Abstract
Carotenoids are essential pigments of the photosynthetic apparatus and an indispensable component of the human diet. In addition to being potent antioxidants, they also provide the vitamin A precursor β-carotene. In tomato (Solanum lycopersicum) fruits, carotenoids accumulate in specialized plastids, the chromoplasts. How the carotenoid biosynthetic pathway is regulated and what limits total carotenoid accumulation in fruit chromoplasts is not well understood. Here, we have introduced the lycopene β-cyclase genes from the eubacterium Erwinia herbicola and the higher plant daffodil (Narcissus pseudonarcissus) into the tomato plastid genome. While expression of the bacterial enzyme did not strongly alter carotenoid composition, expression of the plant enzyme efficiently converted lycopene, the major storage carotenoid of the tomato fruit, into provitamin A (β-carotene). In green leaves of the transplastomic tomato plants, more lycopene was channeled into the β-branch of carotenoid biosynthesis, resulting in increased accumulation of xanthophyll cycle pigments and correspondingly reduced accumulation of the α-branch xanthophyll lutein. In fruits, most of the lycopene was converted into β-carotene with provitamin A levels reaching 1 mg per g dry weight. Unexpectedly, transplastomic tomatoes also showed a >50% increase in total carotenoid accumulation, indicating that lycopene β-cyclase expression enhanced the flux through the pathway in chromoplasts. Our results provide new insights into the regulation of carotenoid biosynthesis and demonstrate the potential of plastids genome engineering for the nutritional enhancement of food crops.
Carotenoids are isoprenoid molecules that are synthesized by all photosynthetic organisms and also by some fungi and nonphotosynthetic bacteria. In plants, they participate in photosynthetic light harvesting and protection against light stress. In addition, carotenoids accumulate to large levels as storage metabolites in chromoplasts of flowers, fruits, and taproots. Carotenoids are also essential to animals, which, however, are unable to synthesize them de novo, and therefore must rely on dietary sources of carotenoids. β-Carotene is the main dietary precursor of vitamin A and therefore also referred to as provitamin A. Vitamin A deficiency in humans represents a global health problem affecting approximately one-third of the countries of the world (Mayer et al., 2008). Presumably due to their antioxidant activity, β-carotene and other carotenoid species also exert protective effects against cardiovascular diseases, certain cancers, and aging-related diseases (Collins, 1999).
While the enzymology of the carotenoid biosynthetic pathways in plants and eubacteria is now reasonably well understood (Armstrong, 1997; Cunningham and Gantt, 1998; Hirschberg, 2001), understanding of the regulation of carotenoid biosynthesis is still rather poor (Bramley, 2002). Mainly using the tomato (Solanum lycopersicum) fruit as model system, the study of pigmentation mutants (Ronen et al., 2000; Isaacson et al., 2002; Galpaz et al., 2006) and transgenic approaches (Giuliano et al., 2000, 2008; Römer and Fraser, 2005; Fraser et al., 2007) have provided first insights into regulatory mechanisms operating in carotenogenesis. For example, constitutive expression of the phytoene desaturase (crtI) gene from the bacterium Erwinia uredovora resulted in elevated β-carotene accumulation in tomatoes, but also led to an unexpected reduction in total carotenoid levels (Römer et al., 2000). The reduction in total carotenoids is believed to be an effect of feedback regulation from β-carotene or one of its downstream metabolites (Bramley, 2002). However, fruit-specific overexpression of the native lycopene β-cyclase resulted in increased β-carotene accumulation, without a concomitant decrease in total carotenoids (Rosati et al., 2000). Why some genetic disturbances of carotenoid biosynthesis negatively affect total carotenoid accumulation and others do not (or even result in an increase; Dharmapuri et al., 2002; Fraser et al., 2002), remains to be established.
Here we have used tomato plastid transformation to address the regulation of carotenoid biosynthesis exerted at the level of lycopene to β-carotene conversion by the enzyme lycopene β-cyclase (Fig. 1A). We show that plastid expression of a plant lycopene β-cyclase does not only trigger efficient conversion of lycopene to β-carotene, but unexpectedly also results in a >50% increase in total carotenoid accumulation. This contrasts moderately increased β-carotene levels and reduced total carotenoid accumulation upon expression of a bacterial lycopene β-cyclase (Wurbs et al., 2007) and suggests lycopene β-cyclase activity as an important regulatory point in plant and microbial carotenoid biosynthesis.
Figure 1.
