Abstract
The plant hormone auxin plays an essential role in plant development. However, only a few auxin biosynthetic genes have been isolated and characterized. Here, we show that the TRANSPORT INHIBITOR RESPONSE2 (TIR2) gene is required for many growth processes. Our studies indicate that the tir2 mutant is hypersensitive to 5-methyl-tryptophan, an inhibitor of tryptophan synthesis. Further, treatment with the proposed auxin biosynthetic intermediate indole-3-pyruvic acid (IPA) and indole-3-acetic acid rescues the tir2 short hypocotyl phenotype, suggesting that tir2 may be affected in the IPA auxin biosynthetic pathway. Molecular characterization revealed that TIR2 is identical to the TAA1 gene encoding a tryptophan aminotransferase. We show that TIR2 is regulated by temperature and is required for temperature-dependent hypocotyl elongation. Further, we find that expression of TIR2 is induced on the lower side of a gravitropically responding root. We propose that TIR2 contributes to a positive regulatory loop required for root gravitropism.
Auxin is known to play an important role in plant development (Davies, 1995). However, many aspects of auxin biology remain poorly understood. Auxin is synthesized primarily in young tissues, such as cotyledons, leaves, and roots (Ljung et al., 2001, 2005), and transported to other tissues where it is perceived by members of the TRANSPORT INHIBITOR RESPONSE1 (TIR1) auxin receptor family. Recent studies have dramatically increased our knowledge of auxin transport and signaling (Quint and Gray, 2006; Vieten et al., 2007). However, the pathways of auxin synthesis and their regulation are still relatively unclear.
Several indole-3-acetic acid (IAA) biosynthetic pathways have been proposed in plants based on research in plant-associated bacteria (Patten and Glick, 1996; Woodward and Bartel, 2005; Spaepen et al., 2007). There are two major types of pathways: the Trp-dependent and Trp-independent pathways. It has been hypothesized that plants have four Trp-dependent pathways that are generally named after an intermediate. In bacteria, the indole-3-pyruvic acid (IPA) pathway, one of the Trp-dependent pathways, has been described in detail (Koga, 1995; Spaepen et al., 2007). The current model for the IPA pathway involves a Trp aminotransferase oxidatively transaminating Trp to IPA. Subsequently, an IPA decarboxylase converts IPA to indole-3-acetaldehyde, and indole-3-acetaldehyde is oxidized to IAA. The IPA pathway is considered a major IAA biosynthetic pathway in plants, since potential intermediates have been isolated from different species (Sheldrake, 1973; Cooney and Nonhebel, 1991; Koga, 1995; Tam and Normanly, 1998). In addition, Trp transamination activity has been found in many plants (Gamborg, 1965; Forest and Wightman, 1972; Truelsen, 1973). Recently, two groups reported the identification of a gene called TAA1. This gene encodes an aminotransferase that converts Trp to IPA and functions in IAA biosynthesis (Stepanova et al., 2008; Tao et al., 2008).
To identify genes that are required for auxin synthesis, transport, and signaling, we previously screened for Arabidopsis (Arabidopsis thaliana) mutants that are resistant to auxin transport inhibitors, such as N-1-napthylpthalamic (NPA; Ruegger et al., 1997). The treatment of seedlings with NPA results in auxin accumulation in the root tip (Ljung et al., 2005). Thus, mutants that are resistant to NPA may have defects in synthesis, transport, or response because roots of these mutants are expected to have lower levels of IAA or reduced sensitivity to IAA. This screen succeeded in isolating mutations in seven genes with weak NPA-resistant phenotypes, including genes related to auxin signaling (TIR1), auxin transport (TIR3), and auxin synthesis (TIR7; Ruegger et al., 1997, 1998; Ljung et al., 2005).
Here, we describe the characterization of TIR2, a gene whose function is required for auxin synthesis. Genetic and physiological analyses of the tir2 mutant suggest that TIR2 is required for the Trp-dependent auxin synthesis pathway and functions as a Trp aminotransferase. Molecular cloning of TIR2 reveals that the gene is identical to TAA1 (Stepanova et al., 2008; Tao et al., 2008). We show that auxin regulates expression of TIR2 in a tissue-specific manner. Furthermore, we show that TIR2 is required for temperature-dependent hypocotyl elongation and that high temperature positively regulates expression of the TIR2 gene, suggesting that temperature regulates hypocotyl elongation directly by stimulating auxin synthesis. Finally, we provide evidence that TIR2 functions in a positive regulatory loop required for root gravitropism.
RESULTS
The tir2 Mutant Exhibits an Altered Response to NPA But Not Auxin
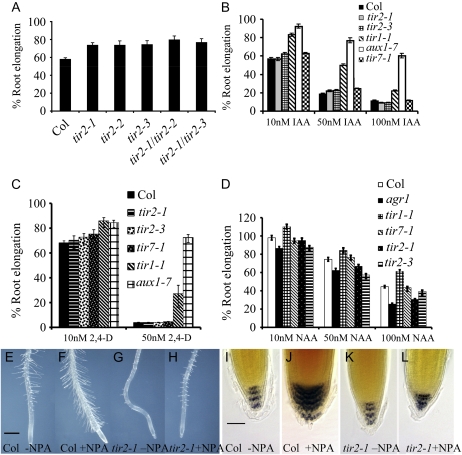
The tir2-1 mutant was isolated in a screen for seedlings that are resistant to the growth inhibiting properties of NPA. The mutant displays an NPA-resistant phenotype similar to other tir mutants, such as tir1 and tir3 (Ruegger et al., 1997). When grown on 5 μm NPA, wild-type seedlings exhibited approximately 60% root elongation after 3 d compared to untreated controls, while tir2-1 seedlings displayed 75% root elongation (Fig. 1A). In addition to root inhibition, NPA treatment induced elongation of root hairs in wild-type (Columbia-0 [Col-0]) roots (Fig. 1, E and F). In tir2-1 plants, this response was clearly reduced (Fig. 1, G and H). Furthermore, wild-type roots produced additional columella cells after treatment with NPA due to auxin accumulation in the root tip (Fig. 1, I and J), while tir2-1 roots had fewer columella cells even after treatment with NPA (Fig. 1, K and L). These results suggest that the roots of the tir2 mutants either have lower levels of auxin or are less sensitive to auxin.
Figure 1.
tir2-1 seedlings are resistant to NPA but not auxin. A, Root elongation on 5 μm NPA. B to D, Root elongation on medium containing IAA (B), 2,4-D (C), and NAA (D). E to H, Root hairs after treatment with 5 μm NPA. Seven-day-old seedlings were grown on ATS medium for 3 d before transfer to medium containing NPA. Bar = 300 μm. I to L, Col-0 and tir2-1 seedlings stained with Lugol stain. Four-day-old seedlings were transferred to medium with or without NPA and stained after 9 d. Bar = 50 μm. [See online article for color version of this figure.]
