Abstract
Plant hormones, cytokinins (CKs), have been for a long time considered to be involved in plant responses to stress. However, their exact roles in processes linked to stress signalization and acclimatization to adverse environmental conditions are unknown. In this study, expression profiles of the entire gene families of CK biosynthetic and degradation genes in maize (Zea mays) during development and stress responses are described. Transcript abundance of particular genes is discussed in relation to the levels of different CK metabolites. Salt and osmotic stresses induce expression of some CK biosynthetic genes in seedlings of maize, leading to a moderate increase of active forms of CKs lasting several days during acclimatization to stress. A direct effect of CKs to mediate activation of stress responses does not seem to be possible due to the slow changes in metabolite levels. However, expression of genes involved in cytokinin signal transduction is uniformly down-regulated within 0.5 h of stress induction by an unknown mechanism. cis-Zeatin and its derivatives were found to be the most abundant CKs in young maize seedlings. We demonstrate that levels of this zeatin isomer are significantly enhanced during early stress response and that it originates independently from de novo biosynthesis in stressed tissues, possibly by elevated specific RNA degradation. By enhancing their CK levels, plants could perhaps undergo a reduction of growth rates maintained by abscisic acid accumulation in stressed tissues. A second role for cytokinin receptors in sensing turgor response is hypothesized besides their documented function in CK signaling.
Abiotic stresses, especially drought and soil salinity, are among the major limiting factors of plant growth and production in the environment. Activation of signaling pathways as a consequence of acquiring tolerance toward stress leads to rapid responses such as stomata closure or long-term changes in metabolism, growth, and development. Modification of gene expression profiles induced by stress response signal transduction can be mediated by phytohormones. Rapid elevation of abscisic acid (ABA) concentration in plant tissues exposed to water deficit is well documented (for review, see Schachtman and Goodger, 2008). ABA in particular reduces transpiration rates and activates at least two downstream signal transduction pathways involving MYB-like and MYC-like transcription factors (Abe et al., 2003). A recent microarray analysis of Arabidopsis cytokinin (CK) receptor mutants clearly showed that CK-mediated signaling can also be involved in stress responses. Knockout lines of two out of three CK receptors were strongly tolerant of drought and salt stress due to up-regulation of many stress-inducible genes (Tran et al., 2007). Alteration in the CK content in plants exposed to various stresses has been frequently reported. For instance, trans-zeatin (tZ) and trans-zeatin riboside (tZR) contents decreased rapidly in the elongation zone of barley (Hordeum vulgare) leaves within several minutes after salinity stress induction. However, concentrations of both CK types increased in the nonelongated part of the leaf blade, indicating a possible reduction of cell division and CK translocation (Fricke et al., 2006). A significant long-term decrease of active isoprenoid CK content was observed in barley roots and shoots after exposure to a higher concentration of NaCl (Kuiper et al., 1990). It was assumed that the water-deficit-induced CK deprivation in leaves is mainly attributed to decelerated transport of root-borne CK via xylem (Davies and Zhang, 1991). Comprehensive CK analysis in wild-type tobacco (Nicotiana tabacum) leaves exposed to drought showed a gradual decrease in bioactive CK levels during stress progress accompanied by elevated activity of CK degradation enzymes. Severe stress, however, led to the accumulation of all CK forms in the roots (Havlová et al., 2008).
Maize (Zea mays) is one of the most important crop species used especially for direct human consumption and animal feed, besides today being a significant plant source for chemical feedstock. Together with model plants Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), maize has been extensively studied on the molecular level, and although the full genome sequence has not been officially published yet, there is enough information accessible in public databases (e.g. The Institute for Genomic Research Maize Database; Chan et al., 2006) to describe coherent gene families. In the Arabidopsis genome, there are more than 50 genes functionally studied that are directly involved in CK signal transduction and metabolism. In 2004, three genes coding for CK-responsive His kinases (CK receptors; HKs) were functionally described in maize (Yonekura-Sakakibara et al., 2004). Surprisingly, in contrast to known CK receptors from Arabidopsis, all three receptors responded to cis-zeatin (cZ), which had for a long time been considered to be inactive. Genetic information about other signal transduction pathway components, His-phosphotransfer proteins and CK primary response regulators (RRs), is accessible from previous works (Sakakibara et al., 1998, 1999; Asakura et al., 2003). One out of 10 recently described rice RR genes was shown to be up-regulated in seedlings exposed to a high concentration of salt (Jain et al., 2006).
CKs are generated in plant tissues by two biosynthetic pathways, de novo synthesis and isoprenylated tRNA degradation. The bulk of isoprenoic CKs are synthesized due to the activity of ATP/ADP isopentenyltransferases (IPTs), whereas the production of cZ in plants seems to be attributed mostly to tRNA degradation (Kasahara et al., 2004; Miyawaki et al., 2006). Several putative ATP/ADP IPTs and one tRNA-IPT gene were mined from a maize bacterial artificial chromosome (BAC) clone library. One of them, ZmIPT2, was recently proved functional and shown to contribute to CK de novo biosynthesis during kernel development (Brugière et al., 2008). CK homeostasis in plants is primarily maintained by different compartmentation of biosynthesis and degradation. While de novo biosynthesis is mostly bound to plastids (Kasahara et al., 2004), CK catabolic processes take place in vacuoles and apoplast (Werner et al., 2003). The irreversible degradation of free CK bases and their derivatives is catalyzed by CK oxidase/dehydrogenases (CKXs), encoded as well by a small gene family. Nevertheless, there is evidence for several maize CKXs (Massonneau et al., 2004); only one member of maize CKX enzymes, ZmCKX1, was studied in detail in connection with plant stress responses (Brugière et al., 2003). Elevated levels of ZmCKX1 transcript were observed in developing kernels during various abiotic stresses. A positive effect on CKX expression in detached leaves was also induced by ABA, which suggests a possible role of this stress hormone in triggering CK degradation machinery.
The goal of this work was to clarify the role of CKs in the responses of maize seedlings to water deficit and salinity stress. The results provide (1) a comprehensive characterization of gene families involved in CK biosynthesis and degradation in maize, (2) a detailed characterization of the transcript abundance in different tissues and organs and their changes following stress treatment, and (3) together with the signaling and response network characterization, support for the concept of succession of gene expression changes that fit the changes in enzyme activities and endogenous contents of phytohormones.
RESULTS
Sequence Analysis of Genes Involved in CK Metabolism and Perception
Homology searches were done by BLASTn software on the maize genome sequencing project database (http://www.maizesequence.org/index.html) with currently known sequences as templates (Massonneau et al., 2004; Brugière et al., 2008) and rice orthologous sequences (Sakamoto et al., 2006; Hirose et al., 2007). BAC clones in the database cover almost 95% of information of the rough draft of the maize genome, as was announced on December 2008. Matched sequences with E values ≤ 2e−7 were checked for IPT and CKX conserved motifs. Contig sequences showing E values significantly below the threshold that contained insertion or deletion in open reading frames (ORFs) were excluded from the study. Eleven putative ORFs showing high similarity to 10 rice IPTs (Sakamoto et al., 2006) were identified. Seven of them are identical to maize IPTs recently annotated (IPT1, IPT2, and IPT4–IPT8; Brugière et al., 2008); hence, the numbering is kept due to the reference. The numbering, contig accession numbers to BAC libraries, locations of exons, as well as chromosome positions are summarized in Supplemental Table S1. Two out of four nonannotated sequences share high mutual homology (90.3%) and therefore were numbered as IPT3 and IPT3b. The third one shows the highest homology to IPT6 (82.4%) and was numbered as IPT9. A putative ortholog to a prokaryotic-origin tRNA-IPT gene (IPT10) highly homologous to rice OsIPT10 and Arabidopsis IPT9 was found (Sakamoto et al., 2006) on chromosome 6.
Two maize CKX genes were proved to encode functional enzymes (CKX1 and CKX3; Houba-Hérin et al., 1999; Morris et al., 1999; Massonneau et al., 2004), and expression profiles of another three genes were analyzed (CKX2, CKX4, and CKX5; Massonneau et al., 2004). Data mining from the maize genome database revealed another eight sequences showing ORFs homologous to annotated CKX proteins. They were numbered upwardly from CKX6 to CKX12 (Supplemental Table S1). The ORF found on chromosome 8 shares 93.4% homology to annotated CKX4 on chromosome 3 and has been annotated as CKX4b. Positions of additional close paralogs CKX2 and CKX3 (93.5% homology) were found on the same chromosome segments as CKX4 and CKX4b, respectively, indicating a common ancestral origin of these genes and the possibility that they segregated during the same duplication event. Patterns of recent chromosome duplication between maize chromosomes 3 and 8 have been reported (Gaut, 2001). Similar duplicates have been identified for gene pairs CKX7/CKX8 (94.2%) and CKX11/CKX12 (91.2%). Recently, the subcellular localization to cytosol and enzymatic activity of CKX10 have been demonstrated (Šmehilová et al., 2009). It seems to be the sole maize CKX isoform missing any signal peptide that contributes to CK depletion in the cytosol. Amino acid alignments of all maize IPTs and CKXs are presented in Supplemental Figures S1 and S2, and phylogenetic trees with rice and Arabidopsis orthologs are shown in Supplemental Figures S3 and S4.