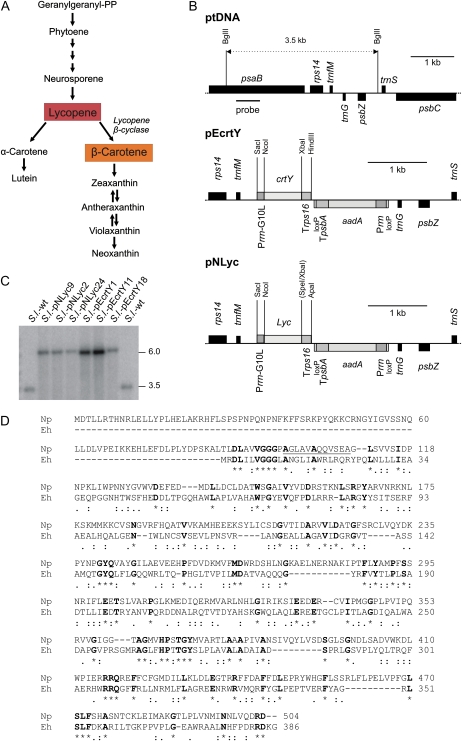
Engineering of the carotenoid biosynthetic pathway by plastid transformation. A, Carotenoid biosynthetic pathway in higher plants. The pathway splits into an α-branch and a β-branch immediately downstream of lycopene, the major storage carotenoid of tomato fruits. The enzyme expressed from the tomato plastid genome in this study, lycopene β-cyclase, leads into the β-branch. B, Physical maps of the targeting region in the plastid genome (ptDNA) and the plastid transformation vectors pEcrtY and pNLyc constructed in this study. Genes above the line are transcribed from the left to the right, genes below the line are transcribed in the opposite direction. The transgenes are targeted to the intergenic region between the trnfM and trnfG genes (Ruf et al., 2001). The selectable marker gene aadA is driven by a chimeric rRNA operon promoter (Prrn; Svab and Maliga, 1993), fused to the 3′-UTR from the psbA gene (TpsbA), and flanked by two loxP sites to allow marker removal by Cre-mediated site-specific recombination (Zhou et al., 2008). The transgene expression cassette consists of the ribosomal RNA operon promoter fused to the 5′ leader from the gene 10 of phage T7 (Prrn-G10L; Kuroda and Maliga, 2001) and the 3′-UTR of the rps16 gene (Trps16). Restriction sites used for cloning or RFLP analysis are indicated, and the psaB-derived hybridization probe is denoted by a horizontal bar. Sites lost due to ligation to heterologous ends are in parentheses. C, Southern-blot analysis of tomato transplastomic lines carrying the lycopene β-cyclase gene from daffodil (S.l.-pNLyc) or from E. herbicola (S.l.-pEcrtY). Total cellular DNA was digested with BglII and hybridized to a radioactively labeled probe detecting the psaB region of the plastid genome, which flanks the transgene insertion site (section B). Fragment sizes are given in kb. wt, Wild type. D, Alignment (produced with ClustalW2) of the amino acid sequences of the lycopin β-cyclases from daffodil (Np) and E. herbicola (Eh). Asterisk (*) denotes residues identical in both sequences (marked in bold), colon (:) indicates conserved substitutions, and a dot indicates semiconserved substitutions. The N-terminal extension of the Np sequence is likely to harbor the transit peptide for protein import into plastids. The amino acids that changed due to correction of the Lyc sequence from daffodil (published sequence: GenBank accession no. X98796.1; corrected sequence: accession no. GQ327929) are underlined. The corrections improve the sequence similarity in the N-terminal domains of the Np and Eh sequences.
RESULTS
Introduction of Lycopene β-Cyclase Genes into the Tomato Plastid Genome
A recent transcriptomics and translatomics analysis of plastid gene expression during tomato fruit ripening revealed that most genes in the plastid genome are drastically down-regulated during fruit development (Kahlau and Bock, 2008). However, this study also identified a small number of plastid expression elements (promoters and translation initiation signals) that remain active in chromoplasts (Kahlau and Bock, 2008). As a previous attempt to express the lycopene β-cyclase gene (crtY) from the carotenoid-producing eubacterium Erwinia herbicola has resulted only in a moderate increase in β-carotene accumulation (to 280 ng/mg dry weight), we wanted to test if transgene expression levels can be improved by using a promoter that retains higher activity in chromoplasts than the previously used atpI promoter (Wurbs et al., 2007). As the ribosomal RNAs represent the most abundant plastid RNA species in chromoplasts (Kahlau and Bock, 2008), we selected the rRNA operon promoter and combined it with the strongest known ribosome-binding site, the Shine-Dalgarno sequence from gene 10 of bacteriophage T7 (Kuroda and Maliga, 2001; Oey et al., 2009; Fig. 1B). In addition to the lycopene β-cyclase gene from E. herbicola, we also cloned the Lyc lycopene β-cyclase gene from the higher plant daffodil (Narcissus pseudonarcissus; Al-Babili et al., 1996; kindly provided by Drs. Peter Beyer and Salim Al-Babili, University of Freiburg, Germany) into the same expression cassette (Fig. 1B). When we resequenced both genes after having finished vector construction, we noted three single-nucleotide deviations from the published Lyc sequence (GenBank accession no. X98796.1). All three deviations were also present in the original Lyc clone and presumably represent errors from band compressions in sequencing gels: a C was missing after position 442 (three Cs instead of two Cs in the published sequence), a G was missing after position 447 (three Gs instead of two Gs in the published sequence), and a C was missing after position 480 (two Cs instead of one C in the published sequence). These insertions change the published protein sequence over a stretch of 12 amino acids (Fig. 1D; as there are altogether three nucleotides missing, the published sequence falls back into the correct reading frame at nucleotide position 481). The corrected Lyc sequence was deposited in the GenBank database (accession no. GQ327929).
