Abstract
Plants have evolved a range of cellular responses to maintain developmental homeostasis and to survive over a range of temperatures. Here, we describe the in vivo and in vitro functions of BOBBER1 (BOB1), a NudC domain containing Arabidopsis (Arabidopsis thaliana) small heat shock protein. BOB1 is an essential gene required for the normal partitioning and patterning of the apical domain of the Arabidopsis embryo. Because BOB1 loss-of-function mutants are embryo lethal, we used a partial loss-of-function allele (bob1-3) to demonstrate that BOB1 is required for organismal thermotolerance and postembryonic development. Recombinant BOB1 protein functions as a molecular chaperone and prevents the aggregation of a model protein substrate in vitro. In plants, BOB1 is cytoplasmic at basal temperatures, but forms heat shock granules containing canonical small heat shock proteins at high temperatures. In addition to thermotolerance defects, bob1-3 exhibits pleiotropic development defects during all phases of development. bob1-3 phenotypes include decreased rates of shoot and root growth as well as patterning defects in leaves, flowers, and inflorescence meristems. Most eukaryotic chaperones play important roles in protein folding either during protein synthesis or during cellular responses to denaturing stress. Our results provide, to our knowledge, the first evidence of a plant small heat shock protein that has both developmental and thermotolerance functions and may play a role in both of these folding networks.
Plants are autotrophic sessile organisms that depend on sunlight for their energetic needs. One consequence of this lifestyle is that plants are often subjected to high temperature stress, especially in dry conditions when transpirational cooling is limited. At a cellular level, elevated temperatures result in changes in protein structure that can result in the exposure of normally buried hydrophobic residues. As a consequence of thermal denaturation, proteins may aggregate and cease to function normally. A universal response to temperature-induced protein unfolding in all living organisms is the production of heat shock proteins (HSPs). HSPs are molecular chaperones that provide organismal thermotolerance by preventing the denaturation and aggregation of target proteins as well as facilitating protein refolding. Highly conserved HSPs are found in all organisms and include the small HSP (sHSP) as well as the Hsp60, Hsp70, Hsp90, and Hsp100 families (Baniwal et al., 2004; Taiz and Zeiger, 2006). Members of the sHSP family are defined by their small size (12–43 kD), their ability to prevent protein aggregation, and by a conserved α-crystallin domain (ACD). Plants are unusual in the large number of ACD-containing sHSPs encoded by their genomes: Arabidopsis (Arabidopsis thaliana) has 19 compared to 10 in humans, four in Drosophila melanogaster, and one or two in bacteria (Haslbeck et al., 2005).
Although the biochemical activity of plant sHSPs has been well characterized (Lee et al., 1995, 1997; Basha et al., 2004; Siddique et al., 2008), little is known about the in vivo functions of plant sHSPs, perhaps due to functional redundancies in this large gene family. Apart from temperature-dependent changes in hypocotyl elongation, which reflects the ability of cells to expand, no developmental roles for a sHSP have been reported in plants (Jenks and Hasegawa, 2005; Dafny-Yelin et al., 2008). In addition to redundancy, a lack of known developmental functions for plant sHSPs may also be a result of the fact that most are only expressed in response to heat or other stresses. Exceptions include a subset of sHSPs expressed during seed and pollen maturation, developmental stages that involve desiccation (Wehmeyer and Vierling, 2000; Dafny-Yelin et al., 2008). However, since most plant sHSPs are not expressed under nonstress conditions, they are unlikely to affect normal growth and development (Swindell et al., 2007).
BOBBER1 (BOB1; At5g53400) is an essential gene required for the normal partitioning and patterning of the apical domain of the Arabidopsis embryo. In bob1-1 and bob1-2 null mutants, meristematic identity is expanded into the portion of the embryo that would normally form the seedling leaves (cotyledons), which in turn are never established. Auxin gradients are never established in bob1 mutant embryos. However, since there are multiple feedback loops involved in auxin signaling and transport, it is unclear whether the lack of auxin maxima in bob1 mutants is a direct or indirect result of a lack of BOB1 activity (Jurkuta et al., 2009). BOB1 encodes a protein with C-terminal homology to NudC, a protein identified in a screen for genes required for nuclear migration in Aspergillus nidulans. Genes with homology to NudC have been shown to interact with dynein microtubule motors. In mammalian tissue culture systems, interference with NudC-like gene function results in defects in chromosome segregation and cytokinesis (Aumais et al., 2003; Nishino et al., 2006; Zhou et al., 2006). The NudC domain has predicted structural homology with the α-crystallin/p23 protein families (Garcia-Ranea et al., 2002), which includes the ACD-containing sHSPs. The ACD, originally identified in the α-crystallin chaperone of the vertebrate eye lens, forms a structure consisting of two antiparallel β-sheets in a sandwich (Scharf et al., 2001; Haslbeck et al., 2005). The NMR structure of the mouse NudC homolog (PDB 1wfi) has the same β-sheet sandwich structure that provides support for the predicted structural homology between NudC domains and ACDs. These observations suggest that NudC domain proteins might share conserved functions with sHSPs. Support for this hypothesis comes from Caenorhabditis elegans where the NudC homolog NUD-1, an essential gene, displays protein chaperone activity in vitro (Faircloth et al., 2009).
Here, we use bob1-3, a partial loss-of-function allele, to show that BOB1 is required for normal development and meristem function after embryogenesis. To determine whether BOB1 functions as a protein chaperone, we characterized the in vitro activity of BOB1 protein. We also investigated the thermotolerance functions of BOB1 using bob1-3 and used a BOB1:GFP line that is biologically active to document that BOB1 protein is incorporated into heat shock granules (HSGs) at high temperatures. All of these data suggest that BOB1 encodes a novel sHSP with dual functions in development and thermotolerance. To our knowledge, this is the first demonstration of a developmental patterning function for a plant sHSP.