Concerning genes involved in CK perception and primary response, the maize genome database screening confirmed loci for all annotated receptor and type A response regulator genes (Sakakibara et al., 1999; Asakura et al., 2003; Yonekura-Sakakibara et al., 2004). Several single nucleotide polymorphisms were detected among the annotated sequences and sequences generated from genome sequencing projects, which can be associated with different cultivars either used by or caused by the inaccuracies in the sequencing data. In the case of the RR3 gene, another locus on chromosome 5 carrying an ORF for a closely homologous gene (83.9%) was found and marked as RR3b. Primers for this response regulator were designed to amplify targets from both sequences. Primers amplifying HK1 were designed according to the annotated sequence NM_001111389 (BAC clone AC195458.3; chromosome 5). However, hybridization with the annotated sequence for receptor HK1a2 (NM_001112387; AC185637.3), which has not been functionally tested, is probable due to shared 92.4% homology. Only one locus was found for the receptor HK3, the corresponding transcript of which was previously shown to be alternatively spliced (Yonekura-Sakakibara et al., 2004). Two alternative versions of HK3 transcripts were described as full-length and functional HK3a and a nonfunctional version missing the third exon. Alignment of the genomic locus (BAC clone AC185417.3) revealed that the third exon (449 bp) is flanked by at least one large intron (third intron, 7,371 bp; sequence of the second intron is interrupted by two not yet arranged subcontigs). The size of the introns probably led to a splicing mistake and exon skipping, resulting in a folding of a nonfunctional receptor without the input domain.
One primer pair was designed to sense the signal of two genes encoding two very homologous cis-zeatin O-glucosyltransferases (cZOGT; Veach et al., 2003). A 9-cis-epoxycarotenoid dioxygenase seems to be the key regulatory enzyme in ABA release from carotenoid structures (Schwartz et al., 2003). The structure of 9-cis-epoxycarotenoid dioxygenase gene as well as its responsiveness to abiotic stress were first described in maize (Tan et al., 1997). The sequence of this gene (VP14, viviparous phenotype; U95953) was used to design a primer pair; however, the amplification was very weak and unspecific. A BLAST search from the Maize Genome Project revealed two homologous ORFs to the VP14 sequence (homology 93.5% and 99.1%). Thus, new sets of primers were designed to avoid all polymorphism, and the most efficient one listed in Supplemental Table S1 was used in this study. A similar approach was employed to design primers for closely related galactinol synthase genes (GALS; Zhao et al., 2004) and betaine aldehyde dehydrogenase (BADH).
Expression Patterns of IPT and CKX Genes in Maize
To determine which of the biosynthetic and degradation enzymes can be involved in stress responses, the IPT and CKX transcript accumulation was analyzed by real-time PCR using cDNA prepared from 7-d-old roots and leaves as well as from various tissues of mature plants along with developing and germinating kernels. The analysis was complemented by genes involved in the CK signal perception and primary CK response described previously (Asakura et al., 2003; Yonekura-Sakakibara et al., 2004).
The absolute transcript levels of all studied genes are summarized in Table I. Values for genes whose transcript level was less than one copy per nanogram of isolated total RNA are not listed, due to the possibility of being unreliable. Concerning IPT genes, two genes for maize tRNA-IPT (IPT1 and IPT10) were highly abundant and constitutively expressed in all tested organs. Other IPT transcripts showed spatial and temporal patterns of expression. Expression of IPT2 is solely bound to developing kernels, as recently published by Brugière et al. (2008). IPT5 is relatively highly present in all vegetative tissues, especially in roots and tassels. Transcripts of IPT4, IPT6, IPT7, and IPT9 were also detected together with IPT5 in roots, and these five IPT enzymes probably mainly contribute to the CK pool rising in the root during maize growth. In leaves, the total adenylate IPT transcript level is almost 1 order of magnitude lower than in roots, and in addition to IPT5, only IPT8 and IPT9 transcripts were determined above the defined threshold.
Table I.
Transcript abundance in different maize organs
Abundance is expressed as gene copy number in 1 ng of total RNA amplified by qPCR, normalized to 18S RNA, and recalculated as primer pair efficiency. Total RNA from each sample was transcribed in two independent reactions, and PCR was performed in duplicate. Mean values with sd are shown.
| Gene | Embryo | Endosperm | Pedicel | Silk | Tassel | Immature Ear | Coleoptile + Radicle | 7-d-Old Root | Mature Root | 7-d-Old Leaf | Mature Leaf |
|---|---|---|---|---|---|---|---|---|---|---|---|
| IPT genes | |||||||||||
| IPT1 | 101.9 ± 33.6 | 110.8 ± 6.6 | 83.4 ± 5.4 | 112.3 ± 12.9 | 111.9 ± 4.8 | 114.2 ± 17.8 | 63.0 ± 8.9 | 71.6 ± 12.0 | 111.2 ± 14.9 | 125.0 ± 17.8 | 38.8 ± 10.6 |
| IPT2 | 10.9 ± 2.2 | 89.4 ± 13.8 | 303.3 ± 17.2 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 |
| IPT3 | <1.0 | <1.0 | 1.1 ± 0.3 | <1.0 | 2.9 ± 0.6 | 1.2 ± 0.2 | <1.0 | <1.0 | 3.5 ± 0.2 | <1.0 | 1.1 ± 0.7 |
| IPT3b | <1.0 | <1.0 | <1.0 | <1.0 | 1.1 ± 0.2 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 |
| IPT4 | 1.9 ± 0.5 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | 1.0 ± 0.4 | 8.3 ± 0.3 | <1.0 | <1.0 |
| IPT5 | 5.1 ± 0.7 | <1.0 | 34.5 ± 6.2 | 32.3 ± 4.2 | 353.7 ± 29.0 | 66.4 ± 13.7 | 3.7 ± 1.0 | 49.7 ± 12.5 | 169.3 ± 7.9 | 14.0 ± 9.3 | 33.2 ± 4.9 |
| IPT6 | <1.0 | 1.7 ± 0.6 | <1.0 | 2.1 ± 0.4 | 8.5 ± 2.1 | 1.6 ± 0.3 | 1.1 ± 0.2 | 8.3 ± 2.7 | 19.2 ± 4.5 | <1.0 | <1.0 |
| IPT7 | <1.0 | <1.0 | 2.1 ± 0.2 | <1.0 | 11.2 ± 2.3 | <1.0 | <1.0 | 1.6 ± 0.4 | <1.0 | <1.0 | <1.0 |
| IPT8 | 8.5 ± 1.1 | 35.8 ± 3.8 | 4.2 ± 1.1 | 3.0 ± 0.5 | 7.3 ± 4.2 | <1.0 | 1.4 ± 0.9 | <1.0 | <1.0 | 6.4 ± 1.4 | 8.8 ± 0.9 |
| IPT9 | 2.2 ± 1.2 | <1.0 | 3.2 ± 0.7 | <1.0 | 7.9 ± 1.2 | <1.0 | <1.0 | 2.7 ± 0.6 | 23.2 ± 3.6 | 2.7 ± 0.3 | <1.0 |
| IPT10 | 51.5 ± 2.3 | 25.1 ± 1.6 | 16.7 ± 1.3 | 8.9 ± 0.6 | 40.9 ± 3.1 | 19.7 ± 2.8 | 48.0 ± 3.4 | 35.9 ± 2.7 | 11.2 ± 3.6 | 47.7 ± 1.0 | 46.9 ± 2.4 |
| CK oxidase/dehydrogenase genes | |||||||||||
| CKX1 | 422.9 ± 13.5 | 37.2 ± 7.4 | 95.2 ± 12.0 | 73.6 ± 14.0 | 190.0 ± 31.7 | 87.1 ± 15.5 | 2.1 ± 0.2 | 54.3 ± 4.5 | 116.7 ± 5.4 | 1.6 ± 0.1 | 1.5 ± 0.1 |
| CKX2 | 23.2 ± 2.7 | 35.8 ± 2.8 | 78.8 ± 8.4 | 19.4 ± 2.5 | 224.6 ± 12.6 | 20.9 ± 1.8 | 6.2 ± 0.7 | 18.7 ± 2.1 | 37.1 ± 1.6 | 27.5 ± 3.7 | 609.4 ± 24.2 |
| CKX3 | 15.3 ± 1.9 | 53.7 ± 0.7 | 38.7 ± 4.2 | 65.2 ± 6.8 | 12.4 ± 1.0 | 36.6 ± 2.2 | 2.9 ± 0.3 | 25.6 ± 0.9 | 5.8 ± 0.2 | 160.4 ± 5.3 | 10.2 ± 0.6 |
| CKX4 | <1.0 | 1.6 ± 0.7 | 57.1 ± 5.1 | 13.9 ± 0.4 | 2.5 ± 0.4 | 2.6 ± 0.1 | <1.0 | 2.6 ± 0.1 | 11.1 ± 0.2 | 1.1 ± 0.1 | 3.8 ± 0.2 |
| CKX4b | 7.9 ± 0.4 | <1.0 | 37.7 ± 6.5 | 21.7 ± 2.0 | 17.4 ± 3.8 | 7.4 ± 1.2 | 2.0 ± 0.2 | 26.3 ± 2.8 | 763.9 ± 96 | 20.3 ± 4.6 | 21.0 ± 1.8 |
