Abstract
Nitrate regulatory mutants (nrg) of Arabidopsis (Arabidopsis thaliana) were sought using a genetic screen that employed a nitrate-inducible promoter fused to the yellow fluorescent protein marker gene YFP. A mutation was identified that impaired nitrate induction, and it was localized to the nitrate regulatory gene NLP7, demonstrating the validity of this screen. A second, independent mutation (nrg1) mapped to a region containing the NRT1.1 (CHL1) nitrate transporter gene on chromosome 1. Sequence analysis of NRT1.1 in the mutant revealed a nonsense mutation that truncated the NRT1.1 protein at amino acid 301. The nrg1 mutation disrupted nitrate regulation of several endogenous genes as induction of three nitrate-responsive genes (NIA1, NiR, and NRT2.1) was dramatically reduced in roots of the mutant after 2-h treatment using nitrate concentrations from 0.25 to 20 mm. Another nrt1.1 mutant (deletion mutant chl1-5) showed a similar phenotype. The loss of nitrate induction in the two nrt1.1 mutants (nrg1 and chl1-5) was not explained by reduced nitrate uptake and was reversed by nitrogen deprivation. Microarray analysis showed that nitrate induction of 111 genes was reduced and of three genes increased 2-fold or more in the nrg1 mutant. Genes involved in nitrate assimilation, energy metabolism, and pentose-phosphate pathway were most affected. These results strongly support the model that NRT1.1 acts as a nitrate regulator or sensor in Arabidopsis.
Inorganic nitrogen is a vital nutrient for plants. Plants take up and assimilate both nitrate and ammonium with nitrate being the predominant form in most agricultural soils (Crawford and Glass, 1998). Nitrate is taken up by roots then transported into cells via transporters from the NRT1 and NRT2 family of nitrate transporters (Forde, 2000; Tsay et al., 2007). Once inside the cell, nitrate is reduced to nitrite by nitrate reductase (NIA) then to ammonium by nitrite reductase (NiR). Ammonium is then assimilated into amino acids.
In addition to serving as a nutrient, nitrate also acts as a signal. When plants are first exposed to nitrate, genes in the nitrate assimilation pathway (NRT, NIA, and NiR) are rapidly induced (Wang et al., 2007). Other genes, which are required for reprogramming carbon metabolism and providing chemical energy for reduction and assimilation, are also induced (Stitt, 1999; Wang et al., 2000, 2003, 2004; Stitt et al., 2002; Scheible et al., 2004; Fritz et al., 2006). Transcriptome analyses have shown that over 1,500 genes are induced or repressed by nitrate within 20 to 180 min of treatment (Wang et al., 2003, 2004, 2007; Scheible et al., 2004; Gutierrez et al., 2007a). Longer-term responses to nitrate include changes in root growth, development and architecture, in root-to-shoot ratios, and in germination rates (Forde, 2002; Alboresi et al., 2005; Filleur et al., 2005; Walch-Liu et al., 2005, 2006; Forde and Walch-Liu, 2009).
The regulatory mechanisms and genes responsible for nitrogen responses in plants have been investigated using genetics (for early examples, see Leydecker et al., 2000; Zhang and Forde, 2000) and systems analysis (Gutierrez et al., 2005, 2007b). The ANR1 MADS-box transcription factor, which controls lateral root branching in response to nitrate and is induced by nitrogen deprivation, was the first to be identified (Zhang and Forde, 1998; Gan et al., 2005). A Dof transcription factor was discovered that improves nitrogen use efficiency at low nitrogen (Yanagisawa et al., 2004). More recent discoveries were the master clock control gene CCA1, which links organic nitrogen regulation and circadian rhythms (Gutierrez et al., 2008), and microRNA167, which mediates cell-specific control of root development in response to nitrogen (Gifford et al., 2008). Most recently, a protein kinase, AtCIPK8, was identified that is needed for nitrate responses at high but not low nitrate concentrations (Hu et al., 2009), and a DNA-binding protein, AtNLP7, was found to function in nitrate regulation of nitrate assimilation (Castaings et al., 2009). AtNLP7 encodes the NIN-like protein 7 (NLP7). NIN (nodule inception) mutants were originally identified in Lotus as being defective in bacterial recognition, infection thread formation, and nodule primordia initiation (Schauser et al., 1999). NIN genes encode nuclear-targeted DNA-binding proteins with bZIP domains containing a signature RWPxRK sequence. The Arabidopsis (Arabidopsis thaliana) NLP7 gene was recently shown to encode a nuclear-targeted protein that is needed for full nitrate induction of several nitrate-responsive genes (Castaings et al., 2009). NLP7 mutants have altered root growth (longer primary roots and more lateral roots) typical of nitrogen-starved plants and are more resistant to water stress.