To understand whether TIR2 is involved in auxin transport, tir2-1 mutant seedlings were treated with the synthetic auxins 2,4-dichlorophenoxyacetic acid (2,4-D) and 1-naphthaleneacetic acid (NAA) as well as the natural auxin IAA. It is known that auxin influx carriers promote uptake of 2,4-D but not NAA (Delbarre et al., 1996), while NAA but not 2,4-D are substrates of the efflux carriers. Thus, the auxin influx transporter mutant (aux1-7; Marchant et al., 1999) is resistant to 2,4-D but not NAA (Fig. 1, C and D). In contrast, the auxin efflux transporter mutant (pin2/agr1; Chen et al., 1998; Luschnig et al., 1998; Muller et al., 1998) is hypersensitive to NAA because the hormone accumulates within the cells of mutant plants (Fig. 1D). If the tir2 mutants are deficient in auxin influx or efflux, we expect that they will be either resistant or hypersensitive to 2,4-D and NAA, respectively. In Figure 1, B to D, we show that tir2-1 seedlings display the same response as Col-0 to all three auxins tested. In contrast, aux1-7 was less sensitive to all concentrations of IAA tested (Fig. 1B). This suggests that tir2-1 can transport auxin effectively. Furthermore, these data indicate that the tir2-1 mutants respond normally to the auxin signal, unlike tir1-1 (Ruegger et al., 1998; Dharmasiri et al., 2005; Fig. 1, B–D). As a control, we tested the tir7-1 mutant, an allele of the ANTHRANILATE SYNTHASE ALPHA SUBUNIT1 (ASA1) gene. This mutant has lower levels of IAA (Ljung et al., 2005). Figure 1, B to D, shows that tir2-1 and tir7-1 have similar responses to the three auxins tested. These data suggest that the TIR2 gene is not involved in auxin transport or signaling but in auxin synthesis.
TIR2 Is Required for Diverse Auxin-Dependent Processes in the Seedling
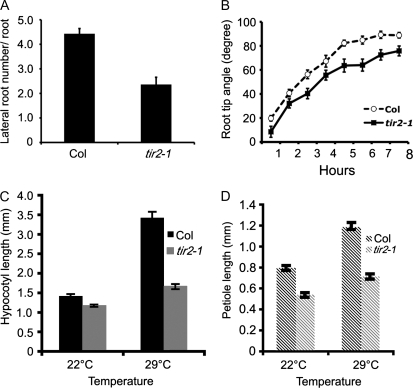
In addition to the NPA-resistant phenotype, the tir2 mutants display many auxin-related defects. Figure 1, E to H, shows that mutant seedlings produce shorter root hairs than the wild type. In addition, the mutant has fewer lateral roots, a reduction in root gravitropism (Fig. 2, A and B), and defects in vascular tissue in cotyledons (data not shown). The diversity of these defects suggests that TIR2-dependent auxin synthesis contributes to many aspects of plant growth.
Figure 2.
Auxin-related defects in the tir2 mutant. A, Number of lateral roots on 10-d-old Col-0 and tir2-1 plants. B, Gravity response of Col-0 and tir2-1 roots. Seedlings were grown in a vertical orientation for 7 d and reoriented at time 0. C and D, Response of hypocotyls (C) and petioles (D) to increased temperature.
Increased temperature has a dramatic effect on Arabidopsis seedling growth. Wild-type seedlings grown at 29°C have higher levels of IAA and as a consequence longer hypocotyls, petioles, and roots, compared to seedlings grown at 22°C (Fig. 2, C and D; Gray et al., 1998). In contrast, the hypocotyls and petioles of tir2-1 seedlings are only slightly longer at the higher temperature, indicating that TIR2 is required for this response.
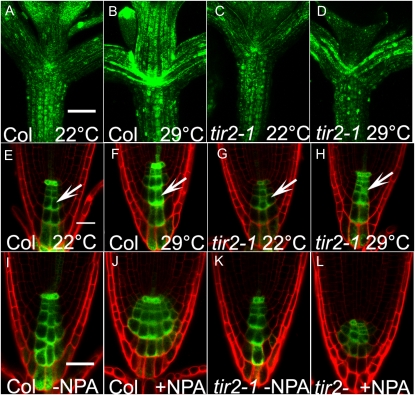
Our data show that tir2 is not resistant to exogenous auxin. However, if the mutant is deficient in IAA synthesis, we expect to see changes in the level of auxin-regulated transcription, particularly at sites of auxin production, such as young leaves, cotyledons, and root tips (Ljung et al., 2001, 2005). To address this question, we crossed the DR5rev:GFP auxin-responsive reporter into the tir2-1 mutant (Benková et al., 2003). The GFP signal was slightly weaker in hypocotyls, petioles, cotyledons, and root tips of tir2-1 compared to Col-0 at 22°C (Fig. 3, A, C, E, and G). At 29°C, the GFP signal increased markedly in the root tips and shoots of wild-type seedlings (Fig. 3, B and F). Furthermore, in wild-type roots the meristem cells were larger at the higher temperature (Fig. 3, E and F, arrow). In tir2-1 seedlings, the GFP signal increased only slightly at 29°C, and the meristem cells were smaller than in wild-type plants (Fig. 3, E–H, arrow). Although the level of DR5rev:GFP activity was affected in tir2-1, the distribution of signal was similar to Col-0, suggesting that tir2-1 is deficient in auxin synthesis but not transport.
Figure 3.
DR5rev:GFP expression responds to high temperature and NPA treatment. A to H, DR5rev:GFP expression in hypocotyls and petioles (A–D) and roots (E–H) after growth at 22°C for 7 d (A, E, C, and G) or for 4 d at 22°C and 3 d at 29°C (B, F, D, and H). Bars = 200 μm in A to D and 50 μm in E to H. Arrows indicate larger cells (E and F) in the root meristem of the wild type at 29°C and smaller cells (G and H) in the root meristem of tir2-1 at 29°C. I to L, DR5rev:GFP expression in 7-d-old seedlings 4 d after treatment with 5 μm NPA. Bar = 30 μm.
We also examined DR5rev:GFP activity in tir2-1 after NPA treatment. In wild-type plants, DR5rev:GFP activity increased in the root tips after treatment with NPA due to accumulation of auxin (Fig. 3, I and J). However, NPA treatment did not significantly alter GFP signal in the mutant (Fig. 3, K and L). These data also suggest that the TIR2 gene is required for auxin synthesis.
