm KNO3 in the medium exceeds the optimal concentration for the NADK2 overexpressors. The growth of the nadk2 mutant was inferior to that of the wild type under any N conditions, and increasing the amount of supplied N did not increase growth. Since the growth of the nadk2 mutant, the wild type, and NADK2 overexpressors was comparable under low-N conditions, NADK2-dependent stimulation of N assimilation would likely only be evident under high-N conditions. The results of the Gln content analysis supported these observations in that Gln was higher in the NADK2 overexpressors than in the wild type grown in 30 or 60 mm KNO3 but was comparable in each of the lines grown in 1 mm KNO3 (Fig. 7C).
Figure 7.
Growth and Gln concentrations in the wild type (WT), the nadk2 mutant, and NADK2 overexpression (OX1 and OX2) lines grown on a low-N medium. A, Plants grown on a modified half-strength MS agar medium containing 1, 10, 30, or 60 mm KNO3 as the sole N source. Bars = 1 cm. B and C, Comparison of growth and Gln concentrations in rosette leaves in plants grown on modified MS medium containing different concentrations of KNO3. Plants were grown on MS medium for 5 d after germination and transferred to MS medium containing 1, 10, 30, or 60 mm KNO3. Plants grown for a further 7 d were compared for growth and Gln contents. Black squares, the wild type; white squares, the nadk2 mutant; black triangles, the NADK2-OX1 line; black diamonds, the NADK2-OX2 line. Values represent means ± sd of at least 25 plants (B) or six independent experiments (C). Significant differences from the wild type are shown (* P < 0.05, ** P < 0.01; t test). FW, Fresh weight.
DISCUSSION
In this report, Arabidopsis plants with enhanced NADK activity were used to investigate the effects of NADP overproduction on plant metabolism, and specifically on C and N assimilation. Although both NAD and NADP are redox regulators of living cells, these compounds likely play overlapping but different roles in other biological processes. NAD is also a substrate for mono- and poly-ADP-ribosylation reactions, which are posttranslational protein modifications. Further analysis of such NAD-degrading reactions in mammals revealed that they are involved in other cellular processes, including energy metabolism and cell death (Hassa et al., 2006; Alvarez-Gonzalez, 2007; Hassa and Hottiger, 2008). It is noteworthy that alteration of ADP-ribosylation activity directly affects intracellular NAD concentrations independently of redox reactions. On the other hand, NADK activity regulates the NAD/NADP ratio and is induced by environmental stresses in higher plants (Zagdańska, 1990; Delumeau et al., 2000; Ruiz et al., 2002). In cyanobacteria, illumination-dependent alterations in the NADP(H)/NAD(H) ratio induce dissociation of the PRK/CP12/GAPDH complex to regulate the activities of PRK and GAPDH (Tamoi et al., 2005). Based on these findings, changes in NAD(P)(H) concentrations would be expected to affect cellular processes in plants independently of the redox state. In this work, we show that transgenic Arabidopsis plants overexpressing NADK2 have elevated levels of NADP(H) in their leaves. However, the total contents and the redox state remained almost unchanged in both the nadk2 mutant and the NADK2 overexpressors. These plants can thus be used for studying the regulation mediated by NADP(H). We found several NADK2 overexpressor phenotypes, including modified C and N metabolism, which are primarily due to the NADP content itself or to the modified NADP/NAD ratio, rather than to variations in redox states. Because the manipulation of NADK2 activity induced pleiotropic effects, NADK2 appears to be critical for proper metabolic regulation in Arabidopsis.
Modification of NADK2 expression also induced several morphological phenotypes in Arabidopsis. Under the growth conditions used, the nadk2 mutant produced small and pale green leaves with decreased chlorophyll content (Chai et al., 2005). Conversely, the chlorophyll content of NADK2 overexpression plants was higher than in the wild type, although overexpression of NADK2 exerted only a limited impact on leaf size when plants were grown in soil. NADK2 deficiency in the nadk2 mutant decreased photosynthetic electron transport in leaves, probably through severe inhibition of zeaxanthin epoxidation (Takahashi et al., 2006b), but NADK2 overexpression did not obviously affect photosynthetic electron transport (data not shown). These observations suggest that the effects of elevated NADP content are not perfectly opposite to the effects generated by its reduction, possibly because some effects might be induced only when the NADP content or the NADP/NAD ratio reaches some threshold. This explanation might be applicable to other phenomena that are inconsistent in the nadk2 mutant and the NADK2 overexpressors.