The nitrate transporter gene NRT1.1 has also been implicated in nitrogen regulation. A transcriptome analysis using serial analysis of gene expression showed that about 300 genes were misregulated in nrt1.1 mutant roots, and in particular, the NRT2.1 high-affinity transporter gene showed reduced ammonium repression in the nrt1.1 mutant (Munos et al., 2004). This result is consistent with the report that NRT1.1 mediates nitrate demand regulation of high-affinity nitrate uptake (Krouk et al., 2006). NRT1.1 also controls root colonization of nitrate-rich patches by a signaling pathway that may include ANR1 as both genes are expressed in similar tissues (especially root tips) and ANR1 derepression requires NRT1.1 function (Remans et al., 2006). A signaling role for NRT1.1 is also supported by the finding that nitrate reversal of Glu inhibition of primary root growth requires NRT1.1 function (Walch-Liu and Forde, 2008; Forde and Walch-Liu, 2009). However, because NRT1.1 functions as a nitrate transporter, making it difficult to distinguish between regulatory and transport functions, it is still controversial whether NRT1.1 is a nitrate sensor or not.
To identify additional nitrate regulatory genes and mechanisms, we performed a forward genetic screen using a nitrate-regulated promoter fused to a yellow fluorescent protein (YFP) marker. Putative mutants that showed reduced nitrate induction of the marker gene were isolated and examined. Two independent mutations were mapped and sequenced and found to reside in the NRT1.1 and the NLP7 genes. Finding the NLP7 mutant demonstrated that this screen could identify nitrate regulatory mutants. The NRT1.1 mutant (nrg1) and the characterization detailed below provide strong support that indeed NRT1.1 is acting as a nitrate regulator.
RESULTS
Identification of Two Nitrate-Nonresponding Mutants
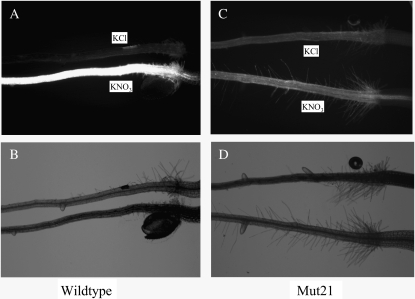
A nitrate-inducible promoter (NRP) was fused to DNA encoding YFP and transformed into Arabidopsis. Homozygous transgenic plants were generated and tested for nitrate-responsive YFP expression using fluorescence microscopy. Seedlings grown 4 d with 2.5 mm ammonium succinate (on agarose plates with no nitrate) were treated with 20 mm KNO3 or 20 mm KCl (both with 2.5 mm ammonium succinate) for 16 h then examined for YFP fluorescence. The nitrate-treated seedlings had much stronger root fluorescence than the chloride-treated controls (Fig. 1A), indicating that YFP expression was induced by nitrate in these plants.
Figure 1.
Nitrate induction of NRP-YFP in wild-type and Mut21 roots. Transgenic seedlings (containing the NRP-YFP construct) grown with ammonium but no nitrate for 4 d were treated with either 20 mm KNO3 or 20 mm KCl in the presence of 2.5 mm ammonium succinate for 16 h. Fluorescent (A and C) and visible light (B and D) images were captured with a fluorescent microscope to visualize YFP expression.
Homozygous transgenic plants were then ethyl methanesulfonate mutagenized to produce M2 seedlings, of which approximately 35,000 were screened for low YFP fluorescence after nitrate treatment. Initially 68 seedlings with low fluorescence were identified. Retesting in the next generation recovered six seedlings. Two mutants, Mut21 (nrg1) and Mut164, were selected for further analysis. An example of the reduced fluorescence phenotype observed in the mutants is shown for Mut21 (Fig. 1, C and D).
Identification of Mut164 as an Allele of NLP7
The Mut164 mutation was mapped to a 55-kb fragment demarcated by the genes At4g23930 and At4g24040. All 15 genes within this region were sequenced from the mutant. This analysis revealed a mutation (C to T) in the second exon of At4g24020 (NLP7) that converted Pro at position 223 to a Ser. Because NLP7 has been identified as a nitrate regulatory gene (Castaings et al., 2009), identification of Mut164 in our screen demonstrated that our strategy for identifying nitrate regulatory mutants was working.