RESULTS
bob1-3 Mutants Exhibit Pleiotropic Postembryonic Developmental Phenotypes
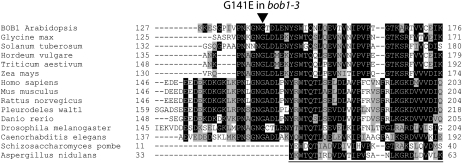
During embryogenesis, BOB1 is required for the establishment of auxin gradients and for patterning the apical domain of the embryo (Jurkuta et al., 2009). A common theme in plant development is that many of the genes involved in embryonic development are subsequently important for vegetative and reproductive development. Expression data generated by the AtGenExpress consortium indicate that BOB1 is expressed in most tissues throughout plant development (Schmid et al., 2005), which suggests that BOB1 has biological functions apart from patterning the apical embryo during embryogenesis. bob1 null alleles are embryo lethal, so we used TILLING (Till et al., 2003) to identify additional alleles in order to investigate the role of BOB1 after embryogenesis. Seven out of 20 ethyl methanesulfonate (EMS) alleles identified were predicted to disrupt BOB1 protein function by SIFT (Ng and Henikoff, 2001). One of these alleles, bob1-3, is a substitution of a Glu for a Gly at the highly conserved amino acid position 141 (G141E), which is just upstream of the NudC domain (Fig. 1). We selected the bob1-3 allele for further characterization because bob1-3 mutants are viable and exhibit pleiotropic developmental phenotypes. The phenotypes observed in homozygous bob1-3 plants can be rescued by either BOB1 or BOB1:GFP transgenes and are exacerbated in bob1-3/bob1-1 trans-heterozygotes (THs), demonstrating that these phenotypes are due to the G141E lesion in bob1-3 as opposed to a tightly linked mutation.
Figure 1.
Alignment of BOB1 homologs. The Gly residue at amino acid position 141 in BOB1 (arrowhead) is invariant among all BOB1 homologs with an N-terminal domain in addition to a NudC domain (the first 30 or 32 amino acids of the NudC domain are underlined).
bob1-3 mutant plants exhibit general growth defects throughout their life cycle. Mutant plants grow more slowly, flower later, and have smaller rosette diameters at flowering compared to wild-type siblings. All of these phenotypes are stronger in bob1 THs than in bob1-3 homozygotes. In addition to shoot defects, bob1-3 mutants have short roots. As in the shoot, the short root phenotype is more severe in TH plants than in bob1-3 homozygotes. When bob1-3 mutants flower they exhibit reduced fertility, which results in short siliques. THs are completely sterile and have very small siliques that do not contain viable seeds. bob1-3 plants also exhibit reduced apical dominance and produce shorter inflorescences with more axillary branches (Table I).
Table I.
BOB1 phenotypes
Rosette diameter was measured in 43-d-old plants. Flowering time was defined as days until floral buds were visible. Root length was measured in 7-d-old seedlings. *, Students t test P value <1 × 10−4 versus the wild type. **, Students t test P value <1 × 10−4 versus both the wild type and bob1-3. ND, Not determined.
| Phenotype | Col-0 | bob1-3/bob1-3 | bob1-3/bob1-1 |
|---|---|---|---|
| Rosette diameter (cm) | 7.7 ± 0.8 (30) | 6.9 ± 0.5 (27)* | 4.4 ± 0.7 (13)** |
| Flowering time (d) | 27.4 ± 2.6 (17) | 32.2 ± 2.6 (17)* | 39.5 ± 1.5 (13)** |
| Root length (cm) | 4.0 ± 0.7 (22) | 2.5 ± 0.3 (17)* | 1.2 ± 0.2 (18)** |
| Plant height (cm) | 45.6 ± 5.3 (20) | 40.1 ± 6.0 (20)* | 14.4 ± 2.8 (23)** |
| Number of axillary inflorescences | 7.1 ± 1.4 (20) | 14.0 ± 2.4 (20)* | ND |
| Silique length (mm) | 13.7±.8 (41) | 11.2 ± 1.2 (44)* | 3.1±.8 (41)** |
bob1-3 leaves are narrower than wild-type leaves and have pronounced serrations on their margins (Fig. 2, A and B). bob1 TH plants also have serrated leaf margins compared to the wild type (Fig. 2C). Because mutations in bob1 result in misexpression of STM, a Knotted-like homeobox (KNOX) gene, during embryogenesis (Jurkuta et al., 2009) and misexpression of KNOX genes can result in the formation of ectopic leaflets on leaf margins (Chuck et al., 1996; Ori et al., 2000), we used reverse transcription (RT)-PCR and KNAT1:GUS and KNAT2:GUS reporters to investigate if the serrations on the margins of bob1-3 leaves are due to ectopic KNOX expression. No ectopic expression of KNAT1, KNAT2, or STM was detected using either approach (Supplemental Fig. S1), demonstrating that the developmental defects observed during embryogenesis and leaf margin patterning are caused by different molecular mechanisms.
Figure 2.
Developmental phenotypes of bob1 mutant plants. bob1-3 mutant plants exhibit pleiotropic developmental phenotypes. Col-0 plants have relatively smooth leaf margins with small serrations (A) compared to bob1-3 (B) or bob1-3;bob1-1 (C) mutant plants. bob1-3 flowers have more sepals, petals, and carpels and fewer stamens than Col-0 flowers. This floral organ phenotype is rescued by a BOB1:GFP transgene (D; n = 33, Student's t test P values versus Col-0 are <0.015* and <4 × 10−7**). Col-0 plants typically have four petals (E), while many bob1-3 flowers have five or more petals (arrowheads, F). Fused stamens were never observed in Col-O plants but are found in 30% of bob1-3 plants (inset, F). Overall plant architecture is changed in bob1 TH plants, which are short and branched. Many of these branches terminate after producing only a small number of flowers (box, G). Terminated branches in this background are due to pin-formed meristems (H). Col-0 (I) and bob1-3 (J) anthers produce pollen, while bob1-3/bob1-1 TH anthers do not (K). TH anthers have patches of stigmatic papillae (arrowheads in K, close-up in L). Bars = 1 cm in A to C and G, 250 μm in H, and 50 μm in I to L.
BOB1 Is Required for Normal Inflorescence and Floral Meristem Function
Arabidopsis inflorescences are produced by the sequential activity of two types of meristems: the inflorescence meristem, which produces cauline leaves and floral meristems; and the floral meristems, which produce floral organs. Floral meristems are normally produced in a spiral phyllotactic pattern on the flanks of the inflorescence meristem. bob1-3/bob1-1 TH plants display defects in inflorescence meristem function. The inflorescence meristem and many of the meristems arising from the axils of cauline leaves stop producing flowers and form pin-like structures reminiscent of pin-formed1 (pin1) and pinoid (pid) mutants (Fig. 2, G and H; Okada et al., 1991; Bennett et al., 1995).
Flower development in Arabidopsis is stereotypical and normally results in flowers with four sepals and petals, six stamens, and two fused carpels (Fig. 2, D and E). This consistency requires correct partitioning of the floral meristem into the four concentric whorls of organs. bob1-3 flowers have aberrant numbers of floral organs, with significantly more sepals, petals, and carpels and fewer stamens (Fig. 2, D and F). In addition to the changes in floral organ numbers, 30% of bob1-3 flowers have at least one set of fused stamen filaments (Fig. 2F, inset). These floral defects are completely rescued by the BOB1:GFP transgene (Fig. 2D). In addition to regulating floral organ number, BOB1 is also required for floral organ identity. bob1 TH plants are completely sterile, and reciprocal crosses and observation of the floral organs suggested that the sterility is due to a defect in pollen production. Wild-type and bob1-3 anthers produce pollen (Fig. 2, I and J), while TH anthers do not (Fig. 2K). Instead, TH anther identity is partially converted into carpel identity and stigmatic papillae grow from the flanks of these anthers (Fig. 2, K and L).