The plastid expression cassettes containing the crtY and Lyc transgenes were inserted into plasmid pKP9 (Zhou et al., 2008), generating the chloroplast transformation vectors pEcrtY and pNLyc, respectively (Fig. 1B). The two constructs were introduced into the plastid genome of the commercial tomato cv IPA-6 by biolistic chloroplast transformation (Svab and Maliga, 1993; Ruf et al., 2001). Eleven transplastomic lines were obtained with pEcrtY (from 400 selection plates with bombarded leaf pieces) and 22 transplastomic lines were obtained with pNLyc (from 350 selection plates). Three independently generated lines per construct were characterized in detail. The lines will be subsequently referred to as S.l.-pNLyc (for Solanum lycopersicum harboring the Narcissus Lyc gene) and S.l.-pEcrtY (for Solanum lycopersicum harboring the Erwinia crtY gene).
Analysis of Transplastomic Tomato Plants Harboring Lycopene β-Cyclase Transgenes
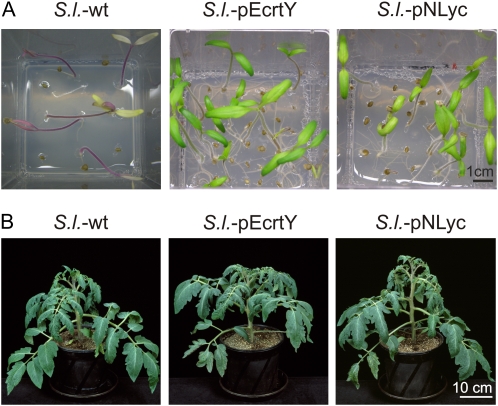
Putative chloroplast transformants were purified to homoplasmy by passing them through additional regeneration cycles under antibiotic selection. Transformation of the chloroplast genome, correct integration of the transgenes via homologous recombination, and homoplasmy of the transplastomic lines (i.e. absence of residual copies of the wild-type chloroplast genome) were assessed by RFLP analysis (Fig. 1C) and homoplasmy was additionally confirmed by seed assays (Fig. 2A). Lack of segregation of the antibiotic resistance in the T1 generation demonstrated homoplasmy (Fig. 2A) and confirmed maternal transgene inheritance, as expected for a plastid-encoded trait in tomato. Homoplasmic T1 plants from S.l.-pNLyc and S.l.-pEcrtY transplastomic lines were indistinguishable from the wild-type control (Fig. 2B), indicating that transgene expression is phenotypically neutral.
Figure 2.
Homoplasmy and phenotypes of transplastomic tomato lines. A, Examples of seed tests to confirm homoplasmy. Seeds from the wild-type (S.l.-wt) and transplastomic plants generated with constructs pEcrtY and pNLyc were germinated on medium with spectinomycin (100 mg/L). Antibiotic resistance and lack of segregation in the T1 generation confirms the homoplasmic state of the transplastomic lines. B, Wild-type-like phenotype of homoplasmic tomato lines expressing lycopene β-cyclase transgenes from their plastid genomes.
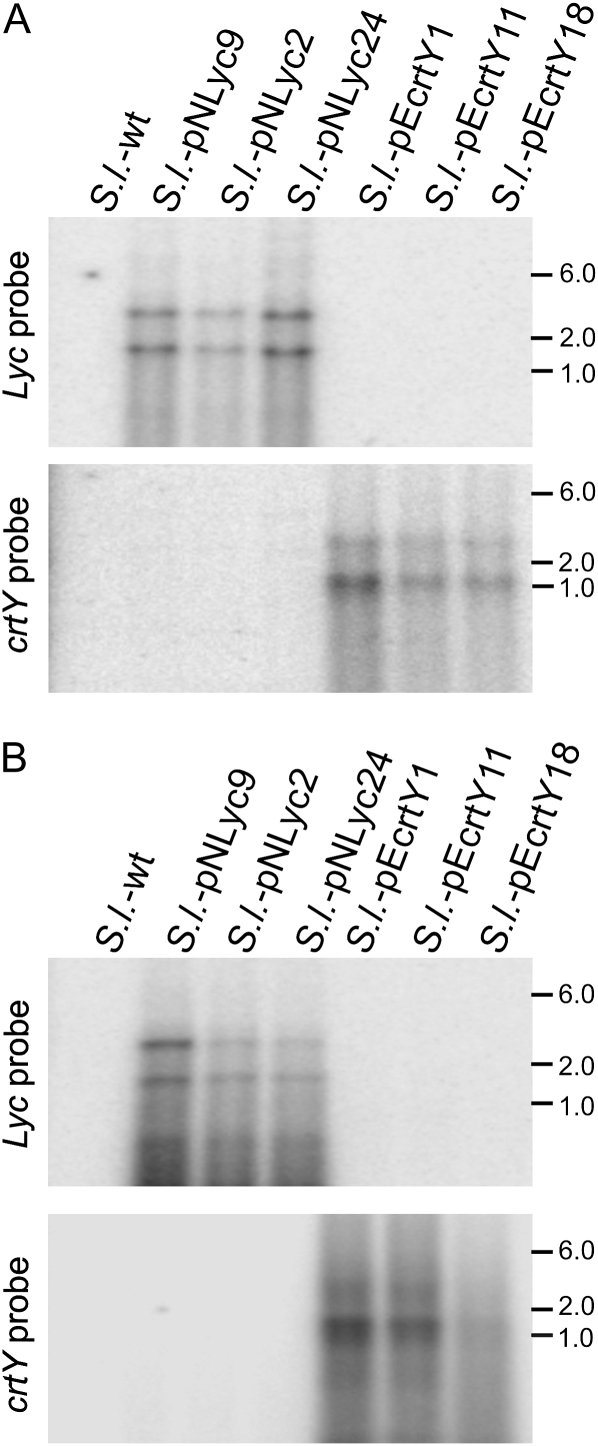
To analyze expression of the introduced carotenoid biosynthesis genes, we first performed a series of RNA gel-blot experiments. Analysis of transgene expression in leaves revealed accumulation of stable monocistronic mRNA and presence of an additional longer transcript species originating from read-through transcription, which had been seen also in previous plastid transformation experiments with vectors that target the same insertion site in the genome (Wurbs et al., 2007; Zhou et al., 2008; Fig. 3A). The same two major RNA species are also detectable in ripe fruits, although presence of a more pronounced smear into the low Mr region of the blot may indicate higher transcript turnover in chromoplasts (Fig. 3B).