Identification of Mut21 as an Allele of NRT1.1
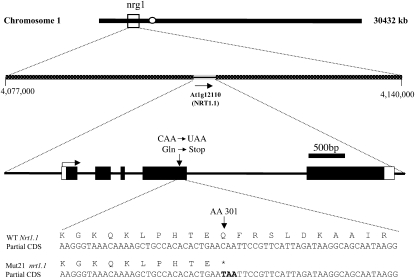
The nrg1 mutation responsible for the Mut21 phenotype was mapped to chromosome 1 in a region encompassed by bacterial artificial chromosome clones F12K11 and F20D23 (Fig. 2). This region contained the NRT1.1 (CHL1) gene. RNA transcript analysis by quantitative PCR (qPCR) using oligonucleotide primers to the 3′ end of the transcript revealed that there was almost no detectable NRT1.1 transcript in the nrg1 mutant (data not shown). NRT1.1 genomic DNA was amplified and sequenced from nrg1. A mutation was found that converted codon Q301 to a stop codon (Fig. 2). Thus, nrg1 is allelic to NRT1.1.
Figure 2.
Mapping of nrg1 (Mut21). Shows schematic diagrams of the Arabidopsis chromosome 1 showing where nrg1 mapped. Exons are shown in large black boxes. Amino acid and nucleotide changes found in Mut21 are also shown. WT, Wild type.
Nitrate Induction of Gene Expression Is Defective in nrg1
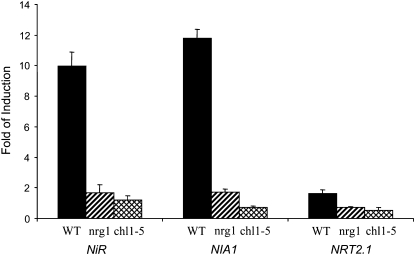
Our analysis of nrg1 showed that nitrate induction of the NRP-YFP transgene was greatly diminished. To determine if regulation of endogenous genes was similarly affected, nitrate regulation of several nitrate-inducible genes (NiR, NIA1, and NRT2.1) was examined. A well-characterized nrt1.1 mutant (deletion mutant chl1-5; Tsay et al., 1993; Munos et al., 2004) was included in these experiments to verify that the Mut21 phenotype was due to the mutation in NRT1.1. Plants were grown for 5 d on agarose plates with 2.5 mm ammonium succinate as the sole nitrogen source, then treated with 20 mm KNO3 or 20 mm KCl in the presence of 2.5 mm ammonium succinate for 2 h. Root mRNA was prepared then analyzed by qPCR. Data in Figure 3 show that nitrate induction of NiR, NIA1, and NRT2.1 in both nrg1 and chl1-5 was significantly reduced (by greater than 80%) compared to wild type. Note that millimolar ammonium was present during these treatments, which explains the low level of nitrate induction of NRT2.1.
Figure 3.
Nitrate induction of endogenous genes. Wild-type (WT) and two nrt1.1 mutant seedlings (nrg1 and chl1-5) were grown on 2.5 mm ammonium succinate for 5 d on agarose plates then treated with either 20 mm KNO3 or 20 mm KCl in the presence of 2.5 mm ammonium succinate for 2 h. Root mRNA levels were determined by qPCR. Error bars represent sd of biological replicates (n = 3).