The TIR2 Gene Is a Trp Aminotransferase
To determine the function of TIR2, we isolated the gene using a positional cloning approach. TIR2 was mapped to an interval on chromosome one that included 30 genes (Supplemental Fig. S1A). The sequence of these genes was determined in the tir2-1 allele, and a mutation was found in the third exon of At1g70560 encoding an alliinase-like protein. The mutation results in the substitution of a Glu at Gly-171. Recently, this gene has been described as TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1; Stepanova et al., 2008; Tao et al., 2008). To confirm that tir2-1 is an allele of TAA1, two T-DNA insertion lines (tir2-2/wei8-3 SALK_127890 and tir2-3/wei8-4 SALK_022743; Alonso et al., 2003; Stepanova et al., 2008) were obtained and analyzed. Both T-DNA insertion lines shared similar defects with tir2-1, including NPA resistance (Fig. 1A), shorter root hairs, and shorter hypocotyls and petioles at 29°C (data not shown). Complementation analysis indicated that tir2-2 and tir2-3 are alleles of tir2-1 (Fig. 1A). Further, the TAA1 cDNA under control of the Cauliflower mosaic virus 35S promoter was introduced into tir2 mutant plants. These lines displayed a wild-type phenotype, including normal NPA response (Supplemental Fig. S2A) and seedling morphology (data not shown), confirming that TIR2 is the same gene as TAA1.
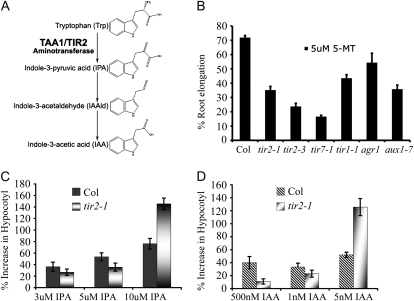
The amino acid sequence of TIR2 is similar to that of garlic (Allium sativum) alliinase (Supplemental Fig. S1B). The function of alliinase has previously been characterized in garlic and onion (Allium cepa; Kuettner et al., 2002). This enzyme catalyzes the conversion of a specific nonprotein sulfur-containing amino acid called alliin to allicin, a compound with antibiotic activity. To understand how TIR2 is related to alliinase-like proteins in other plant species, we performed a phylogenetic study using available sequences from land plants. These proteins fell into two major clades (Supplemental Fig. S3). The garlic and onion alliinases together with a number of uncharacterized proteins define one clade, while TIR2/TAA1 is in the second clade together with two additional Arabidopsis proteins and proteins from diverse species, including the moss Physcomitrella patens. Stepanova et al. (2008) have named the two additional Arabidopsis proteins TAR1 and TAR2 and present evidence indicating that these proteins also function in IAA biosynthesis. Our data show that TIR2/TAA1 and the TAR1/2 proteins are in a separate lineage from garlic and onion alliinase (Supplemental Fig. S3). Further, structural studies show that the garlic alliinase is very similar to aminotransferases (Kuettner et al., 2002). Together with the earlier studies of Stepanova et al. (2008) and Tao et al. (2008), our analysis indicates that TIR2 is a Trp aminotransferase that synthesizes IPA from Trp (Fig. 4A).
Figure 4.
The function of TIR2 in IAA biosynthesis. A, Schematic diagram of the IPA pathway. B, Root elongation after treatment with 5-MT. C and D, Hypocotyl length in seedlings grown on IPA (C) and IAA (D). Seedlings were grown at 22°C for 3 d followed by 4 d in the absence or presence of compound at 29°C. The effect of IPA and IAA on hypocotyl length is expressed as a percentage increase compared to medium without compound.
The TIR2 Gene Is Required for Auxin Synthesis
To further explore the possibility that TIR2 functions in the IPA pathway, we examined the sensitivity of the tir2 mutant to 5-methyl Trp (5-MT), a Trp analog that inhibits Trp synthesis (Hull et al., 2000). If Trp is a substrate of TIR2, the tir2 mutant should be hypersensitive to 5-MT. The data in Figure 4B show that this is the case. As controls, tir1-1, pin2, aux1-7, and tir7-1 mutants were also treated with 5-MT. All of these mutants were more sensitive to 5-MT than wild-type plants (Fig. 4B). However, for tir1, pin2, and aux1, the difference was quite small. In contrast, tir7-1, deficient in Trp synthesis, was even more sensitive than tir2 (Ljung et al., 2005). It has already been shown that the double mutants of the cytochrome P450s (cyp79b2 cyp79b3) are hypersensitive to 5-MT (Zhao et al., 2002). These two genes function in the IAOx pathway, one of the Trp-dependent pathways. Overexpression of TIR2 rescued the 5-MT-sensitive phenotypes of the tir2 mutants (Supplemental Fig. S2B).
If the TIR2 gene encodes a Trp aminotransferase, it may be possible to rescue some aspects of the mutant phenotype by providing additional IAA or IPA. Indeed, exogenous IAA or IPA dramatically increased tir2-1 hypocotyl length at high temperature (Fig. 4, C and D) consistent with the proposed function of TIR2.
The TIR2 Gene Is Expressed at Sites of Auxin Production
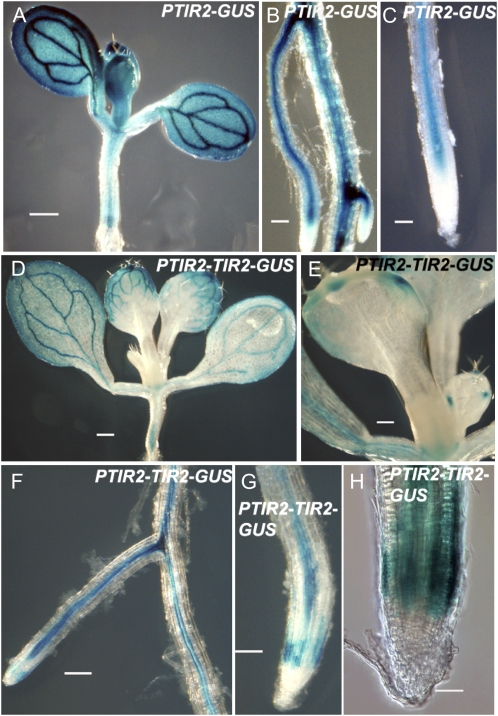
Auxin is produced at high levels in young developing leaves and cotyledons and at lower but still significant levels in roots (Ljung et al., 2001, 2005). To determine if TIR2 is expressed in these tissues, we generated TIR2 transcriptional GUS fusion lines using 3.4 kb of sequence upstream of the TIR2 ATG. GUS expression was detected in young leaves and cotyledons as well as the vascular tissue in shoots, hypocotyls, and roots (Fig. 5, A–C). However, GUS staining was not detected in the root meristem.
Figure 5.
Analysis of TIR2 expression in pTIR2:GUS and pTIR2:TIR2-GUS lines. GUS staining in pTIR2:GUS (A–C) and pTIR2:TIR2-GUS tir2-1 (D–H) seedlings. Bars = 1 mm in A, 125 μm in B, C, F, and G, 500 μm in D, 250 μm in E, and 50 μm in H. Eight-day-old seedlings (A), 6-d-old seedlings (B and C), and 9-d-old seedlings (D–H) are shown.