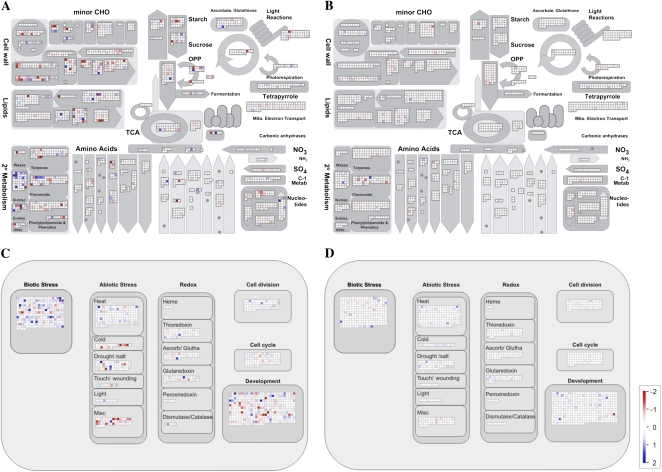
To identify pleiotropic effects caused by modifications of NADP(H) contents, transcriptomes of wild-type Arabidopsis, the nadk2 mutant, and the NADK2 overexpressor were compared by DNA microarray analysis (Fig. 8; Supplemental Tables S1 and S2). Broad effects on gene expression in the nadk2 mutant were revealed. In this mutant, the expression of genes involved in metabolic regulation, stress responses, cellular homeostasis, and morphogenesis was strongly modified. This finding is in accord with results obtained in previous reports, because the mutant shows a severe morphological phenotype and high sensitivity against ROS stress (Chai et al., 2005; Takahashi et al., 2006b). This suggests that decreased levels of NADP(H), which acts as an essential factor in a variety of processes, induces disorders at least partly through modulations of gene expression. On the other hand, limited and comparatively moderate effects on gene expression were found in the NADK2 overexpressor. While the expression levels of 2,878 genes were significantly modified in the nadk2 mutant, only 47 genes were affected in the NADK2 overexpressor (P < 0.01).
Figure 8.
Effects of up- and down-regulation of the NADK2 gene on the gene expression profile in leaves. The transcript levels of genes associated with metabolic activities in the nadk2 mutant (A) or the NADK2-OX2 line (B) were compared with those in the wild type. The transcript levels of genes associated with cellular responses in the nadk2 mutant (C) and the NADK2-OX2 line (D) were also compared with those in the wild type. Genes that exhibited increased expression levels are shown in blue, while those with decreased expression are shown in red. The color scale applies to all panels. CHO, Carbohydrate; OPP, oxidative pentose phosphate.
Importantly, we found that intermediates of the Calvin cycle such as Rib-5-P and RuBP were higher in the NADK2 overexpressors, although NADK2 overexpression had only a limited effect on the expression level of related genes and photosynthetic electron transport. Therefore, some enzyme activity in the Calvin cycle must be different in the NADK2 overexpressors in vitro. Previously, it has been shown that expression of a cyanobacterial dual-function enzyme, FBPase/sedoheptulose-1,7-bisphosphatase, in chloroplasts of transgenic tobacco plants significantly increases Rubisco activity but not the activities of PRK, NADP-GAPDH, and aldolase (Miyagawa et al., 2001). This observation might indicate that there is some mechanism that coordinately regulates Rubisco activity and the activity of Calvin cycle enzymes. One possible mechanism to explain this observation is that the increased NADP(H) content may improve conjugation of NADP(H) to NADP(H)-dependent enzymes or complexes, including the PRK/CP12/GAPDH complex. Indeed, some NAD(P)(H)-dependent enzymes are activated by increased NAD(P)(H) contents (White et al., 1983; Scagliarini et al., 1990; Stewart and Copeland, 1998; Graciet et al., 2002). Therefore, even if there was no change in the activities of PRK and NADP-GAPDH when activity was measured in vitro in the presence of identical concentrations of NAD(P)(H), there is still a possibility that the activity of these enzymes was different in the cells of the wild-type and NADK2 transgenic Arabidopsis plants. This hypothesis is comparable to the case of Rubisco activation, and it has been proposed that increases in RuBP play a role in the activation of Rubisco in vivo. We demonstrate here that significant increases in initial and total Rubisco activities and RuBP contents themselves were induced in the NADK2 overexpressors. However, it is currently unknown whether these changes were induced by activation of the Calvin cycle or triggered the activation of the Calvin cycle. Therefore, further analysis (e.g. of possible modifications of enzyme activity by thiol-mediated regulation) would be necessary to elucidate the relationship between overexpression and modified activity of the Calvin cycle. Discovery of a regulatory facet associated with NADP(H) would then provide a new target for genetic engineering of the Calvin cycle.