Nitrate Induction of Gene Expression Is Restored by Nitrogen Deprivation in nrg1
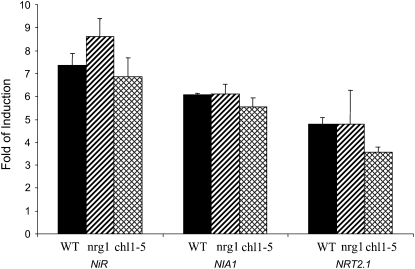
The virtual loss of nitrate-induced gene expression by nrt1.1 mutations was a surprise. We have tested for such phenotypes in the past and found little difference between wild-type and nrt1.1 mutants (R. Wang and N.M. Crawford, unpublished data). Recently, Hu et al. (2009) reported a 1.7 to 2.2 decrease in nitrate-induced levels of NiR, NIA1, and NRT2.1 in chl1-5 mutants compared with wild type (Hu et al., 2009), which is much less than what we observed (see Fig. 3). Upon comparison of experimental protocols, we noticed that our previous conditions included a nitrogen starvation pretreatment to enhance the nitrate response, which was not done in our current experiments with Mut21. To determine if the Mut21 phenotype is affected by nitrogen deprivation, the previous nitrate induction experiment, in which plants were exposed continuously to nitrogen in the form of ammonium (Fig. 3), was repeated except that seedlings were first nitrogen deprived for 24 h before nitrate treatment. The results show almost no loss of nitrate induction in mutant plants (Fig. 4), indicating that nitrogen starvation for 24 h had restored nitrate induction in Mut21 and thus rendered the nitrate response NRT1.1 independent.
Figure 4.
Nitrate induction of endogenous genes after 24 h of nitrogen deprivation. Plants were grown and treated as described in the legend to Figure 3, except at day 4 plants were transferred to nitrogen-free medium for 24 h then treated with 20 mm nitrate or chloride for 2 h with no added ammonium succinate. Root mRNA levels were determined by qPCR. Error bars represent sd (n = 3). WT, Wild type.
To determine how long it takes to lose NRT1.1-dependent induction upon nitrogen starvation, a time-course experiment was performed (Supplemental Figs. S1–S3). Plants were grown hydroponically on 2.5 mm ammonium succinate for 7 d then nitrogen starved by transfer to the same media with no ammonium succinate. Plants were then treated with 1 mm KCl or KNO3 for 30 min. Roots were harvested, and mRNA prepared and analyzed by qPCR. The data show that for all three genes tested (NiR, NIA1, and NRT2.1), nitrate induction began to recover in the mutant after 1 to 2 h of nitrogen starvation. After 24 h, nitrate induction in the mutant was almost as high as for wild-type plants.
The effect of nitrogen starvation on NRT1.1 expression was measured to determine if the loss of the Mut21 phenotype could be accounted for by a loss of NRT1.1 mRNA. Over the first 8 h of nitrogen starvation, the level of NRT1.1 mRNA increased about 1.6-fold (Supplemental Fig. S4). However, after 24 h, the level dropped 4-fold. These results indicate that the loss of NRT1.1-dependent regulation during the first 8 h of nitrogen starvation is not due to the loss of NRT1.1 expression (i.e. mRNA) and may be due to a posttranscriptional modification. At 24 h, the drop in NRT1.1 mRNA was sufficiently large that it should contribute to the loss of the Mut21 phenotype.
The experiments described above cannot determine where it is the nitrogen deprivation in general or the loss of ammonium in particular that is responsible for the loss of the Mut21 phenotype. Including 5 mm ammonium during the 2-h nitrate induction treatment of nitrogen-starved seedlings did not restore the Mut21 phenotype (data not shown). Further experiments are needed to resolve this issue.
Loss of Nitrate Induction in nrg1 Is Not Accounted for by Impaired Nitrate Uptake
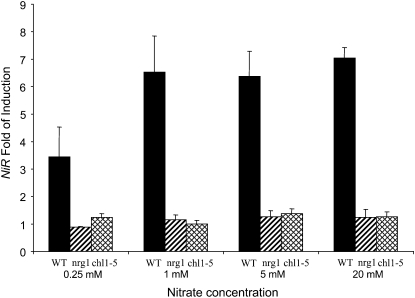
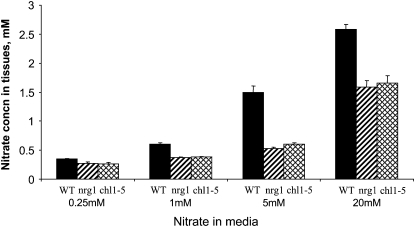
Since NRT1.1 encodes a nitrate transporter, it is possible that the loss of nitrate induction in the nrt1.1 mutants is due to reduced nitrate uptake. To test this idea, nitrate induction of NiR in wild type and the two nrt1.1 mutants were assayed at various concentrations of nitrate (0.25–20 mm) in the presence of ammonium (Fig. 5). Nitrate induction was virtually abolished in both mutants at all nitrate concentrations tested under these conditions. Next, nitrate accumulation in whole seedlings was also measured after the same 2-h treatments (Fig. 6) under the same conditions. Nitrate accumulation was lower in the mutants than the wild type at all the concentrations of nitrate tested; however, the amount of accumulation was still substantial enough in the mutants (36%–77% of wild type) to support nitrate induction. For example, nitrate accumulation at 20 mm nitrate in the mutants is as much or more than in wild-type plants treated with 0.25 mm to 5 mm nitrate, yet nitrate induction is vanishing small in the mutants at 20 mm nitrate (Fig. 6). In fact, the amount of nitrate entering the plants under all concentrations tested is more than sufficient for induction, as uptake from solutions with only 2 to 5 μm nitrate is needed for strong induction (Wang et al., 2007). Thus, reduction in nitrate uptake cannot explain the loss of nitrate induction in the mutants.