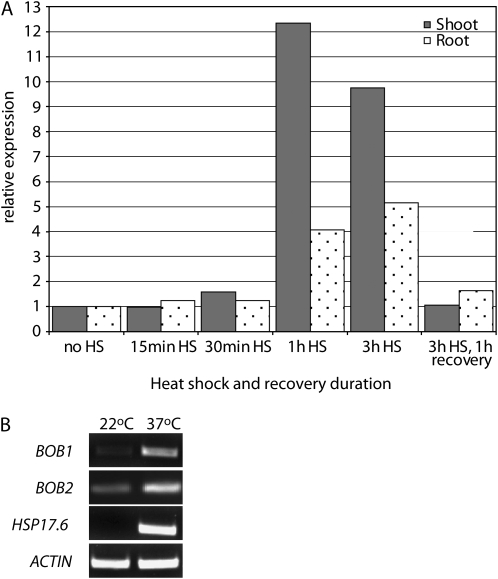
BOB1 Is Induced by High Temperatures
In order to gain further insights into the function of BOB1, we analyzed the AtGenExpress abiotic stress microarray data set, which shows that BOB1 mRNA levels are induced up to 12-fold in the shoots of plants exposed to a 37°C heat shock (Fig. 3A; Kilian et al., 2007). We used RT-PCR to confirm that both BOB1, as well as the duplicated gene BOB2 (At4g27890), are induced by high temperatures (Fig. 3B). Global coexpression analysis performed using ATTED-II (Obayashi et al., 2007) revealed that many BOB1 coexpressed genes are HSPs. Coregulated genes include members of the HSP60, 70, 90, and 100 families as well as canonical sHSPs (Supplemental Fig. S2). This transcriptional profile, coexpression analysis, the structural homology between NudC domains and ACDs (Garcia-Ranea et al., 2002), and the demonstration that the C. elegans NUD-1 protein exhibits chaperone activity in vitro (Faircloth et al., 2009) all suggest that BOB1 encodes a noncanonical sHSP.
Figure 3.
Temperature-dependent BOB1 expression. BOB1 expression in both the shoot and the root increases during a 37°C heat shock (HS) and returns to basal levels after an hour of recovery at 24°C (A; Kilian et al., 2007). Confirmation of this data using RT-PCR shows that BOB1 and BOB2 are induced by elevated temperatures similar to the known sHSP Hsp17.6 (B).
In order to test this hypothesis, we decided to investigate whether BOB1 shares three additional characteristics of plant sHSPs. First, at elevated temperatures, some plant sHSPs have been shown to localize to large cytoplasmic complexes called HSGs (Kirschner et al., 2000; Miroshnichenko et al., 2005). Second, sHSPs exhibit protein chaperone activity in vitro (Lee et al., 1995; Basha et al., 2004). Finally, there is evidence that at least some plant sHSPs are necessary for organismal thermotolerance (Dafny-Yelin et al., 2008).
BOB1 Is Localized to HSGs at High Temperatures
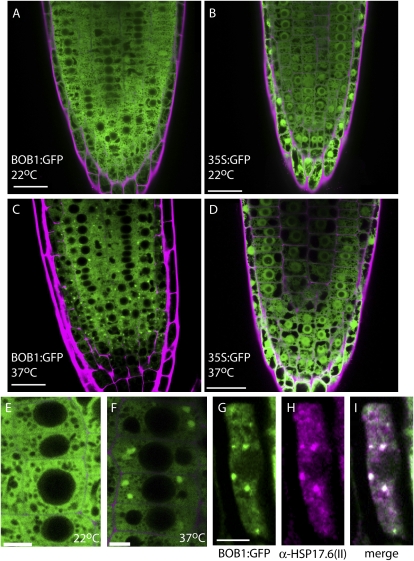
In order to determine the subcellular localization of BOB1, we built a construct containing a translational fusion of a genomic clone of BOB1 with GFP. The genomic clone included the entire 5′ intergenic region (2.4 kb) between BOB1 and At5g53390, the next gene upstream of BOB1. BOB1:GFP transgenic plants developed normally and were crossed to BOB1 null alleles (bob1-1 and bob1-2). Multiple independently transformed lines rescued the embryo lethality of both bob1-1 and bob1-2 and the developmental phenotypes in bob1-3, which demonstrates that BOB1:GFP is a biologically functional allele of BOB1. BOB1:GFP is expressed in all seedling tissues (Supplemental Fig. S3) with highest expression levels at the root tip. In roots grown at 22°C, BOB1:GFP is evenly distributed throughout the cytoplasm and can be detected at low levels in the nucleus, although it appears to be excluded from the nucleolus (Fig. 4, A and E). To determine if BOB1 is incorporated into HSGs similar to other sHSPs, we exposed BOB1:GFP plants to a heat shock. After 1 h at 37°C, BOB1:GFP is incorporated into large (1.5 ± 0.4 μm, n = 31) cytoplasmic granules (Fig. 4, C and F). A control GFP line exhibited an evenly distributed GFP signal in the cytoplasm and nucleus at both 22°C and at 37°C (Fig. 4, B and D), demonstrating that the BOB1:GFP localization is not an artifact caused by the presence of GFP in the BOB1:GFP protein. HSG formation was observed at heat shock temperatures as low as 34°C, and granules could be detected for up to 8 h after plants were returned to 22°C (data not shown).
Figure 4.
Temperature-dependent BOB1 subcellular localization. Both BOB1:GFP (A and E) and 35S:GFP (B) are distributed throughout the cytoplasm of root cells at 22°C. BOB1:GFP forms granules at 37°C (C and F), while 35S:GFP localization at 37°C is unchanged (D). BOB1:GFP (G) and an α-HSP17.6 antibody (H) were used to show that these proteins colocalize in HSGs (white in I). Cell walls stained with propidium iodide are shown in magenta in A to F. Bars = 25 μm in A to D and 5 μm in E to I.