Figure 3.
Analysis of lycopene β-cyclase mRNA accumulation in leaves (A) and ripe fruits (B) of transplastomic tomato plants. Total cellular RNA was hybridized to radiolabeled probes corresponding to the coding regions of crtY or Lyc. With each probe, two major transcript species are detected. While the bottom band represents mature monocistronic lycopene β-cyclase mRNA (1.3 kb for crtY and 1.7 kb for Lyc), the top band most probably represents a stable read-through transcript, as has been observed before with pKP9-derived vectors (Zhou et al., 2008; Oey et al., 2009). Sizes of marker bands are indicated in kb. Note that crtY and Lyc do not cross-hybridize due to insufficient sequence similarity (Fig. 1D). wt, Wild type.
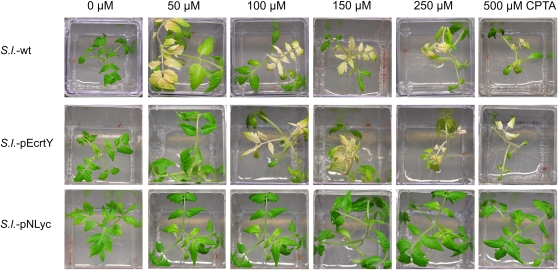
We next wanted to test for elevated lycopene β-cyclase activity in transplastomic tomato plants. To this end, we treated seedlings with the herbicide 2-(4-chlorophenylthio)-triethylamine (CPTA), a specific lycopene cyclase inhibitor (Schuetz and Baldwin, 1958; Wurbs et al., 2007). While S.l.-pEcrtY transplastomic plants showed only moderately increased CPTA tolerance compared to the wild type, S.l.-pNLyc plants displayed an enormous resistance to the herbicide. While the wild type bleaches already at a CPTA concentration of 50 μm, S.l.-pNLyc plants were virtually unaffected by concentrations as high as 500 μm CPTA (Fig. 4). This tentatively suggests that lycopene β-cyclase activity is dramatically increased in S.l.-pNLyc transplastomic plants, but not strongly increased in S.l.-pEcrtY plants.
Figure 4.
Herbicide resistance assays to test for lycopene β-cyclase expression. The herbicide CPTA was used as specific inhibitor of the lycopin β-cyclase activity. Aseptically grown tomato plants were watered with 2.5 mL CPTA solution to obtain the final concentrations indicated (50, 100, 150, 250, or 500 μm). Phenotypic comparison with a water-treated control (0 μm) was done after 7 d. wt, Wild type.
Carotenoid Biosynthesis in Fruits of Transplastomic Tomato Plants
The dark-red carotenoid lycopene represents the main storage carotenoid in tomato fruits. Presence of elevated levels of lycopene β-cyclase activity in fruits should result in enhanced conversion of lycopene into the orange carotenoid β-carotene (provitamin A; Fig. 1A). Inspection of ripe fruits from our transplastomic tomato plants revealed that the S.l.-pNLyc tomatoes were bright orange instead of dark red (Fig. 5). In contrast, S.l.-pEcrtY transplastomic tomatoes were virtually indistinguishable from wild-type tomatoes (Fig. 5). Taken together with the results from the CPTA assays, this suggested that the daffodil cyclase was expressed to high levels and was highly active in tomato plastids.
Figure 5.
Phenotypes of tomato fruits from transplastomic tomato plants expressing lycopene β-cyclase transgenes. Fruits from a wild-type plant (S.l.-wt), an S.l.-pEcrtY line, and an S.l.-pNLyc line were harvested at different ripening stages and photographed from the side (top row) and from the bottom (bottom row). The orange color of ripe S.l.-pNLyc fruits indicates efficient conversion of red lycopene into orange β-carotene (provitamin A).