Figure 5.
Titration of the nitrate induction response. Seedlings were grown 5 d on agarose plates with 2.5 mm ammonium succinate (same as Fig. 3), then treated with various concentrations of KNO3 or KCl for 2 h in the presence of 2.5 mm ammonium succinate before roots were collected for RNA preparation. NiR mRNA levels were determined by qPCR. Error bars represent sd (n = 3). WT, Wild type.
Figure 6.
Nitrate accumulation. Seedlings grown 5 d with 2.5 mm ammonium succinate were treated with various concentrations of KNO3 (same as for Fig. 5) in the presence of 2.5 mm ammonium succinate for 2 h. Whole seedlings were then collected for nitrate assays as described in “Materials and Methods.” Error bars represent sd (n = 3). WT, Wild type.
Microarray Analysis of Nitrate Response in nrt1.1 Mutants
Several transcriptome analyses have been reported for nrt1.1 mutants. In addition to the serial analysis of gene expression experiments for plants grown on ammonium nitrate (Munos et al., 2004), a microarray analysis using ATH1 chips of nitrate-treated (30 min at 25 mm) roots found that 42 genes had absolute transcript levels that were lower in the chl1-5 mutant by 1.7-fold or more (or 17 genes reduced by 2-fold or more) compared with wild type (Hu et al., 2009). We performed microarray analyses in a different way: using both control and nitrate-treated wild-type and nrg1 plants that had been grown without nitrogen starvation (i.e. with continuous ammonium supply) to determine the effect of nrg1 on nitrate induction ratios under these conditions. Seven-day-old plants grown under hydroponic conditions with ammonium were treated with 1 mm KCl or KNO3 in the presence of ammonium for 30 min. Root mRNA was isolated and analyzed using ATH1 chips (total data set is shown in Supplemental Table S1).
The microarray data showed that 111 genes had lower induction ratios of 2-fold or more in nrg1 plants and only three genes had higher induction ratios of 2-fold or more in nrg1 plants (Supplemental Table S2). Many known nitrate-inducible genes including NiR (induction ratio reduced 5.1-fold in mutant), NIA1 (reduced 4.0-fold), UPM1 (reduced 3.8-fold), NIA2 (reduced 2.5-fold), and NRT2.4 (reduced 2.0-fold) showed reduced nitrate induction ratios in the mutant. CIPK1 and CIPK3 were also on this list consistent with the findings of Hu et al. (2009). Biomaps analysis using the Munich Information Center for Protein Sequences database (www.virtualplant.org) revealed that genes most affected by the nrg1 mutation were overrepresented in Gene Ontology groups: energy, photosynthesis, pentose-P pathway, detoxification, and light absorption (Supplemental Table S3).
DISCUSSION
There has been mounting evidence that NRT1.1 functions not only as a nitrate transporter but also as a regulator. NRT1.1 expression is atypical for a root uptake transporter, being targeted to root tips, lateral root primordia, and nascent shoot organs (Guo et al., 2001), and being up-regulated by acidic pH (Tsay et al., 1993) and auxin (Guo et al., 2002). NRT1.1 function is required for high-nitrate repression of NRT2.1 and high-affinity uptake in the presence of high ammonium (Krouk et al., 2006). nrt1.1 mutants are defective in lateral root proliferation in nitrate-rich zones and have reduced expression of the lateral root regulatory gene ANR1 (Remans et al., 2006). nrt1.1 mutants are also defective in the nitrate reversal of Glu inhibition of primary root growth (Walch-Liu and Forde, 2008). It was concluded from these studies that NRT1.1 may be playing a signaling role as a nitrate sensor. Our findings that a nrt1.1 mutant can be captured in a genetic screen for nitrate regulatory mutants and that nrt1.1 mutants are impaired in nitrate regulation of gene expression over a wide variation of nitrate concentrations are certainly consistent with and support this proposal.