| CKX5 | <1.0 | 1.8 ± 0.3 | 7.4 ± 2.0 | 54.6 ± 2.4 | 23.5 ± 1.2 | 2.5 ± 0.6 | <1.0 | <1.0 | 6.1 ± 0.4 | <1.0 | <1.0 |
| CKX6 | 16.4 ± 3.6 | <1.0 | 32.2 ± 3.3 | 4.2 ± 0.3 | 74.0 ± 3.4 | 16.6 ± 1.2 | 38.0 ± 1.7 | 221.2 ± 12.0 | 99.1 ± 11.0 | 93.4 ± 7.0 | 20.4 ± 1.7 |
| CKX7 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 |
| CKX8 | 6.6 ± 0.5 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | 2.1 ± 0.3 | 38.0 ± 19.6 | 2.6 ± 0.1 | 1.1 ± 0.2 | 4.6 ± 0.7 |
| CKX9 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | 1.3 ± 0.1 | <1.0 | <1.0 | <1.0 | 1.8 ± 0.7 | <1.0 |
| CKX10 | 141.6 ± 1.1 | 13.3 ± 0.7 | 229.3 ± 30.2 | 647.2 ± 56.1 | 1,120.1 ± 48.9 | 351.2 ± 28.7 | 219.2 ± 18.4 | 61.2 ± 18.4 | 126.2 ± 10.4 | 310.9 ± 25.1 | 76.6 ± 18.0 |
| CKX11 | 2.0 ± 0.1 | <1.0 | <1.0 | 1.0 ± 0.2 | 3.5 ± 0.3 | 1.3 ± 0.3 | 1.8 ± 0.1 | 2.5 ± 0.3 | 4.6 ± 0.3 | 71.7 ± 4.6 | 2.9 ± 0.7 |
| CKX12 | 9.5 ± 2.7 | 13.4 ± 0.8 | <1.0 | <1.0 | 3.8 ± 1.6 | <1.0 | 1.5 ± 0.5 | 2.4 ± 0.2 | 2.7 ± 0.4 | 10.3 ± 0.6 | 3.5 ± 1.9 |
| CK receptor genes | |||||||||||
| HK1 | 1,712.9 ± 40.9 | 821.7 ± 38.7 | 1,117.1 ± 54.8 | 1,243.1 ± 42.0 | 1,439.8 ± 45.8 | 2,237.4 ± 75.9 | 838.2 ± 45.4 | 2,078.9 ± 45.6 | 763.4 ± 27.5 | 596.3 ± 30.5 | 322.6 ± 17.5 |
| HK2 | 668.2 ± 47.0 | 818.8 ± 61.5 | 573.1 ± 29.5 | 1,849.3 ± 149.2 | 2,344.8 ± 88,3 | 2,233.0 ± 155.0 | 1,166.6 ± 147.0 | 4,812.8 ± 202.2 | 1,095.5 ± 85.7 | 2,394.4 ± 102.5 | 1,322.8 ± 147.8 |
| HK3 | 141.0 ± 9.8 | 340.4 ± 41.5 | 158.0 ± 25.4 | 362.5 ± 36.6 | 273.1 ± 30.0 | 225.1 ± 17.7 | 178.2 ± 18.2 | 641.5 ± 54.4 | 195.6 ± 9.8 | 150.8 ± 7.9 | 240.6 ± 13.2 |
| CK response regulator type A genes | |||||||||||
| RR1 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | <1.0 | 55.5 ± 4.7 | 189.4 ± 32.4 | <1.0 | 449.1 ± 19.7 | <1.0 |
| RR2 | 5.6 ± 1.4 | <1.0 | 29.7 ± 3.8 | <1.0 | 11.2 ± 0.8 | 4.3 ± 0.3 | 38.7 ± 7.2 | 116.4 ± 11.7 | 19.1 ± 3.7 | 66.6 ± 5.8 | 92.9 ± 24.0 |
| RR3 | 16.8 ± 2.7 | <1.0 | <1.0 | <1.0 | <1.0 | 3.3 ± 0.2 | <1.0 | <1.0 | <1.0 | 0.3 ± 0.1 | <1.0 |
| RR4 | 118.2 ± 24.7 | 20.3 ± 3.0 | 201.7 ± 41.0 | 39.2 ± 4.9 | 435.3 ± 33.7 | 112.6 ± 6.7 | 615.6 ± 47.5 | 979.5 ± 54.5 | 190.8 ± 5.6 | 303.5 ± 24.2 | 356.0 ± 26.8 |
| RR5 | 36.8 ± 9.1 | 44.8 ± 4.8 | 67.6 ± 7.5 | 69.2 ± 9.7 | 108.6 ± 9.0 | 28.7 ± 0.9 | 88.0 ± 3.9 | 53.8 ± 7.4 | 76.7 ± 5.4 | 36.2 ± 2.1 | 189.0 ± 27.8 |
| RR6 | 41.2 ± 2.7 | 16.0 ± 1.5 | 1,007.0 ± 71.5 | 102.9 ± 21.5 | 1,693.5 ± 105.6 | 217.6 ± 16.4 | 835.1 ± 121.0 | 4,691.6 ± 350.4 | 920.2 ± 65.5 | 441.4 ± 25.9 | 2,362.1 ± 95.5 |
| RR7 | 35.2 ± 10.5 | 4.7 ± 0.7 | 139.6 ± 18.0 | 37.1 ± 4.3 | 250.0 ± 39.7 | 62.0 ± 4.5 | 1,705.3 ± 254.7 | 4,993.6 ± 387.2 | 846.8 ± 25.7 | 1,693.5 ± 74.0 | 460.1 ± 28.6 |
| Other genes | |||||||||||
| cZOGT | 7.5 ± 1.5 | 29.3 ± 3.8 | 111.1 ± 18.0 | 341.5 ± 23.9 | 806.7 ± 54.2 | 153.9 ± 28.0 | 2,188.7 ± 63.4 | 817.9 ± 29.2 | 279.3 ± 21.4 | 258.8 ± 19.6 | 29.0 ± 2.4 |
| bGLU | 38.3 ± 4.3 | 3.1 ± 1.1 | 11.2 ± 1.5 | 523.1 ± 38.6 | 67.7 ± 4.8 | 2,531.6 ± 89.4 | 150,129 ± 14,771 | 32,448 ± 2,110 | 8,512 ± 1,324 | 11,081 ± 865 | 10.8 ± 2.5 |
| GALS | 54.8 ± 9.7 | <1.0 | 213.1 ± 14.5 | 5,134.0 ± 196.7 | 6,543.6 ± 266.6 | 126.7 ± 9.4 | 28.5 ± 5.8 | 134.0 ± 11.8 | 2,250.2 ± 92.7 | 3,605.2 ± 178.6 | 1,3087.2 ± 489.2 |
| VP14 | 143.6 ± 14.7 | 12.0 ± 2.7 | 605.0 ± 47.8 | 218.4 ± 38.0 | 926.6 ± 72.4 | 568.4 ± 28.4 | 35.3 ± 2.7 | 31.5 ± 5.1 | 607.1 ± 57.1 | 41.1 ± 4.4 | 664.4 ± 29.7 |
The transcript levels of CKX genes seem to be no higher than of those encoding IPTs. The only cytosolic CK degradation enzyme, CKX10, is relatively highly expressed in all tissues with the exception of endosperm. Other CKXs probably have a redundant role or can be expressed in specialized types of cells in different tissues; hence, it is visible from their low presence in all materials tested (e.g. CKX7 or CKX9). There are paralogs predominantly bound to reproductive organs (i.e. CKX4 and CKX5). Among the CKX genes that most likely contribute to the pool of CK degradation activity in tissues exposed to stress conditions are CKX1, CKX6, and CKX8 expressed in young roots and CKX3, CKX6, and CKX11 expressed in young leaves. In the control plants, broad changes were observed in transcript levels of several CKX genes between the first and second weeks of plant development when stress was applied. For example, approximately a 6-fold increase in the copy number of CKX1 between 7 and 10 d of root development or a 3-fold decrease of CKX6 expression in the same tissue was seen. These kinds of fluctuations were also observed in leaves (data not shown). A dramatic increase of CKX2 and CKX4b expression was detected in mature leaves and roots, respectively. However, the level of all CKX transcripts with the exception of cytosolic CKX10 was negligible in coleoptile and radicle (2-d-old plantlet), probably due to the demand for a high concentration of active CKs in these rapidly developing tissues. The above summarized data are in good accord with previously reported expression profiles of CKX1 to CKX5 (Massonneau et al., 2004) obtained by semiquantitative reverse transcription-PCR.
Changes in Expression Profiles during Stress Responses
Relative to expression of all CKXs, IPT was quantified by real-time reverse transcription-PCR in tissues exposed to the two stresses or to an exogenous supply of CK for 0.5 h (Fig. 1, A and C) or 3 d (Fig. 1, B and D). Expression of HK and RR type-A genes was followed at six time points from the initiation of the stress experiment (Fig. 2). Since the expression of IPT and CKX genes is generally low in plant tissues, transcripts of most of them are hard to detect by any convenient hybridization technique. Nevertheless, the changes in expression of several RR type-A genes and the gene responsible for glucosylation of cZ were followed by northern-blot analysis. These data correspond well to those obtained by quantitative PCR (qPCR) analysis (Supplemental Fig. S5; Supplemental Methods S1). Isoforms of the β-actin gene (accession no. U60508) that was selected from two other actin genes and a glyceraldehyde phosphate dehydrogenase gene as the most constantly expressed ones in young leaves and roots were used as references to normalize expression levels among the tested samples.
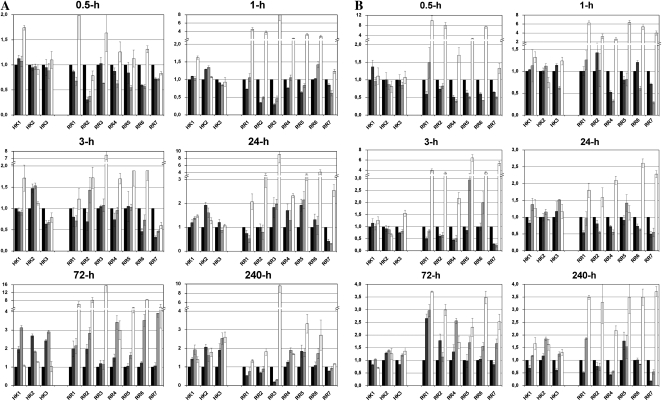
Figure 1.
Expression profiles of genes involved in CK metabolism and perception in maize leaves (A and B) and roots (C and D) exposed to osmotic stress (dark gray bars), salinity stress (light gray bars), and exogenous BAP application (white bars). Total RNA was isolated from tissues exposed to stress and CK stimuli for 0.5 h (A and C) or 72 h (B and D) from at least two independent biological replicates. cDNA from each sample was obtained at least in two independent reactions and run in two separate PCRs. The actin gene was used as an endogenous control, and untreated tissue samples were used as calibrators (black bars) in which expression level was set to 1. Only genes with abundance above one copy per 1 ng of total isolated RNA are included.