To extend these results, we generated a translational fusion of the TIR2 cDNA with GUS under control of the TIR2 promoter and introduced it into the tir2-1 mutant. This construct restored the wild-type phenotype to the mutant, indicating that the fusion protein is functional (Supplemental Fig. S4, A–C). The pattern of expression was similar to that of the transcriptional fusion except that GUS staining was more restricted in cotyledons and young leaves. Strong staining was found in the tips of young leaves and the margins of cotyledons (Fig. 5, D–H). Like the transcriptional fusions, we did not observe staining in the columella or distal meristem. However, a band of strong staining was detected in the proximal meristem. At this point, it is not known why this staining was not observed in the transcriptional fusion line. One possibility is that regulatory sequences are contained within the TIR2 coding region. Stepanova et al. (2008), using a TAA1:TAA1-GFP fusion line, observed the strongest GFP signal in the quiescent center (QC) of etiolated seedlings. Although the significance of these differences is not clear, we note that the transgene we used for our studies was able to complement the mutant phenotype.
Auxin Regulates Expression of the TIR2 Gene
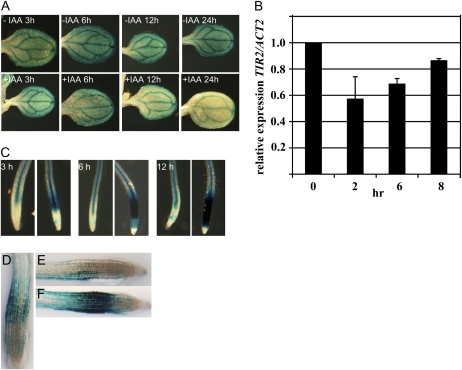
The temporal and spatial distribution of IAA plays an important role in plant development. The concentration of IAA in specific tissues is regulated by changes in auxin synthesis and transport. Although the regulation of IAA synthesis is poorly understood, recent studies have shown that IAA synthesis is regulated by other plant hormones, such as ethylene (Stepanova et al., 2005). In addition, there is evidence that feedback inhibition of IAA biosynthesis contributes to the control of IAA levels (Ljung et al., 2001, 2005). To determine if TIR2 may be part of this feedback mechanism, we examined the effect of auxin treatment on expression of the TIR2:TIR2-GUS transgene. The data in Figure 6A show that GUS staining in the cotyledons is reduced after IAA treatment. However, because GUS is a stable enzyme, GUS staining is not a sensitive indicator of decreased transcription of the reporter gene. Consequently, we measured TIR2 RNA levels in the hypocotyls and cotyledons of auxin-treated Col-0 seedlings using quantitative reverse transcription (RT)-PCR. The results show that TIR2 levels decreased substantially within 2 h of auxin treatment (Fig. 6B). Even after 8 h, TIR2 levels have not recovered to the level of untreated seedlings. These results suggest that IAA negatively regulates TIR2 transcription.
Figure 6.
Regulation of TIR2 by auxin. A, GUS staining of pTIR2:TIR2-GUS seedlings at intervals after treatment with 10 μm IAA. B, Relative TIR2 RNA levels in aerial plant parts after treatment with 50 μm IAA measured by quantitative RT-PCR. TIR2 levels at time 0 were set to 100%. Values are the mean of eight determinations from three independent RNA preparations. Error bars represent sd. C, GUS staining of pTIR2:TIR2-GUS seedlings after treatment with 10 μm IAA for 3, 6, and 12 h. D to F, GUS staining of pTIR2:TIR2-GUS before (D) and 8 h after 90° rotation (E and F). Seedling in F was treated with 50 μm NPA.
In contrast to the cotyledon, we see a dramatic enhancement in GUS staining in the root tips of auxin-treated pTIR2:TIR2-GUS plants (Fig. 6C). At this point, it is not clear if this regulation is transcriptional or posttranscriptional.
Previously, it has been reported that treatment with 1-aminocyclopropane-1-carboxylic acid (ACC) (the precursor of ethylene) induces expression of genes related to auxin synthesis (Stepanova et al., 2005, 2008; Růzicka et al., 2007). Since auxin is known to induce ethylene biosynthesis, it is possible that the effect of auxin on TIR2 expression requires ethylene. To determine if this is the case, we treated pTIR2:GUS and pTIR2:TIR2-GUS seedlings with IAA together with aminoethoxyvinylglycine (AVG; an inhibitor of the ethylene biosynthetic enzyme; Amrhein and Wenker, 1979; Lieberman, 1979). ACC treatment increased GUS staining in DR5:GUS lines in the root meristem as described previously (Stepanova et al., 2005; Růzicka et al., 2007; Supplemental Fig. S5). Also consistent with earlier studies, treatment with 10 μm 5-MT or 10 μm ACC plus 5-MT did not induce DR5:GUS expression, suggesting that ethylene induces DR5:GUS expression in roots by regulating genes in auxin synthesis (Supplemental Fig. S5). In contrast, the addition of AVG did not affect IAA-regulated changes in either the transcriptional or translational fusion lines, suggesting that auxin affects TIR2 expression independently of ethylene (Supplemental Fig. S5).
The phenotype of the tir2 mutant indicates that TIR2-dependent auxin synthesis is required for a normal gravitropic response. Since curvature in the root is dependent on an asymmetry in auxin levels across the root, we wondered if this asymmetry results in differential TIR2 expression on the upper and lower side of the root. To investigate this, we grew pTIR2:TIR2-GUS seedlings in the vertical orientation, turned them 90°, and stained for GUS after 8 h. The results in Figure 6, D and E, show that GUS staining increased in the epidermal cells on the lower side of the root but decreased in the epidermis on the upper side of the root, consistent with preferential auxin transport. Treatment with NPA abolished the gravitropic response and resulted in symmetrical high-level expression of TIR2-GUS, confirming that the gravity response of TIR2 is related to asymmetric auxin transport (Fig. 6F). These results suggest that a positive feedback loop contributes to increased auxin synthesis on the lower side of the root, thus amplifying the gravitropic response.
High Temperature Induces Expression of the TIR2 Gene
In Arabidopsis seedlings, elevated temperature results in an increase in auxin levels. This increase stimulates both hypocotyl and petiole length as well as an increase in cell size in the root meristem (Gray et al., 1998). Because tir2 seedlings do not respond to increased temperature, we reasoned that regulation of TIR2 may be important for the temperature effect on auxin levels. To investigate this, we examined GUS levels in pTIR2:TIR2-GUS and pTIR2:GUS lines at 22°C and 29°C. The data in Figure 7 show that GUS levels increase in the cotyledon, hypocotyls, and root in response to the higher temperature, indicating that TIR2 transcription is regulated by high temperature. Since we have already shown that expression of TIR2 is decreased in much of the seedling in response to auxin, the effect of high temperature is probably independent of auxin signaling.
Figure 7.