Although Arabidopsis HSGs have not been described beyond their identification in electron micrographs, in tobacco (Nicotiana tabacum) protoplasts, HSGs have been shown to consist of protein complexes containing multiple HSPs (Nover et al., 1989; Kirschner et al., 2000). In order to determine if BOB1:GFP granules also contained other sHSPs, we used antibodies raised against Hsp17.6(II) (At5g12020) and performed colocalizations with BOB1:GFP. BOB1:GFP colocalizes with Hsp17.6(II) in HSGs (Fig. 4, G–I), demonstrating that BOB1-containing HSGs also contain canonical sHSPs and do not represent distinct heat-induced structures.
BOB1 Is Required for Organismal Thermotolerance
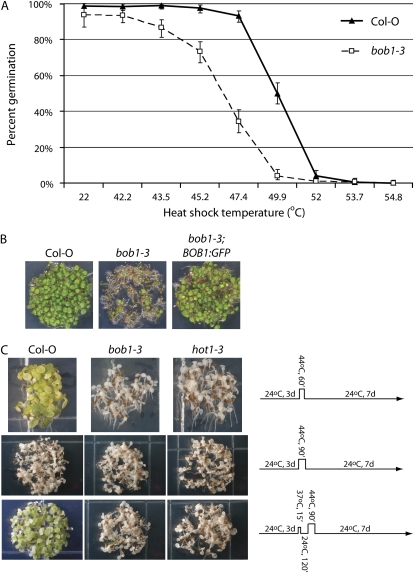
We used bob1-3 to determine whether BOB1 is required for plant survival in response to high temperatures. Germinating plants are often exposed to high soil temperatures, so we investigated if bob1-3 mutant seeds exposed to high temperatures during germination exhibit reduced thermotolerance compared to wild-type seeds. Stratified seeds were incubated for 1 h at temperatures ranging from 42°C to 55°C. The percentage of bob1-3 seeds that germinated at temperatures above 43.5°C was significantly lower than that of wild-type seeds (Fig. 5A). This thermotolerance defect could be rescued by a BOB1:GFP transgene (Fig. 5B), confirming that the phenotype is due to a reduction in BOB1 function. Thermotolerance without an acclimation period at sublethal but elevated temperatures is known as basal thermotolerance. Basal thermotolerance has been shown to decrease during seedling development (Queitsch et al., 2000), so to determine if BOB1 is also required later in development for basal or acquired thermotolerance, we assayed thermotolerance in older seedlings. Three-day-old wild-type, bob1-3, and hot1-3 (an Hsp101 mutant with known thermotolerance defects; Hong and Vierling, 2001) seedlings were exposed to 45°C temperatures for either 60 or 90 min with or without a 15-min acclimation period at 37°C. Wild-type plants can survive a 60-min exposure to 45°C temperatures without acclimation or 90 min at 45°C with previous acclimation (and associated HSP induction) at 37°C. Neither bob1-3 nor hot1-3 mutant seedlings survived these temperature regimens, demonstrating that like Hsp101, normal BOB1 function is required for both basal and acquired thermotolerance after germination (Fig. 5C).
Figure 5.
Thermotolerance of bob1-3 and Col-0 plants. Col-0 and bob1-3 seeds were stratified for 2 d and then exposed to a 1-h heat shock before plating. Seedling germination was scored 7 d after plating (A). The thermotolerance defect with a 46.4°C heat shock during germination can be rescued by the introduction of a BOB1:GFP transgene (B). Survival of 3-d-old Col-0, bob1-3, and hot1-3 seedlings after a 60- or 90-min 44°C heat shock with or without a 15-min 37°C acclimation period was observed 7 d after the heat shock (C). Error bars in A represent 99% binomial confidence intervals.
BOB1 Prevents the Aggregation of Heat-Denatured Proteins in Vitro
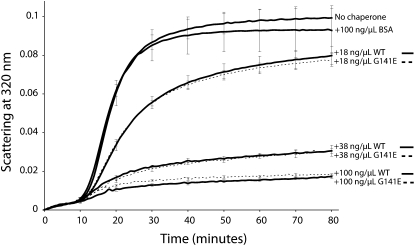
Most plant sHSPs containing ACDs have been shown to prevent aggregation and assist in refolding of heat-denatured model substrates (Lee et al., 1997; Siddique et al., 2008). Similar activity has been demonstrated for NUD-1, the C. elegans BOB1 homolog (Faircloth et al., 2009). In order to determine if BOB1 also possesses this activity, we expressed and purified BOB1 protein and used it in an aggregation assay with the model substrate malate dehydrogenase (MDH) (Lee and Vierling, 1998). Wild-type BOB1 protein was able to prevent the aggregation of MDH nearly completely at 100 ng/μL and to a lesser degree at lower concentrations (Fig. 6). BOB1 protein (100 ng/μL) represents an 8-fold molar excess of BOB1 relative to MDH, suggesting that similar to other sHSPs, BOB1 may act in a multimeric complex. The simplest explanation of the reduced thermotolerance and developmental phenotypes observed in bob1-3 plants is that mutant BOB1 protein has a reduced chaperone activity. In order to test this hypothesis, we repeated the aggregation assays using recombinant G141E (bob1-3) protein. G141E protein had a nearly identical ability to prevent the aggregation of MDH (Fig. 6), demonstrating that the phenotypes observed in bob1-3 are not due to a change in absolute chaperone activity but must be caused by changes in other activities of the protein.
Figure 6.
BOB1 prevents thermal aggregation of MDH. Thermal aggregation of 300 nm MDH at 45°C was monitored every minute for 80 min by measuring scattering at 320 nm in the presence or absence of wild-type (WT) and G141E (bob1-3) His-tagged recombinant BOB1 protein. Bovine serum albumin (BSA) at a concentration equal to the highest BOB1 concentration by weight serves as a negative control. The average of at least three replicates is shown for each condition. Error bars shown at 10-min intervals represent sd.
DISCUSSION
We previously demonstrated that BOB1 is required for partitioning the apical domain of Arabidopsis embryos during embryogenesis. In bob1 mutants, the central meristematic domain is expanded at the expense of the formation of the lateral cotyledons (Jurkuta et al., 2009). In order to gain insights into the mechanistic basis of BOB1-mediated patterning, we took advantage of a partial loss-of-function allele (bob1-3) as well as a BOB1:GFP fusion and an in vitro chaperone assay to establish that BOB1 functions as a sHSP. We also demonstrate that BOB1 is required for a range of postembryonic developmental processes. This sets BOB1 apart from previously characterized plant sHSPs, none of which appear to be required for developmental patterning. Our data also suggest that other NudC domain-containing proteins homologous to BOB1 function as protein chaperones and define a noncanonical family of sHSPs.