Figure 2.
Expression profiles of genes involved in CK perception in maize leaves (A) and roots (B) exposed to osmotic stress (dark gray bars), salinity stress (light gray bars), and exogenous BAP application (white bars). Total RNA was isolated from tissues exposed to stress and CK stimuli at six time points from at least two independent biological replicates. cDNA from each sample was obtained at least in two independent reactions and run in two separate PCRs. The actin gene was used as an endogenous control, and untreated tissue samples were used as calibrators (black bars) in which expression level was set to 1. Only genes with abundance above one copy per 1 ng of total isolated RNA are included.
Generally, CK treatment caused persistent down-regulation of de novo adenylate IPT genes detectable in whole plants 0.5 h after the treatment, whereas the transcript levels of two genes contributing to tRNA-linked CK biosynthesis did not seem to have altered. On the other hand, the levels of CKX genes were up-regulated; however, the extent of up-regulation that was observed 3 d after the treatment differed in intensity. For instance, CKX1 and CKX4b showed extensive up-regulation, while other genes were regulated more moderately. Interestingly, CKX3, which is strongly expressed in leaves, showed a significant down-regulation after the CK treatment. As expected, expression of all type A response regulators were induced in the roots immediately after CK addition to the nutrient solution and then was slightly reduced after 24 h of treatment. However, expression was still higher in comparison with untreated maize until the end of the experiment. Similar up-regulation of RR genes was observed in the leaves, with an approximately 0.5-h delay likely caused by transportation of the CK signal to aerial part of the plant. The levels of receptor transcripts were not significantly altered after exogenous CK application.
Regulation of CK metabolic gene expression in stressed tissues is not straightforward (Fig. 1). There were only two IPT genes induced 0.5 h after osmotic stress application in roots, while the expression of all other genes tested was not immediately influenced in leaves or in whole plant by salinity stress application. The most abundant CKX gene in root tissue, CKX6, and the CKX3 gene were down-regulated 0.5 h after either stress induction. However, other less abundant CKX genes were up-regulated. The situation in leaves appeared likewise: CKX3, CKX6, and CKX11 were slightly down-regulated, whereas the expression of the other genes was not significantly changed. As well, the transcript for the sole cytosolic CKX enzyme does not seem to be regulated in fast response to stress.
Long-term actions of NaCl and water deficit in plant tissues affect transcript levels of most CKX and IPT genes. All abundant IPT genes are up-regulated to some extent. While in leaves salinity causes significantly increased expression levels of both de novo and tRNA-linked synthesis genes, in roots a significant enhancement of two major de novo synthesis gene expressions, IPT5 and IPT6, was observed in osmotically stressed plants. Increase of CKX transcript level was more evident in leaf tissues than in roots. Whereas most of the genes were up-regulated in roots of salt-stressed plants including CKX6, CKX8, and CKX10, long-term osmotic stress preserved elevated expression of CKX6 and two less abundant genes, CKX3 and CKX2. Interestingly, the transcript level of another relatively abundant root gene, CKX1, was found in stressed roots below the levels of control plants. Although the gene is not highly expressed in leaves, its significant up-regulation under stress conditions was observed, indicating a possible tissue specificity of CKX1 promoter activity to the stress response.
Regarding the genes encoding CK receptors, the onset of stress application had no noticeable effect on their regulation. During acclimatization, elevated levels of receptor transcripts appeared only in the leaves of maize seedlings grown under stress conditions. The levels of all type A genes were jointly down-regulated immediately after the stress induction in whole plants. A further stress response slowly increased RR type A levels back to normal or even above the control levels. However, the profile of the up-regulation was not uniform. One hour after the stress treatment, levels of RR5 and RR6 transcripts rose above the control levels in roots exposed to high salinity. On the other hand, expression of the most abundant response regulator gene in maize seedlings, RR7, was significantly down-regulated until the third day of the plant acclimatization.
From a huge family of maize glucosyl transferases and glucosidases, there are only three functionally described enzymes participating in specific glycosylation and deglycosylation of CK molecules (Brzobohatý et al., 1993; Veach et al., 2003). Two genes whose products are responsible for O-glucosylation of cZ derivatives seem to be regulated alternatively in different tissues. While cZOGT transcripts in roots increase in response to stress, their levels in leaves are diminished. Thus, the gene for β-glucosidase Zm-p60.1 (bGLU) encoding the enzyme that catalyzes the reverse reaction shows an opposite expression profile. While water deprivation enhanced its level in leaves, in roots it was slightly down-regulated. As expected, the level of pooled transcripts coding for VP14, the key enzyme in ABA release, was exceedingly up-regulated immediately after the stress application, and this level of transcription was maintained for 3 d, particularly in osmotically stressed tissues. The expression levels of other genes (GALS and BADH) whose products are probably involved in the production of the osmoprotective compounds galactinol and betaine were not found to increase before the late stage of stress.
Changes in CKX Activity during Stress Responses
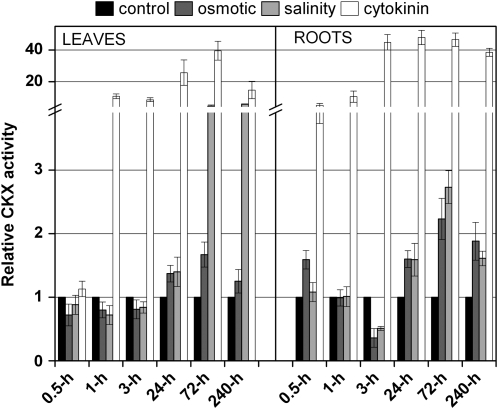
Efficiency of degradation of N6-isopentenyladenosine (iPR) at physiological pH 6.0 and in the presence of quinone electron acceptor was followed for a period of 10 d after stress induction. CKX activity was detected separately in root tissue and upper parts of plants. Total activity was considerably higher in the roots (5.45 pkat mg−1 extracted proteins in 7-d-old control plants) than in leaves (0.43 pkat mg−1). Although the total activity during the stress experiment fluctuated slightly, probably due to diurnal changes (data not shown), generally a slight decrease was observed each day of maize growth (3.75 and 0.28 pkat mg−1 in roots and leaves, respectively, in 17-d-old maize). Activation of CKX enzymes after exogenous CK application was previously well described (Brugière et al., 2003). The increase was detectable in roots 0.5 h after CK addition to the nutrient solution and within 1 h in the leaves (Fig. 3). Stress did not have any immediate effect on CKX activity (within 1 h); however, a significant decrease in the activity was observed in the root tissue 3 h after the application of osmotic or salinity stress. During plant acclimatization to the stress conditions, degradation efficiency gradually grew in the whole plant. A 3- to 4-fold increase was detected in roots exposed to both stresses. In the upper part of the plant, a significant enhancement of CKX activity was detected only in the plants exposed to high salinity, reaching an almost 6-fold increase on the 10th d of acclimatization to stress (Fig. 3).
Figure 3.
Changes in relative CKX activities during stress and CK treatment. Total activity in control leaf and root tissues (0.5 h) was determined as 0.43 and 5.45 pkat mg−1, respectively, with iPR as substrate under the conditions described in “Materials and Methods.” Mean values from free replicate measurements are shown with sd.
CK Content and Changes during Stress Responses
CK content was determined in leaves and roots separately (Supplemental Figs. S6 and S7). All aromatic CKs as well as N7- and N3-glucosides present in all tested tissue samples were under the limit of detection (less then 0.05 pmol g−1 dry weight), with the exception of plants treated by benzyladenine (BAP; Supplemental Fig. S8). Generally, cZ and its derivatives were the most abundant types of CKs in the maize seedlings, followed by tZ and N6-isopentenyladenine (iP) derivatives. Levels of dihydrozeatin derivatives were markedly lower, with only N9-glucoside, riboside-5′-monophosphate, O-glucoside, and riboside O-glucoside detectable in roots. Free bases and ribosides, which are considered to be the active CKs (A-CKs), were less abundant than glucosidic forms. cZ accumulated mainly as its O-glucoside, whereas tZ and iP accumulated as N9-glucosides in roots. Riboside-5′-monophosphates of all three CK types, considered to be the nonactive biosynthetic intermediates, were more abundant than active forms, with the exception of tZR5′MP in leaves.
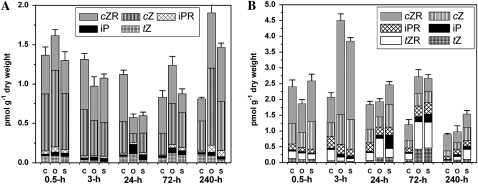
One-half hour after the induction of abiotic stress, few changes in the content of active CKs were observed (Fig. 4). Slight fluctuations among various types of A-CKs appeared, but the total levels of A-CKs in stressed leaves copied those in the control plants still at 3 h after the stress induction. In stressed roots, levels of A-CKs approximately doubled, mainly due to accumulations of cZ and its riboside (cZR), whereas concentrations of other less abundant A-CKs were reduced within the same time interval. After 1 d under the stress, the total level of A-CKs dropped back to normal; however, a significant increase in the levels of iP and tZ types was observed in roots. When plants acclimatized to the stress conditions, the pool of A-CKs remained elevated compared with control plants, which was detectable in roots after the third day of stress treatment. In leaves, there was a noticeable decrease of cZ and its riboside 1 d after the stress induction; however, as in roots, accumulation of tZ and iP types, followed later by cZ and cZR increase, was observed in the course of acclimatization. In the roots, growth of A-CK content was similar or slightly higher than in leaves after 3 d of salinity stress. A higher pool of A-CKs was observed in leaves of osmotically stressed plants.
Figure 4.