Effect of temperature on TIR2 expression. GUS staining in 4-d-old seedlings in pTIR2:TIR2-GUS, pTIR2:GUS, and DR5:GUS lines at 22°C (4 d at 22°C) and at 29°C (2 d at 22°C, 2 d at 29°C). A, B, G, and H, pTIR2:TIR2-GUS. C, D, I, and J, pTIR2:GUS. E, F, K, and L, DR5:GUS. A, C, E, G, I, and K, 22°C. B, D, F, H, J, and L, 29°C. Bars = 1 mm in A to F, 250 μm in G to J, and 125 μm in K and L.
The TIR2 Gene Is Required for Root Meristem Development
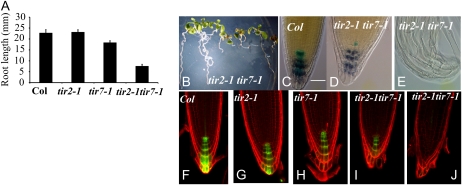
So far we have shown that TIR2 is required for hypocotyl and petiole elongation, lateral root formation, gravitropism, and root hair formation. To learn more about the biological function of TIR2, we generated a tir2-1 tir7-1 double mutant. The double mutant had a much more severe phenotype than the respective single mutants. The root lengths of the tir2-1 and tir7-1 single mutants were similar to the wild type, while the tir2-1 tir7-1 double mutants formed short roots with very diverse phenotypes (Fig. 8, A and B). The more robust seedlings displayed a 60% reduction in root length compared to the wild type. More severely affected seedlings formed extremely short roots. Neither the tir2-1 or tir7-1 single mutants had an obvious root meristem defect, whereas the tir2-1 tir7-1 double mutant had a clear defect (Fig. 8, C–E). To identify which cells are disrupted in the root tips, these double mutants were crossed with a QC25 enhancer-trap line, which expresses the GUS gene in all QC cells (Sabatini et al., 2003). QC cells were detected in these lines by GUS staining, and columella cells were visualized with Lugol staining. In wild-type roots, two QC cells and four layers of columella cells were observed as expected (Fig. 8C). However, in the roots of weakly affected double mutant plants, only one QC cell was detected and the columella was disrupted (Fig. 8D). In severely affected seedlings, QC and columella cells were not observed (Fig. 8E). In addition, the total number of cells in the root meristem decreased in the tir2-1 tir7-1 double mutant compared with the wild type, the tir2-1 mutant, and the tir7-1 mutant (data not shown).
Figure 8.
Phenotype of the tir2-1 tir7-1 double mutants. A, Root length of 7-d-old seedlings. B, Eight-day-old tir2-1 tir7-1 seedlings. C to E, Expression of QC25 (blue) and Lugol staining (dark blue) in Col-0 (C) and tir2-1 tir7-1 (D and E) seedlings. Bar = 50 μm for the 8-d-old seedlings (C–E). F to J, Expression of DR5rev:GFP (green) in Col-0 (F), tir2-1 (G), tir7-1 (H), and tir2-1 tir7-1 (I and J). Propidium iodide staining (red). Bar = 50 μm for the 5-d-old seedlings (F–J).
To examine the distribution of auxin in the mutant lines, we crossed in the DR5:GFP construct. A slight decrease in GFP signal was observed in the single mutants (Fig. 8, G and H). However, in the double mutants, GFP signal was either decreased (Fig. 8I) or absent (Fig. 8J) in the root tips. Surprisingly, GFP was still detected in the vascular tissue of double mutant roots (Fig. 8J; Supplemental Fig. S6). This suggests that genes related to TIR2, such as TAR1 and TAR2, or a different auxin biosynthetic pathway may be more important for auxin synthesis in these tissues.
DISCUSSION
Recent studies confirm that auxin synthesis occurs through multiple pathways in Arabidopsis, including the TAM pathway and the IPA pathway (Cheng et al., 2006; Stepanova et al., 2008; Tao et al., 2008). Although the pathways may be partially redundant, their precise role in various aspects of plant development remains to be fully explored. Here, we show that TIR2/TAA1, encoding a Trp aminotransferase, is required for diverse processes in the seedling, including temperature-dependent hypocotyl elongation and root gravitropism. Our results extend previous studies that demonstrate a role for TIR2/TAA1 in a variety of growth processes during seedling development, including temperature-dependent hypocotyl growth, root gravitropism, root hair formation, and lateral root development.
TIR2 Is Required for the IPA Pathway
Early studies suggested that the IPA pathway is a major source of IAA in plants (Koga, 1995; Spaepen et al., 2007). However, until recently the enzymes in the pathway were unknown. The tir2-1 mutant was recovered in a screen for NPA resistance that identified genes in diverse aspects of auxin biology, including auxin perception (TIR1), response (TIR5/ARF7), transport (TIR3), and synthesis (TIR7/ASA1; Harper et al., 2000; Gil et al., 2001; Dharmasiri et al., 2005; Kepinski and Leyser, 2005; Ljung et al., 2005). Typically, the behavior of NPA-resistant mutants on various auxins provides useful information on the function of the gene product. The tir2-1 mutant responds normally to IAA, 2,4-D, and 1-NAA, suggesting that it is not deficient in auxin response or transport. In contrast, hypersensitivity to 5-MT is consistent with a defect in Trp-dependent IAA synthesis via the IPA pathway. The fact that tir2-1 is rescued by exogenous IPA and IAA also strongly supports this hypothesis.
The TIR2 gene encodes an alliinase-like protein structurally related to aminotansferases. Recently, two groups reported the identification of a Trp aminotransferase named TAA1 and demonstrated a role for this protein in auxin synthesis (Stepanova et al., 2008; Tao et al., 2008). We have confirmed that TIR2 is identical to TAA1. In the tir2-1 allele, Gly-171 is changed to Glu. This residue is conserved among all members of the alliinase/transaminase family and is part of the so-called strained loop involved in pyridoxal phosphate cofactor binding (Kuettner et al., 2002). It is likely that substitution of Glu at this position would disrupt this structure and pyridoxal phosphate binding.
TIR2 Regulation
In bacteria, the IPA decarboxylase is the rate-limiting step in IAA synthesis (Spaepen et al., 2007). Our results suggest that this may be the case in plants as well. Overexpression of TIR2 did not result in growth defects, such as those observed when YUCCA is overexpressed (Zhao et al., 2001). Further, 35S:TIR2 lines displayed normal sensitivity to exogenous Trp, suggesting that increasing endogenous IPA levels does not result in the synthesis of more IAA (Supplemental Fig. S2C). Based on these results, it seems likely that IPA decarboxylase is limiting for auxin biosynthesis, at least under our conditions. The characterization of this enzyme in plants will address this question in the future.
Although TIR2/TAA1 does not appear to be limiting, our results indicate that TIR2 is highly regulated during development. Localized changes in auxin level are known to play a key role in embryogenesis, meristem establishment and maintenance, organogenesis, and tropic growth. The role of auxin transport during these processes is well established, but information on the role of auxin synthesis in the regulation of auxin flux is just beginning to emerge (Cheng et al., 2006; Stepanova et al., 2008; Tao et al., 2008). Expression of the TIR2/TAA1 gene is spatially restricted during embryo development, and seedling development is consistent with existing models (this study; Stepanova et al., 2008; Tao et al., 2008).