Haslbeck et al. (2005) defined several characteristics shared among sHSPs, including induction by stress conditions, ATP-independent protein chaperone activity, a small size, and the presence of an ACD. BOB1 meets all of these criteria. BOB1 is induced by high temperatures, has chaperone activity in vitro, encodes a protein with a predicted mass of 34.5 kD, and contains an α-crystallin like NudC domain. Several results from studies of BOB1 homologs in other systems support the idea that proteins with NudC domains have evolutionarily conserved functions as molecular chaperones. First, NUD-1 in C. elegans exhibits chaperone activity in vitro (Faircloth et al., 2009). Second, in mammalian cells, RNA interference (RNAi) depletion of a NudC-like gene leads to the aggregation and proteasome-mediated degradation of the dynein intermediate chain (Zhou et al., 2006). Both the aggregation and the degradation of presumably misfolded protein are consistent with a chaperone function for NudC proteins. Finally an A. nidulans screen for nuclear migration mutants identified NudC as well as the light and heavy chains of dynein (nudA and nudG) and NudF/Lis1 as being essential for normal nuclear movement (Osmani et al., 1990; Xiang et al., 1994, 1999). Subsequently, NudC proteins were shown to interact with NudF/Lis1, and in A. nidulans, NudC is required to maintain NudF protein levels (Xiang et al., 1995). A role for NudC functioning as a protein chaperone that prevents the degradation of misfolded NudF/Lis1 is consistent with these data.
In addition to the characteristics described above, all sHSPs analyzed to date form large oligomeric complexes in which the subunits rapidly exchange. Subunit exchange and dynamic changes in oligomeric structure seem to be important for sHSP function (Haslbeck et al., 2005). Similar to canonical sHSPs, NUD-1 from C. elegans forms large oligomers (Faircloth et al., 2009), supporting the hypothesis that NudC proteins oligomerize like canonical sHSPs.
A comparison between BOB1 and homologous genes in animal and fungal systems illustrates that an association with microtubules is not an evolutionarily conserved property of NudC proteins. NudC homologs are often described as microtubule-associated proteins in the literature (Lin et al., 2004; Faircloth et al., 2009). They are required for normal microtubule dynamics and have been shown to be associated with dynein complexes, microtubules, and kinetochores in animal and fungal systems (Aumais et al., 2001, 2003; Zhang et al., 2002; Pan et al., 2005; Nishino et al., 2006; Zhou et al., 2006). However, plants do not contains dyneins (Lawrence et al., 2001), and there is no evidence from our localization data that BOB1 is microtubule associated. It seems likely that reported microtubule associations are mediated by interactions between NudCs and dyneins, perhaps mediated by the N termini of these genes, which are relatively conserved among, but not between, animals and plants.
BOB1 protein is expressed throughout growth and development, and this differentiates BOB1 from most other plant sHSPs that are only expressed at elevated temperatures. However, at high temperatures, BOB1 exhibits characteristics similar to canonical sHSPs, such as incorporation into HSG complexes. In order to show that BOB1 HSGs do not define a novel class of granules, we demonstrated that BOB1 colocalizes with Hsp17.6(II). The function(s) and the composition of HSGs (apart from sHSPs, Hsp70s, and HsfA2) is not well understood (Nover et al., 1983; Scharf et al., 1998; Siddique et al., 2008). HSG formation may be required for thermotolerance as immunomodulation of sHSPs results in both thermotolerance defects and an inhibition of HSG formation (Miroshnichenko et al., 2005). To our knowledge, BOB1:GFP is the first reporter described for visualizing HSGs in living plant cells and will be useful for investigating these dynamics.
We used the bob1-3 allele to demonstrate that BOB1 is required not only for thermotolerance but also for normal development. bob1-3 is likely to be a partial loss-of-function allele since all of the phenotypes we measured are more severe when bob1-3 is combined with a null allele (bob1-1 or bob1-2) as a transheterozygote (Muller, 1932). Recombinant protein containing the G141E mutation found in bob1-3 does not exhibit a reduction in its ability to prevent the aggregation of MDH in vitro. This suggests that the defects observed in bob1-3 mutants are unlikely to be due to a reduction in chaperone activity. These results are similar to results from a genetic screen using the single Synechocystis sHSP. A subset of the mutations identified in the screen were clustered in the N terminus and disrupted thermotolerance but not in vitro activity. This demonstrates that in vitro chaperone assays do not capture all aspects of sHSP chaperone function (Giese et al., 2005). Both the α-crystallin and N-terminal domains of sHSPs have been shown to be required for the specificity of interactions with chaperone substrates (Basha et al., 2006). One explanation of the discrepancy between the phenotypes seen in bob1-3 plants and the unchanged MDH chaperone activity of G141E protein is that G141 may not be essential for interactions with the model substrate MDH in vitro even though it is required for interactions with physiologically relevant substrates in planta. Alternatively, the mutation may disrupt interactions with one of the ATP-dependent chaperone systems known to cooperate with sHSPs in refolding bound substrates (Lee and Vierling, 2000; Lee et al., 2005).
bob1 mutants have defects in general growth of the shoot and the root as well as more specific developmental patterning defects. These include narrow serrated leaves, changes in floral organ numbers and identity, and pin-formed inflorescences. These phenotypes could be due to a lack of chaperone activity or due to disruption of an additional uncharacterized activity. If the phenotypes observed in bob1-3 plants grown at normal temperatures are due to decreased BOB1 chaperone activity, it suggests that there is a group of developmentally important proteins that require BOB1 for proper folding during or after protein synthesis in the absence of temperature stress. This would place BOB1 in a small group of protein chaperones with dual roles in de novo protein folding and recovery from denaturing stress (Albanese et al., 2006).
Compared to normal flowers, bob1-3 flowers have increased numbers of sepals, petals, and carpels; however, the average number of organs per flower is not significantly different (16.2 in bob1-3 versus 15.9 in the wild type). This suggests that the floral defect is due to a change in meristematic patterning similar to the phenotype observed in bob1 null mutants (Jurkuta et al., 2009), although we have not ruled out other possibilities, including changes in meristem size. In addition to changes in floral organ number, a third of bob1 flowers have fused anthers. This phenotype is also observed in pid and cuc1;cuc2 mutants (Bennett et al., 1995; Aida et al., 1997). PID is a kinase that regulates the subcellular localization of PIN auxin transporters by directly phosphorylating PIN proteins (Michniewicz et al., 2007; Skirpan et al., 2009). CUC1 and CUC2 are partially redundant transcription factors that regulate organ fusion, and the expression of the CUC genes is regulated by auxin (Aida et al., 2002). bob1 THs develop pin-formed inflorescences. Pin-formed inflorescence phenotypes are observed in a limited set of mutants, most of which are involved in auxin biosynthesis, transport, or signaling (Okada et al., 1991; Bennett et al., 1995; Cheng et al., 2006). These data imply an interaction between BOB1 and auxin-mediated developmental processes and are consistent with the observation that auxin gradients are disrupted in bob1 null mutants (Jurkuta et al., 2009).