Intermediates of the Calvin cycle are utilized in several biosynthetic pathways, including the GS/GOGAT pathway for Glu family amino acid synthesis via 2-oxoglutarate as a C skeleton (Hodges, 2002; Suzuki and Knaff, 2005; Forde and Lea, 2007; Tabuchi et al., 2007). Since 2-oxoglutarate levels were altered proportionally with NADP(H) contents, we hypothesized that Gln and Glu synthesis were also affected by NADK2 overexpression. Indeed, levels of Glu family amino acids were higher in the NADK2 overexpressors. There was no evident activation or repression of gene expression associated with the GS/GOGAT cycle in the nadk2 mutant or the NADK2 overexpressors, although there was a slight modulation of GLT1 and GDH expression proportional to NADK activity. Furthermore, no significant change was observed in GS or GDH activity of the nadk2 mutant or the NADK2 overexpressors, suggesting that other factors may be involved in producing higher Glu family amino acid concentrations in the NADK2 overexpressors. On the other hand, the overexpression of NADK2 significantly affected expression of genes encoding enzymes involved in nitrate reduction. However, NIA and NIR expression was high in the nadk2 mutant but low in the NADK2 overexpressors. This modified gene expression thus likely reflects the altered metabolic balance rather than activation of N assimilation. Despite the reduced levels of NIA and NIR transcripts, no significant difference among the wild type, the nadk2 mutant, and NADK2 overexpressors was found in terms of NR activity and NR activation state. However, nitrate reduction itself appears to be more active in the NADK2 overexpressors, judging from the Gln content in plants that were grown on medium containing nitrate as the sole N source. Thus, NADK2 overexpression appears to stimulate N assimilation not so much through changes in the phosphorylation state of the NR protein but rather through other factors, such as the supply of C skeletons derived from the Calvin cycle, at least in part.
Neither knockout nor overexpression of NADK2 affected Gln contents under low-N (1 mm KNO3) conditions, whereas overexpression of NADK2 induced increases in the Gln content under high-N (30 or 60 mm KNO3) conditions (Fig. 7C). Therefore, we speculate that NADP levels maintained by NADK1 and NADK3 in the cytosol may be sufficient for N assimilation at low levels under low-N conditions, while NADK2 activity in chloroplasts may dictate N assimilation rates under high-N conditions. This hypothesis may be supported by the observation that the growth of the nadk2 mutant is comparable to that of the wild type and the NADK2 overexpressors under low-N conditions but is not as good under high-N conditions (Fig. 7, A and B). Unexpectedly, we observed that the growth of NADK2 overexpressors, but not the wild type, decreased when the concentration of KNO3 in the medium was 60 mm rather than 30 mm. Although the mechanism underlying this phenomenon is currently unknown, it may be that NADK2 is involved in controlling growth in response to exogenous N concentrations in the environment.
In summary, the overexpression of a single gene, NADK2, caused pleiotropic effects on numerous metabolic processes in Arabidopsis leaves. Although the observed effects on metabolism presumably included direct or indirect consequences that are not distinguishable at this stage, our results suggest that modulation of NADP levels would be integrated into the network that controls plant metabolism. Furthermore, our results also highlight the possibility that enhancement of cofactor synthesis is a workable strategy for improving C and N assimilation in planta, because the overexpression of NADK2 improved growth and increased the concentration of Gln, a storage material. More effective improvement of C and N assimilation might be achieved by identification and targeted modifications of key processes that are critical for C and N metabolism.
MATERIALS AND METHODS
Generation of Transgenic Plants
To generate Arabidopsis (Arabidopsis thaliana) plants overexpressing NADK2 under the control of the cauliflower mosaic virus 35S promoter, a cDNA clone containing the complete Arabidopsis NADK2 open reading frame was amplified by PCR using primers with an additional sequence for the attB site. The PCR product obtained was cloned into pDONR201 (Invitrogen) and then transferred into pH2GW7 (Karimi et al., 2002) using the Gateway recombination system. The resultant plasmid (pH2GW7-AtNADK2) was introduced into wild-type Arabidopsis (ecotype Columbia) by Agrobacterium tumefaciens-mediated transformation utilizing the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on 0.8% (w/v) agar plates containing Murashige and Skoog (MS) medium and 40 μg mL−1 hygromycin. Homozygous plants harboring the transgene were used for further analysis.
Plant Materials and Growth Conditions
Plants were grown on MS agar plates or in soil at 23°C under continuous light conditions (60 μmol m−2 s−1). For analysis of metabolite contents and gene expression, leaves of 23-d-old soil-grown plants were frozen in liquid N and stored at −80°C. For evaluation of growth under different N conditions, seeds were germinated on modified half-strength MS medium. Five-day-old seedlings were transferred onto plates containing different concentrations of KNO3 and grown for 7 d. For the analyses shown in Figure 4, the plants were grown with photoperiods of 16 h at 60 μmol m−2 s−1 and 25-d-old plants were used.