In addition, we show that TIR2 promoter activity is dramatically regulated in the seedling in response to auxin, indicating that auxin synthesis is subject to feedback regulation. In the aerial part of the seedling, TIR2 expression is significantly reduced upon auxin treatment. This mechanism may play an important role in the precise regulation of auxin levels during growth processes. In a previous study, Tao et al. (2008) demonstrated that TAA1 expression is reduced in the hypocotyl in response to shade despite the fact that auxin levels increase and hypocotyl elongation is stimulated in shade conditions. These authors also show that most of the auxin in a shade-responding seedling is synthesized in the cotyledons and leaves and is transported into the hypocotyl. Based on our results, we propose that this auxin represses transcription of TIR2 in the hypocotyl.
In previous studies, we showed that Arabidopsis seedlings grown at elevated temperature have higher levels of IAA and that this change results in increased hypocotyl elongation (Gray et al., 1998). The tir2 mutants are deficient in temperature-dependent growth stimulation, suggesting that the IPA pathway may be regulated by temperature. Indeed, we find that transcription of TIR2 is strongly increased at 29°C compared to 22°C. Thus, it seems likely that the temperature effect is at least partly due to increased flux through the IPA pathway. Since TIR2/TAA1 is not limiting, we expect that the IPA decarboxylase is also regulated by temperature. In the future, it will be interesting to determine what other environmental conditions regulate the IPA pathway.
TIR2 and Root Growth
Our results indicate that TIR2 is expressed in a narrow band in the proximal root meristem. Further, TIR2 expression in this region is auxin sensitive, suggesting the existence of a positive regulatory loop that may have an important function during root growth. The structure and function of the root meristem is regulated by an auxin gradient with a maximum at the QC. This gradient is established through the action of members of the PIN family of auxin efflux carriers and maintained by an auxin transport loop that cycles auxin from the QC through the columella to the epidermal layer and back to the QC via the provascular tissue (Blilou et al., 2005; Grieneisen et al., 2007; Scheres, 2007). By itself, the tir2-1 mutation does not confer an obvious defect in meristem structure. However, the phenotype of the tir7-1 tir2-1 double mutant clearly indicates that TIR2 functions in meristem maintenance. The tir7-1 mutation affects ASA1 function and therefore acts upstream of TIR2/TAA1. Because both ASA1 and TIR2/IAA1 function redundantly with other members of their respective families, we infer that the combination of both mutations results in a further reduction in IAA levels in the root tip. We propose that auxin synthesized in the proximal meristem through the IPA pathway contributes to the auxin transport loop. Auxin regulation of TIR2, and perhaps other genes in the pathway, would facilitate stringent regulation of the auxin maximum. Consistent with this, double mutants that are deficient in both TAA1 and TAR2 have a similar meristem defect as the tir2 tir7 mutants, indicating that TAR2 also contributes to this pathway (Stepanova et al., 2008).
It is important to note that the pattern of TIR2 expression we observe with the TIR2:TIR2-GUS line differs slightly from that observed by Stepanova et al. (2008) using a whole gene GFP fusion. In that study, the highest level of GFP signal was observed in the QC, whereas we don't observe any GUS staining in the QC. In addition, cell-type specific array data indicate that TAA1 is broadly expressed in the root, with the highest level in the stele (Birnbaum et al., 2003; Petersson et al., 2009). At this point, the reasons for these differences are not clear. However, we note that the TIR2:TIR2-GUS transgene rescues the tir2 mutant phenotype, indicating that TAA1 function is not required in the QC.
TIR2 also has an important role in root gravitropism. During gravitropic growth, auxin is preferentially transported from the columella into the epidermal cell layer on the lower side of the root, resulting in decreased growth of these cells relative to the cells on the upper side. Our results show that root curvature is associated with increased TIR2 expression in the lower epidermal cells and concomitant loss of expression on the upper side. These changes would have the effect of increasing the asymmetry in auxin concentration and thus amplify the growth response.
Several recent studies have shown that parameters related to auxin transport are sufficient to model the formation and maintenance of the auxin maximum in the root meristem (Blilou et al., 2005; Kramer et al., 2008). Our study, together with other recent studies, suggests that regulation of auxin synthesis may also be an important factor in meristem behavior.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Treatments
All mutants and transgenic lines used in this study were in the Col-0 ecotype. The T-DNA insertion lines tir2-2 (SALK_127890) and tir2-3 (SALK_022743) were obtained from the Arabidopsis Biological Resource Center at Ohio State University. Seeds were surface sterilized in 30% commercial bleach and 0.02% Triton X-100 and plated on ATS media [1% Suc, 5 mm KNO3, 2.5 mm KH2PO4, pH 5.6, 2 mm MgSO4, 2 mm Ca(NO3)2, 50 μm CuSO4, 1 μm ZnSO4, 0.2 μm NaMoO4, 10 μm NaCl, and 0.01 μm CoCl2; Lincoln et al., 1990] containing 7 g agar in 1 L. Seedlings were grown at 21°C to 23°C under continuous light conditions in square petri dishes.
Assay for Root Gravitropism
Root gravitropic response was assayed in a manner similar to the method of Lincoln et al. (1990). Four-day-old seedlings were transferred to new ATS plates and held in an incubator in a vertical orientation for 1 to 2 h. The plates were then rotated 90°, and at 1-h intervals the plates were removed from the incubator and the root tip angles were measured. Each point represents at least 10 seedlings.
Map-Based Cloning
The tir2-1 mutant in Col-0 was crossed to Landsberg erecta. NPA-resistant plants were selected from F2 populations based on root length and root hair phenotypes. DNA was isolated by cetyl-trimethyl-ammonium bromide from these F2 plants. NPA-resistant phenotypes were confirmed in the F3 generation.
For rough mapping, TIR2 was placed in an interval between markers F4N2 (110.0 centimorgans) and nga11 (115.55 centimorgans) on chromosome one. After fine mapping, TIR2 was mapped between base pair 31246 of BAC F24J13 and base pair 35655 of BAC F15H11. The primers for mapping were designed using information from the CEREON collection (http://www.Arabidopsis.org/).
Assays for Root, Hypocotyl, and Petiole Elongation
To perform root elongation assays, 3- or 4-d-old seedlings were grown on ATS plates and transferred to ATS plates containing 5 μm NPA, 5 μm 5-MT, and other compounds. Root length was measured 3 or 4 d after transfer. For hypocotyl elongation assays, seedlings were grown on an ATS plate for 2 to approximately 3 d at 22°C, transferred to new plates, and grown at either 22°C or 29°C and incubated for 4 d. Lengths of roots, hypocotyls, and petioles were measured using a Nikon SMZ1500 dissecting scope and Image SXM software (http://www.liv.ac.uk/∼sdb/ImageSXM/).