Gray et al. (1998) showed that auxin levels as well as the expression of auxin-regulated genes increase dramatically when plants are grown at 29°C as opposed to 20°C. Given auxin's ubiquitous roles in plant development, it is surprising that apart from accelerated growth the only developmental phenotype reported at high temperatures was an auxin-dependent increase in hypocotyl elongation. This suggests that auxin-dependent developmental events are highly buffered against temperature changes. Since BOB1 encodes an sHSP required for establishing auxin maxima during embryogenesis and has phenotypes that are likely to be auxin mediated during vegetative growth, it will be interesting to investigate if BOB1 plays a role in maintaining auxin homeostasis at elevated temperatures.
BOB1 is unusual for a plant sHSP because unlike most plant sHSPs, which are only expressed in response to heat shock, BOB1 is expressed at normal temperatures. Other developmentally regulated sHSPs are expressed during seed maturation and petal development, and sHSP ESTs have been identified in leaves (Wehmeyer and Vierling, 2000; Scharf et al., 2001; Dafny-Yelin et al., 2008). However, sHSP function in these tissues is not well defined. A mechanistic link between BOB1 activity and development probably involves other chaperones since sHSPs function in chaperone networks with other HSPs (Scharf et al., 2001; Haslbeck et al., 2005; Lee et al., 2005). Among the other plant HSP families, only Hsp90s have been shown to play a role in developmental patterning (Jenks and Hasegawa, 2005).
Single mutants of cytoplasmic Hsp90s do not exhibit developmental phenotypes; however, pharmacological inhibition of multiple Hsp90s in plants using the drug geldanamycin results in a wide range of developmental defects (Queitsch et al., 2002). Similarly, inhibition of the cytosolic Hsp90s using RNAi results in stem fasciation and a range of phenotypes similar to those observed in geldanamycin-treated plants (Sangster et al., 2007). Stem fasciation in other mutants is caused by enlarged meristems, suggesting that cytoplasmic Hsp90s are required for normal meristem function (Laufs et al., 1998; Taguchi-Shiobara et al., 2001), similar to BOB1. The BOB1 and Hsp90 examples illustrate that not all plant HSPs function solely in heat stress responses. In yeast, it has been demonstrated that a subset of chaperones are involved in protein folding associated with protein synthesis and that many of these chaperones do not play a role in stress responses (Albanese et al., 2006).
Hsp90s are ATP-dependent protein chaperones that rely on a variety of cochaperones for their activity (Terasawa et al., 2005). In plants, several potential Hsp90 cochaperones have developmental phenotypes, although the role of Hsp90 in these phenotypes has not been established (Sangster and Queitsch, 2005). Using a proteomic approach, Te et al. (2007) identified NudC as a component of the human Hsp90 interactome. Additionally, p23 proteins, which share predicted structural homology with NudC proteins (Garcia-Ranea et al., 2002), are known Hsp90 cochaperones (Felts and Toft, 2003). If NudC/Hsp90 interactions are conserved between Arabidopsis and humans, then the similar phenotypes observed in Hsp90 RNAi lines and bob1 mutants may be caused by disruption of the function of a shared chaperone substrate.
Our results demonstrate that BOB1 is an sHSP with developmental functions at basal temperatures as well as thermotolerance functions at elevated temperatures. Our data in conjunction with previous studies suggest that homologous genes containing NudC domains function as molecular chaperones, and possibly as HSPs, in other organisms. The generation of a functional BOB1:GFP reporter provides a unique opportunity to study the subcellular dynamics of HSGs, which have not been characterized in a live cell system, and the isolation of a partial loss-of-function allele will allow the further characterization of BOB1 functions during plant development. It will be informative to use genetic and biochemical approaches to identify targets of BOB1 chaperone activity with the goal of understanding how this unique sHSP regulates growth and development in plants.
MATERIALS AND METHODS
Plant Growth
Plants grown on soil were grown under standard long-day greenhouse conditions with supplemental lighting. Plants grown on plates were grown on 0.5× Murashige and Skoog (MS) media at 22°C under continuous light in Percival E-30B growth chambers (Percival Scientific).
Plant Stocks
All plants are in a Columbia (Col) background. bob1-1 (EMS) and bob1-2 (T-DNA insertion Salk_001125) are null alleles described by Jurkuta et al. (2009). bob1-1 and bob1-3 were generated using EMS and were backcrossed to Col-0 at least six times before use in these studies.
HSG Visualization
A BOB1 genomic clone, including 2,413 bp 5′ of the start codon (up to the end of the previous gene, At5g53390), was amplified using primers BOB1_attB_F (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCGTCTTCTTGTCTTCACCCATTC-3′) and BOB1_attB_R (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTGTTAAACTTTGCATTTGAGAAGTCCA-3′) to remove the stop codon. The PCR product was recombined into pDONR/Zeo (Invitrogen) and subsequently recombined into pGWB4 (Nakagawa et al., 2007) to create a C-terminal GFP fusion (BOB1:GFP). This construct was transformed into both Col-0 and bob1-3 plants using Agrobacterium tumefaciens strain GV3101 (Clough and Bent, 1998) and subsequently crossed to bob1-1 and bob1-2 to demonstrate the functionality of the transgene. Confocal imaging was performed using a Leica SP5 AOBS confocal microscope (Leica Microsystems). Plants were mounted in 10 μg/mL propidium iodide to allow visualization of cell walls. Immunolocalization of Hsp17.6 using α-Hsp17.6 antibodies (a gift from Elizabeth Vierling) was performed according to the protocol detailed in Sauer et al. (2006), omitting optional steps 4 to 12.
Thermotolerance Experiments
For germination assays, seeds were sterilized and then stratified for 2 to 3 d at 4°C. Stratified seeds were heat shocked for 1 h in thin wall tubes using a PTC-200 gradient PCR machine (MJ Research) and subsequently transferred to 0.5× MS plates for 7 d, at which point germination was scored. Seedling assays were performed by submerging sealed plates in water baths as described by Charng et al. (2006).
Phenotypic Characterization
Plant images were acquired using a Canon Powershot G9 digital camera (Canon USA) and processed using Photoshop (Adobe). Rosette diameter and root lengths were measured from digital images using ImageJ (http://rsbweb.nih.gov/ij/).