Reverse Transcription (RT)-PCR and Real-Time RT-PCR Expression Analysis
Total RNA was extracted from 23-d-old plants with an RNeasy Plant Mini Prep Kit (Qiagen), and 1 μg of total RNA was reverse transcribed with oligo(dT) primer using Ready-To-Go You-Prime First-Strand Beads (GE Healthcare). First-strand cDNA was amplified by PCR with specific primer sets for NADK2 and for the constitutively expressed β-tubulin4 (TUB4) gene as a control. The following primers were used: NADK2 (forward, 5′-GGCTTCTCTGCAGCCCCTATTGCTGTGCC-3′; reverse, 5′-GACTCGTTTGAGGTCTTGCCTGAAGTCCT-3′) and TUB4 (forward, 5′-AGAGGTTGACGAGCAGATGA-3′; reverse, 5′-CCTCTTCTTCCTCCTCGTAC-3′). For real-time RT-PCR, total RNA was treated with RNase-free DNase I (Qiagen) to eliminate genomic DNA contamination according to the manufacturer's instructions. First-strand cDNA was synthesized using a QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer's instructions. Quantitative real-time RT-PCR was performed with the QuantiTect SYBR Green PCR Kit (Qiagen) and an ABI Prism 7500 sequence detection system (Applied Biosystems). Actin8 was used as an internal control in each experiment. The following gene-specific primers were used: GLN1;1 (forward, 5′-TGTCATGTGCGATGCGTACA-3′; reverse, 5′-CCGCAGCGTGTCTTTTGTTA-3′), GLN1;3 (forward, 5′-CACATTGCTGCTTACGGTGAA-3′; reverse, 5′-TGTGTTGATGTCTGCGGTTTC-3′), GLN1;5 (forward, 5′-GGTCGTGACATTGTCGATGCT-3′; reverse, 5′-CACCATTGGCACCACCAATA-3′), GS2 (forward, 5′-TGGTGAAGTTATGCCTGGACAGT-3′; reverse, 5′-CATGATCACCTGCATCAATTCCT-3′), GLU1 (forward, 5′-ATGTCCTCCGTGACCTGCTT-3′; reverse, 5′-AACGGCCGGTTCTACTTTCC-3′), GLU2 (forward, 5′-GCATTTAGCCAACCGTCCTATT-3′; reverse, 5′-CAGGGATAGGGTTACGGTCACT-3′), GLT1 (forward, 5′-GCTATCTCAGAAGGACGTCAAGCT-3′; reverse, 5′-AGCTTGGCGTCTTCGTCATC-3′), GDH1 (forward, 5′-GGGAAGATTGTTGCCGTGAGT-3′; reverse, 5′-TGAGCAAGGCCGGGATATC-3′), GDH2 (forward, 5′-CGAAGCTGATCCACGAGAAAG-3′; reverse, 5′-AGGGTTCCTGATTGCACCAGTA-3′), NIA1 (forward, 5′-TTTGGTCAACCCACGTGAGAA-3′; reverse, 5′-CTTACGAACGTCGTGCGAGAT-3′), NIA2 (forward, 5′-GGCGTGGTTCGTTCTTACAAA-3′; reverse, 5′-TTGGTTCTGGTGCGCCTTA-3′), and NIR (forward, 5′-TCGCAGTTTATCACCGCTAATTC-3′; reverse, 5′-CCACCACACACACATTCCACTT-3′).
Chlorophyll Estimation
Chlorophyll was extracted from leaves as described previously (Oka et al., 2004). Frozen leaves were homogenized with 80% acetone, and homogenates were centrifuged at 1,500 rpm for 5 min. Supernatants were measured at 645 and 663 nm. The chlorophyll content was calculated according to Mackinney (1941) and expressed as mg chlorophyll g−1 fresh weight.