Plant Vectors and Transformations
The TIR2 cDNA with or without stop codons and the TIR2 promoter were cloned using Gateway cloning technology (Invitrogen) and recombined into pMDC32 and pMDC163 (Curtis and Grossniklaus, 2003). For 35S-TIR2, TIR2 cDNA was amplified from cDNAs generated by RT-PCR SuperScript II (Invitrogen) from total RNA from Col-0 seedlings, cloned to pENTR/d-TOPO (Invitrogen), and recombined into pMDC32 containing a 35S promoter. Primers used for 35S-TIR2 were pENTR TIR2 forward (5′-CACCATGGTGAAACTGGAGAACTCG-3′) and TIR2 reverse (5′-CTAAAGGTCAATGCTTTTAATGAG-3′). For pTIR2:GUS, a 3.4-kb region from the start codon of the TIR2 gene was amplified from BAC F24J13 using primers pENTR TIR2 promoter forward (5′-CACCTGAAGATGCTCTTACTTAAATTTA-3′) and TIR2 promoter reverse (5′-CTTCTTCTTCTTGGTTTGGTC-3′), cloned, and recombined into pMDC163. For pTIR2:TIR2-GUS, the TIR2 promoter including 90 bp from the ATG and the TIR2 cDNA without stop codon were cloned and digested by XhoI and AscI (New England Biolabs). The digested fragment of the TIR2 cDNA without the stop codon was ligated into the digested TIR2 promoter in pENTR/d-TOPO and recombined into pMDC 163. Primers used for the TIR2 promoter with 90 bp of the first exon were pENTR TIR2 promoter forward and TIR2 promoter-cDNA reverse (5′-TGATCCAGATTGACCACGAAA-3′). Primers used for the TIR2 cDNA without the stop codon were pENTR TIR2 forward and TIR2 no stop reverse (5′-AAGGTCAATGCTTTTAATGAGC-3′).
Microscopy and Confocal Microscopy
For assays for root and hypocotyl elongation, images were collected using a dissecting microscope (Nikon SMZ1500) with a digital camera (Nikon DMX 1200). For detecting QC cells and columella cells, roots were observed by a Nikon E800 microscope. For detecting GFP signals, seedlings were observed under a Leica TCS SP confocal microscope. Seedlings were mounted in a 0.1 mg/mL propidium iodide (10 mg/mL; Sigma-Aldrich P-4170).
Staining
Seedlings were stained in GUS staining solution in 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-glucuronic acid in N,N-dimethylformamide, 0.5 m sodium phosphate buffer (pH 7.2), 10% Triton X-100, 100 mm potassium ferrocyanide, and 100 mm potassium ferricyanide for 15 to approximately 18 h at 37°C. The orientation of seedlings after gravitropism was marked by removing one cotyledon prior to GUS staining. For Lugol staining, seedlings were stained in Lugol solution (Sigma-Aldrich L-6146) for 1 to approximately 3 h at room temperature.
Treatment of pTIR2:TIR2-GUS, pTIR2:GUS, and DR5:GUS Lines
Six-day-old seedlings were grown on ATS plates and transferred to ATS media on the six-well cell culture plate (Corning) containing 10 μm IAA, 10 μm IAA AVG, 10 μm AVG, 10 μm 5-MT, 10 μm ACC, 10 μm ACC 5-MT, and 10 μm IAA ACC. Seedlings were grown for 2 d under continuous white light at room temperature. To examine the effects of temperature, seedlings were grown on ATS at 22°C for 2 d before transfer to 29°C. Two days later, seedlings were stained for GUS.
Quantitative RT-PCR Analysis
To examine TIR2 expression in response to auxin, total RNA was isolated from the aerial parts of 6-d-old seedlings 2, 6, and 8 h after treatment with 50 μm IAA using Tri-reagent (Sigma-Aldrich). Three micrograms of total RNA were used in reverse transcription reactions using SuperScript III reverse transcriptase (Invitrogen) and 20-mer oligo(dT) primer. Real-time quantitative PCR was performed using the LightCycler 480 Instrument (Roche Diagnostics) with LightCycler 480 Probes Master and Universal ProbeLibrary. Quantification of the TIR2 RNA levels was performed using LightCycler software relative to expression of ACTIN2. Primers were designed using the Universal ProbeLibrary Assay Design Center (https://www.roche-appliedscience.com/sis/rtpcr/upl/adc.jsp; Roche Applied Science). Primers were as follows: tir2f, 5′-GCCGCTCCTTTTTACTCCA-3′; tir2r, 5′-TGTACATACCCGACCGAACA-3′; Actin2f, 5′-CCGCTCTTTCTTTCCAAGC-3′; and Actin2r, 5′-CCGGTACCATTGTCACACAC-3′. The data represent the average fold change in TIR2 transcript levels 2, 6, and 8 h after 50 μm IAA treatment, compared to seedlings treated with Murashige and Skoog alone.
Phylogenetic Analysis
Most protein sequences were extracted from data sets derived from genome sequencing projects: Arabidopsis (Arabidopsis thaliana; The Arabidopsis Information Resource version 8; http://www.arabidopsis.org), Populus trichocarpa (Poptr1_1.Jamboreemodels, http://genome.jgi-psf.org/Poptr1_1), Ricinus comunus (TIGRcastorWGSr0.1, http://castorbean.jcvi.org/), Vitis vinifera (Vitis_vinifera_peptide_v1, http://www.genoscope.cns.fr/spip/Vitis-vinifera-e.html), Medicago truncatula (20080227_imgag_protMAPPED_NO_OVERLAP, http://www.medicago.org/), Oryza sativa (TIGRv5.0, http://rice.plantbiology.msu.edu/), Zea mays (AZM5, http://maize.jcvi.org/), Sorghum bicolor (Sorbi1_Genemodels_Sbi1_4_aa, http://genome.jgi-psf.org/Sorbi1), Selaginella moellendorffii (Selmo1_Genemodels_Filteredmodels2_aa, http://genome.jgi-psf.org/Selmo1), and Physcomitrella patens (Phypa1_1.Filteredmodels3, http://genome.jgi-psf.org//Phypa1_1). Additional sequences were identified in The Institute for Genomic Research Transcript Assemblies from Aquilegia formosa × pubescens, Pinus taeda, Picea glauca, Ceratopteris richardii, and Marchantia polymorpha (http://plantta.jcvi.org/). The remaining sequences were identified in the GenBank Nonredundant protein and High Throughput Genomic Sequence databases. Physcomitrella, Selaginella, and Populus gene models were manually adjusted from the genomic sequences based on homology, splicing predictions, and EST data. The sequences were aligned using T-COFFEE (Notredame et al., 2000) and adjusted using MACCLADE (Maddison and Maddison, 2003). For Bayesian inference, the analysis was run with four runs of four chains for 3,000,000 generations using MRBAYES 3.1.2 with the parameters aamodelpr = mixed, nst = 6, and rates = invgamma on the Indiana University BigRed computing cluster (Huelsenbeck and Ronquist, 2001). Maximum parsimony bootstrap support values were generated using the same protein alignment and PAUP* beta10 for Mac (Swofford, 1998).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Positional cloning of the TIR2 gene.