Because NRT1.1 functions as a nitrate transporter, the signaling defects described above could be explained by reduction of nitrate uptake into cells in which the nitrate sensor resides. In the reports described above, inhibition of nitrate uptake was found not to explain the nrt1.1 mutant phenotypes; however, in the earlier reports where bulk uptake into roots was measured, it was difficult to rule out the possibility that reduced nitrate uptake into select sensing cells in root tips could account for the effects. Results from Walch-Liu and Forde (2008) provide additional insights because they found that a nonphosphorylatable form of NRT1.1, which retains low-affinity but not high-affinity uptake activity (Liu and Tsay, 2003), was not capable of nitrate reversal of Glu inhibition of root growth (Walch-Liu and Forde, 2008). In our experiments, we measured gene expression in whole roots, which is not restricted to a small number of select cells in the root, so that measurements of nitrate uptake into seedlings should be more indicative of nitrate availability for induction. In our system, nitrate uptake in the mutants was more than sufficient to induce a nitrate response, yet induction was clearly impaired.
The most consistent model to explain all the published results and our findings is that NRT1.1 is sensing nitrate directly and thus serves as a nitrate transceptor. Transceptors, which are transporters that also act as sensors, have been described in yeast (Saccharomyces cerevisiae; Holsbeeks et al., 2004). NRT1.1 can transport nitrate yet it appears to play a regulatory role as well. If NRT1.1 is in fact a transceptor, it should be possible to isolate mutants that separate the transport from sensing functions. The T101A mutation that is defective in a sensing function (nitrate reversal of Glu inhibition; Walch-Liu and Forde, 2008) but retains partial transport activity (Liu and Tsay, 2003) provides support for this idea. However, there are many questions still unanswered by this model. Is the regulation of NRT1.1 by nitrate, acidic pH, and auxin, important for controlling nitrate uptake or regulation? How does the switch between high- and low-affinity states by phosphorylation of T101 modulate nitrate regulation? Also, there are several phenotypes for nrt1.1 mutants that are difficult to explain simply by loss of nitrate sensing. nrt1.1 mutants are defective in root growth in young seedlings even in the absence of nitrate in the medium (Guo et al., 2001). The altered regulation of NRT2.1 in the nrt1.1 mutant results from reduced ammonium repression (Munos et al., 2004). Lastly, NRT1.1-dependent regulation is lost during nitrogen deprivation even though nitrate induction of endogenous genes still occurs (Fig. 4). Thus, other nitrate-sensing systems must be present. These questions require further analysis before we can achieve a full understanding of nitrate sensing and the role of NRT1.1.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Mutagenesis
Homozygous backcrossed transgenic seeds containing the NRP-YFP construct (1.2 g in 20 mL of water) were treated with ethyl methanesulfonate (methanesulfonic acid ethyl ester) at 15 mm for 16 h with agitation (30 rpm). M2 seeds were produced and pooled into families for screening. NRP contained promoter fragments from the NIA1 and NiR promoters fused to the 35S minimal promoter (see GenBank accession no. GQ374175).
Mutant Screen
M2 seedlings were screened on vertical 100 × 100 mm square plates containing 25 mL of 0.6% agarose media. Surface-sterilized seeds were aligned horizontally on the plate surface at a density of about 100 seeds per row. Three rows of seeds were placed on each plate. The initial medium (described in Wang et al., 2004) was nitrate free with 2.5 mm ammonium succinate as the nitrogen source. After incubation at 4°C for 2 d, seedlings were grown at 25°C with 24 h of light. Four-day-old seedlings were then flooded with 12.5 mL of medium containing 20 mm KNO3 and 2.5 mm ammonium succinate for 16 h. Seedlings were screened under a fluorescence microscope (Nikon Eclipse TE2000-U) and rescued. Putative mutants were selfed then rescreened. Confirmed mutants were backcrossed to the transgenic wild type and made homozygous before analysis.