Enzyme Assays
Preparation of crude tissue lysates and NAD kinase assays were performed as described previously (Takahashi et al., 2006a). Briefly, frozen leaves were ground in liquid N and further homogenized in a buffer containing 50 mm tripotassium phosphate and 100 mm sodium ascorbate (pH 10.0). After centrifugation at 15,000 rpm for 5 min, the resulting supernatant was used as crude extract. NADK activity was determined in 1 mL of reaction mixture containing 34 mm Tricine-KOH (pH 8.0), 5 mm NAD, 5 mm ATP, 7 mm MgCl2, 6 mm nicotinamide, and 400 to 500 μg of total protein from the crude extract incubated for 30 min at 30°C. NADK activity was expressed as nmol NADP h−1 g−1 fresh weight. For measurement of Rubisco activity, frozen leaf discs were ground with a chilled mortar and pestle and further homogenized in an extraction buffer containing 100 mm HEPES-KOH (pH 8.0), 1 mm MgCl2, 1 mm EDTA, 2 mm dithiothreitol (DTT), and 1 mm phenylmethylsulfonyl fluoride (PMSF). Initial and total activities were measured with a spectrophotometer at 340 nm as described by Sharkey et al. (1991). Stromal FBPase was extracted in 50 mm HEPES-KOH (pH 8.0), 10 mm MgCl2, 1 mm EDTA, and 1 mm PMSF and assayed as described by Hurry et al. (1995). To assay the activity of the other Calvin cycle enzymes, frozen tissues were homogenized in an extraction buffer containing 50 mm HEPES-KOH (pH 8.0), 10 mm MgCl2, 1 mm EDTA, 1 mm DTT, and 1 mm PMSF. Aldolase, PGK, and PRK activities were measured according to the methods described by Haake et al. (1998), Leegood and Walker (1980), and Latzko et al. (1970), respectively. NADP-GAPDH activity was measured in a reaction mixture containing 100 mm Tricine-KOH (pH 8.0), 30 mm MgCl2, 20 mm KCl, 1 mm EDTA, 5 mm DTT, 0.5 mm NADPH, 5 mm ATP, 6 mm phosphoglyceric acid, and 10 units of PGK. Crude extracts were prepared to measure NR activity by the method of Ferrario-Méry et al. (1998). NR activity was assayed in the reaction mixture containing 50 mm HEPES-KOH (pH 7.5), 0.1 mm NADH, 5 mm KNO3, and either 6 mm MgCl2 or 2 mm EDTA. After incubation at room temperature for 15 min, an equal volume of 1% (w/v) sulfanilamide in 3 n HCl and 0.02% (w/v) N-naphthylethylenediamime dehydrochloride were added. Nitrite concentration was measured at 540 nm, and NR activity was expressed as nmol nitrite h−1 g−1 fresh weight. The NR activation state was estimated by calculating the ratio of the activity in the presence of 6 mm MgCl2 (activity of the unphosphorylated form) to the activity in the presence of 2 mm EDTA (total activity) and expressed as a percentage. For measurement of GS and GDH, a crude leaf extract was prepared and GS activity was measured by quantifying produced Gln, according to Ishiyama et al. (2004). GDH activity was determined by quantifying Glu production according to Masclaux et al. (2000) with minor modifications. Protein concentration in the crude extract was measured with a protein assay kit (Bio-Rad Laboratories).
Metabolite Analysis
Samples were ground to fine powder in liquid N and further homogenized with ice-cold 50% (v/v) methanol containing PIPES and Met sulfone as internal standards. Homogenates were then centrifuged at 15,000 rpm for 5 min, the supernatant was filtered through a Millipore 5-kD cutoff filter (Amicon), and the filtrates were used for analysis. Separation and quantification of organic acids, nucleotides, and amino acids were performed by CE-MS (Agilent Technologies), as described previously (Takahashi et al., 2006a, 2006b). For the determination of organic acids and nucleotides, a polyethylene glycol-coated capillary (DB-WAX; J&W Scientific) with 20 mm ammonium acetate (pH 6.8) as a run buffer was used. Amino acids were separated in an uncoated fused-silica capillary using 1 m formic acid (pH 1.9) as a run buffer. Accuracy was verified with known concentrations of reference standard compounds. Nitrate and ammonia contents were measured with an ICS-3000 ion chromatography system (Dionex). Accuracy was established using cation mixed standard solution II and anion mixed standard IV (Kanto Chemical).
Agilent Oligomicroarray Analysis
Triplicate independent biological replicates of microarray experiments were performed on an Agilent Technologies 44K Arabidopsis 3 Oligo Microarray. Total RNA was extracted by the TRIzol method. Total RNA was further purified using the RNeasy Plant Mini Kit (Qiagen). The purified RNA was quantified by 260/280/320-nm UV light absorption measurements with a spectrophotometer (Ultrospec 3000; Pharmacia Biotech) and by an Agilent 2100 bioanalyzer (Agilent Technologies). The purified RNA was used for the preparation of Cy5- and Cy3-labeled cDNA probes. All experiments and data analysis were performed according to the manufacturer's manual and instructions. Feature-extraction and image-analysis software (version 7.5; Agilent Technologies) was used to locate and delineate every spot in the array and to integrate each spot's intensity, using filtering and normalization by the Lowess method. Microarray data were annotated according to the Arabidopsis genome (The Arabidopsis Information Resource 7 version). Genes were classified into functional categories and were visualized using MapMan, mapping version 2.2.0 (http://gabi.rzpd.de/projects/MapMan/; Thimm et al., 2004).