Supplemental Figure S2. Assay for root elongation of p35S:TIR2 lines.
Supplemental Figure S3. TAA1/TIR2-related proteins in plants.
Supplemental Figure S4. The pTIR2:TIR2-GUS gene rescues the tir2 mutant.
Supplemental Figure S5. GUS staining in pTIR2:TIR2-GUS and DR5:GUS seedlings.
Supplemental Figure S6. Expression of DR5rev:GFP in the tir2-1 tir7-1 double mutant.
Supplementary Material
This work was supported by the National Institutes of Health (grant no. GM–43644 to M.E.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Mark Estelle (mestelle@ucsd.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Amrhein N, Wenker D (1979) Novel inhibitors of ethylene production in higher plants. Plant Cell Physiol 20 1635–1642 [Google Scholar]
- Benková EE, Michniewicz MM, Sauer MM, Teichmann TT, Seifertová DD, Jürgens GG, Friml JJ (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302 1956–1960 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433 39–44 [DOI] [PubMed] [Google Scholar]
- Chen R, Hilson P, Sedbrook J, Rosen E, Caspar T, Masson PH (1998) The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc Natl Acad Sci USA 95 15112–15117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney T, Nonhebel H (1991) Biosynthesis of indole-3-acetic acid in tomato shoots: measurement, mass-spectral identification and incorporation of 2H from 2H2O into indole-3-acetic acid, d- and l-tryptophan, indole-3-pyruvate and tryptamine. Planta 184 368–376 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PJ (1995) Plant Hormones: Physiology, Biochemistry, and Molecular Biology. Kluwer Academic, Dordrecht, The Netherlands
- Delbarre A, Muller P, Imhoff V, Guern J (1996) Comparison of mechanisms controlling uptake and accumulation of 2,4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid, and indole-3-acetic acid in suspension-cultured tobacco cells. Planta 198 532–541 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435 441–445 [DOI] [PubMed] [Google Scholar]
- Forest JC, Wightman F (1972) Amino acid metabolism in plants. III. Purification and some properties of a multispecific aminotransferase isolated from bushbean seedlings (Phaseolus vulgaris L.). Can J Biochem 50 813–829 [DOI] [PubMed] [Google Scholar]
- Gamborg OL (1965) Transamination in plants. The specificity of an aminotransferase from mung bean. Can J Biochem 43 723–730 [DOI] [PubMed] [Google Scholar]
- Gil P, Dewey E, Friml J, Zhao Y, Snowden KC, Putterill J, Palme K, Estelle M, Chory J (2001) BIG: a calossin-like protein required for polar auxin transport in Arabidopsis. Genes Dev 15 1985–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Maree AF, Hogeweg P, Scheres B (2007) Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449 1008–1013 [DOI] [PubMed] [Google Scholar]
- Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E (2000) The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12 757–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17 754–755 [DOI] [PubMed] [Google Scholar]
- Hull AK, Vij R, Celenza JL (2000) Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci USA 97 2379–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 446–451 [DOI] [PubMed] [Google Scholar]
- Koga J (1995) Structure and function of indolepyruvate decarboxylase, a key enzyme in indole-3-acetic acid biosynthesis. Biochim Biophys Acta 1249 1–13 [DOI] [PubMed] [Google Scholar]
- Kramer EM, Draye X, Bennett MJ (2008) Modelling root growth and development. SEB Exp Biol Ser 61 195–211 [PubMed] [Google Scholar]
- Kuettner EB, Hilgenfeld R, Weiss MS (2002) The active principle of garlic at atomic resolution. J Biol Chem 277 46402–46407 [DOI] [PubMed] [Google Scholar]
- Lieberman M (1979) Biosynthesis and action of ethylene. Annu Rev Plant Physiol 30 533–591 [Google Scholar]
- Lincoln C, Britton JH, Estelle M (1990) Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K, Bhalerao RP, Sandberg G (2001) Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J 28 465–474 [DOI] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Celenza J, Yamada M, Estelle M, Normanly J, Sandberg G (2005) Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17 1090–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR (1998) EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev 12 2175–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison DR, Maddison WP (2003) MacClade 4: Analysis of Phylogeny and Character Evolution. Sinauer Associates, Sunderland, MA
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ (1999) AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J 18 2066–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A, Guan C, Galweiler L, Tanzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K (1998) AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J 17 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302 205–217 [DOI] [PubMed] [Google Scholar]
- Patten CL, Glick BR (1996) Bacterial biosynthesis of indole-3-acetic acid. Can J Microbiol 42 207–220 [DOI] [PubMed] [Google Scholar]
- Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, Moritz T, Grebe M, Benfey PN, Sandberg G, Ljung K (2009) An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Gray WM (2006) Auxin signaling. Curr Opin Plant Biol 9 448–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M (1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev 12 198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Hobbie L, Brown D, Bernasconi P, Turner J, Muday G, Estelle M (1997) Reduced naphthylphthalamic acid binding in the tir3 mutant of Arabidopsis is associated with a reduction in polar auxin transport and diverse morphological defects. Plant Cell 9 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růzicka K, Ljung K, Vanneste S, Podhorská R, Beeckman T, Friml J, Benková E (2007) Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19 2197–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B (2007) Stem-cell niches: nursery rhymes across kingdoms. Nat Rev Mol Cell Biol 8 345–354 [DOI] [PubMed] [Google Scholar]
- Sheldrake AR (1973) The production of hormones in higher plants. Biol Rev Camb Philos Soc 48 509–559 [Google Scholar]
- Spaepen S, Vanderleyden J, Remans R (2007) Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev 31 425–448 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM (2005) A Link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17 2230–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jurgens G, Alonso JM (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133 177–191 [DOI] [PubMed] [Google Scholar]
- Swofford DL (1998) PAUP. Phylogenetic Analysis Using Parsimony (and Other Methods). Sinauer Associates, Sunderland, MA
- Tam YY, Normanly J (1998) Determination of indole-3-pyruvic acid levels in Arabidopsis thaliana by gas chromatography-selected ion monitoring-mass spectrometry. J Chromatogr A 800 101–108 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truelsen TA (1973) Indole-3-pyruvic acid as an intermediate in the conversion of tryptophan to indole-3-acetic acid. II. Distribution of tryptophan transaminase activity in plants. Physiol Plant 28 67–70 [Google Scholar]
- Vieten A, Sauer M, Brewer PB, Friml J (2007) Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci 12 160–168 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291 306–309 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR, Normanly J, Chory J, Celenza JL (2002) Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev 16 3100–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.