Changes in active CK content in maize leaves (A) and roots (B) during responses to osmotic (O) and salinity (S) stress compared with untreated plants (C). All values are derived from at least two biological replicates that were determined in at least two technical replicates. Error bars show the sums of sd values of all represented CKs. Levels of A-CKs not shown were under the detection limit of the method.
A rapid elevation of cZ and its riboside in roots occurred within 3 h of stress and was accompanied by a 2- to 3-fold increase of its riboside-5′-monophosphate (cZR5′MP), which was observed in roots that were salt stressed for 0.5 h. Furthermore, levels of N6-isopentenyladenosine-5′-monophosphate (iPR5′MP) and tZR5′MP in the stressed samples diminished after a 3-h interval but started to elevate quickly within the first day of stress response. Thus, tZR5′MP level in roots was 4- to 5-fold higher than the control after 72 h. In leaves, iPR5′MP was similarly elevated in the same time interval and tZR5′MP was below the detection limit. Regarding the glucosides of isoprenoid CKs, the levels in stressed tissues compared with control plants started to distinctly rise after an extended period of stress, with noticeable change on the third day in both roots and leaves. Only cZ did not preferably accumulate in stressed roots as N- or O-glucoside (cZOG, cZROG, cZ9G). In the leaves of osmotically and salt-stressed plants, however, increasing accumulation of cZOG and cZROG was observed on the third day of plant acclimatization or later.
ABA and Its Metabolites in Stressed Tissues
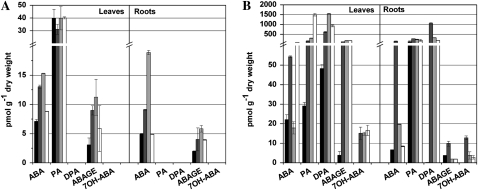
As expected, levels of free ABA were elevated in whole plants 30 min after stress application (Fig. 5). In leaves, levels increased approximately 2-fold, while a more robust 4-fold ABA accumulation was detected in salt-stressed roots. Interestingly, free ABA accumulation returned to control levels in salt-stressed plants 72 h after the stress treatment while being kept significantly elevated in osmotically stressed plants. However, high levels of ABA catabolic products such as phaseic acid and dihydrophaseic acid in salt stress samples, which are biologically inactive, indicate a massive degradation of released ABA within the first 3 d. Free ABA level reduction back to the control level is likely related to long-term acclimatization to salt stress. On the other hand, levels of ABA glucosyl ester (ABAGE) were constantly elevated in leaves under both stress treatments. However, ABAGE concentration was insignificant in roots. Neophaseic acid, 9′-OH-ABA, ABA alcohol, and ABA aldehyde were below detection limits in all analyzed samples.
Figure 5.
Endogenous levels of ABA and its derivatives in untreated maize roots and leaves (black bars) and tissues exposed to osmotic stress (dark gray bars), salinity stress (light gray bars), and exogenous BAP application (white bars) for 0.5 h (A) and 72 h (B). All bars show mean values of two replicates with indicated sd. DPA, Dihydrophaseic acid; PA, phaseic acid.
Exogenously applied 10 μm BAP to unstressed maize plants dramatically increased the concentration of ABA and related compounds (Fig. 5). A massive accumulation of free ABA and its degradation products was detected in leaves after 3 d of growing in a nutrient solution with CK. The concentrations of ABA and phaseic acid were even 2- and 10-fold higher, respectively, compared with osmotically stressed tissues. Increased levels of dihydrophaseic acid indicate increased metabolism of ABA in roots, despite the fact that the levels of ABA and its glucosyl ester were comparable with the control plants. Hence, the accumulation of ABA metabolites matches the observed reduction in leaf size of plants exposed to an oversupply of CK. The influence of exogenously applied CK on ABA levels in plants was formerly tested only in the halophyte Mesembryanthemum crystallinum without any positive correlation (Thomas et al., 1992). Nevertheless, those authors did not specify which parts of the plant were used for analysis.
DISCUSSION
Molecular mechanisms regulating responses to abiotic stresses have been extensively studied during the last decade. For instance, the essential role of mitogen-activated protein and salt overly sensitive kinase cascades and the function of transcription factors containing ABA-responsive elements are undoubtedly well established. CKs are an important signal traveling from roots to the shoots in response to nutrient insufficiency (Takei et al., 2004); however, conclusions concerning their role in drought, osmotic, and salinity responses or signalization are derived from inferences with no cause/effect data (Kuiper et al., 1990; Davies and Zhang, 1991; Kudoyarova et al., 2007; Tran et al., 2007). In this article, we, to our knowledge for the first time, describe complete gene families of CK biosynthetic and degradation genes in maize and regulation of their expression in a short-term response to osmotic and salinity stress as well as during the plant acclimatization to these adverse environmental stimuli. We also present the expression of CK signaling components with the aim to pursue all of the expression profiles in the context of endogenous CK and ABA levels and in particular the levels of the active forms.
Seedlings of 7-d-old maize were studied due to the fact that CK metabolism and CK responsiveness in young tissues begin extensively on the seventh day of development. This is supported by relatively high expression levels of many genes encoding biosynthetic and degradation enzymes as well as CK signaling components (Table I). During germination and the early phase of seedling development, CKs are probably still supplied in the form of glucosides from the kernel, as becomes evident from the massive expression of the β-glucosidase gene and the marginal expression of all IPT and CKX genes detected in 2-d-old radicles and coleoptiles. On the other hand, in developing kernels, where the CK role in response to water stress was studied previously (Yang et al., 2001; Brugière et al., 2003), only specific genes for de novo biosynthesis (IPT2), degradation (CKX1 and CKX4), and signal response (RR3) are active. Total content of active CKs reaches a maximum in 1-week-old seedlings (Fig. 4) and later slowly declines as CKs are converted to nonactive glycosylated forms. The observed peak of active CKs in 7-d-old seedlings is probably due to meristematic activity and rapid elongation promoted by induction of de novo biosynthesis and depletion of kernel-stored CK forms. Recently, similar accumulations of active CK forms were observed in 6-d-old pea (Pisum sativum) seedlings, while in 9-d-old seedlings the levels started to decline (Stirk et al., 2008).
Immediately after the stress induction, we did not observe any significant decline or increase in CK content, with the exception of cZR5′MP, as was previously demonstrated in several studies (Mustafina et al., 1998; Fricke et al., 2006). A constant level of active CKs 30 min after stress induction is in accordance with unchanged activities of degradation enzymes. Nevertheless, expression of CK metabolic genes is already influenced, especially in osmotically stressed roots. Genes for two de novo IPTs (IPT6 and IPT7) were up-regulated, while the main root-expressed degradation gene, CKX6, was down-regulated, resulting in an approximately 2-fold decrease in CKX activity and accumulation of active CKs in stressed roots during first 24 h of stress application. A more pronounced positive effect of the stress on CK biosynthesis was observed after 3 d, when also the level of the most abundant transcript, IPT5, was significantly elevated in whole seedlings. Regulation mechanisms of the expression of CKX gene family members are not uniform, as is evident from the expression profiles under stress conditions and BAP treatment. For instance, most CKX genes are significantly up-regulated after exogenous application of BAP (CKX1, CKX2, CKX4b); however, the expression of CKX3 is approximately 10-fold less than the levels in untreated tissue. Accordingly, two out of 11 rice CKX genes were down-regulated 2 h after tZ application on rice seedlings, whereas the others were up-regulated (Hirose et al., 2007). Tissue-dependent variations in CKX gene regulation are apparent as well; whereas the stress status induces CKX1 expression in leaves, it has an opposite effect in roots. The direct effect of ABA on the amount of CKX1 transcripts in maize leaves, as was demonstrated by Brugière et al. (2003), is not likely in roots, where higher ABA levels stimulated by stress do not induce CKX1 expression. On the other hand, significant elevation of CKX1 transcripts was observed later, 42 d after pollination, and could be attributed to the accumulation of ABA in the desiccating kernels, as the total level of active CKs is distinctively decreased in this stage of development (Brugière et al., 2003). No correlation is evident in the leaves where ABA as well as CKX1 transcript levels are kept high under long-term osmotic stress, although in salt-exposed plants at the same moment active ABA concentration returns to normal level but CKX1 is still up-regulated. This suggests that no short-term feedback mechanism exists to return CKX1 transcript level back to normal after induction by ABA. Alternatively, degradation products of ABA, which are still highly accumulated in salt-stressed tissues, might to some extent keep ABA responsiveness, as was demonstrated for 7′-OH-ABA in barley (Hill et al., 1995). Hence, levels of free ABA correlate with growth rates of acclimatized plants, while in NaCl-treated seedlings, elongation growth is to some extent restored and polyethylene glycol (PEG)-treated seedlings display significantly less elongation of leaves within a 7-d period of stress action, probably due to the permanent increase in free ABA level.