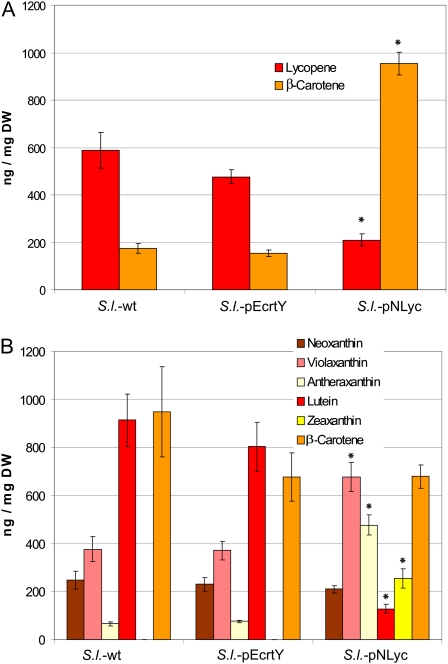
To determine to what extent plastid expression of the lycopene β-cyclase transgenes led to altered carotenoid contents or composition in tomatoes, we measured carotenoid accumulation in fruits by HPLC. While the carotenoid composition in S.l.-pEcrtY tomatoes was not significantly different from that in wild-type tomatoes, expression of the daffodil cyclase resulted in a dramatic increase in β-carotene accumulation (reaching 1 mg per g dry weight) and a concomitant reduction in lycopene accumulation (Fig. 6A). However, unexpectedly, these changes were not proportional in that the increase in β-carotene greatly exceeded the decrease in lycopene. Consequently, S.l.-pNLyc tomatoes had a much higher total carotenoid content (52% increase compared to the wild type; Fig. 6A). This suggests that plastid expression of the daffodil cyclase does not only trigger efficient lycopene-to-β-carotene conversion, it also results in a general enhancement of the carotenoid biosynthetic pathway in the fruit.
Figure 6.
HPLC analysis of pigment accumulation in fruits and leaves from wild-type plants (S.l.-wt) and transplastomic S.l.-pEcrtY and S.l.-pNLyc plants. A, Comparison of lycopene and β-carotene contents (in ng/mg dry weight [DW]) in ripe fruits as determined by HPLC. Values represent means from 12 measurements: three fruits harvested from three to four independent plant lines per construct. B, Comparison of carotenoid contents in leaves. Values represent means from nine measurements, which included leaves harvested from at least four independent transplastomic lines per construct. The sd is shown as error bar. Asterisks indicate significant differences compared to the wild type (P value < 0.05).
Carotenoid Biosynthesis in Leaves of Transplastomic Tomato Plants
We next wanted to determine whether or not lycopene β-cyclase overexpression alters carotenoid biosynthesis also in leaves. When leaf tissue from the transplastomic plants expressing the Erwinia cyclase gene was analyzed, carotenoid composition was found to be unaltered (Fig. 6B). In contrast, leaves from S.l.-pNLyc plants displayed pronounced changes in pigment composition. While chlorophyll contents were identical to the wild type (data not shown), the carotenoid spectra showed significantly elevated levels of the xanthophyll cycle carotenoids violaxanthin, antheraxanthin, and zeaxanthin and a significant reduction in lutein accumulation (Fig. 6B). These changes are proportional in that they do not entail a significant alteration of total leaf carotenoid contents. This suggests that in S.l.-pNLyc plants, the flux through the β-branch of the carotenoid biosynthetic pathway (Fig. 1A) is enhanced and the flux through the α-branch is proportionately reduced.
DISCUSSION
In this work, we have expressed two lycopene β-cyclase genes from the plastid genome of tomato. While the gene from the eubacterium Erwinia did not result in a significant change in carotenoid accumulation, expression of the lycopene β-cyclase from the higher plant daffodil did not only trigger efficient lycopene-to-provitamin A conversion, but also led to a massive increase in total fruit carotenoid content.
The lack of efficient expression of the Erwinia gene was surprising, because an earlier attempt to convert lycopene to β-carotene by expression of this gene from the plastid genome had resulted in at least a moderate increase in β-carotene accumulation (to 286 μg/g dry weight), although total carotenoid levels declined by >10% (Wurbs et al., 2007). The lycopene β-cyclases from Erwinia and Narcissus share significant sequence similarity at the protein level (Fig. 1D), but the similarity at the DNA level is very low. The low CPTA tolerance of the S.l.-pEcrtY plants (Fig. 4) clearly points to a problem with protein accumulation. The promoter and 5′-untranslated region (UTR) used here should have triggered stronger expression than the previously used atpI promoter (Kahlau and Bock, 2008; Oey et al., 2009) and it is not entirely clear why this was not the case. One possible reason could be that accessibility of the Shine-Dalgarno sequence to the plastid ribosomes is impaired in the Prrn-G10L sequence context, for example, by aberrant secondary structure formation between the 5′-UTR and the crtY coding region. Masking of the Shine-Dalgarno sequence by RNA secondary structure formation has been demonstrated to effectively prevent translation in both bacteria (Hall et al., 1982) and plastids (Hirose and Sugiura, 1997). As aberrant RNA folding is notoriously difficult to predict and nearly impossible to analyze in vivo, its possible involvement in the lack of crtY expression from the Prrn-G10L cassette was not investigated further.
Interestingly, plastid expression of the daffodil lycopene β-cyclase led to a strong elevation of total fruit carotenoid content. This was surprising, because previous expression of the lycopene β-cyclase from Erwinia had resulted in a lower total carotenoid content (Wurbs et al., 2007). A plausible explanation could be that the daffodil enzyme is less susceptible to negative feedback regulation by β-carotene than the bacterial enzyme. This would also be compatible with the high β-carotene (and total carotenoid) accumulation that the daffodil enzyme triggers in flower chromoplasts. The daffodil lycopene β-cyclase, therefore, may provide a valuable tool for enhancing the carotenoid metabolic pathway in fruits.