Reference Reagents
Analytical-grade dihydroxyacetone phosphate, Fru-1,6-bisphosphate, ribulose-5-phosphate, Rib-5-P, RuBP, Glc-6-P, 3PGA, phosphoenolpyruvate, pyruvate, citrate, 2-oxoglutarate, and amino acids were used as reference reagents. Standard solutions were prepared by adjusting these compounds to 50 μm.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Selected metabolism-related genes that differentially expressed between nadk2 and wild-type plant.
Supplemental Table S2. Selected metabolism-related genes that differentially expressed between NADK2-OX and wild-type plant.
Supplementary Material
Acknowledgments
We thank Masahiro Tamoi (Department of Food and Nutrition, Faculty of Agriculture, Kinki University) for helpful discussion and evaluation of CO2 uptake ratio of the Arabidopsis plants.
This work was supported by a Grant-in-Aid for Young Scientists Start-up (grant no. 19870031), by a Research Fellowship for Young Scientists of the Japan Society for the Promotion of Science (to S.-n.H.), and by Core Research for Evolutional Science and Technology, Japan Science and Technology Agency, Japan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hideyuki Takahashi (h-takahashi@ibrc.or.jp).
The online version of this article contains Web-only data.
References
- Alvarez-Gonzalez R (2007) Genomic maintenance: the p53 poly(ADP-ribosyl)ation connection. Sci STKE 2007 68. [DOI] [PubMed] [Google Scholar]
- Berrin JG, Pierrugues O, Brutesco C, Alonso B, Montillet JL, Roby D, Kazmaier M (2005) Stress induces the expression of AtNADK-1, a gene encoding a NAD(H) kinase in Arabidopsis thaliana. Mol Genet Genomics 273 10–19 [DOI] [PubMed] [Google Scholar]
- Chai MF, Chen QJ, An R, Chen YM, Chen J, Wang XC (2005) NADK2, an Arabidopsis chloroplastic NAD kinase, plays a vital role in both chlorophyll synthesis and chloroplast protection. Plant Mol Biol 59 553–564 [DOI] [PubMed] [Google Scholar]
- Chai MF, Wei PC, Chen QJ, An R, Chen J, Yang S, Wang XC (2006) NADK3, a novel cytoplasmic source of NADPH, is required under conditions of oxidative stress and modulates abscisic acid responses in Arabidopsis. Plant J 47 665–674 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Coruzzi G, Bush DR (2001) Nitrogen and carbon nutrient and metabolite signaling in plants. Plant Physiol 125 61–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi GM, Zhou L (2001) Carbon and nitrogen sensing and signaling in plants: emerging ‘matrix effects.’ Curr Opin Plant Biol 4 247–253 [DOI] [PubMed] [Google Scholar]
- Delumeau O, Morére-Le Paven MC, Montrichard F, Laval-Martin DL (2000) Effects of short-term NaCl stress on calmodulin transcript levels and calmodulin-dependent NAD kinase activity in two species of tomato. Plant Cell Environ 23 329–336 [Google Scholar]
- Ferrario-Méry S, Valadier MH, Foyer CH (1998) Overexpression of nitrate reductase in tobacco delays drought-induced decrease in nitrate reductase activity and mRNA. Plant Physiol 117 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG, Lea PJ (2007) Glutamate in plants: metabolism, regulation, and signalling. J Exp Bot 58 2339–2358 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Parry M, Noctor G (2003) Markers and signals associated with nitrogen assimilation in higher plants. J Exp Bot 54 585–593 [DOI] [PubMed] [Google Scholar]
- Graciet E, Lebreton S, Camadro JM, Gontero B (2002) Thermodynamic analysis of the emergence of new regulatory properties in phosphoribulokinase-glyceraldehyde 3-phosphate dehydrogenase complex. J Biol Chem 277 12697–12702 [DOI] [PubMed] [Google Scholar]
- Haake V, Zrenner R, Sonnewald U, Stitt M (1998) A moderate decrease of plastid aldolase activity inhibits photosynthesis, alters the levels of sugars and starch, and inhibits growth of potato plants. Plant J 14 147–157 [DOI] [PubMed] [Google Scholar]
- Harding SA, Oh SH, Roberts DM (1997) Transgenic tobacco expressing a foreign calmodulin gene shows an enhanced production of active oxygen species. EMBO J 16 1137–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashida SN, Takahashi H, Uchimiya H (2009) The role of NAD biosynthesis in plant development and stress responses. Ann Bot (Lond) 103 819–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa PO, Haenni SS, Elser M, Hottiger MO (2006) Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev 70 789–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa PO, Hottiger MO (2008) The diverse biological roles of mammalian PARPs, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci 13 3046–3082 [DOI] [PubMed] [Google Scholar]