Levels of cZR5′MP were significantly increased in roots exposed to a high NaCl concentration 0.5 h after the application (Supplemental Fig. S7). A similar increase was also observed later in osmotically stressed roots. Dephosphorylation and deglycosylation of cZR5′MP resulted afterward in an increased pool of cZ and its riboside in stressed plants detectable 3 h after application. Interestingly, the elevated level of cZR5′MP is accompanied by a significant up-regulation of a gene encoding cZ O-glucosyltransferase 0.5 h after the stress induction (Fig. 1C), resulting in cZOG and cZROG accumulation detected after 3 h in roots of both stressed samples. This indicates that the expression of the enzyme is activated rapidly by its substrate or its precursor. Thus, O-glucosylation is a mechanism by which the plant cell could react very fast to the disturbed hormone homeostasis induced by physiological stimuli. Since expression of IPT genes was not initially up-regulated in salt-stressed samples, the 3-fold elevated level of cZR5′MP well supports previous studies where the origin of cZ-type CKs is attributed to prenyl-tRNA degradation (for review, see Sakakibara, 2006). Recently, a massive and rapid endonucleolytic cleavage of different types of mature tRNAs as well as rRNA has been observed in yeast and Arabidopsis exposed to oxidative and some other stresses (Thompson et al., 2008). Interestingly, enhanced tRNA cleavage does not significantly reduce the pool of mature tRNAs in yeast exposed to hydrogen peroxide. Although the authors focused only on specific fragmentation of tRNA within the anticodon loop, prenylation of tRNA right at this tRNA structure indicates that the described stress-induced fragmentation may lead to a release of free modified nucleotides from destabilized small RNA structures or they may be released by postulated cytotoxic tRNAses (Tomita et al., 2000). Processes connected with the release of prenylated nucleotides from RNA molecules could be linked to the specific cell compartments called stress granules or RNA granules, which rapidly form in all kinds of eukaryotic cells within 15 to 30 min after various stress stimuli. Although the primary function of these granules is thought to be the degradation of specific mRNAs, the granules also contain other enzymes linked to various forms of RNA decay (for review, see Anderson and Kedersha, 2006). The contribution of tRNA-released CKs to their pool in stressed tissues is well demonstrated in earlier work on NaCl-stressed maize (Atanassova et al., 1997). Hydrolyzed RNA from long-term stressed samples contained more CK bases than the control. In accordance, the expression of both tRNA-IPTs was elevated 72 h after stress induction; however, these genes seem to be more or less constitutively expressed during maize development. Nevertheless, further research needs to be done (e.g. stress responses of tRNA-IPT mutants) to confirm such a rapid release of modified nucleotides from degraded RNA. The dynamics of stress-induced CK changes needs to be viewed also from the point of cellular compartmentation. As the only known CK receptors are situated on the cytoplasmic membrane (Kim et al., 2006), the efflux rates of CKs out of the cell that were not characterized so far might be the limiting factor of CK-mediated transduction.
From our study, it is evident that stress response followed by acclimatization caused CK imbalance by increased rates of metabolism. Up-regulation of the majority of CKX genes leads to higher CK degradation activities in plants acclimatized to osmotic and salinity stress. Nevertheless, the enhanced catabolism is accompanied by similarly accelerated de novo biosynthesis and, most likely together with CK release from RNA and other CK-linked metabolism, such as β-glucosidase activity in leaves, contributes to moderate accumulation of active CKs in stressed plants. Apparent CK accumulation in 72-h stressed roots is subsequently transposed to the upper part of plants, being promoted by increased root levels of either tZR or O-glucosides, which are thought to be a transport form of CKs (Bano et al., 1993; Hansen and Dörffling, 2003). Root-to-shoot trafficking of CKs could also explain certain discrepancies in this study regarding a later increase in cZOG in stressed leaves that contrast with down-regulation of cZOGT and up-regulation of bGLU genes. Thus, the supply of this form from roots is most likely. Hence, it is premature to discuss all changes in expression profiles, since almost nothing is known about substrate specificities and turnover rates of determined gene products. For instance, our recent work clearly shows that diverse Arabidopsis isoforms of CKX significantly differ in their preference for cZ and tZ isoforms (Pertry et al., 2009) and that the degradation of differently glycosylated CKs is pH dependent (Galuszka et al., 2007). Alkalinization of xylem sap and apoplast observed under water-deficient conditions (for review, see Schachtman and Goodger, 2008) might therefore induce significant changes in CKX substrate preferences. It will be a question in subsequent studies to describe substrate specificities and cellular compartmentation of maize CKX and IPT enzymes. As visible from the phylograms (Supplemental Figs. S3 and S4), maize proteins cluster independently of Arabidopsis orthologs. Thus, CK homeostasis in cereal plants can be functionally diverse based on details emerging from the study of the dicot model Arabidopsis (Galuszka et al., 2007).
In the early phase of stress treatment, when plants were apparently wilted, expression of all CK response regulator genes was markedly down-regulated (Fig. 2), although the levels of active CKs were not altered. Complementation studies with Arabidopsis CK receptors previously showed that variations in osmotic pressure can involve changes in the activity of these receptors and subsequent regulation of downstream phosphorylation in yeast in the presence of CKs (Reiser et al., 2003). However, it is not clear whether the reduction in CK sensitivity is directly caused by double antagonistic responsiveness of CK receptors to CK molecules and turgor changes or the CK signaling transduction is affected by other relative turgor receptors and/or signaling pathways. Recently, the antagonistic function of CK receptors and another related His kinase (AHK1) in abiotic stress signaling has been demonstrated by studying gain- and loss-of function mutants in Arabidopsis (Tran et al., 2007).
The dynamics of RR gene expression observed within the first day of stress action could contribute not only to increased turgor sensing but also to fluctuation of specific CK forms. Active cZ forms reached 3-fold higher levels in 3-h stressed roots than in the control. The primary increase in cZR5′MP detected in salt-stressed roots is accompanied by RR1 transcript elevation followed by RR5 and RR6 expression increase detected at 1 and 3 h after the stress application, respectively, while the other RR genes, especially RR4 and RR7, are still expressed below the control level. Transcript levels of these response regulators decreased as the levels of cZ types declined 24 h after the stress application. At the same time point, however, expression of most RR genes in the leaves returned to control levels or started to be slightly up-regulated, despite the fact that the levels of cZ types dropped 2-fold. Such an enhancement can be attributed to the increase of iP and the lower amount of tZ in the stressed leaves. Thus, the responsiveness of the CK transduction pathway in maize seems to be uniformly influenced by the changes in turgor as well as specifically (at least to some extent) by different types of CKs.
Generating data from full-genome transcriptional profile screening exploiting Affymetrix chips is easy via Genevestigator software. Although there are no accessible results from experiments done on Affymetrix maize genome array so far, data from 166 Affymetrix Rice Genome arrays, including several providing expression information from abiotic stress treatments, were recently released (Zimmermann et al., 2008). Since there is a high degree of genetic synteny between maize and rice genomes, we were able to unambiguously locate nearly identical rice homologs to almost all studied genes from maize (Supplemental Figs. S3 and S4). Beyond sequence similarity, transcripts of these orthologs demonstrate comparable levels of abundance. Expression profiles of all CK-related genes from the experiment where 7-d-old rice seedlings were exposed for 3 h to drought and salinity stress induced by 200 mm NaCl (Jain et al., 2007) are summarized in Supplemental Table S2. Interestingly, changes in the expression of CK metabolic and perception genes in stressed rice are closely comparable to those we described in maize 0.5 h after stress induction. Thus, all genes for type A response regulators were down-regulated 3 h after salinity stress application and almost all when seedlings were dried. Transcripts of rice CKX genes, showing highest homology to ZmCKX3 and ZmCKX6, generally the most abundant maize seedling orthologs, were as well significantly below control levels in both stress samples. These CKXs probably contribute to the decrease in CKX activity observed 3 h after stress application in maize. Concerning the genes for de novo IPTs, two of them showing highest signals on the chip were slightly up-regulated. This documented agreement in expression profiling between our results obtained from maize and published rice microarray data demonstrates that initial fluctuation and later accumulation of CKs induced by stress stimuli found here in maize seedlings can be a general response to abiotic stress at least in crop plants from the Poaceae family.
In conclusion, we demonstrated that CKs do not have a direct function in stress signaling similar to ABA due to a slow response of metabolic genes to stress induction. Thus, CK levels start to significantly alter in comparison with unstressed tissues as late as several hours after stress action, with significant accumulation after plant acclimatization to the adverse conditions. Therefore, restriction of growth rate under abiotic stress does not seem to be caused by the limited amount of active CKs due to accelerated degradation, as was assumed previously. On the other hand, at least in stressed maize seedlings, reduction of growth is linked only to the level of active ABA, which directly regulates stress responses and can have an impact on ethylene production that was shown to have a direct negative effect on shoot growth as well (for review, see Sharp and LeNoble, 2002). Hereafter, accumulation of active CKs among other processes is just the way plants try to overcome stress status and release themselves from growth inhibition. Hence, crop plants with enhanced endogenous levels of active CKs, engineered for instance by knockout of particular CKX genes, can be tested in the future as new transgenic cultivars with increased drought and salinity tolerance.
MATERIALS AND METHODS
Plant Material
Maize seeds (Zea mays ‘Cellux’; Morseva) were imbibed in tap water and germinated in the dark on wetted filter paper. After 2 d, the germinated seedlings were transferred to aerated hydroponic tanks filled with Hoagland nutrient solution. The plants were grown in a growth chamber with a 16-h light period (250 μE m−2 s−1) at 27°C and an 8-h dark period at 20°C. During 1 week of growth, the nutrient solution was exchanged two times and finally supplied with 175 mm NaCl and 25 mm CaCl2, 20% PEG 6000, or 10 μm BAP. Concentrations of salt and PEG were set up to be osmotically equivalent to cause a similar decline in leaf water potential (Ueda et al., 2003). PEG lowers the osmotic potential of the external medium and reduces water availability for root tissues, unlike NaCl, which crosses plant membranes and has a direct toxic effect on plant cells. Calcium chloride was added to the NaCl solution to eliminate severe sodium-induced calcium deficiency, which leads to extensive membrane destabilization and appeared to be more limiting to shoot growth than Na toxicity per se (Grieve and Fujiyama, 1987). Stress treatment was set up 3 h after turning on the light. Root and leaf tissues of 10 plants were harvested at 0.5, 1, 3, 8, 24, 72, and 240 h after the stress induction, immediately frozen in liquid nitrogen, and lyophilized for hormone analysis. Control plants were grown in parallel and harvested at the same time points to exclude hormonal variation due to circadian rhythms. The experiment was carried out in three period-independent replicates, and the plant material from each replicate collected at each time point was processed separately with the exception of 0.5-h time point collection, which was run in four replicates. CK content at each time point was statistically evaluated from three biological replicates, where each replicate sample was measured twice. Changes in expression levels were determined from two biological replicates each done at least in four technical replicates with the exception of 0.5-h time point, which was done in four biological replicates.