Expression of the daffodil lycopene β-cyclase also resulted in altered carotenoid composition in leaves. Accumulation of the xanthophyll cycle pigments zeaxanthin, antheraxanthin, and violaxanthin was increased and accumulation of the α-branch xanthophyll lutein was correspondingly reduced (Fig. 1A). In leaves, conversion of lycopene to β-carotene and α-carotene is usually complete in that no lycopene is detectable. Obviously, there is substrate competition between lycopene β-cyclase and lycopene ε-cyclase and presence of elevated amounts of lycopene β-cyclase is, therefore, sufficient to channel more lycopene into the β-branch of carotenoid biosynthesis. This alteration in flux through the pathway did not result in any phenotypic effect, which is not unexpected, because lutein, the pigment species reduced in S.l.-pNLyc plants, was shown previously to be dispensable for photosynthesis in higher plants (Pogson et al., 1996).
Both elevating the provitamin A content and increasing the total carotenoid content of tomatoes represent important goals of breeding and genetic engineering efforts (Ye et al., 2000; Giuliano et al., 2008; Mayer et al., 2008). Having obtained, by plastid genome engineering in a commercial tomato variety, provitamin A levels approaching 1 mg/g dry weight and, at the same time, a >50% increase in total fruit carotenoid content, therefore, is also an encouraging step forward in metabolic engineering of the carotenoid pathway in tomato. Given that approximately 10 mg per day of β-carotene provides the adult recommended dietary allowance for vitamin A (http://www.nlm.nih.gov/medlineplus/druginfo/natural/patient-vitamina.html), S.l.-pNLyc tomatoes provide a very rich source of provitamin A. Expression of the daffodil lycopene β-cyclase represents only the second (and the to date most successful) example of metabolic engineering by plastid transformation in a food crop (Wurbs et al., 2007). This is because tobacco (Nicotiana tabacum) is still the only plant in which plastid transformation is routine. Generation of transplastomic plants in the few other species that can be transformed is much more difficult, laborious, and time consuming (Ruf et al., 2001; Dufourmantel et al., 2005). However, as the transplastomic technology offers unique advantages, including high-level foreign protein accumulation (of up to >70% of the plant's total soluble protein; Oey et al., 2009), absence of epigenetic effects, convenient transgene stacking in operons, and greatly increased transgene containment due to the maternal inheritance of plastid genes (Ruf et al., 2007), there is clearly a persuasive case for the further development of plastid transformation technology and its application in agriculture and biotechnology. Our data presented here demonstrate that if appropriate combinations of transgenes and expression elements are chosen, efficient metabolic engineering via plastid transformation is also possible in non-green tissues.
MATERIALS AND METHODS
Plant Material
Aseptically grown tomato (Solanum lycopersicum ‘IPA-6’) plants were obtained from surface-sterilized seeds germinated and grown on agar-solidified Murashige and Skoog medium (Murashige and Skoog, 1962) with 20 g/L Suc. Homoplasmic transplastomic lines were rooted and propagated on the same media. Rooted homoplasmic plants were transferred to soil and grown to maturity under standard greenhouse conditions (250 μmol quanta m−2 s−1). Fruits were harvested at different stages of ripening.
Cloning Procedures
The coding region of the crtY gene from Erwinia herbicola was excised as NcoI/XbaI fragment from a previously described plasmid (Wurbs et al., 2007), cloned into a Prrn-G10L-driven expression cassette (Kuroda and Maliga, 2001; Oey et al., 2009), and then integrated as SacI/HindIII fragment into the previously described plastid transformation vector pKP9 (Zhou et al., 2008), generating vector pEcrtY (Fig. 1C). A cloned lycopene β-cyclase gene (Lyc) from daffodil (Narcissus pseudonarcissus; kindly provided by Drs. Peter Beyer and Salim Al-Babili, University of Freiburg, Germany) was amplified by PCR with primers PNlyc5′ (5′-TTTTCCATGGATACTCTATTGAGAACCCA-3′) and PNlyc3′ (5′-TTTTACTAGTCCCTATCTTGAACTAAGTTA-3′). With the primer sequences, a 5′ NcoI restriction site and a 3′ SpeI site were introduced (underlined). The entire coding region of the Lyc gene (including the putative transit peptide sequence) was cloned into the same Prrn-G10L expression cassette (as NcoI/SpeI fragment into the NcoI/XbaI-digested vector) and subsequently integrated into pKP9 as SacI/ApaI fragment, generating transformation vector pNLyc (Fig. 1C).
Transformation of Tomato Chloroplasts
Plastid transformation was carried out using a biolistic protocol (Svab and Maliga, 1993; Ruf et al., 2001). Young leaves from tomato plants grown under aseptic conditions were bombarded with plasmid DNA-coated 0.6-μm gold particles using a biolistic gun with a Hepta adaptor (PDS1000He; Bio-Rad). Spectinomycin-resistant tomato lines were selected on a modified Murashige and Skoog medium (Wurbs et al., 2007) containing spectinomycin (500 mg/L). For each construct, several independent transplastomic tomato lines were subjected to three additional rounds of regeneration on spectinomycin-containing medium to obtain homoplasmic tissue. Homoplasmy was confirmed by inheritance assays (Bock, 2001) in which seeds were germinated on Murashige and Skoog medium containing spectinomycin (100 mg/L).