- Heineke D, Riens B, Grosse H, Hoferichter P, Peter U, Flugge UI, Heldt HW (1991) Redox transfer across the inner chloroplast envelope membrane. Plant Physiol 95 1131–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges M (2002) Enzyme redundancy and the importance of 2-oxoglutarate in plant ammonium assimilation. J Exp Bot 53 905–916 [DOI] [PubMed] [Google Scholar]
- Hurry VM, Keerberg O, Parnik T, Gardestrom P, Oquist G (1995) Cold-hardening results in increased activity of enzymes involved in carbon metabolism in leaves of winter rye (Secale cereale L.). Planta 195 554–562 [Google Scholar]
- Ishiyama K, Inoue E, Watanabe-Takahashi A, Obara M, Yamaya T, Takahashi H (2004) Kinetics properties and ammonium-dependent regulation of cytosolic isoenzymes of glutamine synthetase in Arabidopsis. J Biol Chem 279 16598–16605 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7 193–195 [DOI] [PubMed] [Google Scholar]
- Kawai S, Mori S, Mukai T, Hashimoto W, Murata K (2001. a) Molecular characterization of Escherichia coli NAD kinase. Eur J Biochem 268 4359–4365 [DOI] [PubMed] [Google Scholar]
- Kawai S, Mori S, Mukai T, Suzuki S, Yamada T, Hashimoto T, Murata K (2000) Inorganic polyphosphate/ATP-NAD kinase of Micrococcus flavus and Mycobacterium tuberculosis H37Rv. Biochem Biophys Res Commun 276 57–63 [DOI] [PubMed] [Google Scholar]
- Kawai S, Suzuki S, Mori S, Murata K (2001. b) Molecular cloning and identification of UTR1 of a yeast Saccharomyces cerevisiae as a gene encoding an NAD kinase. FEMS Microbiol Lett 200 181–184 [DOI] [PubMed] [Google Scholar]
- Lam HM, Coschigano KT, Oliveira IC, Melo-Oliveira R, Coruzzi GM (1996) The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu Rev Plant Physiol Plant Mol Biol 47 569–593 [DOI] [PubMed] [Google Scholar]
- Latzko E, Garnier RV, Gibbs M (1970) Effects of photosynthesis, photosynthetic inhibitors and oxygen on the activity of ribulose 5-phosphate kinase. Biochem Biophys Res Commun 39 1140–1144 [DOI] [PubMed] [Google Scholar]
- Lea US, Leydecker MT, Quilleré I, Meyer C, Lillo C (2006) Posttranslational regulation of nitrate reductase strongly affects the levels of free amino acids and nitrate, whereas transcriptional regulation has only minor influence. Plant Physiol 140 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegood RC, Walker DA (1980) Autocatalysis and light activation of enzymes in relation to photosynthetic induction in wheat chloroplasts. Arch Biochem Biophys 200 575–582 [DOI] [PubMed] [Google Scholar]
- Lerner F, Niere M, Ludwig A, Ziegler M (2001) Structural and functional characterization of human NAD kinase. Biochem Biophys Res Commun 288 69–74 [DOI] [PubMed] [Google Scholar]
- Lin Y, Hwang CF, Brown JB, Cheng CL (1994) 5′ proximal regions of Arabidopsis nitrate reductase genes direct nitrate-induced transcription in transgenic tobacco. Plant Physiol 106 477–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinney G (1941) Absorption of light by chlorophyll solutions. J Biol Chem 140 315–322 [Google Scholar]
- Masclaux C, Valadier MH, Brugière N, Morot-Gaudry JF, Hirel B (2000) Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta 211 510–518 [DOI] [PubMed] [Google Scholar]
- Miyagawa Y, Tamoi M, Shigeoka S (2001) Overexpression of a cyanobacterial fructose-1,6-/sedoheptulose-1,7-bisphosphatase in tobacco enhances photosynthesis and growth. Nat Biotechnol 19 965–969 [DOI] [PubMed] [Google Scholar]
- Oka E, Tagami Y, Oohashi T, Kondo N (2004) A physiological and morphological study on the injury caused by exposure to the air pollutant, peroxyacetyl nitrate (PAN), based on the quantitative assessment of the injury. J Plant Res 117 27–36 [DOI] [PubMed] [Google Scholar]
- Outten CE, Culotta VC (2003) A novel NADH kinase is the mitochondrial source of NADPH in Saccharomyces cerevisiae. EMBO J 22 2015–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouteau S, Cherel I, Vaucheret H, Caboche M (1989) Nitrate reductase mRNA regulation in Nicotiana plumbaginifolia nitrate reductase-deficient mutants. Plant Cell 1 1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaelli N, Finaurini L, Mazzola F, Pucci L, Sorci L, Amici A, Magni G (2004) Characterization of Mycobacterium tuberculosis NAD kinase: functional analysis of the full-length enzyme by site-directed mutagenesis. Biochemistry 43 7610–7617 [DOI] [PubMed] [Google Scholar]