Tissues for screening IPT and CKX expression profiles were collected from maize plants grown in a greenhouse during summer.
CKX Activity Assay
Plant material was powdered with liquid nitrogen using a hand mortar and pestle and extracted with a 2-fold excess (v/w) of 0.2 m Tris/HCl buffer, pH 8.0, containing 1 mm phenylmethylsulfonyl fluoride and 0.3% Triton X-100. Cell debris was removed by centrifugation at 12,000g for 10 min.
The assay was performed according to the method described previously (Frébort et al., 2002). Samples were incubated in a reaction mixture composed of 100 mm McIlvaine buffer, pH 6.0, 0.5 mm electron acceptor 2,3-dimethoxy-5-methyl-1,4-benzoquinone, and 0.25 mm substrate iPR for 2 to 10 h at 37°C. For determination of specific activities, protein contents in the samples were assayed according to Bradford (1976) with bovine serum albumin as a standard.
qPCR Analysis
Total RNA for reverse transcription was isolated using the RNAqueous kit and Plant RNA Isolation Aid solutions (Ambion). To minimize bias in qPCR data, isolated RNA was treated twice by Ambion's TURBO DNase-free kit to remove all traces of genomic DNA contamination. First-strand cDNA was synthesized by RevertAid H Minus Moloney murine leukemia virus reverse transcriptase and oligo(dT) or random hexamer primers (Fermentas). Diluted cDNA samples were used as templates in real-time PCRs containing POWER SYBR Green PCR Master Mix or TaqMan Gene Expression Master Mix (Applied Biosystems), 300 to 900 nm of each primer, and 250 nm specific 5′ 6-carboxyfluorescein- and 3# nonfluorescent quencher-labeled minor groove binder probe, respectively. Primers for all genes were mostly designed to cover the 3′ end of the ORF to minimize inaccuracy caused by possible partial degradation of mRNA usually starting from the 5′ end. However, some gene family members share high mutual homology, which does not allow this kind of design. Moreover, some primer combinations caused strong unspecific amplification, disturbing correct fluorescence reading. In these cases, amplification with specific TaqMan probes was exploited (i.e. IPT5, IPT6, CKX1, CKX4, CKX5, and CKX8) or primers were designed to the nonhomologous 5′ end sequences (i.e. CKX2, CKX3, and RR2) to avoid cross-reactivity of primer pairs to highly homologous paralogs. The primers for PCR exploiting SYBR Green chemistry were designed using Primer Express 3.0 software (Applied Biosystems). TaqMan probes together with corresponding primers were designed by Applied Biosystems customer service. The primer and probe sequences are listed in Supplemental Table S1. RNA from every biological replicate was at least transcribed in two independent reactions, and each cDNA sample was run in at least two technical replications on the StepOnePlus Real-Time PCR System in a default program (Applied Biosystems). To ensure that primers amplified the desired gene sequence, amplicons for every used primer pair were produced by standard Taq polymerase, cloned into pDRIVE vector, and sequenced by a commercial sequencing service. For each pair of primers, plasmid DNA was used afterward as template to generate a calibration curve for determining the efficiency of PCR. Cycle threshold values were normalized with respect to the 18S small RNA subunit gene (expression during development and in different organs) or the β-actin gene (expression in stressed samples) and the efficiency of amplification.
CK Analysis
The procedure used for CK purification was a modification of the method described by Faiss et al. (1997). Three fractions were obtained by this procedure: the first one contained free bases, ribosides, and N9-glucosides; the second fraction was enriched by nucleotides; and the third fraction contained O-glucosides. Deuterium-labeled CK internal standards (OlChemIm) were added, each at 1 pmol per sample, to check the recovery during the purification and to validate the determination (Novák et al., 2008). The samples were purified using immunoaffinity chromatography based on wide-range specific monoclonal antibodies against CKs (Novák et al., 2003). The methanolic eluates from immunoaffinity columns were evaporated to dryness, and the obtained solids were dissolved in 20 μL of the mobile phase used for quantitative analysis. The samples were analyzed with an ultraperformance liquid chromatograph (Acquity UPLC; Waters) coupled to a Quatro micro API (Waters) triple quadrupole mass spectrometer equipped with an electrospray interface. The purified samples were injected onto a C18 reverse-phase column (BEH C18, 1.7 μm, 2.1 × 50 mm; Waters). The column was eluted with a linear gradient (0 min, 10% B; 0–8 min, 50% B; flow rate of 0.25 mL min−1; column temperature of 40°C) of 15 mm ammonium formate (pH 4.0; A) and methanol (B). Quantification was achieved by multiple reaction monitoring of [M+H]+ and the appropriate product ion. The quantification was performed by Masslynx software using a standard isotope dilution method. The ratio of endogenous CK to the appropriate labeled standard was determined and further used to quantify the level of endogenous compounds in the original extract, according to the known quantity of the added internal standard.
Analysis of ABA and Its Metabolites
Approximately 50-mg aliquots of plant tissue were homogenized and extracted for 1 h in 750 μL of ice-cold methanol:water:acetic acid (10:89:1, v/v) containing sodium diethyldithiocarbamate (400 μg g−1 dry weight) as an antioxidant. Internal standard mixtures, containing 50 pmol each of [2H2]ABA alcohol, [2H2]ABA aldehyde, (−)-7′,7′,7′[2H3]phaseic acid, (−)-7′,7′,7′[2H3]dihydrophaseic acid, (−)-8′,8′,8′[2H3]neophaseic acid, (+)-4,5,8′,8′,8′[2H5]ABAGE, (−)-5,8′,8′,8′[2H4]7′-OH-ABA, and (+)-3′,5′,5′,7′,7′,7′[2H6]ABA, were added to the samples. The homogenates were centrifuged (21,000g, 10 min, 4°C) after 1 h of extraction, and the pellets were then reextracted in the same way for 30 min. The combined extracts were purified by solid-phase extraction on Oasis HLB cartridges (60 mg, 3 mL; Waters), evaporated to dryness, and finally analyzed by ultraperformance liquid chromatograph-electrospray ionization (−/+)-tandem mass spectrometry.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of all maize putative IPT proteins.
Supplemental Figure S2. Alignment of all maize putative CKX proteins.
Supplemental Figure S3. Phylogenetic tree of all ZmIPT, OsIPT, and AtIPT proteins.
Supplemental Figure S4. Phylogenetic tree of all ZmCKX, OsCKX, and AtCKX proteins.
Supplemental Figure S5. Northern blots of selected genes in tissues exposed to stress and CK.
Supplemental Figure S6. Fluctuation of CK content in maize leaves during exposure to osmotic and salinity stress.
Supplemental Figure S7. Fluctuation of CK content in maize roots during exposure to osmotic and salinity stress.
Supplemental Figure S8. CK content in maize exposed to BAP compared with untreated plants.
Supplemental Table S1. Sequences of primers and TaqMan probes used for qPCR.
Supplemental Table S2. Expression profiles of genes involved in CK metabolism and perception in rice seedlings exposed to drought and salinity stress.
Supplemental Methods S1. Northern-blot analysis.
Supplementary Material
Acknowledgments
We thank Petra Amakorová, Katarína Mrízová, Kateřina Závodná, and Kateřina Slováková for skillful technical assistance and Miroslav Strnad for critical reading of the manuscript.
This work was supported by the Grant Agency of the Czech Republic (grant nos. 522/06/0703 and 522/08/0920) and the Ministry of Education, Youth, and Sports of the Czech Republic (grant nos. MSM6198959216, ME861, and 1M06030).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Petr Galuszka (petr.galuszka@upol.cz).