Growth and Treatment Conditions
For qPCR analyses and nitrate accumulation assays, seedlings were grown on vertical agarose plates as described above for 5 d with 2.5 mm ammonium succinate as the sole nitrogen source in plant growth medium (Wang et al., 2004). The seedlings were then flooded with 12.5 mL of plant growth medium (with 2.5 mm ammonium succinate) plus KNO3 at various concentrations for 2 h with agitation (60 rpm) under light. Roots were then collected for total RNA preparation (as described Wang et al., 2003). Control samples were prepared at the same time with the same concentration of KCl in place of KNO3.
For nitrate treatments without ammonia, seedlings were grown on plant growth medium with 2.5 mm ammonium succinate for 4 d then transferred to fresh agarose plates without nitrogen for 24 h, followed by flooding with nitrate-containing plant growth media as described above except that there was no ammonium succinate in the liquid medium.
For the microarray analysis, plants were grown in aseptic hydroponics as described (Wang et al., 2007) for 7 d with modifications as follows: Seedlings were transferred to 100 mL of fresh medium with 2.5 mm ammonium succinate after 6 d of growth and continued incubation for 24 h. Nitrate and control chloride treatments were initiated by adding KNO3 or KCl to the growth media to yield 1 mm concentration then incubated for 30 min before harvesting roots.
Gene Expression and Nitrate Analysis
qPCR Analysis
RNA samples were prepared from roots as described (Wang et al., 2007). Real-time qPCR was performed as described (Wang et al., 2004). Relative expression levels of NRT1.1 were compared with the internal reference gene, ubiquitin-associated protein gene (At5g12120).
Microarray Analysis
Roots were collected for total RNA preparation as described (Wang et al., 2007). Experiments were done in duplicate then averaged to generate induction ratios using the Affymetrix software as described (Wang et al., 2003). In all cases, data were filtered to require that signal levels were detectable in both replicates for at least one of the treatments (indicated as P by the Affymetrix software) and had an absolute value of 100 or more.
Nitrate Accumulation
Nitrate in seedlings of wild type and mutants were measured using the hydrazine-sulfate method as described (Wang et al., 2004).
Positional cloning of nrg1 was performed on individual F2 recombinants using simple sequence length polymorphisms as described (Lukowitz et al., 2000).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number GQ374175.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. NiR induction after N starvation.
Supplemental Figure S2. NIA1 induction after N starvation.
Supplemental Figure S3. NRT2.1 induction after N starvation.
Supplemental Figure S4. NRT1.1 mRNA levels after N starvation.
Supplemental Table S1. Complete microarray data set.
Supplemental Table S2. Genes affected by nrg1 mutation.
Supplemental Table S3. Biomaps analysis.
Supplementary Material
Acknowledgments
We thank Dr. Mamoru Okamoto and Kati Wu for valuable assistance and advice.
This work was supported by a grant from the National Science Foundation (grant no. IOB–0519985).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Nigel M. Crawford (ncrawford@ucsd.edu).
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Alboresi A, Gestin C, Leydecker MT, Bedu M, Meyer C, Truong HN (2005) Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ 28 500–512 [DOI] [PubMed] [Google Scholar]
- Castaings L, Camargo A, Pocholle D, Gaudon V, Texier Y, Boutet-Mercey S, Taconnat L, Renou JP, Daniel-Vedele F, Fernandez E, et al (2009) The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J 57 426–435 [DOI] [PubMed] [Google Scholar]
- Crawford NM, Glass ADM (1998) Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci 3 389–395 [Google Scholar]
- Filleur S, Walch-Liu P, Gan Y, Forde BG (2005) Nitrate and glutamate sensing by plant roots. Biochem Soc Trans 33 283–286 [DOI] [PubMed] [Google Scholar]
- Forde BG (2000) Nitrate transporters in plants: structure, function and regulation. Biochim Biophys Acta 1465 219–235 [DOI] [PubMed] [Google Scholar]
- Forde BG (2002) Local and long-range signaling pathways regulating plant responses to nitrate. Annu Rev Plant Biol 53 203–224 [DOI] [PubMed] [Google Scholar]
- Forde BG, Walch-Liu P (2009) Nitrate and glutamate as environmental cues for behavioural responses in plant roots. Plant Cell Environ 32 682–693 [DOI] [PubMed] [Google Scholar]
- Fritz C, Palacios-Rojas N, Feil R, Stitt M (2006) Regulation of secondary metabolism by the carbon-nitrogen status in tobacco: nitrate inhibits large sectors of phenylpropanoid metabolism. Plant J 46 533–548 [DOI] [PubMed] [Google Scholar]