Isolation of Nucleic Acids and Gel-Blot Analyses
Total plant DNAs were isolated from fresh leaf tissue samples by a cetyltrimethylammoniumbromide-based method (Doyle and Doyle, 1990). Total cellular RNA was extracted using the peqGOLD TriFast reagent (Peqlab GmbH). For Southern-blot analysis, samples of 5 μg total DNA were digested with the restriction enzyme BglII, separated by gel electrophoresis on 1% agarose gels, and transferred onto Hybond nylon membranes (GE Healthcare) by capillary blotting. Total cellular RNA samples were electrophoresed in formaldehyde-containing 1% agarose gels and blotted onto Hybond nylon membranes. Hybridizations were performed at 65°C in Church buffer (Church and Gilbert, 1984). Hybridization probes were purified by agarose gel electrophoresis following extraction of the DNA fragments of interest from excised gel slices using the GFX PCR (DNA and gel band purification) kit (GE Healthcare). A 550 bp PCR product generated by amplification of a portion of the psaB coding region using primers P7247 and P7244 (Wurbs et al., 2007) was used as an RFLP probe to verify plastid transformation and assess homoplasmy. Transgene-specific probes for northern-blot analyses were generated by excising the Erwinia and Narcissus lycopene β-cyclase coding regions from plasmid clones with NcoI and XbaI.
Herbicide Tolerance Assays
Synthesis of the chemical CPTA, as a specific lycopene cyclase inhibitor, was described previously (Schuetz and Baldwin, 1958; Wurbs et al., 2007). Tolerance of transplastomic tomato plants to CPTA was assessed by adding 2.5 mL CPTA solution to plants grown under aseptical conditions in boxes on 2× Murashige and Skoog medium (to obtain final herbicide concentrations of 50, 100, 150, 250, or 500 μm) followed by phenotypic comparison with a water-treated control after 7 d.
HPLC Analyses of Pigments
Carotenoids and chlorophylls were isolated from leaf tissue (that was frozen, ground, and lyophilized) by extraction with 80% acetone followed by two additional extractions with 100% acetone and combination of the three extracts. Tomato fruit tissue harvested at the onset of fruit softening was frozen, ground, lyophilized, and extracted as described above. Separation, identification, and quantification of carotenoids were performed by HPLC using an Agilent 1100 Series HPLC system with a diode array detection unit (Agilent). For all separations, a YMC ODS-A 250 × 4.6 mm column + precolumn was used. Separation was performed as described previously (Wurbs et al., 2007). All pigment species were identified and quantified by comparison with known amounts of pure standards.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number GQ327929.
Acknowledgments
We are grateful to Drs. Peter Beyer and Salim Al-Babili (University of Freiburg, Germany) for providing a daffodil lycopene cyclase clone. We thank Dr. Stephanie Ruf and Steffi Seeger (Max-Planck-Institut für Molekulare Pflanzenphysiologie) for help with plant transformation, Helga Kulka (Max-Planck-Institut für Molekulare Pflanzenphysiologie Green Team) for plant care and cultivation, Josef Bergstein (Max-Planck-Institut für Molekulare Pflanzenphysiologie) for photography, and Yossi Hirschberg (The Hebrew University of Jerusalem) for helpful discussion and critically reading the manuscript.
This work was supported by the Max Planck Society and by the Deutsche Forschungsgemeinschaft (grant no. BO 1482/11–1 to R.B.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ralph Bock (rbock@mpimp-golm.mpg.de).
Open access articles can be viewed online without a subscription.