RT-PCR
RNA from seedlings grown in 0.5× MS liquid media was prepared using RNeasy plant mini kits (Qiagen), quantified using a Nanodrop ND-1000 (Thermo Scientific), and reverse transcribed using the ProtoScript II RT-PCR kit (NEB). The following primers were used for 25-cycle PCR reactions: ACTIN-F, 5′-GAAGAACTATGAATTACCCGATGGGC-3′; ACTIN-R, 5′-CCCGGGTTAGAAACATTTTCTGTGAACG-3′; HSP17.6-F, 5′-CGATCCGTTCTCGCTGGATG-3′; HSP17.6-R, 5′-GGGCACGGTAACCGACAACA-3′; BOB2-F, 5′-GGTCCCATCGTTCCTAACAAAG-3′; BOB2-R, 5′-TTGATCAAACATCATCTTCTCAACG-3′; BOB1-F, 5′-TGGGACTAAAGCACGGACTGTTG-3′; and BOB1-R, 5′- AGTTAAACTTTGCATTTGAGAAGTCCA-3′.
In Vitro Aggregation Assays
In order to add an N-terminal His tag to BOB1, a BOB1 cDNA was cloned into pET15b (Novagen). This construct (pAJ1) was transformed into Escherichia coli strain BL21(DE3). Site-directed mutagenesis was used to introduce the G141E mutation into pAJ1, resulting in pAJ2. Expressed protein was purified using Ni-NTA resin (Qiagen). Protein purity was determined using SDS-PAGE, and protein concentration was determined using calculated extinction coefficients (Pace et al., 1995) and the Bio-Rad protein reagent. Aggregation assays were performed as described (Lee et al., 1995, 1997) using porcine heart MDH (Sigma-Aldrich) and methacrylate cuvettes in a Cary 50 spectrophotometer equipped with a temperature-controlled 18-cuvette holder (Varian).
Scanning Electron Microscopy
Fresh tissue was imaged using low vacuum mode on an FEI Quanta 200 scanning electron microscope equipped with a cooled stage.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analysis of KNOX gene expression in bob1-3.
Supplemental Figure S2. List of BOB1 coregulated genes.
Supplemental Figure S3. BOB1:GFP expression at normal temperatures.
Supplementary Material
Acknowledgments
We thank Elizabeth Vierling for her generous gift of α-Hsp antibodies and hot1-3 seeds. Greenhouse care was provided by Bill Pinder. We also thank Jennifer Pfluger, Damon Lisch, Pablo Jenik, Sarah Hake, and Kathy Barton for helpful discussions and comments on this manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Nicholas J. Kaplinsky (nkaplin1@swarthmore.edu).
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Vernoux T, Furutani M, Traas J, Tasaka M (2002) Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development 129 3965–3974 [DOI] [PubMed] [Google Scholar]
- Albanese V, Yam AY, Baughman J, Parnot C, Frydman J (2006) Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell 124 75–88 [DOI] [PubMed] [Google Scholar]
- Aumais JP, Tunstead JR, McNeil RS, Schaar BT, McConnell SK, Lin SH, Clark GD, Yu-Lee LY (2001) NudC associates with Lis1 and the dynein motor at the leading pole of neurons. J Neurosci 21 RC187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumais JP, Williams SN, Luo W, Nishino M, Caldwell KA, Caldwell GA, Lin SH, Yu-Lee LY (2003) Role for NudC, a dynein-associated nuclear movement protein, in mitosis and cytokinesis. J Cell Sci 116 1991–2003 [DOI] [PubMed] [Google Scholar]
- Baniwal SK, Bharti K, Chan KY, Fauth M, Ganguli A, Kotak S, Mishra SK, Nover L, Port M, Scharf KD, et al (2004) Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J Biosci 29 471–487 [DOI] [PubMed] [Google Scholar]
- Basha E, Friedrich KL, Vierling E (2006) The N-terminal arm of small heat shock proteins is important for both chaperone activity and substrate specificity. J Biol Chem 281 39943–39952 [DOI] [PubMed] [Google Scholar]
- Basha E, Lee GJ, Demeler B, Vierling E (2004) Chaperone activity of cytosolic small heat shock proteins from wheat. Eur J Biochem 271 1426–1436 [DOI] [PubMed] [Google Scholar]
- Bennett SRM, Alvarez J, Bossinger G, Smyth DR (1995) Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J 8 505–520 [Google Scholar]
- Charng YY, Liu HC, Liu NY, Hsu FC, Ko SS (2006) Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol 140 1297–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Lincoln C, Hake S (1996) KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8 1277–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Dafny-Yelin M, Tzfira T, Vainstein A, Adam Z (2008) Non-redundant functions of sHSP-CIs in acquired thermotolerance and their role in early seed development in Arabidopsis. Plant Mol Biol 67 363–373 [DOI] [PubMed] [Google Scholar]
- Faircloth L, Churchill P, Caldwell G, Caldwell K (2009) The microtubule-associated protein, NUD-1, exhibits chaperone activity in vitro. Cell Stress Chaperones 14 95–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felts SJ, Toft DO (2003) p23, a simple protein with complex activities. Cell Stress Chaperones 8 108–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ranea JA, Mirey G, Camonis J, Valencia A (2002) p23 and HSP20/alpha-crystallin proteins define a conserved sequence domain present in other eukaryotic protein families. FEBS Lett 529 162–167 [DOI] [PubMed] [Google Scholar]
- Giese KC, Basha E, Catague BY, Vierling E (2005) Evidence for an essential function of the N terminus of a small heat shock protein in vivo, independent of in vitro chaperone activity. Proc Natl Acad Sci USA 102 18896–18901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Franzmann T, Weinfurtner D, Buchner J (2005) Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol Biol 12 842–846 [DOI] [PubMed] [Google Scholar]
- Hong SW, Vierling E (2001) Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. Plant J 27 25–35 [DOI] [PubMed] [Google Scholar]
- Jenks M, Hasegawa P (2005) Plant Abiotic Stress. Wiley-Blackwell, Oxford
- Jurkuta RJ, Kaplinsky NJ, Spindel JE, Barton MK (2009) Partitioning the apical domain of the Arabidopsis embryo requires the BOBBER1 NudC domain protein. Plant Cell 21 1957–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D'Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50 347–363 [DOI] [PubMed] [Google Scholar]
- Kirschner M, Winkelhaus S, Thierfelder JM, Nover L (2000) Transient expression and heat-stress-induced co-aggregation of endogenous and heterologous small heat-stress proteins in tobacco protoplasts. Plant J 24 397–411 [DOI] [PubMed] [Google Scholar]
- Laufs P, Dockx J, Kronenberger J, Traas J (1998) MGOUN1 and MGOUN2: two genes required for primordium initiation at the shoot apical and floral meristems in Arabidopsis thaliana. Development 125 1253–1260 [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Morris NR, Meagher RB, Dawe RK (2001) Dyneins have run their course in plant lineage. Traffic 2 362–363 [DOI] [PubMed] [Google Scholar]
- Lee GJ, Pokala N, Vierling E (1995) Structure and in vitro molecular chaperone activity of cytosolic small heat shock proteins from pea. J Biol Chem 270 10432–10438 [DOI] [PubMed] [Google Scholar]