- Gan Y, Filleur S, Rahman A, Gotensparre S, Forde BG (2005) Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana. Planta 222 730–742 [DOI] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD (2008) Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA 105 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo FQ, Wang R, Chen M, Crawford NM (2001) The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is activated and functions in nascent organ development during vegetative and reproductive growth. Plant Cell 13 1761–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo FQ, Wang R, Crawford NM (2002) The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is regulated by auxin in both shoots and roots. J Exp Bot 53 835–844 [DOI] [PubMed] [Google Scholar]
- Gutierrez RA, Gifford ML, Poultney C, Wang R, Shasha DE, Coruzzi GM, Crawford NM (2007. a) Insights into the genomic nitrate response using genetics and the Sungear Software System. J Exp Bot 58 2359–2367 [DOI] [PubMed] [Google Scholar]
- Gutierrez RA, Lejay LV, Dean A, Chiaromonte F, Shasha DE, Coruzzi GM (2007. b) Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol 8 R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RA, Shasha DE, Coruzzi GM (2005) Systems biology for the virtual plant. Plant Physiol 138 550–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, et al (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA 105 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsbeeks I, Lagatie O, Van Nuland A, Van de Velde S, Thevelein JM (2004) The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem Sci 29 556–564 [DOI] [PubMed] [Google Scholar]
- Hu HC, Wang YY, Tsay YF (2009) AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J 57 264–278 [DOI] [PubMed] [Google Scholar]
- Krouk G, Tillard P, Gojon A (2006) Regulation of the high-affinity NO3- uptake system by NRT1.1-mediated NO3- demand signaling in Arabidopsis. Plant Physiol 142 1075–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leydecker MT, Camus I, Daniel-Vedele F, Truong HN (2000) Screening for Arabidopsis mutants affected in the Nii gene expression using the Gus reporter gene. Physiol Plant 108 161–170 [Google Scholar]
- Liu KH, Tsay YF (2003) Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J 22 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR (2000) Positional cloning in Arabidopsis: why it feels good to have a genome initiative working for you. Plant Physiol 123 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munos S, Cazettes C, Fizames C, Gaymard F, Tillard P, Lepetit M, Lejay L, Gojon A (2004) Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell 16 2433–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Filleur S, Diatloff E, Mounier E, Tillard P, Forde BG, Gojon A (2006) The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc Natl Acad Sci USA 103 19206–19211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402 191–195 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M (1999) Nitrate regulation of metabolism and growth. Curr Opin Plant Biol 2 178–186 [DOI] [PubMed] [Google Scholar]
- Stitt M, Muller C, Matt P, Gibon Y, Carillo P, Morcuende R, Scheible WR, Krapp A (2002) Steps towards an integrated view of nitrogen metabolism. J Exp Bot 53 959–970 [DOI] [PubMed] [Google Scholar]
- Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK (2007) Nitrate transporters and peptide transporters. FEBS Lett 581 2290–2300 [DOI] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA, Crawford NM (1993) A herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72 705–713 [DOI] [PubMed] [Google Scholar]
- Walch-Liu P, Filleur S, Gan Y, Forde BG (2005) Signaling mechanisms integrating root and shoot responses to changes in the nitrogen supply. Photosynth Res 83 239–250 [DOI] [PubMed] [Google Scholar]
- Walch-Liu P, Forde BG (2008) Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises L-glutamate-induced changes in root architecture. Plant J 54 820–828 [DOI] [PubMed] [Google Scholar]
- Walch-Liu P, Ivanov II, Filleur S, Gan Y, Remans T, Forde BG (2006) Nitrogen regulation of root branching. Ann Bot (Lond) 97 875–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Guegler K, LaBrie ST, Crawford NM (2000) Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12 1491–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Tischner R, Gutierrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford NM (2004) Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol 136 2512–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Xing X, Crawford N (2007) Nitrite acts as a transcriptome signal at micromolar concentrations in Arabidopsis roots. Plant Physiol 145 1735–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Akiyama A, Kisaka H, Uchimiya H, Miwa T (2004) Metabolic engineering with Dof1 transcription factor in plants: improved nitrogen assimilation and growth under low-nitrogen conditions. Proc Natl Acad Sci USA 101 7833–7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Forde BG (2000) Regulation of Arabidopsis root development by nitrate availability. J Exp Bot 51 51–59 [PubMed] [Google Scholar]
- Zhang HM, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279 407–409 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.