References
- Al-Babili S, Hobeika E, Beyer P (1996) A cDNA encoding lycopene cyclase from Narcissus pseudonarcissus L. (PGR96-107). Plant Physiol 112 1398 [Google Scholar]
- Armstrong GA (1997) Genetics of eubacterial carotenoid biosynthesis: a colorful tale. Annu Rev Microbiol 51 629–659 [DOI] [PubMed] [Google Scholar]
- Bock R (2001) Transgenic chloroplasts in basic research and plant biotechnology. J Mol Biol 312 425–438 [DOI] [PubMed] [Google Scholar]
- Bramley PM (2002) Regulation of carotenoid formation during tomato fruit ripening and development. J Exp Bot 53 2107–2113 [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AR (1999) Oxidative DNA damage, antioxidants, and cancer. Bioessays 21 238–246 [DOI] [PubMed] [Google Scholar]
- Cunningham FX, Gantt E (1998) Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol 49 557–583 [DOI] [PubMed] [Google Scholar]
- Dharmapuri S, Rosati C, Pallara P, Aquilani R, Bouvier F, Camara B, Giuliano G (2002) Metabolic engineering of xanthophyll content in tomato fruits. FEBS Lett 519 30–34 [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12 13–15 [Google Scholar]
- Dufourmantel N, Tissot G, Goutorbe F, Garcon F, Muhr C, Jansens S, Pelissier B, Peltier G, Dubald M (2005) Generation and analysis of soybean plastid transformants expressing Bacillus thuringiensis Cry1Ab protoxin. Plant Mol Biol 58 659–668 [DOI] [PubMed] [Google Scholar]
- Fraser PD, Enfissi EMA, Halket JM, Truesdale MR, Yu D, Gerrish C, Bramley PM (2007) Manipulation of phytoene levels in tomato fruit: effects on isoprenoids, plastids, and intermediary metabolism. Plant Cell 19 3194–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Romer S, Shipton CA, Mills PB, Kiano JW, Misawa N, Drake RG, Schuch W, Bramley PM (2002) Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. Proc Natl Acad Sci USA 99 1092–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galpaz N, Ronen G, Khalfa Z, Zamir D, Hirschberg J (2006) A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus. Plant Cell 18 1947–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Aquailani R, Dharmapuri S (2000) Metabolic engineering of plant carotenoids. Trends Plant Sci 5 406–409 [DOI] [PubMed] [Google Scholar]
- Giuliano G, Tavazza R, Diretto G, Beyer P, Taylor MA (2008) Metabolic engineering of carotenoid biosynthesis in plants. Trends Biotechnol 26 139–145 [DOI] [PubMed] [Google Scholar]
- Hall MN, Gabay J, Debarbouille M, Schwartz M (1982) A role for mRNA secondary structure in the control of translation initiation. Nature 295 616–618 [DOI] [PubMed] [Google Scholar]
- Hirose T, Sugiura M (1997) Both RNA editing and RNA cleavage are required for translation of tobacco chloroplast ndhD mRNA: a possible regulatory mechanism for the expression of a chloroplast operon consisting of functionally unrelated genes. EMBO J 16 6804–6811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg J (2001) Carotenoid biosynthesis in flowering plants. Curr Opin Plant Biol 4 210–218 [DOI] [PubMed] [Google Scholar]
- Isaacson T, Ronen G, Zamir D, Hirschberg J (2002) Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of β-carotene and xanthophylls in plants. Plant Cell 14 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlau S, Bock R (2008) Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: Chromoplast gene expression largely serves the production of a single protein. Plant Cell 20 856–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Maliga P (2001) Complementarity of the 16S rRNA penultimate stem with sequences downstream of the AUG destabilizes the plastid mRNAs. Nucleic Acids Res 29 970–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer JE, Pfeiffer WH, Beyer P (2008) Biofortified crops to alleviate micronutrient malnutrition. Curr Opin Plant Biol 11 166–170 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue culture. Physiol Plant 15 473–497 [Google Scholar]
- Oey M, Lohse M, Kreikemeyer B, Bock R (2009) Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J 57 436–445 [DOI] [PubMed] [Google Scholar]
- Pogson B, McDonald KA, Truong M, Britton G, DellaPenna D (1996) Arabidopsis carotenoid mutants demonstrate that lutein is not essential for photosynthesis in higher plants. Plant Cell 8 1627–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer S, Fraser PD (2005) Recent advances in carotenoid biosynthesis, regulation and manipulation. Planta 221 305–308 [DOI] [PubMed] [Google Scholar]
- Römer S, Fraser PD, Kiano JW, Shipton CA, Misawa N, Schuch W, Bramley PM (2000) Elevation of the provitamin A content of transgenic tomato plants. Nat Biotechnol 18 666–669 [DOI] [PubMed] [Google Scholar]
- Ronen G, Carmel-Goren L, Zamir D, Hirschberg J (2000) An alternative pathway to β-carotene formation in plant chromoplasts discovered by map-based cloning of beta and old-gold color mutations in tomato. Proc Natl Acad Sci USA 97 11102–11107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati C, Aquilani R, Dharmapuri S, Pallara P, Marusic C, Tavazza R, Bouvier F, Camara B, Giuliano G (2000) Metabolic engineering of beta-carotene and lycopene content in tomato fruit. Plant J 24 413–419 [DOI] [PubMed] [Google Scholar]
- Ruf S, Hermann M, Berger IJ, Carrer H, Bock R (2001) Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat Biotechnol 19 870–875 [DOI] [PubMed] [Google Scholar]
- Ruf S, Karcher D, Bock R (2007) Determining the transgene containment level provided by chloroplast transformation. Proc Natl Acad Sci USA 104 6998–7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz RD, Baldwin RA (1958) The synthesis and properties of some substituted phenyl ω-(N,N-dialkylamino)-alkyl sulfides. J Am Chem Soc 80 162–164 [Google Scholar]
- Svab Z, Maliga P (1993) High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc Natl Acad Sci USA 90 913–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurbs D, Ruf S, Bock R (2007) Contained metabolic engineering in tomatoes by expression of carotenoid biosynthesis genes from the plastid genome. Plant J 49 276–288 [DOI] [PubMed] [Google Scholar]
- Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, Beyer P, Potrykus I (2000) Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287 303–305 [DOI] [PubMed] [Google Scholar]
- Zhou F, Badillo-Corona JA, Karcher D, Gonzalez-Rabade N, Piepenburg K, Borchers A-MI, Maloney AP, Kavanagh TA, Gray JC, Bock R (2008) High-level expression of HIV antigens from the tobacco and tomato plastid genomes. Plant Biotechnol J 6 897–913 [DOI] [PubMed] [Google Scholar]