- Lee GJ, Roseman AM, Saibil HR, Vierling E (1997) A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J 16 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GJ, Vierling E (1998) Expression, purification, and molecular chaperone activity of plant recombinant small heat shock proteins. Methods Enzymol 290 350–365 [DOI] [PubMed] [Google Scholar]
- Lee GJ, Vierling E (2000) A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol 122 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee U, Wie C, Escobar M, Williams B, Hong S-W, Vierling E (2005) Genetic analysis reveals domain interactions of Arabidopsis Hsp100/ClpB and cooperation with the small heat shock protein chaperone system. Plant Cell 17 559–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SH, Nishino M, Luo W, Aumais JP, Galfione M, Kuang J, Yu-Lee LY (2004) Inhibition of prostate tumor growth by overexpression of NudC, a microtubule motor-associated protein. Oncogene 23 2499–2506 [DOI] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130 1044–1056 [DOI] [PubMed] [Google Scholar]
- Miroshnichenko S, Tripp J, Nieden U, Neumann D, Conrad U, Manteuffel R (2005) Immunomodulation of function of small heat shock proteins prevents their assembly into heat stress granules and results in cell death at sublethal temperatures. Plant J 41 269–281 [DOI] [PubMed] [Google Scholar]
- Muller H (1932) Further studies on the nature and causes of gene mutations. In DF Jones, ed, Proceedings of the Sixth International Congress of Genetics, Vol 1. Brooklyn Botanic Garden, Brooklyn, NY, pp 213–255
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104 34–41 [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S (2001) Predicting deleterious amino acid substitutions. Genome Res 11 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino M, Kurasawa Y, Evans R, Lin SH, Brinkley BR, Yu-Lee LY (2006) NudC is required for Plk1 targeting to the kinetochore and chromosome congression. Curr Biol 16 1414–1421 [DOI] [PubMed] [Google Scholar]
- Nover L, Neumann D, Scharf KD (1989) Heat Shock and Other Stress Response Systems of Plants, Vol 16. Springer-Verlag, Berlin [PubMed]
- Nover L, Scharf KD, Neumann D (1983) Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol Cell Biol 3 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H (2007) ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res 35 D863–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y (1991) Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S (2000) Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127 5523–5532 [DOI] [PubMed] [Google Scholar]
- Osmani AH, Osmani SA, Morris NR (1990) The molecular cloning and identification of a gene product specifically required for nuclear movement in Aspergillus nidulans. J Cell Biol 111 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace CN, Vajdos F, Fee L, Grimsley G, Gray T (1995) How to measure and predict the molar absorption coefficient of a protein. Protein Sci 4 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan RM, Yang Y, Wei MX, Yu XB, Ge YC, Xu P (2005) A microtubule associated protein (hNUDC) binds to the extracellular domain of thrombopoietin receptor (Mpl). J Cell Biochem 96 741–750 [DOI] [PubMed] [Google Scholar]
- Queitsch C, Hong S-W, Vierling E, Lindquist S (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S (2002) Hsp90 as a capacitor of phenotypic variation. Nature 417 618–624 [DOI] [PubMed] [Google Scholar]
- Sangster TA, Bahrami A, Wilczek A, Watanabe E, Schellenberg K, McLellan C, Kelley A, Kong SW, Queitsch C, Lindquist S (2007) Phenotypic diversity and altered environmental plasticity in Arabidopsis thaliana with reduced Hsp90 levels. PLoS One 2 e648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster TA, Queitsch C (2005) The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Curr Opin Plant Biol 8 86–92 [DOI] [PubMed] [Google Scholar]
- Sauer M, Paciorek T, Benkova E, Friml J (2006) Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat Protocols 1 98–103 [DOI] [PubMed] [Google Scholar]
- Scharf KD, Heider H, Hohfeld I, Lyck R, Schmidt E, Nover L (1998) The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol Cell Biol 18 2240–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Siddique M, Vierling E (2001) The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (Acd proteins). Cell Stress Chaperones 6 225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Siddique M, Gernhard S, von Koskull-Doring P, Vierling E, Scharf KD (2008) The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones 13 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirpan A, Culler AH, Gallavotti A, Jackson D, Cohen JD, McSteen P (2009) BARREN INFLORESCENCE2 interaction with ZmPIN1a suggests a role in auxin transport during maize inflorescence development. Plant Cell Physiol 50 652–657 [DOI] [PubMed] [Google Scholar]
- Swindell WR, Huebner M, Weber AP (2007) Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D (2001) The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev 15 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz L, Zeiger E (2006) Plant Physiology, Ed 4. Sinauer Associates, Sunderland, MA
- Te J, Jia L, Rogers J, Miller A, Hartson SD (2007) Novel subunits of the mammalian Hsp90 signal transduction chaperone. J Proteome Res 6 1963–1973 [DOI] [PubMed] [Google Scholar]
- Terasawa K, Minami M, Minami Y (2005) Constantly updated knowledge of Hsp90. J Biochem 137 443–447 [DOI] [PubMed] [Google Scholar]
- Till BJ, Reynolds SH, Greene EA, Codomo CA, Enns LC, Johnson JE, Burtner C, Odden AR, Young K, Taylor NE, et al (2003) Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res 13 524–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehmeyer N, Vierling E (2000) The expression of small heat shock proteins in seeds responds to discrete developmental signals and suggests a general protective role in desiccation tolerance. Plant Physiol 122 1099–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Beckwith SM, Morris NR (1994) Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc Natl Acad Sci USA 91 2100–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Osmani AH, Osmani SA, Xin M, Morris NR (1995) NudF, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol Biol Cell 6 297–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Zuo W, Efimov VP, Morris NR (1999) Isolation of a new set of Aspergillus nidulans mutants defective in nuclear migration. Curr Genet 35 626–630 [DOI] [PubMed] [Google Scholar]
- Zhang MY, Huang NN, Clawson GA, Osmani SA, Pan W, Xin P, Razzaque MS, Miller BA (2002) Involvement of the fungal nuclear migration gene nudC human homolog in cell proliferation and mitotic spindle formation. Exp Cell Res 273 73–84 [DOI] [PubMed] [Google Scholar]
- Zhou T, Zimmerman W, Liu X, Erikson RL (2006) A mammalian NudC-like protein essential for dynein stability and cell viability. Proc Natl Acad Sci USA 103 9039–9044 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.