- Ruiz JM, Sánchez E, Garcia PC, López-Lefebre LR, Rivero RM, Romero L (2002) Proline metabolism and NAD kinase activity in greenbean plants subjected to cold-shock. Phytochemistry 59 473–478 [DOI] [PubMed] [Google Scholar]
- Sakuraba H, Kawakami R, Ohshima T (2005) First archaeal inorganic polyphosphate/ATP-dependent NAD kinase, from hyperthermophilic archaeon Pyrococcus horikoshi: cloning, expression, and characterization. Appl Environ Microbiol 71 4352–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scagliarini S, Trost P, Valenti V, Pupillo P (1990) Glyceraldehyde 3-phosphate:NADP+ reductase of spinach leaves. Plant Physiol 94 1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibe R (2004) Malate valves to balance cellular energy supply. Physiol Plant 120 21–26 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Gonzales-Fontes A, Lauerer M, Müller-Röber B, Caboche M, Stitt M (1997) Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9 783–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey TD, Savitch LV, Butz ZD (1991) Photometric method for routine determination of k-cat and carbamylation of Rubisco. Photosynth Res 28 41–48 [DOI] [PubMed] [Google Scholar]
- Sinclair TM, Purcell LC, Sneller CH (2004) Crop transformation and the challenge to increase yield potential. Trends Plant Sci 9 70–75 [DOI] [PubMed] [Google Scholar]
- Stewart D, Copeland L (1998) Uridine 5′-diphosphate-glucose dehydrogenase from soybean nodules. Plant Physiol 116 349–355 [Google Scholar]
- Stitt M, Schulze D (1994) Does Rubisco control the rate of photosynthesis and plant growth? An exercise in molecular ecophysiology. Plant Cell Environ 17 465–487 [Google Scholar]
- Suzuki A, Knaff DB (2005) Glutamate synthase: structural, mechanistic and regulatory properties, and role in the amino acid metabolism. Photosynth Res 83 191–217 [DOI] [PubMed] [Google Scholar]
- Tabuchi M, Abiko T, Yamaya T (2007) Assimilation of ammonium ions and reutilization of nitrogen in rice (Oryza sativa L.). J Exp Bot 58 2319–2327 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Hayashi M, Goto F, Sato S, Soga S, Nishioka T, Tomita M, Kawai-Yamada M, Uchimiya H (2006. a) Evaluation of metabolic alteration in transgenic rice overexpressing dihydroflavonol-4-reductase. Ann Bot (Lond) 98 819–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Watanabe A, Tanaka A, Hashida SN, Kawai-Yamada M, Sonoike K, Uchimiya H (2006. b) Chloroplast NAD kinase is essential for energy transduction through the xanthophyll cycle in photosynthesis. Plant Cell Physiol 47 1678–1682 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Sasaki Y, Iba S, Morikawa H (2001) Nitrite reductase gene enrichment improves assimilation of NO2 in Arabidopsis. Plant Physiol 126 731–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamoi M, Miyazaki T, Fukamizo T, Shigeoka S (2005) The Calvin cycle in cyanobacteria is regulated by CP12 via the NAD(H)/NADP(H) ratio under light/dark conditions. Plant J 42 504–513 [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37 914–939 [DOI] [PubMed] [Google Scholar]
- Turner WL, Waller JC, Snedden WA (2005) Identification, molecular cloning and functional characterization of a novel NADH kinase from Arabidopsis thaliana (thale cress). Biochem J 385 217–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz M, Moureaux T, Leydecker MT, Vaucheret H, Caboche M (1993) Regulation of nitrate and nitrite reductase expression in Nicotiana plumbaginifolia leaves by nitrogen and carbon metabolites. Plant J 3 315–324 [DOI] [PubMed] [Google Scholar]
- White BA, Paone DAM, Cacciapuoti AF, Fricke RJ, Mosbach EH, Hylemon PB (1983) Regulation of bile acid 7-dehydroxylase activity by NAD+ and NADH in cell extracts of Eubacterium species V.P.I. 12708. J Lipid Res 24 20–27 [PubMed] [Google Scholar]
- Yanagisawa S, Akiyama A, Kisaka H, Uchimiya H, Miwa T (2004) Metabolic engineering with Dof1 transcription factor in plants: improved nitrogen assimilation and growth under low-nitrogen conditions. Proc Natl Acad Sci USA 101 7833–7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagdańska B (1990) NAD kinase activity in wheat leaves under water deficit. Acta Biochim Pol 37 385–389 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.