The online version of this article contains Web-only data.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N (2006) RNA granules. J Cell Biol 172 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Hagino T, Ohta Y, Aoki K, Yonekura-Sakakibara K, Deji A, Yamaya T, Sugiyama T, Sakakibara H (2003) Molecular characterization of His-Asp phosphorelay signaling factors in maize leaves: implications of the signal divergence by cytokinin-inducible response regulators in the cytosol and the nuclei. Plant Mol Biol 52 331–341 [DOI] [PubMed] [Google Scholar]
- Atanassova L, Stojanov I, Pissarska M, Valkova C (1997) Salt stress-induced changes of cytokinins in maize and pea plants RNA. Bulg J Plant Physiol 23 33–39 [Google Scholar]
- Bano A, Dörffling K, Bettin D, Hahn H (1993) Abscisic acid and cytokinins as possible root-to-shoot signals in xylem sap of rice plants in drying soil. Aust J Plant Physiol 20 109–115 [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Brugière N, Humbert S, Rizzo N, Bohn J, Habben JE (2008) A member of the maize isopentenyltransferase gene family, Zea mays isopentenyl transferase 2 (ZmIPT2), encodes a cytokinin biosynthetic enzyme expressed during kernel development: cytokinin biosynthesis in maize. Plant Mol Biol 67 215–229 [DOI] [PubMed] [Google Scholar]
- Brugière N, Jiao S, Hantke S, Zinselmeier C, Roessler JA, Niu X, Jones RJ, Habben JE (2003) Cytokinin oxidase gene expression in maize is localized to the vasculature, and is induced by cytokinins, abscisic acid, and abiotic stress. Plant Physiol 132 1228–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzobohatý B, Moore I, Kristoffersen P, Bako L, Campos N, Schell J, Palme K (1993) Release of active cytokinin by a β-glucosidase localized to the maize root meristem. Science 262 1051–1054 [DOI] [PubMed] [Google Scholar]
- Chan AP, Pertea G, Cheung F, Lee D, Zheng L, Whitelaw C, Pontaroli AC, SanMiguel P, Yuan Y, Bennetzen J, et al (2006) The TIGR maize database. Nucleic Acids Res 34 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies WJ, Zhang J (1991) Root signals and the regulation of growth and development of plants in drying soil. Annu Rev Plant Physiol Plant Mol Biol 42 55–76 [Google Scholar]
- Faiss M, Zalubilová J, Strnad M, Schmülling T (1997) Conditional transgenic expression of the ipt gene indicates a function of cytokinins in paracrine signaling in whole tobacco plants. Plant J 12 401–415 [DOI] [PubMed] [Google Scholar]
- Frébort I, Šebela M, Galuszka P, Werner T, Schmülling T, Peč P (2002) Cytokinin oxidase/dehydrogenase assay: optimized procedures and applications. Anal Biochem 306 1–7 [DOI] [PubMed] [Google Scholar]
- Fricke W, Akhiyarova G, Wei W, Alexandersson E, Miller A, Kjellbom PO, Richardson A Wojciechowski T, Schreiber L, Veselov D, et al (2006) The short-term growth response to salt of the developing barley leaf. J Exp Bot 57 1079–1095 [DOI] [PubMed] [Google Scholar]
- Galuszka P, Popelková H, Werner T, Frébortová J, Pospíšilová H, Mik V, Köllmer I, Schmülling T, Frébort I (2007) Biochemical characterization of cytokinin oxidases/dehydrogenases from Arabidopsis thaliana expressed in Nicotiana tabacum L. J Plant Growth Regul 26 255–267 [Google Scholar]
- Gaut BS (2001) Patterns of chromosomal duplication in maize and their implications for comparative maps of the grasses. Genome Res 11 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieve GM, Fujiyama H (1987) The response of two rice cultivars to external Na/Ca ratio. Plant Soil 103 245–250 [Google Scholar]
- Hansen H, Dörffling K (2003) Root-derived trans-zeatin riboside and abscisic acid in drought-stressed and re-watered sunflower plants: interaction in the control of leaf diffusive resistance? Funct Plant Biol 30 365–375 [DOI] [PubMed] [Google Scholar]
- Havlová M, Dobrev PI, Motyka V, Štorchová H, Libus J, Dobrá J, Malbeck J, Gaudinová A, Vaňková R (2008) The role of cytokinins in responses to water deficit in tobacco plants over-expressing trans-zeatin O-glucosyltransferase gene under 35S or SAG12 promoters. Plant Cell Environ 31 341–353 [DOI] [PubMed] [Google Scholar]
- Hill RD, Liu JH, Durnin D, Lamb N, Shaw A, Abrams SR (1995) Abscisic acid structure-activity relationships in barley aleurone layers and protoplasts. Plant Physiol 108 573–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N, Makita N, Kojima M, Kamada-Nobusada T, Sakakibara H (2007) Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol 48 523–539 [DOI] [PubMed] [Google Scholar]
- Houba-Hérin N, Pethe C, d'Alayer J, Laloue M (1999) Cytokinin oxidase from Zea mays: purification, cDNA cloning and expression in moss protoplasts. Plant J 17 615–626 [DOI] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP (2007) F-box proteins in rice: genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143 1467–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP (2006) Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa). BMC Plant Biol 6 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara H, Takei K, Ueda N, Hishiyama S, Yamaya T, Kamiya Y, Yamaguchi S, Sakakibara H (2004) Distinct isoprenoid origins of cis- and trans-zeatin biosyntheses in Arabidopsis. J Biol Chem 279 14049–14054 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I (2006) Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci USA 103 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoyarova GR, Vysotskaya LB, Cherkozyanova A, Dodd IC (2007) Effect of partial rootzone drying on the concentration of zeatin-type cytokinins in tomato (Solanum lycopersicum L.) xylem sap and leaves. J Exp Bot 58 161–168 [DOI] [PubMed] [Google Scholar]
- Kuiper D, Schuit J, Kuiper PJC (1990) Actual cytokinin concentrations in plant tissue as an indicator for salt resistance in cereals. Plant Soil 123 243–250 [Google Scholar]
- Massonneau A, Houba-Hérin N, Pethe C, Madzak C, Falque M, Mercy M, Kopečný D, Majira A, Rogowsky P, Laloue M (2004) Maize cytokinin oxidase genes: differential expression and cloning of two new cDNAs. J Exp Bot 55 2549–2557 [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Tarkowski P, Matsumoto-Kitano M, Kato T, Sato S, Tarkowská D, Tabata S, Sandberg G, Kakimoto T (2006) Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc Natl Acad Sci USA 103 16598–16603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RO, Bilyeu KD, Laskey JG, Cheikh N (1999) Isolation of gene encoding a glycosylated cytokinin oxidase from maize. Biochem Biophys Res Commun 255 328–333 [DOI] [PubMed] [Google Scholar]
- Mustafina A, Veselov S, Valcke R, Kudoyarova G (1998) Contents of abscisic acid and cytokinins in shoots during dehydration of wheat seedlings. Biol Plant 40 291–293 [Google Scholar]
- Novák O, Hauserová E, Amakorová P, Doležal K, Strnad M (2008) Cytokinin profiling in plant tissues using ultra-performance liquid chromatography-electrospray tandem mass spectrometry. Phytochemistry 69 2214–2224 [DOI] [PubMed] [Google Scholar]
- Novák O, Tarkowski P, Lenobel R, Doležal K, Strnad M (2003) Quantitative analysis of cytokinins in plants by liquid chromatography/single-quadrupole mass spectrometry. Anal Chim Acta 480 207–218 [Google Scholar]
- Pertry I, Václavíková K, Depuydt S, Galuszka P, Spíchal L, Temmerman W, Stes E, Schmülling T, Kakimoto T, van Montagu M, et al (2009) Identification of Rhodococcus fascians cytokinins and their modus operandi to reshape the plant. Proc Natl Acad Sci USA 106 929–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser V, Raitt DC, Saito H (2003) Yeast osmosensor Sln1 and plant cytokinin receptor Cre1 respond to changes in turgor pressure. J Cell Biol 161 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H (2006) Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57 431–449 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Hayakawa A, Deji A, Gawronski SW, Sugiyama T (1999) His-Asp phosphotransfer possibly involved in the nitrogen signal transduction mediated by cytokinin in maize: molecular cloning of cDNAs for two-component regulatory factors and demonstration of phosphotransfer activity in vitro. Plant Mol Biol 41 563–573 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Suzuki M, Takei K, Deji A, Taniguchi M, Sugiyama T (1998) A response-regulator homologue possibly involved in nitrogen signal transduction mediated by cytokinin in maize. Plant J 14 337–344 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Sakakibara H, Kojima M, Yamamoto Y, Nagasaki H, Inukai Y, Sato Y, Matsuoka M (2006) Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol 142 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Goodger JQD (2008) Chemical root to shoot signaling under drought. Trends Plant Sci 13 281–287 [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Zeevaart JAD (2003) Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol 131 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp RE, LeNoble ME (2002) ABA, ethylene and the control of shoot and root growth under water stress. J Exp Bot 53 33–37 [PubMed] [Google Scholar]
- Šmehilová M, Galuszka P, Bilyeu KD, Jaworek P, Kowalska M, Šebela M, Sedlářová M, English JT, Frébort I (2009) Subcellular localization and biochemical comparison of cytosolic and secreted cytokinin dehydrogenase enzymes from maize. J Exp Bot 60 2701–2712 [DOI] [PubMed] [Google Scholar]
- Stirk WA, Novák O, Václavíková K, Tarkowski P, Strnad M, van Staden J (2008) Spatial and temporal changes in endogenous cytokinins in developing pea roots. Planta 227 1279–1289 [DOI] [PubMed] [Google Scholar]
- Takei K, Ueda N, Aoki K, Kuromori T, Hirayama T, Shinozaki K, Yamaya T, Sakakibara H (2004) AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol 45 1053–1062 [DOI] [PubMed] [Google Scholar]
- Tan BC, Schwartz SH, Zeevaart JAD, McCarty DR (1997) Genetic control of abscisic acid biosynthesis in maize. Proc Natl Acad Sci USA 94 12235–12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JC, McElwain EF, Bohnert HJ (1992) Convergent induction of osmotic stress-responses: abscisic acid, cytokinin, and the effect of NaCl. Plant Physiol 100 416–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Lu C, Green PJ, Parker R (2008) tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14 2095–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Ogawa T, Uozumi T, Watanabe K, Masaki H (2000) A cytotoxic ribonuclease which specifically cleaves four isoaccepting arginine tRNAs at their anticodon loops. Proc Natl Acad Sci USA 97 8278–8283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci USA 104 20623–20628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A, Kathiresan A, Inada M, Narita Y, Nakamura T, Shi W, Takabe T, Bennett J (2003) Osmotic stress in barley regulates expression of a different set of genes than salt stress does. J Exp Bot 55 2213–2218 [DOI] [PubMed] [Google Scholar]
- Veach YK, Martin RC, Mok DWS, Malbeck J, Vaňková R, Mok MC (2003) O-Glucosylation of cis-zeatin in maize: characterization of genes, enzymes, and endogenous cytokinins. Plant Physiol 131 1374–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang J, Wang Z, Zhu Q, Wang W (2001) Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol 127 315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Kojima M, Yamaya T, Sakakibara H (2004) Molecular characterization of cytokinin-responsive histidine kinases in maize: differential ligand preferences and response to cis-zeatin. Plant Physiol 134 1654–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao TY, Martin D, Meeley RB, Downie B (2004) Expression of the maize GALACTINOL SYNTHASE gene family. II. Kernel abscission, environmental stress and myo-inositol influences accumulation of transcript in developing seeds and callus cells. Physiol Plant 121 647–655 [Google Scholar]
- Zimmermann P, Laule O, Schmitz J, Hruz T, Bleuler S, Gruissem W (2008) Genevestigator transcriptome meta-analysis and biomarker search using rice and barley gene expression databases. Mol Plant 1 851–857 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.